Abstract
Plant responses mediated by phytochrome A display a first phase saturated by transient light signals and a second phase requiring sustained excitation with far-red light (FR). These discrete outcomes, respectively so-called very-low-fluence response (VLFR) and high-irradiance response (HIR), are appropriate in different environmental and developmental contexts but the mechanisms that regulate the switch remain unexplored. Promoter analysis of a light-responsive target gene revealed a motif necessary for HIR but not for VLFR. This motif is required for binding of the Bell-like homeodomain 1 (BLH1) to the promoter in in vitro and in yeast 1-hybrid experiments. Promoter substitutions that increased BLH1 binding also enhanced HIR. blh1 mutants showed reduced responses to continuous FR and to deep canopy shadelight, but they retained normal responses to pulsed FR or red light and unfiltered sunlight. BLH1 enhanced BLH1 expression and its promotion by FR. We conclude that BLH1 specifically regulates HIR and not VLFR of phytochrome A.
Keywords: Arabidopsis thaliana, promoter, TALE
When dark-grown seedlings are exposed to light, stem extension is arrested while foliage growth, pigmentation, and photosynthetic capacity are promoted. This developmental transition is largely mediated by phytochrome B (phyB) when the seedlings are exposed to red light (R) and by phytochrome A (phyA) when the seedlings are exposed to far-red light (FR) (1, 2). phyA initiates 2 photobiologically discrete response phases (3). One, often called very-low-fluence response (VLFR), saturates with infrequent excitation of phyA (e.g., 1 brief pulse every hour) with very low light fluence rates. The other phase, called high-irradiance response (HIR), operates above a threshold of excitation frequency and requires higher fluence rates of FR (3–5).
Processing of the incoming information by the cellular signaling circuits results in a wide range of input-output relationships that serve specific functional purposes (6, 7). The duality of phyA-mediated photobiology adds tools to achieve a versatile control of development by light. The VLFR to brief light exposures, such us those experienced by seeds during soil tillage, stimulates germination under favorable conditions (8). The HIR initiates the synthesis of photoprotective pigments only when the seedlings experience prolonged exposures to higher fluence rates (9). The expression of light-harvesting chlorophyll a-b binding protein (Lhcb) genes can exhibit both VLFR and HIR, providing a rapid reaction in response to the first photons when the seedling begins to emerge from the soil (VLFR) and a stronger promotion when excitation becomes persistent (HIR) (10). The transition between VLFR and HIR provides a suitable and functionally meaningful model to investigate mechanisms of regulation of input-output relationships in multicellular organisms.
The phyA-302 allele, harboring a Glu-777 to Lys substitution in the PAS2 domain of the C terminus, retains near-normal VLFR and lacks HIR (11). Under continuous FR, the formation of nuclear speckles containing phyA, and phyA degradation are reduced in phyA-302 providing correlative evidence for a role of these processes in HIR. EID1 is an F-box protein that negatively regulates the HIR of phyA and has little effect on VLFR (12, 13). During the early steps of de-etiolation under continuous FR, phyA targets a relatively large number of transcription factors (14). However, no transcription factor is known to affect selectively the HIR of its target genes. Previously we observed that a target gene promoter contained cis-acting elements, required for both VLFR and HIR, and a 42-bp region between −176 and −134 specifically required for HIR (10). Here, we refine the definition of this HIR-specific motif and use this information to identify a transcription factor by using yeast 1-hybrid screens. We also show that this factor, the Bell-like homeodomain 1 (BLH1) protein, is specifically involved in the HIR but not in VLFR.
Results
Identification of a Sequence Specifically Required for HIR in the Lhcb1*2 Promoter.
