Abstract
In budding yeast, telomeres and the mating type (HM) loci are found in a heterochromatin-like silent structure initiated by Rap1 and extended by the interaction of Silencing Information Regulator (Sir) proteins with histones. Binding data demonstrate that both the H3 and H4 N-terminal domains required for silencing in vivo interact directly with Sir3 and Sir4 in vitro. The role of H4 lysine 16 deacetylation is well established in Sir3 protein recruitment; however, that of the H3 N-terminal tail has remained unclear. To characterize the role of H3 in silent chromatin formation and compare it to H4 we have generated comprehensive high resolution genome-wide binding maps of heterochromatin proteins. We found that H4 lysine 16 deacetylation is required for the recruitment and spreading of heterochromatin proteins at all telomeres and HM loci. In contrast, the H3 N terminus is required for neither recruitment nor spreading of Sir proteins. Instead, deletion of the H3 tail leads to increased accessibility within heterochromatin of an ectopic bacterial dam methylase and the decreased mobility of an HML heterochromatic fragment in sucrose gradients. These findings indicate an altered chromatin structure. We propose that Sir proteins recruited by the H4 tail then interact with the H3 tail to form a higher order silent chromatin structure.
Keywords: hml and hmr, silencing, sir proteins, telomere
Large segments of the eukaryotic genome, often near centromeres and telomeres, are found in constitutively condensed domains termed heterochromatin (1). This structure is important for both the repression of genes and the faithful segregation of chromosomes, and defects in heterochromatin function can lead to cancer. However, the precise mechanism by which heterochromatin forms and silences genes is not well understood.
In the budding yeast, Sacharomyces cerevisiae, telomeric chromatin and that of the silent mating type loci (HML and HMR) are found in heterochromatin-like domains of silent chromatin. Formation of these domains requires several protein components and key among them are Repressor/Activator Protein 1 (Rap1), the Silencing Information Regulator (Sir) proteins, and the core histones H3 and H4 (2). Silent chromatin formation is thought to involve 3 discrete events: initiation, spreading, and the barrier to spreading. Genetic and biochemical work in a number of labs has produced a model for silent chromatin establishment at the naturally truncated telomere at the right end of chromosome VIR. Silencing is initiated by the sequence specific DNA binding protein Rap1 (3). After recruitment of Sir4 to the initiation site by Rap1 (4), Sir4 recruits Sir2 that de-acetylates the histone H4 N-terminal lysine 16 (H4 K16) (5, 6). This provides a high affinity binding site for Sir3 (7–9). Successive rounds of Sir protein binding to and deacetylation of the histones are thought to allow the silencing complex to spread inwards along the chromosome. Once the Sir complex has spread, the chromatin folds back on itself (10–12) to form a structure capable of repressing genes located at subtelomeric loci. Excessive continued spreading of heterochromatin is prevented by the action of the histone acetyltransferase (HAT) Sas2, which acetylates H4 K16 in adjacent euchromatin, thus preventing the promiscuous spread of Sir3 and the rest of the Sir protein complex (13, 14). Heterochromatin is further tethered to the nuclear periphery by yKu70/80 telomeric DNA binding proteins and Sir4, a subcellular localization that can influence silencing (15, 16).
Although there is a fairly good understanding of the role of the H4 N terminus in the formation of heterochromatin the role of H3 has been much less clear. Like H4 K16, the H3 N terminus and K56 in the core of H3 are deacetylated by Sir2 in vitro (6, 17). Also, the H3 N-terminal domain (residues 1–20) is genetically important for silencing at telomere VIIL and the HM loci and the H3 N-terminal sequence containing this same domain interacts in vitro with Sir3 and Sir4 (7, 9, 18). Significantly, there is a strong synergism between mutants in the H3 and H4 tail at the silent mating type locus HMR leading to a complete loss of silencing and therefore mating only in the double mutant (18). This suggests that the H3 tail acts with H4 to bind to Sir proteins in silent chromatin. However, it is not known whether the H3 N terminus does indeed recruit Sir proteins for their spreading along heterochromatin or whether it interacts with Sir proteins at a subsequent stage.
To address the role of H3 we started with a genome wide approach. Most studies of silent chromatin in yeast have focused on a handful of silent regions, such as the HM loci or telomeres that are naturally (VIR) or synthetically (VIIL) truncated (19). This removes subtelomeric repeated elements that promote the spreading of heterochromatin. However, there are clear differences between silent loci in regards to silencing strength, the factors involved in silencing establishment (e.g., Sir1 at the HM loci), and effects of histone mutants (18, 20) Therefore, the extent of variation in spreading among heterochromatic loci in yeast is best addressed using high resolution whole-genome approaches. The recent development of Affymetrix high-density DNA tiling arrays allows for the detection of chromatin binding proteins at very high resolution. These arrays also include telomeric and repeated sequence elements that are absent from older arrays designed primarily to study coding sequences and their associated regulatory elements (21).
