Abstract
Mammalian calreticulin (CRT) is a multifunctional Ca2+-binding protein involved in more than 40 cellular processes in various subcellular compartments, such as Ca2+ storage and protein folding in the endoplasmic reticulum (ER). CRT homologues were discovered in plants almost 15 years ago, and recent studies revealed that many plant species contain 2 or more CRTs that are members of 2 distinct families, the CRT1/2 family and the plant-specific CRT3 family. However, little is known about their physiological functions. Here we report ebs2 (EMS-mutagenized bri1 suppressor 2) as an allele-specific suppressor of bri1–9, a dwarf Arabidopsis mutant caused by retention of a defective brassinosteroid receptor in the ER. EBS2 encodes the Arabidopsis CRT3 that interacts with ER-localized bri1–9 in a glycan-dependent manner. Loss-of-function ebs2 mutations compromise ER retention of bri1–9 and suppress its dwarfism, whereas EBS2 over-expression enhances its dwarf phenotype. In contrast, mutations of 2 other CRTs or their membrane-localized homologues calnexins had little effect on bri1–9. A domain-swapping experiment revealed that the positively charged C-terminal tail of CRT3 is crucial for its “bri1–9-retainer” function. Our study revealed not only a functional role for a plant-specific CRT, but also functional diversity among the 3 Arabidopsis CRT paralogues.
Keywords: BRI1, calnexin, ER chaperone, ER quality control, UGGT
Calreticulin (CRT) is a highly conserved Ca2+-binding protein in eukaryotes that consists of 3 distinct domains: a globular N-domain, a proline-rich middle (P) domain, and a highly acidic C-terminal domain with a (K/H)DEL endoplasmic reticulum (ER) retrieval signal (1). Mammalian CRT has been implicated in more than 40 cellular processes in different subcellular compartments, including Ca2+ homeostasis and protein folding in the ER (1). CRT and its membrane-bound homologue calnexin (CNX) also contain a unique lectin site in their globular domains that specifically binds a mono-glucosylated glycan on newly synthesized glycoproteins, thus establishing the so-called CRT/CNX cycle. This system also includes glucosidase II and UDP-glucose:glycoprotein glucosyltransferase (UGGT) that catalyze removal and addition of the terminal glucose residue of Asn-linked glycans, respectively, to facilitate folding and quality control of glycoproteins in the ER (2). Deletion of CRT in the mouse leads to embryonic lethality along with abnormal cardiac development as a result of impaired Ca2+ homeostasis (3, 4).
Many plant species contain 2 or more CRTs that are members of 2 distinct CRT families: CRT1/2 and CRT3, which were thought to be duplicated before the splitting of monocots and dicots (5). Despite the fact that plant CRTs were discovered 15 years ago (6), their physiological functions remain largely unknown. Experiments using heterologous tissue cultures or CRT-over-expression transgenic plants implicated plant CRTs in multiple physiological processes such as defense against virus (7), plasmodesmata-mediated cellular transport (8, 9), tissue regeneration (10, 11), ER Ca2+ buffering (12, 13), and stress tolerance (14, 15). A recent study using a T-DNA insertional mutant suggested that the Arabidopsis CRT1, which can substitute the mammalian CRT function in cell culture, plays a role in ER stress response (16).
An Arabidopsis dwarf mutant, bri1–9, is an excellent genetic system in which to study plant ER quality control (17). BRI1 is a leucine-rich repeat receptor-like kinase that functions as a cell surface receptor for the plant steroid hormone brassinosteroids (BRs) (18). We have recently discovered that bri1–9, which carries a S662F mutation in its ligand-binding motif, is retained in the ER by an over-vigilant ER quality control system in Arabidopsis (17). Loss-of-function mutations in the Arabidopsis EBS1 gene, which encodes a homologue of the mammalian UGGT thought to function as a protein folding sensor (19), significantly compromise this quality control system and suppress the dwarf phenotype of bri1–9.
To identify additional components involved in the ER quality control of the mutated BR receptor, we studied ebs2 (EMS-mutagenized bri1 suppressor 2), which also suppresses the bri1–9 mutation in an allele-specific manner. By using the map-based cloning approach, we discovered that EBS2 encodes the Arabidopsis CRT3. Our biochemical experiments showed that CRT3 interacts with bri1–9 in a mono-glucosylation-dependent manner to retain the mutated BR receptor in the ER. Loss of CRT3 function allows substantial amount of mis-folded bri1–9 to move to plasma membrane to initiate BR signaling despite the presence of 2 other CRT paralogues and their membrane-localized homologues, CNXs, which were known to interact with bri1–9 in vivo. We therefore conclude that CRT3 functions as a key retention factor to keep bri1–9 in the folding compartment.
Results
The ebs2 Mutation Partially Suppresses the Dwarf Phenotype of bri1–9.