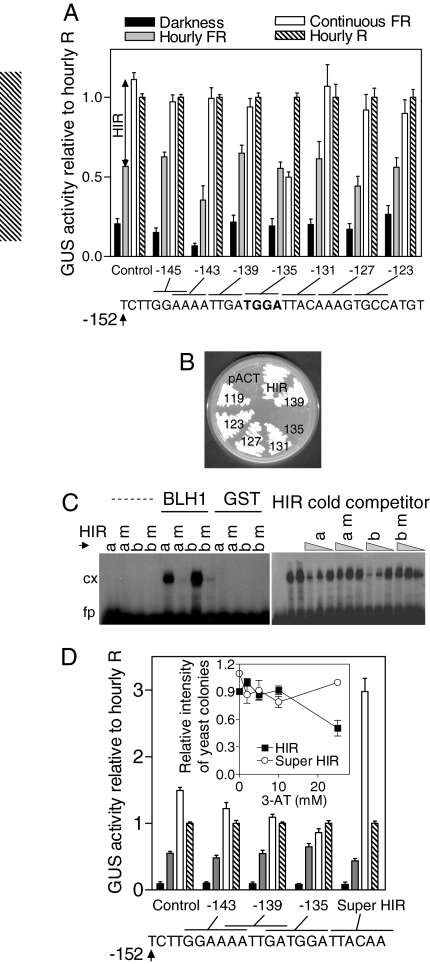
Hourly pulses of FR (3 min, 60 μmol m−2 s−1) saturate the VLFR phase. The difference between the effects of hourly and continuous FR defines the HIR, which operates above a threshold of excitation frequency. We first narrowed down the sequence containing a cis-acting motif specifically required for HIR to the −152 to −134 region of the Lhcb1*2 promoter (Fig. S1A) (10). Subsequent substitution analysis carried out in the −152 to +67 Lhcb1*2 promoter context shows that none of the mutations affected GUS activity in seedlings incubated in darkness or under hourly R or FR. Substitution of TGGA between −138 and −135 bp by CCCC abrogated the HIR because it reduced GUS activity under continuous FR to the level observed under hourly FR and severely impaired the fluence-rate dependency typical of the HIR (Fig. 1A and Fig. S1 B and C). The TGGA sequence is found in Lhcb genes of different species and shows a conserved position relatively to other motifs, including AAAATCT and GATA (15) (Fig. S2A). Substitution of GATA motifs and of ATCT within the AAAATCT motif in the context of the −453 to +67 Lhcb1*2 promoter caused reduced GUS activity in transgenic Arabidopsis grown under continuous FR, hourly pulses of R or FR, or darkness (Fig. S2B). Thus, while TGGA is specifically required for HIR, other motifs are required for multiple phytochrome-mediated responses.
Fig. 1.
BLH1 binds the HIR box of the Lhcb1*2 promoter of Nicotiana plumbaginifolia, which is required for HIR but not for VLFR of phyA or for phyB-mediated responses. (A) Etiolated seedlings were exposed to hourly pulses of FR, continuous FR (60 μmol m−2 s−1), or hourly pulses of R or remained as dark controls. Substitutions to CCCC affecting the indicated 4 bp in the context of the −152 to +67 promoter define the sequence required for HIR as TGGA between −138 and −135 bp (i.e., −135, see sequences in Fig. S1B). Data are means and SE of 5 independent transgenic lines. For each line, activity was normalized to the values under R pulses, which was unaffected by the substitutions (see SI Text). The interaction between light and promoter is significant at P < 0.0001 (2-way ANOVA), and the difference between hourly and continuous FR is significant for all promoters except −135 (P < 0.01, Bonferroni post-tests). (B) BLH1 protein binds the HIR box in a yeast 1-hybrid system in a sequence-specific manner. Yeast strains transformed with pACT-BLH1 fail to grow in the absence of histidine and the presence of 2 mM 3-AT if the bait reporter construct carries a TGGA to CCCC substitution between −138 and −135 bp. (C) BLH1 protein binds the HIR box in vitro in a sequence-specific manner. Left: Electrophoretic mobility shift assay with recombinant BLH1 (5 μg) protein and labeled probes (10 femtomol) containing the TGGA motif (HIRa or HIRb) or the CCCC substitution (HIRa m or HIRb m). Right: Competition experiment where BLH1 (5 μg, except first lane with 0 μg and third lane with 10 μg) was incubated with labeled HIRb in the presence of 1000-, 500-, or 100-fold excess of the indicated unlabeled probe. cx, complex; fp, free probe. (D) Super HIR Lhcb1*2 promoter shows enhanced HIR and binding by BLH1. At least 5 independent transgenic lines were used for each 6-bp substitution in the −453 Lhcb1*2 promoter context. The inset shows enhanced growth of yeast strains transformed with pACT-BLH1 in the absence of HIS and the presence of 25 mM 3-AT if the bait reporter construct carries a TGGAGGT sequence (Super HIR) repeated 4 times compared to the WT HIR sequence repeated 4 times. Relative intensity of yeast colonies is the brightness intensity of yeast colonies photographed against a dark background and scanned with Photoshop software, expressed relative to the brightness with 0 mM 3-AT (means and SE of 3 experiments, P < 0.001 with 25 mM 3-AT).
The Homeodomain Transcription BLH1 Factor Binds the TGGA Motif In Vivo.
We conducted a yeast 1-hybrid screening of an Arabidopsis thaliana cDNA library from dark-grown seedlings to identify proteins binding the motif required for HIR. As bait we used a tandem of 4 repeats of a 23-bp sequence composed of the −141 to −129 sequence of the promoter followed by 10 bp complementary to the −141 to −132 fragment in inverted order. The yeast reporter strain transformed with an empty HIS3 reporter vector was able to grow on medium without histidine, but not if it contained 2 mM 3-amino-1,2,4 triazole (3-AT) (Fig. S3). A 1.5-kb clone (pACT-BLH1) was identified on 2 independent occasions, its sequence corresponds to the BLH1 (At2g35940) protein lacking the 50 aa at the amino terminal end and 168 aa at the carboxy terminal end, but retaining the Bell domain, the homeodomain, and extended sequences of poor similarity to other BLH proteins (16).
