Abstract
α-Crystallins are molecular chaperones that protect vertebrate eye lens proteins from detrimental protein aggregation. αB-Crystallin, 1 of the 2 α-crystallin isoforms, is also associated with myopathies and neuropathological diseases. Despite the importance of α-crystallins in protein homeostasis, only little is known about their quaternary structures because of their seemingly polydisperse nature. Here, we analyzed the structures of recombinant α-crystallins using biophysical methods. In contrast to previous reports, we show that αB-crystallin assembles into defined oligomers consisting of 24 subunits. The 3-dimensional (3D) reconstruction of αB-crystallin by electron microscopy reveals a sphere-like structure with large openings to the interior of the protein. αA-Crystallin forms, in addition to complexes of 24 subunits, also smaller oligomers and large clusters consisting of individual oligomers. This propensity might explain the previously reported polydisperse nature of α-crystallin.
Keywords: electron microscopy, molecular chaperone, protein aggregation, small heat shock protein, stress response
Molecular chaperones called α-crystallins, which belong to the small heat-shock protein (sHsp) family, constitute an important component of the stress response (1–3). Their cellular function is to bind to partially unfolded polypeptides and maintain them in a refolding-competent state (4–6). Thus, they act as molecular traps that protect the cell from the consequences of irreversible protein aggregation. This holdase function of sHsps enables to separate the binding of unfolded polypeptides from the refolding process thereby sustaining protein homeostasis (4, 7–9).
There are 2 α-crystallin genes, αA and αB, that encode for proteins that share ∼60% sequence identity (10). In the eye lens, α-crystallins account for ≈40% of the total protein content (11). The hetero-oligomeric mixtures of the 2 isoforms cannot be separated under native conditions (12). Whereas αA-crystallin expression is mainly limited to the lens and only trace amounts are found in few other tissues (13), αB-crystallin is ubiquitously expressed and is abundant in brain, heart, and muscle (14). The importance of α-crystallins as molecular chaperones was demonstrated by knock-out mice, in which the deletion of αA-crystallin led to young onset cataract formation (15). The functional importance of αB-crystallin is not restricted to the eye lens, because it was shown to be associated with a number of neuropathological protein folding diseases, including Alzheimer's, Parkinson's, and Creutzfeldt-Jakob disease (16, 17). Additionally, αB-crystallin is the most abundant gene transcript present in early active multiple sclerosis lesions (18), and it appears to exhibit anti-apoptotic as well as neuroprotective functions (19, 20).
sHsps share common structural features. The most distinctive characteristic is the structurally conserved “α-crystallin” domain (2), which is flanked by a genetically diverse N-terminal region that differs in sequence and length, and a moderately conserved C-terminal extension (21). sHsps assemble into large oligomers with mostly 12 or 24 subunits (22). The dynamic nature of these assemblies renders their structure determination challenging, and only few sHsp crystal structures and reconstructions based on electron microscopy (EM) are available (23–28). These revealed similar spherical or toroidal structures with internal cavities.
α-Crystallins were reported to form large oligomers that display great heterogeneity. The current opinion is that α-crystallins cannot be obtained in homogeneous form. Neither the structure of monomeric α-crystallin nor the subunit arrangement within the oligomer are yet known. Based on studies using scattering methods, several structural models have been proposed that differed substantially from each other (29–33). Transmission electron microscopy (TEM) has also been used to elucidate the oligomer structure of α-crystallin (23, 34). Recombinant human αB-crystallin was suggested to be an asymmetric and roughly spherical assembly of 32 subunits with a large central cavity (23). In addition, the presence of multiple oligomer assemblies and a highly variable quaternary structure was reported (23). Authentic α-crystallin from bovine eye lens was also shown to form asymmetric oligomers with mean diameters larger than those found for recombinant human αB-crystallin and with an even higher degree of heterogeneity (34). More recently, a mass spectrometric (MS) analysis suggested that α-crystallins form a continuum of assemblies with an average mass corresponding to 28 subunits (35).
To re-evaluate the degree of heterogeneity and the quaternary structure of α-crystallins, we analyzed the structures of recombinant human αA- and αB-crystallin and compared them to authentic α-crystallin purified from bovine eye lenses (αL-crystallin).
Results
Chaperone Activity of α-Crystallins.
Human αA- and αB-crystallins were recombinantly expressed in Escherichia coli and purified to homogeneity (for details see Materials and Methods and Fig. S1 A–D). For both proteins, MS revealed the correct monomer masses (Fig. S1 A and B). In addition, authentic αL-crystallin was purified from bovine eye lenses. MS analysis identified the correct isoforms and known posttranslational modifications (Fig. S1C). The secondary and tertiary structures of the preparations were assessed by circular dichroism spectroscopy (Fig. S1 E and F) and found to correspond to published data (36).