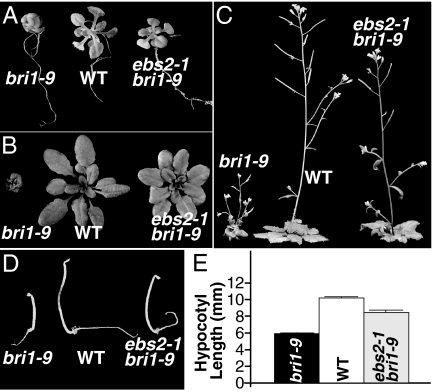
The same genetic screening that led to identification of EBS1 uncovered the second bri1–9 suppressor, ebs2. Similar to ebs1, ebs2 partially suppresses multiple defects of bri1–9 phenotypes throughout its life cycle. Compared with bri1–9, ebs2–1 bri1–9 has elongated petioles and expanded rosette leaves at both seedling (Fig. 1A) and rosette (Fig. 1B) stages. At maturity, ebs2–1 bri1–9 has an intermediate height compared with bri1–9 and WT (Fig. 1C). In addition, ebs2–1 partially rescues the short hypocotyl phenotype of bri1–9 in the dark (Fig. 1 D and E).
Fig. 1.
The ebs2 mutation partially suppresses the bri1–9 mutant. Images of 7-d-old bri1–9, WT, and ebs2–1 bri1–9 grown on half-strength Murashige and Skoog medium(A); 5-week-old soil-grown plants of bri1–9, WT, and ebs2–1 bri1–9 (B); 7-week-old mature plants of bri1–9, WT, and ebs2–1 bri1–9 grown in soil (C); and 4-d-old dark-grown seedlings of bri1–9, WT, and ebs2–1 bri1–9 (D). (E) Average hypocotyl length of 4-d-old etiolated seedlings of bri1–9, WT, and ebs2–1 bri1–9. Each bar represents the average of more than 40 seedlings of duplicated experiments. Error bar denotes SE.
ebs2 Allele-Specifically Suppresses the bri1–9 Mutation.
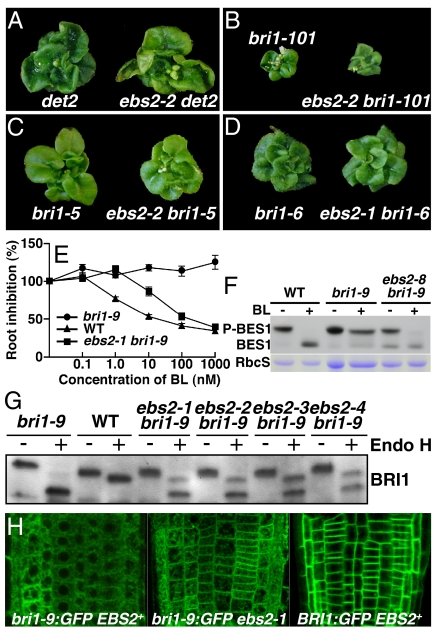
To investigate the underlying mechanism by which ebs2 suppresses the bri1–9 phenotype, we studied the genetic interaction between ebs2 and a BR-deficient mutant det2 (20). As shown in Fig. 2A, ebs2–2 det2 is morphologically similar to det2, suggesting that ebs2 does not lead to constitutive activation of BR signaling. We also crossed ebs2 with 3 bri1 mutants: bri1–101, bri1–5, and bri1–6. The bri1–101 contains a missense mutation in the kinase domain of BRI1 whereas bri1–5 and bri1–6 mutants contain mutations in their extracellular domain (21). It should be noted that bri1–5 is also retained in the ER by at least 3 retention mechanisms (22). As shown in Fig. 2 B-D, none of these bri1 mutations was suppressed by ebs2, indicating that ebs2 is an allele-specific suppressor of the bri1–9 mutation. We suspected that ebs2 might directly act on bri1–9 to restore its BR receptor function. Indeed, a BR-induced root inhibition (23) and BES1-dephosphorylation assays revealed that ebs2 partially restores BR sensitivity to bri1–9. As shown in Fig. 2E, treatment of bri1–9 with increasing concentrations of brassinolide (BL), the most active BR, had little effect on root growth, whereas similar treatments clearly inhibited root growth of ebs2–1 bri1–9 and WT. Fig. 2F revealed that 1 h BL treatment resulted in complete and near-complete dephosphorylation of BES1, a robust biochemical marker for BR signaling (24), in WT and ebs2–8 bri1–9, respectively, whereas significant amounts of phosphorylated BES1 were still present in bri1–9.
Fig. 2.