Our data demonstrate different functions for the H3 and H4 N-terminal regions involved in silencing. We have found that H4 lysine 16 deacetylation is indeed required for the recruitment and spreading of Sir proteins along all heterochromatin. However, our experiments using genetic epistasis, bacterial dam methylase access and sucrose gradient sedimentation all indicate a role for the H3 N terminus at a subsequent stage, promoting the formation of a silent higher order chromatin structure once Sir proteins have already spread.
Results
Genome-Wide Binding Map of Silencing Proteins.
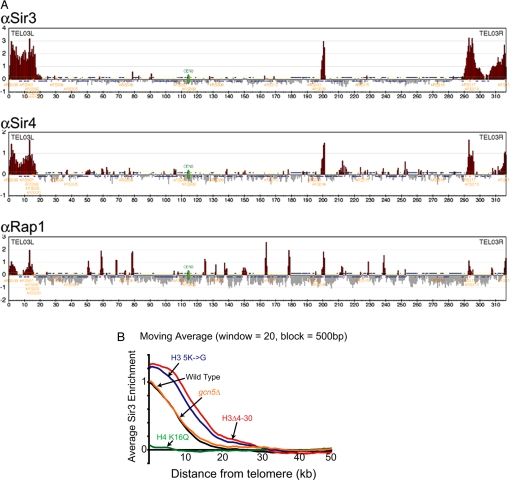
We wished to apply high resolution genome wide chromatin immunoprecipitation in combination with Affymetrix DNA microarrays (ChIP-on-chip) to determine a detailed map of heterochromatin initiation and spreading in yeast and to generate a baseline for the study of silencing factors. Polyclonal antibodies to various silencing factors were used to immunoprecipitate cross-linked chromatin and the associated DNA was purified, labeled, and hybridized to Affymetrix microarrays. We mapped sites of heterochromatin initiation and spreading by determining the genome-wide distribution of the DNA binding protein Rap1 in addition to the silencing factors Sir3 and Sir4 (Fig. 1A and Fig. S1 a–c). Rap1, Sir3 and Sir4 bind to all 32 yeast telomere proximal regions and the silent mating type loci. We observe several euchromatic sites with minor peaks of Sir3 or Sir4 binding (e.g., at ChrX near YJR137C and ChrXIII near YMR314W in Fig. S1a). However, ORFs adjacent to these sites are not derepressed in a sir3Δ mutant (Fig. S1d), and we chose not to pursue this further. We conclude that the binding of silencing proteins, and therefore the presence of heterochromatin, is limited to telomeric regions and the silent mating type loci.
Fig. 1.
A high resolution map of silencing proteins reaveals that the H4 tail but not that of H3 is required for recruitment and spreading of Sir3. ChIP DNA of Sir3, Sir4, and Rap1 and input from wild-type cells were amplified, fragmented, labeled and hybridized to Affymetrix Tiling 1.0R arrays. (A) Chromosome III is shown along with ORFs (blue), ARSs (orange), and the centromere (green). Data for chromosomes I through XVI can be found in Fig. S1. Binding data have been divided into 500 bp bins and values >1 SD above the average (yellow line) have been colored in red. Enrichment is measured as the Log2 score of IP versus input. (B) The moving average (block = 500 bp, window = 20) of Sir3 binding at all 32 yeast telomeres was plotted as a function of distance from the chromosome end. Sir3 enrichment is measured as the Log2 score of IP versus input. Data for Sir3 binding in wild type (black), H4 K16Q (green), H3Δ4–30 (red), H3 K9, 14, 18, 23, 27G (5K→G, blue), and GCN5 deletion (gcn5Δ, orange).
Deacetylation of lysine 16 on the H4 tail acts as a key determinant in the spreading of silent chromatin (8, 9, 22). To verify that this, in fact, occurs at all silent loci and to generate a baseline for comparison to the H3 tail lesion, we examined the role of H4 K16 acetylation on Sir3 binding at a genome-wide level. Substitutions of H4 K16 with glutamine (Q) or glycine (G) mimic the hyperacetylated uncharged state and result in the disruption of telomeric heterochromatin and that of HML (14, 22). We carried out ChIP-on-chip for Sir3 in this strain and plotted Sir3 enrichment as a moving average of distance from the telomere. We found that on average K16Q leads to a substantial loss of Sir3 spreading at telomeric heterochromatin (Fig. 1B). We conclude that the acetylation state of lysine 16 on the H4 tail is a major determinant of Sir protein spreading throughout telomeric heterochromatin.
Loss of the H3 Tail Leads to Increased Levels of Subtelomeric Sir3.