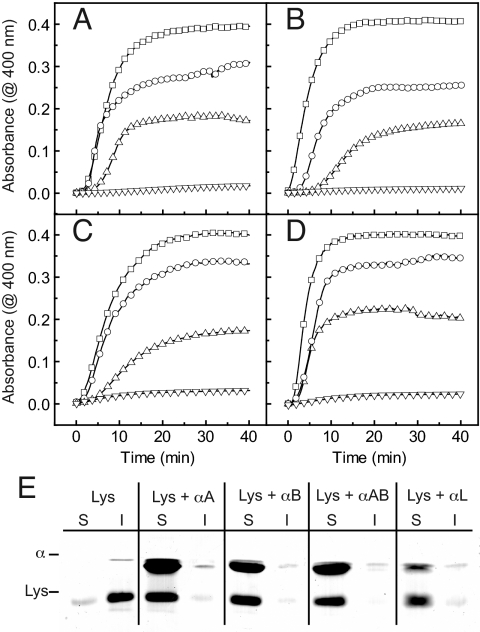
To test the functional properties of the purified proteins, we analyzed their chaperone activity in aggregation assays using lysozyme as substrate (37). In the presence of equimolar concentrations of either recombinant α-crystallin species, the aggregation signal was reduced by approximately half, and when a 2-fold molar excess over lysozyme was used, the signal was entirely suppressed (Fig. 1 A and B). Analysis of the supernatant and pellet fractions of the samples by SDS/PAGE confirmed this picture (Fig. 1E). Also, αAB-crystallin (an equimolar mixture of recombinant αA- and αB-crystallin) (Fig. 1 C and E) and authentic αL-crystallin (Fig. 1 D and E) suppressed lysozyme aggregation in a concentration-dependent manner with efficacies similar to those observed for the isolated recombinant isoforms.
Fig. 1.
Chaperone activity of α-crystallin. The chaperone function of α-crystallin was assessed at 37 °C by its ability to suppress the aggregation of 10 μM lysozyme. Lysozyme aggregation was monitored at 400 nm in a UV/Vis spectrophotometer in the absence (□) and presence of 5 μM (○), 10 μM (▵), and 20 μM (▿) α-crystallin. (A) αA-Crystallin, (B) αB-crystallin, (C) αAB-crystallin (equimolar mixture of recombinant αA- and αB-crystallin), and (D) authentic αL-crystallin. (E) The solubility of denatured lysozyme (Lys) in the absence and presence of a 2-fold molar excess of respective α-crystallin (α) was analyzed by comparing the soluble supernatant fraction (S) and the insoluble sediment fraction (I) using SDS/PAGE.
Hydrodynamic Size of α-Crystallins.
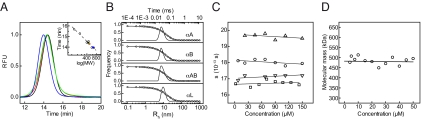
Most sHsps studied so far form oligomers consisting of 12 or 24 subunits (22). For α-crystallin, however, structural data in the literature suggest a heterogeneous assembly with a continuum of different oligomeric states (34, 35). To address this issue, we subjected the recombinant α-crystallin isoforms as well as authentic αL-crystallin to analytical size exclusion chromatography (SEC) and dynamic light scattering (DLS). Elution profiles of α-crystallins from an analytical SEC column are shown in Fig. 2A. αB-crystallin eluted in a single narrow and symmetric peak with a full-width at half maximum (FWHM) of 1.45 min. According to linear calibration, the elution time corresponds to a molecular mass of ∼475 kDa consistent with a complex of 24 subunits (20.2 kDa monomeric mass). Similarly, αA-crystallin eluted in a single and symmetric peak with a slightly smaller apparent molecular mass of 440 kDa (19.9 kDa monomeric mass). Although the FWHM of αL-crystallin was very similar to that observed for αA- and αB-crystallin, the elution time was smaller, indicating that its hydrodynamic properties differ from that of recombinant αA- and αB-crystallin. Interestingly, αAB-crystallin eluted at times similar to those of the isolated recombinant proteins, suggesting that the formation of αA-αB-hetero-oligomers per se is not responsible for the different hydrodynamic behavior of αL-crystallin. For comparison, Hsp26 from yeast and Hsp16.5 from Methanococcus jannaschii, which form distinct 24-mers (25, 28, 38), were also subjected to SEC. Their FWHMs were very similar to those obtained for our α-crystallin preparations.
Fig. 2.
Hydrodynamic analysis. (A) The SEC elution profiles of recombinant human αA- (black line), αB-crystallin (red line), an equimolar mixture of both isoforms (αAB-crystallin) (green line), and authentic bovine αL-crystallin (blue line) were obtained using a Tosoh Bioscience TSK 4000 PW column. All species eluted from the SEC column within a single, narrow, and symmetric peak with a width at half-height in the range of 1.4 min. Inset: Linear calibration revealed a molecular mass of 441 kDa for αA-crystallin, ≈475 kDa for αB-crystallin as well as for the mixture of both isoforms, and 628 kDa for αL-crystallin. (B) Plots of the DLS autocorrelation function (G(1)(t)) of αA-, αB-, αAB-, and αL-crystallin and the corresponding particle size distributions. Frequency refers to the observed molecule size distribution. The hydrodynamic radii are 8.04 ± 0.23 nm for αA- (peak weight 99.88%), 7.26 ± 0.18 nm for αB- (peak weight 99.80%), 8.83 ± 0.29 nm for αAB- (peak weight 100%), and 10.19 ± 0.27 nm for αL-crystallin (peak weight 99.31%). (C) Concentration-dependent analysis of the sedimentation coefficient (s) by SV AUC as a function of the concentration of αA-crystallin (○), αB-crystallin (□), αAB-crystallin (▿), and αL-crystallin (▵). (D) Apparent molecular masses for αB-crystallin determined from SE AUC experiments as a function of the protein concentration. Data were fitted to a molecular mass of 482 ± 12 kDa, which is independent of protein concentration.