To further characterize the binding of BLH1, we designed another set of HIS3 reporters based on the −141 to −119 bp sequence of the Lhcb1*2 promoter. This resulted in 6 additional plasmids carrying various 4-bp substitutions equivalent to those used in Fig. 1A. When transformed with pACT-BLH1, all strains were able to grow in the absence of histidine (Fig. S3), but those carrying the TGGA to CCCC substitution between −138 and −135 bp failed to grow in the presence of 2 mM 3-AT (135 in Fig. 1B and Fig. S3).
BLH1 Binds the TGGA Motif in Vitro.
Recombinant BLH1 derived from the 1.5-kb clone identified in yeast screenings was purified from Escherichia coli as a GST-BLH1 protein fusion. Gel shift analysis of recombinant BLH1 with 2 radioactively labeled HIR probe variants resulted in complex formation (HIRa and HIRb in Fig. 1C, Left). The same TGGA to CCCC substitution that abolished activation in a yeast 1-hybrid assay strongly reduced in vitro binding of BLH1 (HIRa m and HIRb m in Fig. 1C, Left). Lanes with control GST extracts did not show any complexes. The HIR probes containing a TGGA to CCCC substitution did not compete with the HIRb fragment for BLH1 binding (compare HIRa versus HIRa m and HIRb versus HIRb m in Fig. 1C, Right).
Super HIR Promoter Shows Enhanced Binding by BLH1.
Before using the promoter-GUS fusions carrying 4-bp substitutions, we had constructed fusions bearing 6-bp substitutions in the −453 bp promoter context. Disruption of the TGGA core motif in the −135 substitution reduced the HIR, but the effect was not complete, very likely because it created a TGGT in a close location (−135 in Fig. 1D and SI Materials and Methods). Noteworthy, the −129 substitution showed a HIR that more than doubled the HIR of the control promoter (Fig. 1D). The −129 substitution incorporated GGTACC between −134 and −129 bp, and this created a TGGAGGT sequence between −137 and −132, which could account for the enhanced HIR via stronger binding of a transcription factor. Consistently with this interpretation, a combination of pACT-BLH1 with a construct containing the TGGAGGT sequence (super HIR box) allowed enhanced growth of yeast under increasing 3-AT concentrations when compared to the construct containing the wild-type (WT) promoter (Fig. 1D, Inset, and Fig. S3F).
Normal HIR Require BLH1.
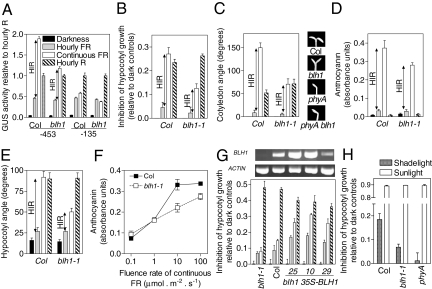
To investigate the physiological significance of BLH1, we obtained homozygous blh1-1, blh1-4, and blh1-5 mutant seedlings, which show reduced BLH1 expression, and the corresponding WT BLH1 siblings (Fig. S4 A and B). In blh1 hypocotyl length, hypocotyl angle, cotyledon angle, anthocyanin levels, and Lhcb1*2 promoter activity were WT in darkness and under hourly pulses of R or FR. However, blh1 showed significantly reduced responses to continuous FR (Fig. 2 and Fig. S4 C and D). The phyA and phyA blh1 mutants were indistinguishable and blind to FR (Fig. 2). The promotion of the endogenous Lhcb expression measured by real-time PCR was also reduced in blh1 under continuous FR (Darkness: WT = 1 ± 0; blh1-1 = 1 ± 0; hourly FR: WT = 138 ± 19; blh1-1 = 153 ± 33; continuous FR: WT = 467 ± 98; blh1-1 = 246 ± 39; P < 0.05). The induction of seed germination by a single pulse of long-wavelength FR, which is a typical VLFR (17, 18), was normal in blh1 (Fig. S4E). This demonstrates that blh1 mutations have specific effects on the HIR without altering the VLFR or phyB-mediated responses.
Fig. 2.