It has been shown that the H3 tail can bind to Sir proteins in vitro (7, 9) and is required for efficient silencing of a telomeric URA3 transgene (18). Therefore, it was reasonable to postulate that the H3 tail might also be involved in recruiting Sir proteins to silent chromatin. To investigate how the H3 tail promotes the formation of silent chromatin, we used ChIP-on-chip to examine Sir3 binding across the entire yeast genome in a mutant lacking H3 residues 4–30. Surprisingly, loss of the H3 tail leads to a dramatic increase in subtelomeric Sir3 occupancy at half of all telomeres (data to be deposited) and a significant increase in the genome average binding (Fig. 1B). This increase in spreading occurs largely without a concomitant decrease in Sir3 occupancy at normally bound regions (Fig. 1B). The ectopic chromatin resembles heterochromatin in that its core histones are hypoacetylated at all sites examined as shown for telomere VIR (Fig. S2). Thus, the H3 tail is not required for recruitment of Sir proteins to silent chromatin as its deletion leads to, on average, increased association of Sir proteins with subtelomeric heterochromatin and adjacent regions. We were concerned that this effect might be indirect, due to altered expression of silencing factors, because deletion of the H3 tail produces global transcriptional defects (23) and overexpression of Sir3 is sufficient to induce ectopic spreading and silencing (24, 25). However, we have examined published microarray data and none of the known core silencing factors show a change in expression when the H3 tail is deleted (23). Therefore, the role of the H3 tail in limiting the spreading of Sir proteins is likely to be a direct one.
Acetylation Sites of the H3 N Terminus Help Prevent Ectopic Spreading of Sir3 at Telomeric and HM Heterochromatin.
To localize more precisely which residues within the H3 tail prevent ectopic spreading we first examined Sir3 spreading at telomere VIR in strains carrying smaller H3 tail deletions, encompassing either residues 4–10 or 4–20. The 4–10 deletion has only a small effect on Sir3 spreading, whereas the 4–20 mutant has an effect approximately half that of the 4–30 deletion strain (Fig. S3). Therefore, the H3 N-terminal sequence involved in blocking the ectopic spread of Sir3 protein is found in most, if not all, of the domain 4–30. To determine whether the acetylatable lysines in the H3 tail might be responsible for the spreading defect, we examined Sir3 spreading genome wide in a strain containing substitutions in all 5 acetylatable lysines (K9, 14, 18, 23 and 27) to glycine (H3 5K to G). As shown in Fig. 1B the H3 5K to G mutation led to an increase in Sir3 spreading almost equivalent to that seen in the Δ4–30 mutant. Deletion of the H3/H2B specific HAT, GCN5, which would preserve positive charges at its target acetylation sites, leads to little change in Sir3 spreading on average although there is increased spreading of Sir3 at only 5 of 32 telomeres (including Chr. VIR) as opposed to 16 of 32 in the H3Δ4–30 mutant (Fig. 1B). We conclude that, although the acetylatable lysines in the H3 tail are important for limiting Sir spreading, the Gcn5 HAT is not solely or perhaps even primarily responsible for this activity.
H4 K16 Substitution Suppresses the Increased Spreading Caused by Deletion of the H3 Tail.
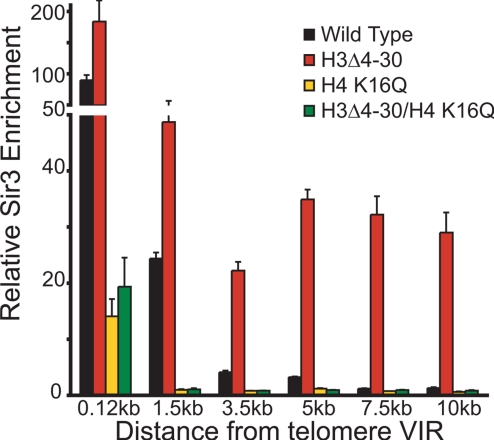
The opposing effects of H4 K16Q and H3Δ4–30 on the spreading of Sir proteins (Fig. 1) makes it unlikely that both the H3 and H4 N termini are involved in recruiting Sir proteins to heterochromatin. It is therefore possible that the H3 tail acts at a stage subsequent to that of the H4 N terminus after Sir proteins have bound and de-acetylated H4 K16. If that is the case, then one might expect that H4 K16Q effects on Sir3 spreading at a telomere would be epistatic to the effects of H3Δ4–30. As shown in Fig. 2 the H3Δ4–30/H4 K16Q mutant has an effect similar to that of H4 K16Q alone on Sir3 spreading. Our data are consistent with a role for the H3 N terminus in Sir complex formation downstream of H4 K16 in Sir3 recruitment and spreading.
Fig. 2.
H3 tail-mediated ectopic spreading is suppressed by amino acid substitution of H4 K16. Sir3 enrichment at telomere VIR as determined by ChIP in mutant strains containing either single mutations in H3 and H4 or the double mutants of the H3 tail deletion in combination with H4 K16Q. Values are averages of at least 3 independent experiments. Error bars represent the SEM.
H3 N-Terminal Tail Is Involved in Silencing HMR Independent of Sir Protein Recruitment.