To further address the issue of solution quaternary structure, we subjected the 4 α-crystallin samples to DLS (Fig. 2B). The respective particle size distributions indicated hydrodynamic radii of 8.04 ± 0.23 nm for αA-, 7.26 ± 0.18 nm for αB-, 8.83 ± 0.29 nm for αAB-, and 10.19 ± 0.27 nm for αL-crystallin (Fig. 2B), which is in good agreement with the SEC analyses.
Quaternary Structure of α-Crystallins.
As SEC and DLS measurements alone do not suffice for an unequivocal judgement of the homogeneity of protein samples, we subjected our α-crystallin preparations to analytical ultracentrifugation (AUC) and TEM.
Sedimentation velocity (SV) AUC of αB-crystallin generated a single sedimentation boundary, indicating the absence of significant fractions of differently sized particles. Data analysis using the time derivative (dC/dT) method (39) revealed a narrow distribution of apparent sedimentation coefficients (s*) with the highest frequency at ≈16.5 S (Fig. S2B). αA-Crystallin and αAB-crystallin, however, showed larger sedimentation coefficients (s) of 18 S and 17 S, respectively, with broader distributions (Fig. S2 A and C). The authentic αL-crystallin exhibited the largest s value of 20 S (Fig. S2D). The s values of all 4 species were shown to be independent of concentration within the range of 2–150 μM (Fig. 2C). Because αB-crystallin was found to lack significant polydispersity, we performed sedimentation equilibrium (SE) AUC to more precisely analyze its quaternary structure. We determined a concentration-independent molecular mass of 482 ± 12 kDa, which is in agreement with the theoretical mass of 484 kDa for a 24-mer (Fig. 2D).
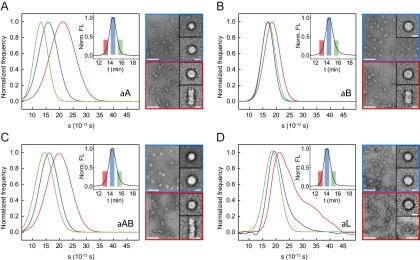
To further investigate the nature of the different assemblies, we separated α-crystallin preparations by SEC into an early, a medium, and a late fraction (Fig. 3 A–D, Insets) and analyzed these fractions separately by SV AUC and TEM. Interestingly, the sedimentation coefficients of the αA-crystallin fractions differed substantially from each other with an inverse correlation between elution time in SEC and s values (Fig. 3A, Left). TEM revealed the presence of small (11.5 nm diameter) and large (13.5 nm) assemblies in the medium αA fraction (Fig. 3A, Right, Top). In the fraction at early elution times, large oligomers (13.5 nm) and elongated assemblies resulting from oligomer-oligomer associations were detected (Fig. 3A, Right, Bottom).
Fig. 3.
Heterogeneity of α-crystallins. (A–D) Analysis of fractions obtained from SEC (see Materials and Methods) by SV analytical ultracentrifugation (Left) and TEM (Right). s distributions for the early (red), medium (blue), and late (green) fraction of the elution peaks (Insets) were determined using the dC/dT method. Representative electron micrographs of negatively stained [ammonium molybdate, 1.5% (wt/vol), pH 5.5] α-crystallin samples obtained from the medium (blue frame) and the early SEC fraction (red frame) are depicted (Scale bars, 50 nm). For each sample, average images (Scale bars, 10 nm) are included for every population with >10% abundance. (A) αA-crystallin. (B) αB-crystallin. Note the much higher degree of homogeneity. (C) αAB-crystallin. (D) Authentic αL-crystallin.
In contrast, αB-crystallin exhibited a much higher degree of homogeneity. The medium and late fractions after SEC (Fig. 3B, Inset) yielded very similar s distributions with the highest frequency at 16.7 S, indicating the absence of smaller oligomeric species (Fig. 3B, Left). TEM analysis confirmed this finding; only globular oligomers (13.5 nm), similar to the large ones in αA-crystallin samples, were identified (Fig. 3B, Right, Top). The early SEC fraction contained a population of oligomers sedimenting with 18.5 S. TEM revealed the presence of large assemblies in this fraction, which appear to be a population of interacting oligomers (Fig. 3B, Right, Bottom). However, the amount of clustered oligomers was lower than in the respective αA-crystallin samples.
Interestingly, αAB-crystallin fractions (Fig. 3C) turned out to be similar to those of αA-crystallin; whereas the early fraction contained larger oligomers and oligomer clusters (Fig. 3C, Right, Bottom), the medium fraction was characterized by the presence of both larger and smaller oligomers (Fig. 3C, Right, Top), suggesting that smaller species are an intrinsic property of αA-crystallin.