EBS2 functions in quality control of bri1–9. Images of 6-week-old soil-grown det2 and ebs2–2 det2 mutants (A); 4-week-old soil-grown plants of bri1–101 and ebs2–2 bri1–101 (B); 5-week-old soil-grown bri1–5 and ebs2–2 bri1–5 mutants (C); and 5-week-old soil grown bri1–6 and ebs2–1 bri1–6 mutants (D). (E) Quantitative analysis of BR sensitivity. Root lengths of 7-d-old seedlings grown on BL-containing medium were measured and presented as the relative value of the average root length of BL-treated seedlings to that of mock-treated seedlings of the same genotype. Each data point represents the average of approximately 30 seedlings of duplicated experiments. Error bar denotes SE. (F) BR treatment leads to BES1 dephosphorylation: 2-week-old seedlings were treated with 1 μM BL for 1 h. Total proteins were extracted in 2× SDS buffer, separated by 10% SDS/PAGE, and analyzed by immunoblotting using anti-BES1 antibody (24). Coomassie blue staining of RbcS served as a loading control. (G) Endo H sensitivity of BRI1 and bri1–9. Equal amounts of proteins of 4-week-old leaves from different samples were treated with 1 μL of Endo Hf for 1 h at 37°, followed by Western blot analysis with anti-BRI1 antibody. (H) Confocal examination of subcellular localization of bri1–9:GFP and BRI1:GFP in root tips of 6-d-old light-grown seedlings.
ebs2 Mutations Compromise the ER Retention of bri1–9.
These genetic and physiological behaviors of ebs2 were strikingly similar to those of ebs1 (17), implying that EBS2 might also be involved in the quality control of bri1–9. To directly test this possibility, we examined endoglycosidase H (Endo H) sensitivity of bri1–9 protein from ebs2–1 bri1–9 and 3 other allelic ebs2 bri1–9 mutants. Endo H removes high-mannose-type glycans of ER-localized glycoproteins but cannot cleave Golgi-processed glycans (25), thus providing a convenient way to examine the subcellular distribution of bri1–9. As shown in Fig. 2G, bri1–9 is sensitive whereas BRI1 is largely resistant to Endo H. Consistent with our hypothesis, a significant pool of Endo H-digested bri1–9 exhibits the same mobility as the Endo-H-processed BRI1 on Western blot filter, suggesting that ebs2 mutations reduce the stringency of quality control of bri1–9. Indeed, confocal microscopy analysis of bri1–9:GFP revealed the presence of bri1–9:GFP at the cell surface in the ebs2–1 mutant (Fig. 2H). We thus concluded that EBS2 plays a key role in retaining bri1–9 in the ER.
EBS2 Encodes Arabidopsis CRT3.
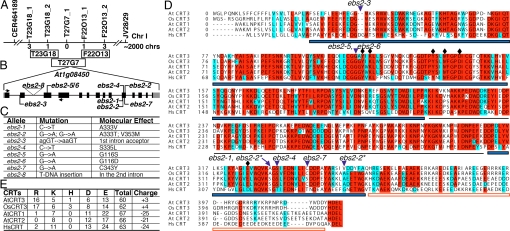
To understand the biochemical role of EBS2 in ER retention of bri1–9, we cloned the EBS2 gene by chromosomal walking. PCR-based genetic mapping delimited the EBS2 locus to a 150-kb region on the top of chromosome I (Fig. 3A). This region contains 3 annotated genes encoding putative ER chaperones, including CRT3 (At1g08450), BiP3 (At1g09080), and CRT2 (At1g09210). Sequencing of these genes from ebs2–1 bri1–9 identified a single-bp substitution in At1g08450, which changes Ala-333 to Val in CRT3 (Fig. 3 B and C). The identity of At1g08450 as EBS2 gene was confirmed by sequencing 6 other potential ebs2 alleles isolated from the same genetic screen, each containing a single nucleotide change in At1g08450 except ebs2–2, which carries 2 mutations, A333T and V353M (Fig. 3 B and C). In addition, a double mutant between bri1–9 and a null T-DNA insertional mutation of At1g08450, named as ebs2–8, exhibited a suppressed-bri1–9 phenotype that can be complemented by expression of a CRT3 transgene containing its native promoter [supporting information (SI) Fig. S1].
Fig. 3.
Molecular cloning of EBS2. (A) EBS2 was mapped to a 150-kb genomic region between markers T23G18_2 and F22O13_1 on the top of chromosome I. The line represents genomic DNA, and markers and numbers of recombinants are shown above and below the line, respectively. (B) The gene structure of EBS2. EBS2 contains 14 exons (black bar) and 13 introns (line). Gray boxes denote untranslated regions, lines indicate positions of ebs2 mutations, and triangle denotes T-DNA insertion. (C) Nucleotide change and molecular defects of 8 ebs2 alleles. (D) Alignment of CRT sequences from Arabidopsis thaliana (At), Oryza sativa (Os), and Homo sapiens (Hs). Comparison of AtCRT3 (NP_563816), OsCRT3 (BAC06263), AtCRT1 (NP_176030), AtCRT2 (NP_172392), and HsCRT (AAB51176) was performed using the ClustalW program at Network Protein Sequence analysis (http://npsa-pbil.ibcp.fr) (39). Aligned sequences were shaded using the BoxShade web server (http://bioweb.pasteur.fr). Residues identical in more than 4 sequences are shaded in red and similar ones are shaded in cyan. Each CRT contains a signal peptide, a globular domain (marked by thick blue lines), a P domain (indicated by a thin gray line), and a C-terminal domain (marked by a red open line). The positions of ebs2 mutations are indicated by blue triangles. Stars denote 2 substitutions in ebs2–2, and diamonds indicate 6 residues thought to be involved in glycan binding. (E) Number of basic and acidic residues in C-termini [the ER retention signal (H/K)DEL was not counted] of AtCRT3, OsCRT3, AtCRT1/2, and HsCRT. The total number of amino acid residues and the net charge of these C-termini are also listed.