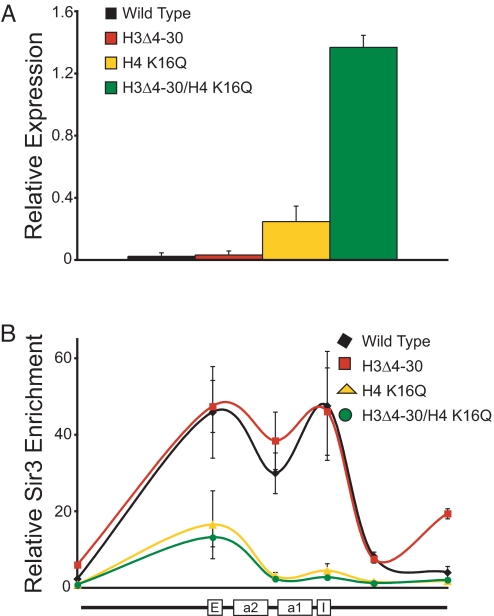
The acetylation site mutants H4 K16G or K16Q disrupt silencing of telomere adjacent regions (18) and HML (22). However, we have observed that in an H4 K16Q mutant there is only a minor decrease in silencing at HMR as measured by quantitative mating assays (18). We confirmed that HMR is efficiently silenced in the H4 K16Q mutant using quantitative RT-PCR (Fig. 3A). We have shown that H4 K16Q mutation leads to a severe silencing defect at HMR only when combined with removal of residues 4–30 from histone H3 (18). We confirmed this result using quantitative RT-PCR (Fig. 3A). Loss of the H3 tail alone has no obvious effect on HMRa1 gene expression. However, in the H3Δ4–30/H4 K16Q double mutant HMRa1 expression increases by >4-fold. Because Sir3 and Sir4 can directly bind the H3 tail in vitro we asked whether the H3 tail is required redundantly with H4 K16 for recruitment of Sir proteins at HMR. Therefore, we examined the binding of Sir3 in both H3Δ4–30 and the double mutant containing H3Δ4–30 and H4 K16Q (Fig. 3B). Loss of the H3 tail alone leads to little change in Sir protein binding across HMR. However, Sir protein binding in the H3/H4 double mutant is indistinguishable from that seen in the H4 K16Q mutant. We conclude that the H3 tail is important for silencing but not for recruitment of Sir proteins at HMR.
Fig. 3.
The H3 and H4 tails cooperate to silence HMR but not to recruit Sir proteins. (A) Expression of HMRa1 in MATα cells carrying the following histone mutants: wild type, H3Δ4–30, H4 K16Q, and the H3/H4 double mutant. Relative level of HMRa1 mRNA was calculated by normalizing to the SCR1 gene, which does not change in the H3Δ4–30 mutant (23). (B) ChIP of Sir3 enrichment across HMR in wild type (black diamond), H3Δ4–30 (red square), H4 K16Q (yellow triangle) and the H3/H4 (green circle) double mutant. Position and orientation of the HMR locus are shown under the graph. Values are averages of 3 independent ChIP experiments. Error bars represent the SEM.
H3 Tail Is Important for Restricting Access to Silent Chromatin at the Telomere and HMR.
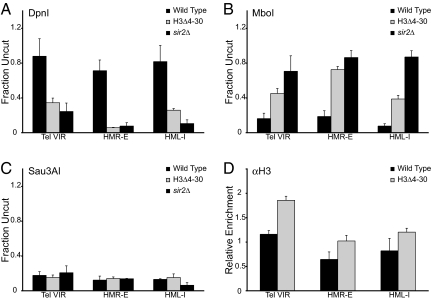
Because the H3 tail does not appear to be necessary for recruitment of Sir proteins, we wished to probe how it exerts its effects on silencing. The epistasis data above suggest that the H3 tail may function after Sir proteins have bound and spread along the chromatin to promote the formation of a specialized chromatin structure important for silencing. To test this possibility we sought to determine whether deletion of the H3 tail might allow increased access to other factors using an in vivo bacterial dam DNA methylase accessibility assay (26). We harvested genomic DNA from either wild type or H3Δ4–30 yeast containing an integrated dam methylase gene and subjected it to DpnI, MboI, and Sau3AI digestion, which recognize methylated (DpnI), unmethylated (MboI), or both methylated and unmethlated (Sau3AI) GATC sites. We then measured the degree of digestion at the heterochromatic domains of telomere VIR, HMR-E and HML-I using quantitative real-time PCR with primers that cross the digest site. It has been shown that loss of Sir proteins leads to a dramatic change in dam methylase accessibility at subtelomeric heterochromatin (26). As shown in Fig. 4A, >70% of the wild type DNA at all 3 loci is inaccessible to the dam methylase and fails to be cut by DpnI. In contrast, only 10%–30% of the DNA in the H3Δ4–30 mutant strain remains inaccessible at telomere VIR, HMR-E and HML-I. The degree of increased accessibility is comparable with that seen in a sir2Δ deletion in which spreading and silencing are abolished (Fig. 4A). In contrast, these same samples were more efficiently cut by MboI (Fig. 4B) as compared with wild type, and in all 3 strains we observed cleavage of >90% by Sau3AI (Fig. 4C). It is unlikely that these effects are due to a decrease in nucleosome density as H3 levels increase slightly at all 3 loci in H3Δ4–30 as measured by ChIP with an H3 C-terminal antibody (Fig. 4D). Our data suggest that, although the H3 tail is not required for Sir protein recruitment it interacts with Sir proteins that have spread to form a heterochromatic structure inaccessible to complexes of the transcription apparatus and dam methylase.