The authentic αL-crystallin exhibited the highest degree of heterogeneity within our analyses (Fig. 3D). In general, s values were larger compared with the recombinant preparations (Fig. 3D, Left), which corresponds well to SEC data. TEM analysis of the medium fraction revealed the existence of populations of smaller and larger oligomers (Fig. 3D, Right, Top). The early fraction produced a broad and asymmetric s distribution ranging from 15–45 S (Fig. 3D, Left). In accordance, a significantly higher degree of oligomer-oligomer association as well as larger oligomer clusters were observed by TEM (Fig. 3D, Right, Bottom) when compared with the other recombinant α-crystallin samples.
Electron Microscopy and 3D Reconstruction of αB-Crystallin.
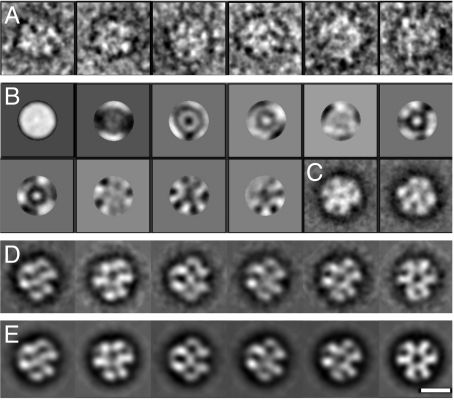
Because of the homogeneity of recombinant αB-crystallin, we focused on analyzing its 3-dimensional (3D) structure by EM and image processing (for details see Materials and Methods and Fig. S3). For this purpose, 2565 single particle images (Fig. 4A) were selected from electron micrographs of the negatively stained samples and subjected to multivariate statistical analysis (MSA) and classification. Eigenimages, i.e., images representing the most important differences in the data set, obtained after a first cycle of translational alignment and MSA (Fig. 4B) as well as preliminary class averages (Fig. 4C) already indicated the presence of projection images with 3-fold symmetry. Thus, and because of the 24-meric assembly of αB-crystallin oligomers as determined by SEC (Fig. 2A) and SE AUC (Fig. 2C), compatible point group symmetries, i.e., C3, D3, tetrahedral, and octahedral, were imposed for the calculation of the respective preliminary 3D models by angular reconstitution based on selected class averages (see Materials and Methods and Fig. S3). For each imposed symmetry, the preliminary model was refined in iterative cycles at which the reprojections of the preceding model were used as new references for multi-reference alignment (MRA) of the whole data set followed by MSA and classification. In each cycle, the quality of the resulting model was checked by projection matching (40), i.e., by comparing the consistency between the class averages and reprojections of the model in the orientations found for the class averages. For C3, D3, and octahedral symmetries, the class averages and corresponding reprojections did not match sufficiently, and the refinement procedure did not lead to a significant resolution improvement (Fig. S4). Tetrahedral symmetry, however, gave consistent results and led to a final 3D model at ≈20 Å resolution as judged by the 0.5 Fourier shell correlation (FSC) criterion (41). Selected class averages and corresponding reprojections of the final 3D model are shown in Fig. 4 D and E, respectively. For the calculation of the model, 40 classes were used.
Fig. 4.
Image analysis of negatively stained αB-crystallin oligomers. (A) Gallery of single αB-crystallin oligomers. (B) First 10 eigenimages (statistical difference images) obtained after translational alignment of the data set. (C) Two class averages with apparent 3-fold and mirror-symmetries resulting from the MSA of the translationally aligned data set. (D) Characteristic final class averages. (E) 2D reprojections of the 3D model of αB-crystallin in the orientations found for the class averages shown in D (Scale bar, 10 nm).
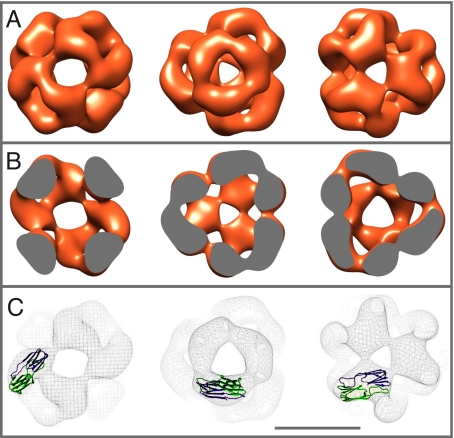
The 3D reconstruction shows that human αB-crystallin oligomer is roughly spherical with a diameter of 13.5 nm (Fig. 5A and Movie S1). The oligomer accommodates a large central cavity of ≈8.5 nm diameter that is surrounded by a symmetrical protein shell with a mean thickness varying from 2.5 to 4 nm (Fig. 5B). The protein shell contains openings at the positions of the 2- and 3-fold axes, with diameters of ≈3.5 nm for the 2-fold axes and 3 and 2 nm for the 2 unequal 3-fold axes. The overall quaternary structure of human recombinant αB-crystallin shows similarities to the structures of Hsp20.2 from Archaeoglobus fulgidus (24) and Hsp16.5 from M. jannaschii (25), which both form spherical, 24-meric complexes with octahedral symmetry. As shown in Fig. 5C, the dimeric α-crystallin domain of Hsp16.5 from M. jannaschii (25) is well-accommodated in the corresponding position of the αB-crystallin model (see also Movie S1).