Impaired HIR and normal VLFR in blh1 mutants. (A) Lhcb1*2 promoter activity. Plants of the blh1-1 mutant and of the BLH1 siblings were transformed with the −453 Lhcb1*2 promoter or the −453 Lhcb1*2 promoter where TGGA was substituted by CCCC (−135). Etiolated seedlings were exposed to hourly pulses of FR, continuous FR (60 μmol m−2 s−1), or hourly pulses of R or remained as dark controls. Data are means and SE of 5 independent transgenic lines for each transgene/background combination. For each line, activity was normalized to the values under R pulses, unaffected by the −135 substitution or blh1 mutation (SI Text). (B–E) Hypocotyl growth inhibition relative to the length in dark controls (length in darkness, mm: WT = 13.4 ± 0.1, blh1 = 13.1 ± 0.2, P > 0.1), cotyledon angle (photographs under continuous FR), anthocyanin levels, and hypocotyl angle. One-day-old seedlings were grown for 3 days under hourly pulses of FR, continuous FR, or hourly pulses of R, or remained as dark controls before measurements. The fluence rate of continuous FR was 0.3 (B), 0.5 (C), 10 (D), or 0.5 (E) μmol m−2 s−1. Data are means and SE of at least 3 replicate boxes. ANOVA followed by Bonferroni post-tests indicate significant effects of blh1 (P < 0.001) under continuous FR but not under any other light condition. The blh1 mutation had no effects in the phyA-211 mutant background under continuous FR (dark control data included in brackets). Inhibition of hypocotyl growth: phyA = 0.02 ± 0.02 (0.00 ± 0.02); phyA blh1 = 0.02 ± 0.03 (0.00 ± 0.01); cotyledon angle (degrees): phyA = 1 ± 0 (1 ± 1); phyA blh1 = 1 ± 0 (1 ± 0); anthocyanin (absorbance units): phyA = 0.01 ± 0.00 (0.01 ± 0.00); phyA blh1 = 0.01 ± 0.00 (0.01 ± 0.00); hypocotyl angle (randomization of growth orientation) (degrees): phyA = 6 ± 1 (8 ± 1); phyA blh1 = 6 ± 1 (8 ± 1). (F) Fluence rate-response curve of anthocyanin levels. Data are means and SE of 3 replicate boxes. ANOVA followed by Bonferroni post-tests indicate significant effects of blh1 under 10 (P < 0.001) and 100 (P < 0.05) μmol m−2 s−1. (G) When expressed in the blh1-1 background under the control of the CaMV 35S promoter, BLH1 inhibits hypocotyl growth even under pulsed FR. One-day-old seedlings were grown for 3 days in darkness or under hourly FR or continuous FR (0.1 μmol m−2 s−1) before measurements of hypocotyl length. Data are means and SE of at least 3 replicate boxes. Each transgenic line is significantly different (P < 0.05) from blh1 under hourly FR and under continuous FR (symbols as in A). BLH1 expression levels in blh1-1, WT, and different transgenic lines are shown at the top. (H) Phenotype of the blh1-1 mutant under a dense canopy. One-day-old seedlings of the WT Col, the phyA-201 mutant, and the blh1-1 mutant were grown for 3 days under a dense canopy of Cynodon dactylon before measurements. At a clear midday, the FR irradiance beneath the canopy was in average 1 μmol m−2 s−1, and the R/FR ratio was 0.01. The seedlings were grown in the same plastic boxes used for indoor experiments. Dark controls were placed beneath the canopy but wrapped in black plastic and aluminum foil, and unfiltered sunlight controls were grown outside the canopy. Under the canopy, the inhibition observed in blh1-1 is significantly smaller (P < 0.01) than in the WT according to ANOVA and a Bonferroni post-test. Data are means and SE of 6 replicate boxes from 2 independent experiments.
We produced independent transgenic lines of the blh1-1 background expressing the BLH1 gene under the control of the CaMV 35S promoter (construct 35S-BLH1) at higher levels than the WT (lines 25 and 10) or at a lower overall level than the WT (but with a different tissue distribution predicted by the use of the CaMV 35S promoter) (Fig. 2G). These lines showed HIR (difference between hourly and continuous FR) even at a fluence rate (0.1 μmol m−2 s−1), where HIR was weak in the WT and fully absent in blh1-1, indicating that the BLH1 transgene rescued the mutant phenotype (Fig. 2G). More importantly, although the blh1 mutation did not affect the response to hourly pulses of FR, the transgenic lines ectopically expressing BLH1 showed enhanced responses to hourly pulses of FR (Fig. 2G and Fig. S5). This indicates that overexpression of BLH1 bypasses the need of continuous FR to make BLH1 physiologically effective. This gain-of-function phenotype of the BLH1 transgenics was absent in the phyA mutant background (Fig. S5B), indicating that the enhanced response to hourly FR induced by overexpressed BLH1 is mediated by phyA. Overexpression of BLH1 did not affect phyB-mediated signaling under hourly R (Fig. S5C).