Fig. 4.
Deletion of the H3 tail leads to increased accessibility of an ectopic dam methylase within silent chromatin. (A–C) Yeast genomic DNA was isolated from wild type, H3Δ4–30, and sir2Δ cells expressing the E. coli dam methylase, and digested with DpnI (A), MboI (B), or Sau3AI (C). Percentage digestion was measured by quantitative real-time PCR with primers that cross cut sites at either HMR-E, telomere VIR (≈1.5 kb from the telomere), or HML-I, and normalized to an uncut DNA fragment at the ACT1 locus. (D) ChIP of H3 using a C-terminal specific antibody in wild type and H3Δ4–30 cells at HMR, telomere VIR, and HML using the same primer sets as in A. H3 ChIP data are normalized to an intergenic region on chromosome I and to input. Values are averages of 3 independent experiments. Error bars represent the SEM.
H3 Tail Is Important for Maintaining the Higher Order Structure of Silent Chromatin.
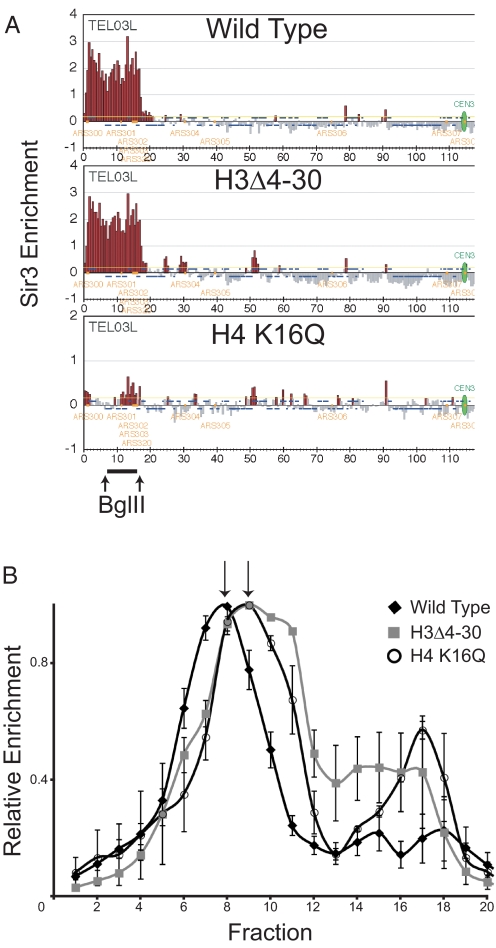
Increased accessibility of heterochromatin in the H3Δ4–30 mutant to the dam methylase could be caused by sliding of proteins normally bound over the GATC site or by a change in chromatin higher order structure that normally excludes macromolecules. To distinguish between these two possibilities we have assayed the hydrodynamic properties of silent chromatin in wild-type and mutant cells. The structural differences between active, repressed, and heterochromatic loci have been analyzed by sucrose gradient sedimentation in higher eukaryotes (27, 28). In these experiments, defined chromatin segments are liberated from nuclei by digestion with endonucleases and the resultant soluble chromatin is then analyzed by sucrose gradient ultracentrifugation (29). Mobility in the sucrose gradient is determined by the mass and shape of a particle; chromatin that is less condensed sediments more slowly than a compact chromatin fragment with a similar mass. We have adapted this technique to study yeast silent chromatin. We chose to examine HML because it is derepressed (18), shows relatively little change in Sir3 enrichment (Fig. 5A), and undergoes increased dam accessibility when the H3 tail is deleted. Importantly, it exists in a relatively large block of contiguous silent chromatin that is flanked by a common restriction endonuclease site. Yeast nuclei from wild-type and mutant strains were prepared and digested with BglII enzyme to release an ≈9 kb fragment from the HML locus. This soluble chromatin was then separated over a 20–40% sucrose gradient and the relative position of the HML fragment was followed by quantitative real-time PCR. As shown in Fig. 5B the HML fragment from H3Δ4–30 cells migrates more slowly in the sucrose gradient than does the same fragment isolated from wild-type cells. Chromatin isolated from H4 K16Q cells, in which Sir protein binding and spreading is disrupted, shows a similar shift upwards in the gradient. We conclude that deletion of the H3 tail or mutation of H4 K16 leads to a similar change in the higher order structure of chromatin that includes the silent mating type locus HML.