Fig. 5.
3D reconstruction of human recombinant αB-crystallin. (A) Surface representations of the 3D model of human recombinant αB-crystallin. The density threshold applied for the surface representation has been chosen such that the corresponding molecular mass was in agreement with a 24-meric αB-crystallin. The molecules are viewed along the 2-fold axis (first row) and the 2 unequal 3-fold axes (second and third row). (B) Density cross-sections through the 3D model of αB-crystallin. (C) Superposition of the ribbon representation of the dimeric α-crystallin domain of Hsp16.5 from M. jannaschii (25) with the meshed surface representations of the 3D model of αB-crystallin (Scale bar, 10 nm).
Discussion
The contribution of sHsps to the cellular chaperone network as efficient molecular traps for non-native proteins is a rather recent concept. A characteristic trait conserved from prokaryotes to eukaryotes and archaea is their elaborate quaternary structure in which 12 or 24 subunits assemble to barrel- or sphere-shaped complexes. In this context, the eye lens protein α-crystallin was considered to be rather unique because it was reported to possess a highly variable quaternary structure (34, 35). However, we demonstrate that both αA- and αB-crystallin assemble into distinct oligomers with a well-defined quaternary structure. The 2 proteins differ in their propensities to form further associations leading to heterogeneous populations.
According to our results, human recombinant αB-crystallin forms distinct complexes consisting of 24 subunits. By negative stain electron microscopy and image processing, we were able to reconstruct a 3D model of the αB-crystallin assembly at 20 Å resolution that allows a detailed look at its structural characteristics. In line with the oligomer architectures observed for other sHsps, αB-crystallin oligomer is a hollow, sphere-like assembly. The positioning of the α-crystallin domain from M. jannaschii Hsp16.5 in the 3D model of αB-crystallin suggests that the subunit topography within the oligomer is conserved. A characteristic feature of sHsps, large openings in the protein shell leading to the hollow interior, is also present in αB-crystallin oligomer, albeit more pronounced as in the structures of other sHsps (24, 25, 28). In contrast to 24-meric sHsps complexes from yeast and archaea (24, 28), αB-crystallin oligomers do not show detectable size variations.
Whereas αB-crystallin is characterized by a well-defined quaternary structure, our combined SEC/AUC/TEM analyses clearly show that αA-crystallin forms, besides complexes of 24 subunits, also smaller oligomers and large clusters consisting of individual oligomers. This heterogeneity also dominates in mixed complexes consisting of both αA- and αB-crystallin. The propensity of single αA-crystallin oligomers to further associate to larger particles may result in a continuum of assemblies that could explain the previously suggested polydisperse character of α-crystallin, which is composed of both αA- and αB-crystallin. Interestingly, authentic α-crystallin seems to possess, as compared with the mixture of recombinant αA- and αB-crystallin, a larger degree of heterogeneity including large oligomer clusters, giving rise to the assumption that the formation of oligomer clusters might be influenced by the posttranslational modifications of the α-crystallins in the eye lens. Whether these modifications are important for maintaining the transparency of the eye lens or are merely the result of aging remains to be elucidated. It is noteworthy that the deletion of αB-crystallin has no lens phenotype, whereas the deletion of αA-crystallin leads to severe cataract, i.e., the formation of large aggregates of eye lens proteins, as shown by gene deletion studies in mice (15, 42). Thus, it is tempting to speculate that the 2 α-crystallins differ not only in their structural but also in their functional properties in lenticular cells, with αA-crystallin being the more active chaperone toward lens proteins such as the β- and γ-crystallins. Furthermore, the polydispersity introduced by αA-crystallin might be essential to abolish higher order structures of α-crystallins under the extremely high protein concentrations (∼450 mg/mL) in the eye lens. Another aspect to consider is that α-crystallins have to be permanently active in the eye lens for several decades—in contrast to regulable sHsps, such as Hsp26. The latter is activated by a temperature-induced structural rearrangement exposing the chaperone binding site (38, 43). In the case of α-crystallin, multiple binding sites may be permanently exposed on the surface of the oligomer and interact with each other, leading to the formation of larger assemblies. A similar tendency has also been reported for Hsp70, which forms dimers mediated by their substrate binding domains (44, 45), and Hsp90, which can form larger oligomers (46).
At present, it is still premature to reconcile the observed differences in association behaviors and polydispersity of the 2 α-crystallin isoforms with specific functional differences. The elucidation of their 3D structures and their assembly properties should allow addressing this question in the near future.
Materials and Methods
Cloning and Protein Purification.