The blh1 mutant showed reduced hypocotyl growth inhibition under a dense canopy, a condition where phyA is required for de-etiolation (Fig. 2H). The latter observation highlights the biological significance of BLH1 control of Arabidopsis photomorphogenesis.
Transcriptome of blh1.
To investigate the effect of blh1 on the transcriptome, WT and blh1-1 seedlings were grown in darkness for 2 days and then transferred to continuous FR (60 μmol m−2 s−1) for 6 h before harvest. Dark controls were harvested simultaneously without exposure to FR. The effects of FR were consistent with previous reports (Fig. S6A). On average, the blh1 mutation caused a 60% reduction of the response to FR (Fig. S6B). We used factorial ANOVA followed by clustering to select the genes showing little difference between the WT and blh1 in darkness and reduced response to FR in the mutant (Table S1). Three clusters (125 genes) grouped genes with reduced promotion of expression by FR in blh1. Plastid-related genes were highly overrepresented [normed frequency ± bootstrap SD (19): 9.7 ± 2.1] in these clusters. Two clusters (89 genes) grouped genes with reduced inhibition of expression by FR in blh1. Genes related to endoplasmic reticulum (8.3 ± 3.8) and receptor binding or activity (4.0 ± 2.3) were highly overrepresented in these clusters. Among the genes significantly affected in blh1, each 1 of the 3 possible 6-mer promoter sequences of the Lhcb1*2 promoter containing the functional TGGA motif was overrepresented (GATGGA: P < 0.006, TGGAAT: P < 0.03, and ATGGAT: P < 0.03).
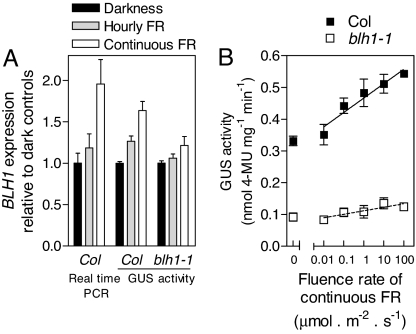
BLH1 Promotes BLH1 Promoter Activity.
The BLH1 gene showed expression in darkness, and this is consistent with the fact that it was found in a library from dark-grown Arabidopsis. BLH1 promoter activity was reduced in dark-grown seedlings of the blh1 mutant compared to the WT (Fig. S7A). In the WT, continuous FR increased BLH1 expression (20) to a larger extent than hourly pulses of FR (Fig. 3A). The blh1 mutant showed reduced response to continuous FR (Fig. 3A and Fig. S7B). GUS activity driven by the BLH1 promoter showed FR fluence rate dependency over a wide range of fluence rates, and this response was severely reduced in the blh1 background (Fig. 3B).
Fig. 3.
BHL1 promotes its own expression. (A) BLH1 enhances the expression of BLH1 under FR. Transgenic seedlings bearing BLH1::GUS were grown in darkness for 2 days and then exposed to hourly FR, continuous FR (60 μmol m−2 s−1), or no light for 24 h before harvest. BLH1 expression was also analyzed by real-time PCR in WT seedlings exposed for 6 h to the different light/dark treatments. Data are means and SE of 3 biological replicates (real-time PCR) or of 5 (WT) or 3 (blh1) independent transgenic lines bearing the BLH1 promoter fused to GUS. The effect of continuous FR is significant (P < 0.001) in the WT but not (P > 0.05) in blh1. (B) BLH1 enhances BLH1::GUS activity in darkness and its response to continuous FR. Data are means and SE of 6 biological replicates from representative transgenic lines in the WT or blh1-1 background. The slopes are significantly different (P < 0.002).