Fig. 5.
Silent chromatin shows reduced mobility in a sucrose gradient when the H3 tail is deleted. (A) ChIP-on-chip of Sir3 at chromosome IIIL in wild type, H3Δ4–30, and H4 K16Q strains to demonstrate the change in Sir3 binding at HML in these mutants. Data are displayed as in Fig. 1A. The bar represents HML fragment liberated by digestion with BglII enzyme. (B) Quantitative real-time PCR analysis of fractions from 20–40% sucrose gradient containing digested chromatin isolated from wild type (diamond), H3 tail deletion (square), and H4 K16Q (open circle) strains. Fractions were collected from the bottom. The peak value for each experiment was set to 1. Arrows indicate peak fractions. Error bars represent the SEM.
Discussion
H3 Tail Is Not Required for Recruitment of Sir Proteins.
Sir proteins are recruited and spread by virtue of their direct interaction with the H4 N-terminal residues between amino acids 16–29. In the absence of these residues or even in a mutant containing nonconservative changes in K16 (e.g., K16Q, K16G) Sir proteins are no longer efficiently recruited and so cannot spread along telomeric heterochromatin.
However, Sir3 and Sir4 also interact directly with the N terminus of H3 (7, 9). Surprisingly, our data show that the H3 tail is not only unnecessary for recruitment of Sir proteins, but that removal of much of the H3 tail (residues 4–30) or substitution of all 5 of the H3 N-terminal acetylatable lysines with nonconservative glycine residues, results in an average increase in Sir protein association with heterochromatin and with adjacent euchromatin. However, this mutant heterochromatin, although enriched for Sir proteins, is defective in silencing, shows increased access to protein probes such as dam methylase and has sedimentation properties similar to that of H4 K16Q mutant heterochromatin that is almost completely devoid of Sir proteins. Therefore, we propose that the H3 N terminus functions after Sir proteins have been recruited, to enable a higher order heterochromatin structure that promotes silencing and which is prevented from spreading into adjacent euchromatin by H3 N terminus-Sir protein interactions.
Role of the H3 N Terminus in Silencing Telomeric and HM Heterochromatin.
Heterochromatin containing Sir proteins that spread as a result of H4 K16 deacetylation (sas2Δ) or conservative substitution (K16R) is able to effectively silence adjacent genes (13, 14). However, the subtelomeric Sir protein complex formed in the presence of the H3 tail lesions, although apparently plentiful, is defective in silencing. This is most evident when examining expression of a reporter inserted in the truncated and sensitized telomere VIIL (18) but is also seen more subtly genome wide at genes in natural telomeres in the same H3 N-terminal deletion strains (23, 30). Similarly, the effects of the H3 lesions on HMR silencing are only apparent, as measured by quantitative mating assay, when examined in a genetic background containing the nonconservative mutation H4 K16Q that by itself has only a weak effect on silencing HMR (18). HML silencing is partially defective in the H3 N-terminal deletion strain but is enhanced by deletion of SIR1, which shows no effect on silencing when mutated on its own (18). The varying effects on silencing at the different loci likely reflect their intrinsic silencing strength (2, 20). Therefore, although deletion of the H3 tail produces varying defects in silencing at telomeres, HML and HMR, it clearly plays an important role in silencing at all 3 heterochromatic loci.
In the H3 tail deletion strain, loss of silencing at both HM loci and the telomere occurs despite the fact that our data show that there is either no change or an increase in Sir3 binding when compared with the silencing competent state. There is also no increase in methylation of H3 K79 (31) or in histone acetylation of H4, H2A and H2B sites at telomere VIR in the H3Δ4–30 mutant. This suggests that Sir occupancy, K79 hypomethylation and hypoacetylation of heterochromatin, although necessary, are not sufficient to allow transcriptional silencing, and is consistent with other published reports of Sir protein binding and spreading in the absence of efficient silencing (17, 32). Our data indicate that there must be an additional key step required for the formation of silent chromatin that occurs after Sir3 has spread but before it is capable of silencing.
Higher-Order Structure of Silent Chromatin.
How might the H3 tail promote silencing when it is not required for Sir recruitment or spreading? At both the telomere and HM loci, removal of the H3 tail leads to an increase in accessibility of both an ectopic bacterial dam methylase and active RNA polymerase complex suggesting that in the absence of the H3 tail silent chromatin exists in a more open chromatin conformation. In addition, when either the H3 tail is deleted or H4 K16 is replaced with glutamine, chromatin fragments that include HML show reduced mobility in sucrose gradients when compared with wild type, consistent with an increase in the frictional coefficient due to a less compact higher order structure. In addition, the mutant histone complexes contain some chromatin that sediments more slowly than the peak fractions, which may result from increased accessibility of more open chromatin to endogenous nucleases. A trivial explanation for this result is that deletion of the H3 tail leads to destabilization of the silencing complex and Sir protein dissociation from the chromatin during sedimentation. However, we observe no loss of Sir3 and core histones in vivo, as assayed by ChIP when the H3 tail is deleted, despite increased in vivo dam methylase accessibility. Therefore, our in vivo and in vitro results are consistent with the H3 tail being important for higher order heterochromatin structure after Sir proteins have bound to and spread along H4 N termini deacetylated at residue K16.