Recombinant αA- and αB-crystallin were cloned from plasmid DNA containing the respective genes (kind gift by Prof. N.H. Lubsen, Nijmegen, The Netherlands) by PCR amplification using primers carrying the NcoI and NotI restriction sites. PCR fragments were subcloned into the vector pET28b+ and verified by sequencing. For gene expression, E. coli BL21(DE3) was transformed with the respective plasmid, cells were grown at 30 °C, and expression was induced by 1 mM IPTG. After cell disruption, the cleared lysate was applied on a Q Sepharose column (Amersham Bioscience) equilibrated with TE buffer (50 mM Tris-HCl, 2 mM EDTA, pH 9.0). Elution was carried out with a linear NaCl gradient. Fractions containing the target protein were pooled, concentrated, and loaded onto a Superdex 200-pg column (Amersham Bioscience) run in TE, 100 mM NaCl. Fractions containing the target protein were applied to a Resource Q column (Amersham Bioscience) in TE buffer, and the protein was eluted with a linear NaCl gradient. Recombinant αA-crystallin and αB-crystallin were dialyzed against 10 mM NaP, pH 7.5, and stored at −80 °C.
Authentic α-crystallin (αL-crystallin) was isolated from bovine calf lenses. The tissue was homogenized, and the soluble αL-crystallin was purified to homogeneity as described above. SDS/PAGE analysis and matrix-assisted laser desorption ionization (MALDI) mass spectrometry demonstrated that the protein preparations were homogeneous.
Lysozyme Aggregation Assay.
The aggregation of 10 μM lysozyme (Sigma-Aldrich) was initiated by 1 mM Tris(2-carboxyethyl)phosphine (TCEP) (Sigma-Aldrich). All lysozyme assays were carried out at 37 °C in PBS buffer. The aggregation process was followed at 400 nm by recording absorbance changes in a Cary 50 UV/VIS spectrophotometer (Varian) equipped with a temperature-adjustable cuvet holder. To determine the chaperone activity of α-crystallins, increasing concentrations of each α-crystallin species were added before reduction. The solubility of denatured lysozyme in the absence and presence of α-crystallin was determined by SDS/PAGE and Coomassie staining, analyzing the sediment and supernatant fraction after centrifugation at 10,000 rpm for 2 min in a table-top centrifuge.
Analytical Gel-Filtration.
A TosoHaas TSK G4000PW (30 × 0.75 cm) gel filtration column with a separation range from 10 to 1,500 kDa (Tosoh Bioscience) was used for gel filtration. All experiments were performed at room temperature using PBS buffer and a flow rate of 0.75 mL/min. Detection was performed with a FP 920 fluorescence detector (Jasco), using an excitation wavelength of 280 nm and an emission wavelength of 340 nm. The following proteins were used for calibration: Thyroglobulin (669 kDa), ferritin (450 kDa), catalase (240 kDa), and aldolase (158 kDa).
Analytical Ultracentrifugation.
Analytical ultracentrifugation was carried out in Beckman XL-A and XL-I machines, equipped with UV and interference detection systems (Beckman Coulter). For SV, 400 μL sample and 410 μL reference buffer were loaded into sector-shaped double-channel centerpieces and spun at 25,000 rpm at 20 °C in a Ti60 rotor. Scans were recorded continuously at 280 nm. Data analysis was carried out using the dC/dT method (39). For SE, 100 μL sample mixed with 15 μL FC40 oil and 120 μL reference buffer were loaded into 6-channel centerpieces and spun at 8,000 rpm at 20 °C. The molecular masses for each concentration were calculated from liner fitting of plots of the natural logarithm of the concentration versus the square of the radius. Buffer density was taken from tables and the partial specific volume calculated on the basis of the primary sequence. The true molecular weight (MW) was derived from linear regression of the calculated molecular masses.
Dynamic Light Scattering.
Light scattering measurements were performed with an ALV/CGS-8 apparatus equipped with an ALV/5000/6010 digital autocorrelator operated in the single cross-correlation mode. A Uniphase (power >35 mW) He-Ne laser operating at a wavelength of 632.8 nm was used as a light source. Scattering was monitored using a pair of avalanche photon counting modules (SPCM-CD2969; PerkinElmer). A fraction of the incoming laser beam was monitored continuously by 2 quadrant photodiodes placed after a precision attenuator. The readings of the photodiodes were used to normalize the total scattered intensities and to exclude artifacts stemming from long-term fluctuations or changes in the pointing stability of the laser beam. Data were analyzed using the inverse Laplace transformation algorithm CONTIN (47).
Electron Microscopy.
α-Crystallin samples were adsorbed for 2 min onto carbon-coated grids that were glow discharged in air before the application of 5 μL protein solution at a concentration of 0.05 mg/mL. Excess protein solution was blotted off, and the samples were negatively stained for 30 s using 5 μL ammonium molybdate solution [1.5% (wt/vol), pH 5.5]. Electron micrographs were recorded at a nominal magnification of 50,000 and at defocus values of 900–1200 nm using a JEOL JEM 100CX electron microscope operated at 100 kV. Suitable micrographs were digitized at a step size of 8.5 μm using a FlexTight Precision II array scanner, resulting in a pixel size of 1.69 Å at the specimen level.
Image Processing.