CRT3 Is a Land Plant-Specific ER Lectin with a Unique Expression Pattern.
The Arabidopsis contains 3 CRTs: CRT1, CRT2, and CRT3, which share similar domain organization with the rice CRT3 and the human CRT (Fig. 3D). Sequence comparison of the 5 CRTs revealed that, whereas they exhibit high sequence identity in the globular and P domains critical for chaperone function, the C domain of CRT3 is significantly diverged from those of CRT1 and CRT2. Unlike the C-termini of CRT1/CRT2 that are rich in acidic residues and predicted to be critical for Ca2+ storage (26, 27), the C-domains of 2 CRT3s are positively charged and contain short stretches of basic residues (Fig. 3 D and E). Gene co-expression analysis using ATTED-II (28) revealed that, although CRT1 and CRT2 share a similar expression profile with other ER chaperones, CRT3 is co-expressed with genes involved in plant defense and other unknown processes (Fig. S2), suggesting that the 3 Arabidopsis CRTs are involved in different physiological processes.
BLAST search and phylogenetic analysis (Fig. S3) identified CRT3s from 3 lower plant species, Marchantia polymorpha, Physcomitrella patens, and Selaginella moellendorffii. Marchantia species are members of liverworts closely related to the earliest land plants, Physcomitrella species are mosses thought to be the sister to all land plants except liverworts, and Selaginella species are considered to be the oldest living vascular plants, at approximately 420 million years old (29). Given the fact that all known CRTs from non-land-plant eukaryotes carry acidic C-termini (see http://pfam.sanger.ac.uk/family?acc=PF00262), including Mesostigma viride (Fig. S4), a scaly green flagellate thought to be the common ancestor of all land plants plus charophycean algae (30), our analyses suggested that the members of the CRT1/CRT2 family are more closely related to the ancestral plant CRT than the CRT3 members and that the CRT1/2-CRT3 duplication likely occurred before or during the rise of the first land plant.
CRT3 Interacts with bri1–9.
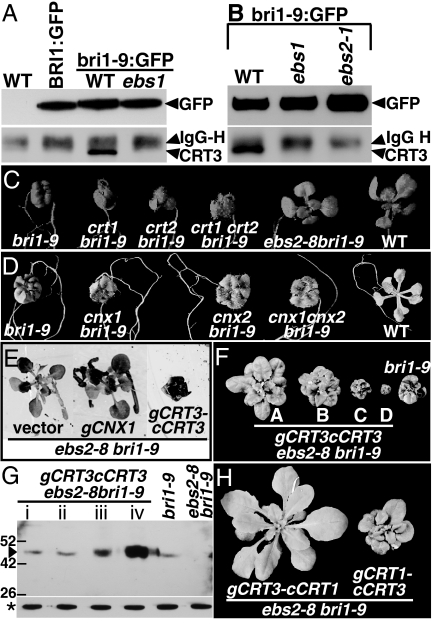
The facts that mutations in CRT3 rescue the bri1–9 phenotype and permit export of a significant amount of bri1–9 proteins out of the ER suggest that CRT3 is a major retention factor that keeps bri1–9 in the ER. However, our previous study showed that bri1–9 interacts only with CNXs but not with any of the CRTs (17) when analyzed with an anti-maize CRT antibody thought to detect all Arabidopsis CRTs and CNXs (5). We suspected that this antibody failed to detect the Arabidopsis CRT3. Indeed, Western blot analysis of T-DNA insertional mutants of 2 CNXs and 3 CRTs revealed that none of the detected CNX/CRT bands corresponds to CRT3 (Fig. S5). We therefore generated a CRT3-specific antibody (Fig. S6) to test CRT3-bri1–9 interaction with anti-GFP immunoprecipitates from transgenic plants expressing BRI1:GFP or bri1–9:GFP. As shown in Fig. 4A, a band corresponding to CRT3 was detected from only bri1–9:GFP transgenic lines, not from BRI1:GFP transgenic plants or WT control, indicating a CRT3-bri1–9 interaction in vivo. Such a CRT3-bri1–9 interaction likely depends on mono-glucosylation of bri1–9, as bri1–9:GFP failed to interact with CRT3 when the bri1–9:GFP transgene was crossed into ebs1 lacking a functional UGGT that is responsible for re-glucosylating glycans on bri1–9 (17) (Fig. 4A).
Fig. 4.