Discussion
We have narrowed-down the HIR-specific motif of Lhcb1*2, which is not required for VLFR or phyB-mediated responses, to the 4-bp core sequence TGGA (Fig. 1). “Phylogenetic footprinting” indicates that the TGGA motif is present at relatively conserved distance from CCAAT and GATA motifs (15) in a number of Lhcb genes from different species (Fig. S2A), suggesting that it might be a binding site conserved by natural selection (21). Using a bait based on this sequence in combination with a library from dark-grown A. thaliana seedlings in the yeast 1-hybrid system we identified the homeodomain transcription factor BLH1. BLH1 belongs to the 3-amino acid loop extension (TALE) superclass of homeobox genes. In vivo and in vitro binding experiments confirmed the specific interaction between BLH1 and TGGA (Fig. 1 B and C). The TGGA core is in the TGATGGATTA sequence of the tobacco Lhcb1*2 promoter, which (i) differs only 2 bases from the central sequence of the ATGATAGATAAT segment from the nitrate reductase 2 gene of Arabidopsis, previously shown to bind in vitro BLH1 protein obtained from nuclear extracts of Arabidopsis roots (22); (ii) contains ATTA, the canonical binding site for most homeodomains, preceded by GG, which provides binding specificity for homeodomains containing a Lys residue at the position 50 in the recognition helix (23); and (iii) presents a full match to TGATXXATTA, the consensus binding site of heterodimers of PBC/Hox TALE proteins (24). Noteworthy, when we replaced the bases following the TGGA motif to obtain TGGAGGT, the promoter showed both stronger binding of BLH1 in yeast and stronger HIR (Fig. 1D), providing gain-of-function evidence for the role of BLH1 binding to the promoter in HIR. The human homeodomain CCAAT-displacement protein (CDP)/Cux binds ATGGAT (25), the same core motif reported here for BLH1. In yeast and in vitro binding experiments, we used the originally identified BLH1 clone, which lacks some amino acids at both ends. The full-length BLH1 clone failed to form complexes in in vitro binding assays and yeast 1-hybrid experiments, and this resembles the behavior of CDP, a well-established case of auto-inhibition involving the N-terminal end of the protein (25, 26). The full-length maize BLH KIP does not bid DNA in the absence of maize Knotted-1 (27). Since BLH1 in nuclear extracts does form complexes with DNA sequences similar to those reported here, and these complexes contain other (unidentified) homeodomain proteins (22), these proteins could be required for cooperative binding of full-length BLH1 to DNA (28). The presence of the ATGGAT sequence in the 500-bp upstream region of genes with altered expression in the blh1 mutant is higher than expected by chance.
In blh1 seedlings, hypocotyl length, hypocotyl angle, cotyledon angle, anthocyanin levels, and Lhcb1*2 promoter activity were normal in darkness and showed normal responses to hourly pulses of FR or R, but reduced responses to continuous FR (Fig. 2). Gene expression responses to 6 h of high-fluence continuous FR were reduced in average in a 60% (Fig. S6B). The VLFR of seed germination was also normal in blh1 (Fig. S4E). Therefore, blh1 mutations and mutations of a DNA motif required for BLH1 binding had specific effects on the HIR without altering the VLFR or phyB-mediated responses. The partial effect of blh1 on HIR could reflect redundancy, which is not uncommon among TALE proteins (29, 30). BLH6 shows strong promotion of expression in response to continuous FR (20) and BLH5 shows the strongest similarity to BLH1. Both blh6 and blh5 mutations reduce HIR (Fig. S8). The expression of BLH1 and its promotion by continuous FR depend strongly on BLH1 (Fig. 3 A and B, and Fig. S7).
The TALE superclass of homeobox genes is conserved among animals, fungi, and plants, where they are required for meristem maintenance and proper regulation of shoot and leaf development in response to endogenous cues (31, 32). Both, TALE proteins and the HIR of phyA have been implicated in plant evolution, the former in the evolution of land plants from green algal ancestors and the diversification of the plant body (32), the latter in the adaptation to the dimly lit understory habitats experienced by the earliest angiosperms (33). Here, we relate BLH1 to HIR, placing a TALE protein as a regulator of plant responses to the specific environmental cue experienced when a seedling emerges from the soil under a deep canopy.
Materials and Methods
Transgenic Lines Carrying Fusions Containing Different Fragments of the Lhcb1*2 Promoter.
The transgenic lines of A. thaliana accession Columbia carrying the −453, −176, and −134 to +67 fragments of the Lhcb1*2 promoter of Nicotiana plumbaginifolia fused to the coding region of gusA have been described previously (10). We continued to use this tobacco promoter to take advantage of previous studies and because the distribution of motifs important for promoter activity is very similar to that of Lhcb1*1 and Lhcb1*2 of A. thaliana (Fig. S2). The procedures to obtain transgenic lines were basically as described and are presented in further detail in SI Text.
blh1 T-DNA Mutant and BLH1 OX Transgenic Lines.
The blh1-1 allele in Columbia [Salk-089095 (34)] containing a T-DNA insertion in exon 6 of BLH1 was obtained from Arabidopsis Biological Resource Center (ABRC) through The Arabidopsis Information Resource at www.arabidopsis.org. To rescue blh1-1 seedling with the BLH1 gene, the A. thaliana full-length cDNA (ABRC stock # U18537) (35), cloned into the the KpnI-SalI sites of the pPZP221 and pPZP212-based binary vectors CHF1 and CHF3 (36) under the control of the CaMV 35S promoter, were introduced in Agrobacterium tumefaciens strain GV3101, and used to transform blh1-1 plants using the floral dip method (37). Homozygous lines were selected by kanamycin or gentamycin resistance. The blh1-4 allele in Landsberg erecta (CSHL_GT9784) containing a Ds insertion in exon 3 was obtained from Gene Trap CSHL (http://genetrap.cshl.org). The blh1-5 allele in Columbia (SM_3_16405) containing a T-DNA insertion in exon 3 was obtained from ABRC. The primers used to identify homozygous mutants and WT siblings are provided in SI Text.