Well-separated particles on electron micrographs were semimanually selected using Boxer from the Eman software package (48). This package was also used for the determination of the defoci and the correction of contrast transfer function (CTF) by phase flipping. The following image processing procedures and the 3D reconstruction (Fig. S3) were carried out using the Imagic suite (49).
In a first step, CTF-corrected molecular images from each negative were pooled, band-pass filtered (between 12 and 170 Å), and normalized. Afterward, a first translational alignment was performed by “self-alignment,” i.e., the input image was iteratively centered relative to a rotationally symmetrized version of itself. Upon identification of eigenimages, MSA, and the following classification, only structurally well-defined class averages were used as references for multi-reference rotational and translational alignment (MRA) of the data set. Once the number and quality of the class averages have been improved sufficiently in iterative cycles of MRA and MSA (Fig. S3, loop 1), 4 preliminary 3D models differing in their symmetries, i.e., models with C3, D3, tetrahedral, and octahedral point group symmetry, were reconstructed based on representative class averages with well-assigned projection angles by the method of angular reconstitution. The respective symmetries were taken into account for the choice of “common lines” (50). For each symmetry imposed, the preliminary model was subjected to iterative cycles of refinement (Fig. S3, loop 2), at which the reprojections of the preceding model were used as new references for MRA of the data set followed by MSA and classification. In each cycle, the quality of the resulting model was checked.
Supplementary Material
Acknowledgments.
We thank Andreas Kastenmüller and Marianne Hanzlik for helping us at various stages of the project; Prof. T. Kiefhaber for making available the light scattering apparatus; and Karin Joder for help in our initial attempts. This work was supported by the Deutsche Forschungsgemeinschaft SFB594 (to J.B., M.H., and S.W.) and the Fonds der chemischen Industrie (J.B. and M.H.). J.P. acknowledges a scholarship from the Studienstiftung des deutschen Volkes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902651106/DCSupplemental.
References
- 1.Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingolia TD, Craig EA. Four small Drosophila heat shock proteins are related to each other and to mammalian alpha-crystallin. Proc Natl Acad Sci USA. 1982;79:2360–2364. doi: 10.1073/pnas.79.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klemenz R, et al. Alpha B-crystallin is a small heat shock protein. Proc Natl Acad Sci USA. 1991;88:3652–3656. doi: 10.1073/pnas.88.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 6.Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakob U, Buchner J. Assisting spontaneity: The role of Hsp90 and small Hsps as molecular chaperones. Trends Biochem Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 8.Kampinga HH, et al. Cells overexpressing Hsp27 show accelerated recovery from heat-induced nuclear protein aggregation. Biochem Biophys Res Commun. 1994;204:1170–1177. doi: 10.1006/bbrc.1994.2586. [DOI] [PubMed] [Google Scholar]
- 9.Ehrnsperger M, et al. Stabilization of proteins and peptides in diagnostic immunological assays by the molecular chaperone Hsp25. Anal Biochem. 1998;259:218–225. doi: 10.1006/abio.1998.2630. [DOI] [PubMed] [Google Scholar]
- 10.Horwitz J. Alpha-crystallin. Exp Eye Res. 2003;76:145–153. doi: 10.1016/s0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 11.Bloemendal H, et al. Ageing and vision: Structure, stability, and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Bloemendal H, Groenewoud G. One-step separation of the subunits of alpha-crystallin by chromatofocusing in 6 M urea. Anal Biochem. 1981;117:327–329. doi: 10.1016/0003-2697(81)90787-9. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasan AN, Nagineni CN, Bhat SP. alpha A-crystallin is expressed in non-ocular tissues. J Biol Chem. 1992;267:23337–23341. [PubMed] [Google Scholar]
- 14.Iwaki T, Kume-Iwaki A, Goldman JE. Cellular distribution of alpha B-crystallin in non-lenticular tissues. J Histochem Cytochem. 1990;38:31–39. doi: 10.1177/38.1.2294148. [DOI] [PubMed] [Google Scholar]
- 15.Brady JP, et al. Targeted disruption of the mouse alpha A-crystallin gene induces cataract and cytoplasmic inclusion bodies containing the small heat shock protein alpha B-crystallin. Proc Natl Acad Sci USA. 1997;94:884–889. doi: 10.1073/pnas.94.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renkawek K, et al. alpha B-crystallin is present in reactive glia in Creutzfeldt-Jakob disease. Acta Neuropathol (Berl) 1992;83:324–327. doi: 10.1007/BF00296796. [DOI] [PubMed] [Google Scholar]
- 17.Renkawek K, et al. Expression of alpha B-crystallin in Alzheimer's disease. Acta Neuropathol. 1994;87:155–160. doi: 10.1007/BF00296185. [DOI] [PubMed] [Google Scholar]
- 18.Chabas D, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294:1731–1735. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- 19.Andley UP, et al. Lens epithelial cells derived from alphaB-crystallin knockout mice demonstrate hyperproliferation and genomic instability. FASEB J. 2001;15:221–229. doi: 10.1096/fj.00-0296com. [DOI] [PubMed] [Google Scholar]