CRT3 is the major lectin that retains bri1–9 in the ER. (A) CRT3 interacts with bri1–9 in an UGGT-dependent manner. (B) The CRT3-bri1–9 interaction is abolished in ebs2–1. For A and B, anti-GFP immunoprecipitates from 18-d-old whole seedlings of WT, or transgenic lines were separated by 10% SDS/PAGE and analyzed with anti-GFP and anti-CRT3 antibodies. IgG heavy-chain interference is indicated. (C) Shown here are 18-d-old seedlings of bri1–9, crt1 bri1–9, crt2 bri1–9, crt1 crt2 bri1–9, ebs2–8 bri1–9, and WT grown on half-strength Murashige and Skoog medium. (D) Shown here are 18-d-old seedlings of bri1–9, cnx1 bri1–9, cnx2 bri1–9, cnx1 cnx2 bri1–9, and WT grown on half-strength Murashige and Skoog medium. (E) Three-week-old transgenic ebs2–8 bri1–9 seedlings containing pPZP222 vector, a gCNX1 transgene, or a gCRT3-cCRT3 transgene. (F) Shown here are 6-week-old soil-grown bri1–9 and 4 gCRT3-cCRT3 ebs2–8 bri1–9 transgenic plants. (G) Analysis of CRT3 protein abundance in gCRT3-cCRT3 ebs2–8 bri1–9 transgenic lines shown in F. Protein extracts of 3-week-old transgenic plants were separated by 10% SDS/PAGE and analyzed by immunoblotting with an anti-CRT3 antibody. After stripping off bound IgGs, the filter was re-probed with anti-BiP antibody. The numbers indicate molecular weight (in kDa), triangle represents CRT3, and star denotes BiP for loading control. (H) The C terminus is crucial for the “retainer” function of CRT3. Shown here are 6-week-old soil-grown transgenic ebs2–8 bri1–9 mutants expressing the pCRT1:gCRT1N-CRT3C or pCRT3:gCRT3N-CRT1C chimeric transgene.
Consistent with our finding that the suppression phenotype of ebs2–1 bri1–9 is caused by loss of CRT3 function, the CRT3-bri1–9 interaction is abolished in bri1–9:GFP ebs2–1 transgenic plants (Fig. 4B). This result suggests that Ala-333, which is changed to Val in ebs2–1, is important for the lectin function or proper folding of CRT3. Interestingly, the same residue is changed to Thr in ebs2–2 that contains a second-site mutation, Val353Met. It is worth to note that most of our analyzed CRT3 sequences contain Ala at this position (Fig. S7) whereas almost all other CRTs contain a Ser residue instead (see http://pfam.sanger.ac.uk/family?acc=PF00262).
CRT3 Is the Major Lectin That Retains bri1–9 in the ER.
Our discovery that CRT3 interacts with bri1–9 is quite surprising, as our previous study revealed no interaction between bri1–9 and 2 other CRTs (17) that share a highly similar globular domain involved in mono-glucosylated glycan binding (Fig. 3D). To eliminate the possibility that the lack of CRT1/CRT2-bri1–9 interaction might be caused by our assay condition, we crossed T-DNA insertional mutations of crt1 and crt2 into bri1–9. As shown in Fig. 4C, neither single nor double mutations of crt1 and crt2 suppresses the bri1–9 phenotype, indicating that CRT1 and CRT2 play no role in retaining bri1–9.
Our finding that lack of CRT3 alone is sufficient to suppress the bri1–9 phenotype to a similar extent as loss-of-function UGGT mutations is also quite puzzling. We have previously detected a strong interaction between bri1–9 and CNXs, which largely depends on mono-glucosylation of bri1–9 by UGGT (17). To test if observed CNX/bri1–9 interaction plays a role in retaining bri1–9 in the ER, we generated double and triple mutants of bri1–9 with T-DNA insertional mutations of 2 CNXs (22). As shown in Fig. 4D, neither single nor double mutations of the 2 CNXs suppressed the bri1–9 mutant. Consistent with this result, over-expression of CNX1 by its native promoter had little effect on bri1–9 (Fig. 4E). By contrast, over-expression of a CRT3 transgene with its native promoter not only rescued the T-DNA insertional mutation of CRT3 in ebs2–8 bri1–9 but also resulted in a stronger dwarf phenotype when the CRT3 abundance was very high (Fig. 4 F and G). Taken together, our data revealed that, among the 5 CRT/CNX-type ER lectins, CRT3 plays a key role in retaining bri1–9 in the ER.
The C-Terminal Tail of CRT3 Is Critical for Its “bri1–9 Retainer” Function.
Based on our observation that CRT3, but not CRT1/CRT2, retains bri1–9 in the ER, we suspected that the CRT3-C terminus might be important for the bri1–9 retainer function. To directly test this hypothesis, we swapped the C-termini of CRT1 and CRT3 and generated 2 chimeric transgenic constructs, pCRT1:CRT1N-CRT3C and pCRT3:CRT3N-CRT1C, driven by the native promoters of CRT1 and CRT3, respectively. As shown in Fig. 4H, when transformed into ebs2–8 bri1–9, only CRT1N-CRT3C but not CRT3N-CRT1C was able to rescue the ebs2–8 mutation. This result indicates that, although the globular and P domains of CRT1 and CRT3 are interchangeable, the CRT3 C terminus is crucial for retaining bri1–9 in the ER.