Yeast 1-Hybrid.
Yeast 1-hybrid assays were conducted according to the method described (38) using yeast strain Y187 (Clontech), containing the bait sequence cloned as a NotI-XbaI fragment in the pINTI-HIS3NB integrative reporter gene system (GenBank accession no. AY061966) integrated at the nonessential PDC6 locus (39). We cloned the bait sequence at the NotI-XbaI restriction sites of the integrative vector pINTI-HIS3NB. Yeast cells Y187 were transformed by a lithium acetate procedure, and transformants were selected on YAPD plates with 150 μg/mL G418 for the presence of the dominant antibiotic APTl marker gene present in vector pINT1-HIS3NB. The following sequence was used as baits in cDNA library screenings: 5′-GGCCGCTGATGGATTACAATAATCCATCATGATGGATTACAATAATCCATCATGATGGATTACAATAATCCATCATGATGGATTACAATAATCCATCAAGATCTT-3′ Details of yeast transformation and library description are provided in SI Text.
Electrophoretic Mobility Shift Assays.
Electrophoretic mobility shift assays (EMSA) were carried out essentially as described by (40). Details are provided in SI Text.
Physiological Experiments.
For the experiments to measure GUS activity driven by the Lhcb1*2 promoter constructs, inhibition of hypocotyl growth, cotyledon angle, anthocyanin levels, and hypocotyl angle (randomization of growth orientation), the seedlings were grown in plastic boxes (10, 11). Details are provided in SI Text.
Analysis of BLH1 and Endogenous Lhcb Expression.
We analyzed endogenous BLH1 and Lhcb (combined Lhcb1*1 and Lhcb1*2) expression by real-time PCR. We also constructed transgenic plants bearing the full-length BLH1 promoter fused to the GUS reporter (SI Text).
Microarray Experiment.
Seedlings grown in darkness for 2 days after the induction of seed germination were either exposed to 6 h continuous FR or left as dark controls. Two independent samples were harvested in liquid nitrogen for each genotype by FR/dark combination, and total RNA was extracted by using the RNEASY Plant mini kit (Qiagen). cDNA and cRNA synthesis and hybridization to 22 K (ATH1) Affymetrix Gene Chips were performed according to Affymetrix instructions. The scaling tab of the Affymetrix microarray suite in the mode “all probe sets” was used to standardize the trimmed mean signal of each array to the “target signal” according to the manufacturer's instructions. Details of the statistical analysis of the data, the functional classification of the genes, and the analysis of upstream promoter sequences are given in SI Text.
Supplementary Material
Acknowledgments.
We thank the Arabidopsis Biological Resource Center (ABRC) for their provision of seeds carrying the Salk-089095 insert, seeds of the line SM_3_16405, the Arabidopsis library CD4–22, and the clone U18537 and Cold Spring Harbor Lab for the provision of the CSHL_GT9784 line. This work was financially supported by Agencia Nacional de Promoción Cientìfica y Tecnológica (Argentina) grant nos. PME54 and PICT 11631 and 32492 to J.J.C., University of Buenos Aires grant no. AG021 to J.J.C., Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina) grants 5958 to J.J.C. and 06082 to R.J.S. A.H.M. was supported by the EU FP5 Project TF-STRESS (QLK3–2000-00328). A.A. and P.B.F.O. were supported by the EU FP6 INCO-MPC2 project CEDROME (INCO-CT-2005–015468).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906598106/DCSupplemental.