- 20.Ousman SS, et al. Protective and therapeutic role for alphaB-crystallin in autoimmune demyelination. Nature. 2007;448:474–479. doi: 10.1038/nature05935. [DOI] [PubMed] [Google Scholar]
- 21.De Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha-crystallin—small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 22.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 23.Haley DA, Horwitz J, Stewart PL. The small heat-shock protein, alphaB-crystallin, has a variable quaternary structure. J Mol Biol. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- 24.Haslbeck M, et al. Structural dynamics of archaeal small heat shock proteins. J Mol Biol. 2008;378:362–374. doi: 10.1016/j.jmb.2008.01.095. [DOI] [PubMed] [Google Scholar]
- 25.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 26.van Montfort RL, et al. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 27.Stamler R, Kappe G, Boelens W, Slingsby C. Wrapping the alpha-crystallin domain fold in a chaperone assembly. J Mol Biol. 2005;353:68–79. doi: 10.1016/j.jmb.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 28.White HE, et al. Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26. Structure. 2006;14:1197–1204. doi: 10.1016/j.str.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 29.Augusteyn RC, Koretz JF. A possible structure for alpha-crystallin. FEBS Lett. 1987;222:1–5. doi: 10.1016/0014-5793(87)80180-1. [DOI] [PubMed] [Google Scholar]
- 30.Carver JA, Aquilina JA, Truscott RJ. A possible chaperone-like quaternary structure for alpha-crystallin. Exp Eye Res. 1994;59:231–234. doi: 10.1006/exer.1994.1101. [DOI] [PubMed] [Google Scholar]
- 31.Tardieu A, et al. Calf lens alpha-crystallin quaternary structure. A three-layer tetrahedral model. J Mol Biol. 1986;192:711–724. doi: 10.1016/0022-2836(86)90023-9. [DOI] [PubMed] [Google Scholar]
- 32.Vanhoudt J, Abgar S, Aerts T, Clauwaert J. A small-angle X-ray solution scattering study of bovine alpha-crystallin. Eur J Biochem. 2000;267:3848–3858. doi: 10.1046/j.1432-1327.2000.01423.x. [DOI] [PubMed] [Google Scholar]
- 33.Wistow G. Possible tetramer-based quaternary structure for alpha-crystallins and small heat shock proteins. Exp Eye Res. 1993;56:729–732. doi: 10.1006/exer.1993.1090. [DOI] [PubMed] [Google Scholar]
- 34.Haley DA, et al. Small heat-shock protein structures reveal a continuum from symmetric to variable assemblies. J Mol Biol. 2000;298:261–272. doi: 10.1006/jmbi.2000.3657. [DOI] [PubMed] [Google Scholar]
- 35.Aquilina JA, et al. Polydispersity of a mammalian chaperone: Mass spectrometry reveals the population of oligomers in alphaB-crystallin. Proc Natl Acad Sci USA. 2003;100:10611–10616. doi: 10.1073/pnas.1932958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun TX, Das BK, Liang JJ. Conformational and functional differences between recombinant human lens alphaA- and alphaB-crystallin. J Biol Chem. 1997;272:6220–6225. doi: 10.1074/jbc.272.10.6220. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg ME, Rudolph R, Jaenicke R. A kinetic study of the competition between renaturation and aggregation during the refolding of denatured-reduced egg white lysozyme. Biochemistry. 1991;30:2790–2797. doi: 10.1021/bi00225a008. [DOI] [PubMed] [Google Scholar]
- 38.Haslbeck M, et al. Hsp26: A temperature-regulated chaperone. EMBO J. 1999;18:6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stafford WFI. Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 40.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 41.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Brady JP, et al. AlphaB-crystallin in lens development and muscle integrity: A gene knockout approach. Invest Ophthalmol Vis Sci. 2001;42:2924–2934. [PubMed] [Google Scholar]
- 43.Franzmann TM, Menhorn P, Walter S, Buchner J. Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol Cell. 2008;29:207–216. doi: 10.1016/j.molcel.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Angelidis CE, Lazaridis I, Pagoulatos GN. Aggregation of hsp70 and hsc70 in vivo is distinct and temperature-dependent and their chaperone function is directly related to non-aggregated forms. Eur J Biochem. 1999;259:505–512. doi: 10.1046/j.1432-1327.1999.00078.x. [DOI] [PubMed] [Google Scholar]
- 45.Benaroudj N, et al. Overexpression in Escherichia coli, purification and characterization of the molecular chaperone HSC70. Eur J Biochem. 1994;221:121–128. doi: 10.1111/j.1432-1033.1994.tb18720.x. [DOI] [PubMed] [Google Scholar]
- 46.Jakob U, et al. Structural organization of procaryotic and eucaryotic Hsp90. Influence of divalent cations on structure and function. J Biol Chem. 1995;270:14412–14419. doi: 10.1074/jbc.270.24.14412. [DOI] [PubMed] [Google Scholar]
- 47.Provencher SW. Contin—a general-purpose constrained regularization program for inverting noisy linear algebraic and integral-equations. Comput Phys Commun. 1982;27:229–242. [Google Scholar]
- 48.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 49.van Heel M, et al. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 50.van Heel M, et al. Single-particle electron cryo-microscopy: Towards atomic resolution. Q Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.