The H3 tail could be involved in the formation of at least 3 distinct higher order structures within heterochromatin: The Sir-mediated fold-back of the telomere (10, 11) and HMR (12), the clustering of silent loci at the nuclear periphery (15), and compaction of the chromatin fiber itself. The fold-back is capable of bringing distant elements into close proximity, and is thought to be responsible for the observation of Rap1 binding at sites internal to the silencer (10, 12). However, we see an increase in internal Rap1 enrichment in the H3Δ4–30 mutant strain as compared with wild type, mirroring the increase in Sir protein spreading, which suggests that the telomeres remain folded (Fig. S4). Although perinuclear anchoring of telomeres has been reported to be at least partially disrupted when the H3 tail is deleted (7), we do not favor the idea that the silencing and structural phenotypes we observe are a consequence of reduced telomeric anchoring. Mutations that decrease perinuclear tethering tend to display decreased Sir3 enrichment at telomeric loci (4), because this nuclear organization is believed to be important for maintaining a high local concentration of silencing factors. In addition, our hydrodynamic mobility results cannot be accounted for by a loss of telomeric anchoring as the chromatin fragments have been purified away from the nucleus and its associated matrix. Instead, our data are most consistent with the H3 tail promoting silencing by helping the chromatin fiber form a higher order structure, possibly compaction. There is considerable controversy as to exactly which step in transcriptional activation is prevented by silent chromatin (33, 34). However, our results do not directly address this as higher order chromatin structure could inhibit transcription by either excluding large complexes such as the polymerase holoenzyme altogether or by restricting the transition to elongation.
In conclusion, although initiation, spreading, and the barrier to spreading are all key events in the formation of heterochromatin in yeast, it is becoming clear that transcriptional silencing, itself, is a discrete event that occurs once Sir proteins have spread. Our data argue that the H3 N terminus mediates the chromatin structure that promotes this latter silencing function of heterochromatin.
Methods
Yeast Strains and Plasmids.
Lists of yeast strains and plasmids used in this study are provided in Tables S1 and S2. Gene disruption and tagging were performed using standard techniques. Primer sequences for these experiments and those described below are available upon request.
ChIP.
ChIP was performed essentially as described in ref. 24. Antibodies against individual histone acetylation sites are described in ref. 35. Antibodies against Rap1 no. 477, Sir2 no. 480, Sir3 no. 347, and Sir4 no. 482 were previously generated in house and used at 1, 2, 1, and 5 μL per 50 μL of lysate, respectively. H3 ab1791 (Abcam) was used at 1 μL per 100 μL of lysate in a final volume of 900 μL. ChIP DNA was assayed by quantitative PCR and is expressed as fold enrichment over an internal control (ACT1 or an intergenic region on chromosome I) and is normalized to input.
Genome-Wide Binding Microarrays.
Sample amplification of ChIP DNA was performed essentially as described in ref. 36 except that each reaction was done in duplicate and the final product was combined in a final Qiaquick PCR purification (Qiagen) step to produce sufficient DNA concentration. Five micrograms sample DNA was fragmented, labeled, and hybridized to Affymetrix 1.0R tiling arrays at the UCLA DNA Microarray Core facility according to the Affymetrix 500K protocol. Data were normalized to input DNA (a total of 13 samples was used) using Affymetrix Tiling Analysis Software (TAS) with the following parameters: quantile normalization, PM only, Bandwidth = 50. Enrichment is represented as the log2 fold change between the treatment and control signals.
RNA Extraction and RT-PCR.
RNA was extracted using the hot phenol method (17). RT reaction was used for subsequent analysis by quantitative real-time PCR with IQ SYBR Green Supermix (Bio-Rad) according to manufacturer's instructions on an Applied Biosystems 7500 device.
dam DNA Methylase Accessibility Assay.
Appropriate yeast strains expressing the E. coli dam methylase were grown to stationary phase and genomic DNA was purified using a Genomic DNA purification kit (Qiagen). Samples were subjected to DpnI, MboI or Sau3AI digestion overnight at 37 °C and analyzed by quantitative real-time PCR. Primers were designed to cross restriction sites at HMR, HML or telomere VIR. Results were normalized to DNA content by comparing to a primer set amplifying an uncut sequence at the ACT1 gene.
Sucrose Gradient Fractionation of Chromatin.