Discussion
In this study we demonstrated that the Arabidopsis CRT3, the founding member of the plant-specific CRT family, functions as a key retention factor in keeping a defective BR receptor in the ER. Results from our previous and current studies have shown that bri1–9 interacts with CNXs and CRT3 in a mono-glucosylation-dependent manner (17), yet loss-of-function mutations in CRT3 alone are sufficient to compromise the quality control of a defective BR receptor. A simple explanation for our finding is that CRT3 and CNX both retain bri1–9 in the ER, but CRT3 plays a dominant role. However, over-expression of CNX1 failed to suppress the T-DNA insertional mutation of CRT3 in the ebs2–8 bri1–9 mutant. We thus hypothesize that CNXs and CRT3 are involved in different cellular processes, with the 2 CNXs functioning as molecular chaperones in facilitating bri1–9 folding while CRT3 serves as an obligate retention factor to keep the mis-folded bri1–9 in the ER; and that the retainer function of CRT3 can not be accomplished by the 2 bri1–9-interacting CNXs. A similar case was reported for the folding of influenza virus hemagglutinin in mammalian cells that employ CNX for folding but use CRT for retention (31). The exact functional consequence of CNXs-bri1–9 interaction needs further investigation, and a quadruple mutant of bri1–9 crt3 cnx1 cnx2 might be useful for such an investigation.
The most surprising discovery of our study is the functional diversification of the 3 CRT paralogues in Arabidopsis. Despite sharing a highly similar lectin-binding domain with CRT3, neither CRT1 nor CRT2 interacts with bri1–9 in our co-immunoprecipitation assay. One possible explanation is that CRT1 and CRT2 have different client preference than CRT3. Previous studies on mammalian CRT/CNX seem to support this explanation. It is well known that mammalian CRT and CNX share the same binding specificity toward mono-glucosylated glycan in vitro but have overlapping yet distinct client proteins in vivo (32). Such client specificity is thought to be caused largely by different sub-organelle localizations of the 2 ER lectins, with CNX sitting on the ER membrane and CRT being an ER luminal protein. It was shown that ER membrane-localized CRT exhibits a similar client preference as CNX, whereas a luminal CNX shares a similar pool of client substrates with CRT (33, 34). The positively charged C-terminal tail of CRT3 might be important for a unique sub-organelle localization pattern for CRT3 to facilitate CRT-bri1–9 interaction. Consistent with this hypothesis, a CRT1-CRT3 chimeric lectin is able to complement the ebs2–8 mutation. It is also possible that plants have evolved different CRT isoforms to participate in distinct cellular processes while the animal CRT had successfully evolved into a truly multifunctional protein. Members of the CRT1/2 family might be important for ER Ca2+ homeostasis as a result of their acidic C-terminal tails containing low-affinity/high-capacity Ca2+-binding sites whereas CRT3 is mainly responsible for retaining mis-folded glycoproteins in the folding compartment, especially when plants are under various stress conditions in a terrestrial habitat. Consistent with this hypothesis, plant CRTs were known to affect ER Ca2+ levels both in vitro and in vivo and that the Arabidopsis CRT1 was recently shown to rescue Ca2+ deficiency of the mouse CRT-depleted embryonic fibroblasts (16). In addition, gene co-expression analysis revealed that, whereas CRT1/2 are co-expressed with many other ER chaperones, CRT3 is clustered with genes involved in stress resistance (Fig. S2). Further investigation using T-DNA insertional mutants of these ER-localized lectins could lead to a better understanding of physiological functions of plant CRT paralogues.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis ecotype Columbia-0 (Col-0) is the parental line for mutants and transgenic plants with the following exceptions: bri1–9 [Wassilewskija (Ws-2)] used for cloning EBS2 and bri1–5 (Ws-2) and bri1–6 (Enkheim) used for genetic analysis. The T-DNA insertional mutants of CRT3 (SALK_051336), CNX1 (SALK_083600), CNX2 (SALK_044381), CRT1 (SALK_142821), and CRT2 (SALK_062083) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Methods for seed sterilization and conditions for plant growth were described previously (35).
Plasmid Construction and Generation of Transgenic Plants.