References
- 1.Quail PH. Phytochrome photosensory signalling networks. Nature Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 3.Casal JJ, Yanovsky MJ, Luppi JP. Two photobiological pathways of phytochrome A activity, only one of which shows dominant negative suppression by phytochrome B. Photochem Photobiol. 2000;71:481–486. doi: 10.1562/0031-8655(2000)071<0481:tppopa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Schäfer E, Beggs CJ, Fukshansky L, Holmes MG, Jabben M. A comparative study of the responsivity of Sinapis alba L. seedlings to pulsed and continuous irradiation. Planta. 1981;153:258–261. doi: 10.1007/BF00383896. [DOI] [PubMed] [Google Scholar]
- 5.Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc Natl Acad Sci USA. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi H, et al. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scopel AL, Ballaré CL, Sánchez RA. Induction of extreme light sensitivity in buried weed seeds and its role in the perception of soil cultivations. Plant Cell Environ. 1991;14:501–508. [Google Scholar]
- 9.Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- 10.Cerdán PD, et al. Sustained but not transient phytochrome A signaling targets a region of a Lhcb1*2 promoter not necessary for phytochrome B action. Plant Cell. 2000;12:1203–1211. [PMC free article] [PubMed] [Google Scholar]
- 11.Yanovsky MJ, et al. Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell. 2002;14:1591–1603. doi: 10.1105/tpc.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieterle M, Zhou Y-C, Schäfer E, Funk M, Kretsch T. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 2001;15:939–944. doi: 10.1101/gad.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y-C, Dieterle M, Buche C, Kretsch T. The negatively acting factors EID1 and SPA1 have distinct functions in phytochrome A-specific light signaling. Plant Physiol. 2002;128:1098–1108. doi: 10.1104/pp.010811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tepperman JM, Zhu T, Chang H-S, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- 16.Becker A, Bey M, Bürglin TR, Saedler H, Theissen G. Ancestry and diversity of BEL1-like homeobox genes revealed by gymnosperm (Gnetum gnemon) homologs. Dev Genes Evol. 2002;212:452–457. doi: 10.1007/s00427-002-0259-7. [DOI] [PubMed] [Google Scholar]
- 17.Botto JF, Sánchez RA, Whitelam GC, Casal JJ. Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 1996;110:439–444. doi: 10.1104/pp.110.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinomura T, et al. Action spectra for phytochrome A-and phytochrome B-specific photoinduction od seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Provart N, Zhu T. A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr Comput Mol Biol. 2003;2003:271–272. [Google Scholar]
- 20.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 22.Sherameti I, et al. The endophytic fungus Piriformospora indica stimulates the expression of nitrate reductase and starch-degrading enzyme glukan-water dikinase in tobacco and Arabidopsis roots through a homeodomain transcription factor that binds to a conserved motif in their promoters. J Biol Chem. 2005;280:26241–26247. doi: 10.1074/jbc.M500447200. [DOI] [PubMed] [Google Scholar]
- 23.Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi taruzu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- 24.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 25.Huang C-J, Chang J-G, Wu S-C, Choo K-B. Negative transcriptional modulation and silencing of the Bi-exonic Rnf35 gene in the preimplantation embryo. Binding of the CCAAT-displacement protein/Cux to the untranslated exon 1 sequence. J Biol Chem. 2005;280:30681–30688. doi: 10.1074/jbc.M413144200. [DOI] [PubMed] [Google Scholar]
- 26.Truscott M, et al. The N-terminal region of the CCAAT displacement protein (CDP)/Cux transcription factor functions as an autoinhibitory domain that modulates DNA binding. J Biol Chem. 2004;279:49787–49794. doi: 10.1074/jbc.M409484200. [DOI] [PubMed] [Google Scholar]
- 27.Smith HM, Boschke I, Hake S. Selective interaction of plant homeodomain protein mediates high DNA-binding affinity. Proc Natl Acad Sci USA. 2002;99:9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Banerjee AK, Hannapel DJ. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 2004;38:276–284. doi: 10.1111/j.1365-313X.2004.02048.x. [DOI] [PubMed] [Google Scholar]
- 29.Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Devel. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- 30.Selleri L, et al. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol Cell Biol. 2004;24:5324–5331. doi: 10.1128/MCB.24.12.5324-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hake S, et al. The role of KNOX genes in plant development. Annu Rev Cell Dev Biol. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- 32.Dolan L. Plant evolution: TALES of development. Cell. 2008;133:771–773. doi: 10.1016/j.cell.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Mathews S, Burleigh JG, Donoghue MJ. Adaptive evolution in the photosensory domain of phytochrome A in early angiosperms. Mol Biol Evol. 2003;20:1087–1097. doi: 10.1093/molbev/msg123. [DOI] [PubMed] [Google Scholar]
- 34.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 35.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 36.Staiger D, et al. The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 2003;17:256–268. doi: 10.1101/gad.244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 38.Ouwerkerk PBF, Meijer AH. Current Protocols in Molecular Biology. Supplement 55. Vol. John Wiley & Sons; 2001. [Google Scholar]
- 39.Meijer AH, Ouwerkerk PBF, Hoge JH. Vectors for transcription factor isolation and target gene identification by means of genetic selection in yeast. Yeast. 1998;14:1407–1416. doi: 10.1002/(SICI)1097-0061(199811)14:15<1407::AID-YEA325>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 40.Ouwerkerk PBF, Trimborn TO, Hilliou F, Memelink J. Nuclear factors GT-1 and 3AF1 interact with multiple sequences within the promoter of the Tdc gene from Madagascar periwinkle: GT-1 is involved in UV light-induced expression. Mol Gen Genet. 1999;261:610–622. doi: 10.1007/s004380050003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.