Yeast nuclei were prepared from 2 L of cells grown to an OD600 of ≈2. Cells were spheroplasted in 50 mL of YPD/1.2 M Sorbitol by adding 1 mL of 1 M DTT and 10 mg of Zymolyase 100-T and incubating at 30 °C for 40–60 min. Spheroplasts were washed in YPD/1.2 M Sorbitol and then 1.2 M Sorbitol before being resuspended in 50 mL of cold Buffer N (25 mM K2SO4, 30 mM Hepes pH 7.6, 5 mM MgSO4, 1 mM EDTA, 10% Glycerol, 0.5% Nonidet P-40, 7.2 mM Spermidine, 3 mM DTT, protease inhibitors). Spheroplasts were lysed by passage twice through a Yamato homogenizer at 500 rpm in the cold. Cellular debris were removed (2,000 rpm for 10 min) and nuclei were collected (6,000 rpm for 20 min) by centrifugation at 4 °C in a JA-20 rotor (Beckman). Nuclei were resuspended in 1.5 mL of Buffer N and the DNA concentration was measured on a Nanodrop spec by diluting 5 μL of nuclei into 200 μL of 2 M NaCl/5 M Urea. Nuclei digests with 2,500U BglII enzyme were performed with ≈80–100 μg of nuclei as described in ref. 29. Twenty to forty percent sucrose gradients were poured, and 200 μL of sample was overlayed on the gradient. Gradients were centrifuged at 36,000 rpm for 6.5 h at 4 °C in an SW40Ti rotor (Beckman). The 500-μL fractions were collected from the bottom by needle puncture. The relative position of the HML fragment was followed by quantitative real-time PCR.
Supplementary Material
Acknowledgments.
We thank members of the Grunstein laboratory for helpful comments and discussion during the course of this project and in particular Catherine Millar for plasmids and Graham Davies, Wei Xie, and Tasuku Kitada for computational assistance; John Wyrick (Washington State University, Pullman, WA) for providing the pYJ031 plasmid; Rodolfo Ghirlando and Gary Felsenfeld (National Institutes of Health, Bethesda, MD) for assistance with the sucrose sedimentation experiments; and the University of California, Los Angeles microarray core facility for array services. This work was supported by grants from the National Institutes of Health Medical Scientist Training Program and a Jonsson Comprehensive Cancer Center Foundation Fellowship (to A.S.S.) and National Institutes of Health Grant GM42421 (to M.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expresion Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE16742).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906866106/DCSupplemental.
References
- 1.Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 2.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 3.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 4.Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoppe GJ, et al. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 7.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 8.Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 9.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 10.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin D, Zaman Z, Liberatore RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- 12.Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT. Long-range communication between the silencers of HMR. Mol Cell Biol. 2008;28:1924–1935. doi: 10.1128/MCB.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 14.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 15.Gotta M, et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 17.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 19.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 20.van Leeuwen F, Gottschling DE. Assays for gene silencing in yeast. Methods Enzymol. 2002;350:165–186. doi: 10.1016/s0076-6879(02)50962-9. [DOI] [PubMed] [Google Scholar]
- 21.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 22.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin AM, Pouchnik DJ, Walker JL, Wyrick JJ. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics. 2004;167:1123–1132. doi: 10.1534/genetics.104.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 25.Renauld H, et al. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 26.Gottschling DE. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert N, Allan J. Distinctive higher-order chromatin structure at mammalian centromeres. Proc Natl Acad Sci USA. 2001;98:11949–11954. doi: 10.1073/pnas.211322798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher EA, Felsenfeld G. Comparison of the folding of beta-globin and ovalbumin gene containing chromatin isolated from chicken oviduct and erythrocytes. Biochemistry. 1986;25:8010–8016. doi: 10.1021/bi00372a033. [DOI] [PubMed] [Google Scholar]
- 29.Ghirlando R, Litt MD, Prioleau MN, Recillas-Targa F, Felsenfeld G. Physical properties of a genomic condensed chromatin fragment. J Mol Biol. 2004;336:597–605. doi: 10.1016/j.jmb.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 30.Jin Y, Rodriguez AM, Wyrick JJ. Genetic and genomewide analysis of simultaneous mutations in acetylated and methylated lysine residues in histone H3 in Saccharomyces cerevisiae. Genetics. 2009;181:461–472. doi: 10.1534/genetics.108.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altaf M, et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchmaier AL, Rine J. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:852–862. doi: 10.1128/MCB.26.3.852-862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Widom J. Mechanism of transcriptional silencing in yeast. Cell. 2005;120:37–48. doi: 10.1016/j.cell.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Gao L, Gross DS. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol Cell Biol. 2008;28:3979–3994. doi: 10.1128/MCB.00019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 36.Robyr D, Grunstein M. Genomewide histone acetylation microarrays. Methods. 2003;31:83–89. doi: 10.1016/s1046-2023(03)00091-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.