Transgenic plants expressing BRI1:GFP and bri1–9:GFP were previously described (17). The CNX1 gene was amplified from genomic DNA of WT plants and cloned into pPZP222 (36) to generate pPZP222-gCNX1. A 3.5-kb genomic fragment containing a 900-bp native promoter and a 400-bp cDNA fragment of CRT3 were cloned into a modified pPZP222-E9 vector that carries a terminator sequence of the pea RbcS E9 gene to create pPZP222-pCRT3∷gCRT3-cCRT3 plasmid. PCR amplified genomic fragments of CRT3 and CRT1 carrying their promoter sequence were fused with cDNA fragments of CRT1 and CRT3 encoding their C-termini, respectively (see Table S1 for primer sequences), into pZPZ222-E9 to generate pPZP222-pCRT3∷gCRT3-cCRT1 and pPZP222-pCRT1∷gCRT1-cCRT3 plasmids. These plasmids were then transformed into ebs2–8 bri1–9 by Agrobacterium species-mediated vacuum infiltration (37).
Map-Based Cloning of EBS2.
The ebs2–1 bri1–9 was crossed with bri1–9 (Ws-2) and the resulting F1 plants were self-fertilized. PCR-based genetic mapping with DNAs of approximately 1,000 F2 ebs2–1 bri1–9 seedlings and molecular markers listed in Table S1 located the EBS2 locus to a 150-kb region on the top of chromosome I. Three ORFs (At1g08450, At1g09080, and At1g09210) were independently amplified from ebs2–1 bri1–9, sequenced, and compared with the published sequences of WT Col-0. Several other ebs mutants similar to ebs2–1 bri1–9 were crossed with bri1–9 (Ws) and approximately 50 to 100 F2 ebs2 bri1–9-like seedlings for each cross were used to estimate the position of each EBS locus. Genomic DNAs of ebs mutants with their EBS loci mapped near EBS2 were used to amplify At1g08450 and the resulting PCR fragments were sequenced to determine if they carry single-nucleotide change in EBS2.
Endo H Treatment.
Leaves from 4-week-old plants were homogenized in “Tris, NaCl, EDTA and Triton X (TSEX) buffer” (50 mM Tris pH 8.0, 100 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 10% glycerol, and protease inhibitors). After centrifugation, the protein extracts were denatured at 95 °C for 10 min in 1× denaturing buffer, and incubated with or without 1,000 U of Endo Hf (New England Biolabs) in the G5 buffer for 1 h at 37 °C. The treated proteins were separated by 7% SDS/PAGE and analyzed by Western blotting with anti-BRI1 serum.
Anti-CRT3 Antibody Generation.
A 123-bp cDNA fragment encoding the C-terminal 41 aa of Arabidopsis CRT3 (AtCRT3) was inserted into pGEX-KG (38). The plasmid was expressed in Rosetta cells (Novagen) and the GST-CRT3C41 fusion protein was purified using glutathione Sepharose 4B-based affinity chromatography (Amersham Biosciences). The eluent was separated by SDS/PAGE and the GST-CRT3C41 band was excised and sent to Pacific Immunology for antiserum production. The anti-CRT3 specific antibody was purified using proteins bound to nitrocellulose filters modified from the procedure of Chin-Sang (http://130.15.90.245/antibody_purification.htm).
Co-Immunoprecipitation and Western Blot Analysis.
A total of 0.2 g of 3-week-old seedlings were ground in liquid N2 and extracted with the TSEX buffer. The entire procedure was conducted at 4 °C. After centrifugation at 5,000 × g for 10 min, the supernatant was incubated with anti-GFP antibody (TP401; Torrey Pines Biolabs) for 1 h and precipitated with protein A agarose beads (Invitrogen) for 1 h. The immunoprecipitates were washed 3 times with the TSEX buffer, separated by 7.5% SDS/PAGE, and analyzed by Western blotting using anti-GFP (MMS-118P; Covance), anti-maize CRT, or anti-CRT3 antibody. For the latter 2 antibodies, the HRP-conjugated anti-rabbit IgG light-chain antibody (Jackson Immunology) was used as the secondary antibody to avoid interference from IgG heavy chain. For other Western blot experiments, total proteins of 3-week-old seedlings were extracted with 2× SDS buffer, separated by 10% SDS/PAGE, and analyzed with anti-BES1 (24), anti-CRT3, or anti-BiP (SPA-818; Stressgen) antibody.
Bioinformatics and Phylogeny Analysis.
A total of 38 unique protein sequences were aligned using a ClustalW program on the Network Protein Sequence analysis Web site (http://npsa-pbil.ibcp.fr) (39). The human CRT sequence (HsCRT) was used as the out group to root the tree. The phylogeny was constructed by using the Phylogeny Inference Package tools (http://evolution.genetics.washington.edu/phylip.html). Briefly, ClustalW alignment was analyzed using PRODIST and NEIGHBOR programs. Bootstrap assessment of tree topology derived from neighbor-joining analysis was performed 100 bootstraps with the SEQBOOT program. The derived consensus tree was displayed with the TreeView program. Bootstrap values were shown at the nodes, indicating how many times the sequences to the right of the node occurred in the same group, among 100 trees.
Confocal Microscopy.
The sub-cellular localization of BRI1:GFP and bri1–9:GFP was examined by imaging root tips of 6-d-old seedlings of BRI1:GFP, bri1–9:GFP, and bri1–9:GFP ebs2–1 transgenic lines using a Zeiss LSM510 confocal microscope filtered with FITC10 set (excitation of 488 nm with emissions of 505–530 and 530–560 nm).
Supplementary Material
Acknowledgments.
We are grateful to the RIKEN BioResource Center for supplying a CRT3 cDNA plasmid, the Arabidopsis Biological Resource Center at Ohio State University for supplying cDNA/genomic clones and T-DNA insertional mutants of Arabidopsis CRTs/CNXs, F. Tax for seeds of bri1–9 (Ws-2) and bri1–5, J. Chory for anti-BRI1 antibody, Y. Yin for anti-BES1 antibody, and R. Boston for anti-maize CRT serum. We thank Drs. A. Chang, S. Clark, C. Duan, M. Uhler, Y. Wang, and Y.-L. Qiu, plus members of the Li lab, for helpful discussions. This work was supported by National Institutes of Health Grant GM060519 (to J.L.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 13151.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906144106/DCSupplemental.
References
- 1.Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem J. 2009;417:651–666. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- 2.Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo L, et al. Cardiac-specific expression of calcineurin reverses embryonic lethality in calreticulin-deficient mouse. J Biol Chem. 2002;277:50776–50779. doi: 10.1074/jbc.M209900200. [DOI] [PubMed] [Google Scholar]
- 4.Mesaeli N, et al. Calreticulin is essential for cardiac development. J Cell Biol. 1999;144:857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persson S, et al. Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 2003;133:1385–1396. doi: 10.1104/pp.103.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crofts AJ, Denecke J. Calreticulin and calnexin in plants. Trends Plants Sci. 1998;3:396–399. [Google Scholar]
- 7.Chen MH, Tian GW, Gafni Y, Citovsky V. Effects of calreticulin on viral cell-to-cell movement. Plant Physiol. 2005;138:1866–1876. doi: 10.1104/pp.105.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baluska F, Samaj J, Napier R, Volkmann D. Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J. 1999;19:481–488. doi: 10.1046/j.1365-313x.1999.00530.x. [DOI] [PubMed] [Google Scholar]
- 9.Laporte C, et al. Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell. 2003;15:2058–2075. doi: 10.1105/tpc.013896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin ZL, et al. Over-expression of Chinese cabbage calreticulin 1, BrCRT1, enhances shoot and root regeneration, but retards plant growth in transgenic tobacco. Transgenic Res. 2005;14:619–626. doi: 10.1007/s11248-005-5694-6. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Komatsu S. Molecular cloning and characterization of calreticulin, a calcium-binding protein involved in the regeneration of rice cultured suspension cells. Eur J Biochem. 2000;267:737–745. doi: 10.1046/j.1432-1327.2000.01052.x. [DOI] [PubMed] [Google Scholar]
- 12.Persson S, et al. The Ca(2+) status of the endoplasmic reticulum is altered by induction of calreticulin expression in transgenic plants. Plant Physiol. 2001;126:1092–1104. doi: 10.1104/pp.126.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyatt SE, Tsou PL, Robertson D. Expression of the high capacity calcium-binding domain of calreticulin increases bioavailable calcium stores in plants. Transgenic Res. 2002;11:1–10. doi: 10.1023/a:1013917701701. [DOI] [PubMed] [Google Scholar]
- 14.Jia XY, et al. Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in drought-stressed responses. J Exp Bot. 2008;59:739–751. doi: 10.1093/jxb/erm369. [DOI] [PubMed] [Google Scholar]
- 15.Sharma A, et al. A novel interaction between calreticulin and ubiquitin-like nuclear protein in rice. Plant Cell Physiol. 2004;45:684–692. doi: 10.1093/pcp/pch077. [DOI] [PubMed] [Google Scholar]
- 16.Christensen A, et al. Functional characterization of Arabidopsis calreticulin1a: A key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 2008;49:912–924. doi: 10.1093/pcp/pcn065. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 19.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 21.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 22.Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mora-Garcia S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 26.Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: One protein, one gene, many functions. Biochem J. 1999;344:281–292. [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura K, et al. Complete heart block and sudden death in mice overexpressing calreticulin. J Clin Invest. 2001;107:1245–1253. doi: 10.1172/JCI12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obayashi T, et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu YL, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann Bot. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molinari M, et al. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell. 2004;13:125–135. doi: 10.1016/s1097-2765(03)00494-5. [DOI] [PubMed] [Google Scholar]
- 32.Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 33.Danilczyk UG, Cohen-Doyle MF, Williams DB. Functional relationship between calreticulin, calnexin, and the endoplasmic reticulum luminal domain of calnexin. J Biol Chem. 2000;275:13089–13097. doi: 10.1074/jbc.275.17.13089. [DOI] [PubMed] [Google Scholar]
- 34.Wada I. Calnexin is involved in the quality-control mechanism of the ER. Seikagaku. 1995;67:1133–1137. [PubMed] [Google Scholar]
- 35.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 37.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 38.Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 39.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: Network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.