Abstract
The majority of studies on autophagy, a cytoplasmic homeostatis pathway of broad biological and medical significance, have been hitherto focused on the phosphatidylinositol 3-kinases as the regulators of autophagy. Here, we addressed the reverse process driven by phosphoinositide phosphatases and uncovered a key negative regulatory role in autophagy of a phosphatidylinositol 3-phosphate (PI3P) phosphatase Jumpy (MTMR14). Jumpy associated with autophagic isolation membranes and early autophagosomes, defined by the key factor Atg16 necessary for proper localization and development of autophagic organelles. Jumpy orchestrated orderly succession of Atg factors by controlling recruitment to autophagic membranes of the sole mammalian Atg factor that interacts with PI3P, WIPI-1 (Atg18), and by affecting the distribution of Atg9 and LC3, the two Atg factors controlling organization and growth of autophagic membranes. A catalytically inactive Jumpy mutant, R336Q, found in congenital disease centronuclear myopathy, lost the ability to negatively regulate autophagy. This work reports for the first time that initiation of autophagy is controlled not only by the forward reaction of generating PI3P through a lipid kinase but that its levels are controlled by a specific PI3P phosphatase, which when defective can lead to human disease.
Keywords: Atg18, autophagy, LC3, myopathy, PI3P phosphatase
Introduction
Autophagy is an ancient, highly conserved eukaryotic intracellular homeostatic process carrying out degradation of cytoplasmic components including damaged or superfluous organelles, toxic protein aggregates and intracellular pathogens (Levine and Deretic, 2007; Levine and Kroemer, 2008; Mizushima et al, 2008). Autophagy takes place at basal levels in all eukaryotic cells, turning over long-lived macromolecules and large supra-molecular structures including whole organelles. In addition to its housekeeping role, autophagy can be upregulated during metabolic, genotoxic or hypoxic stress conditions and acts as an adaptive mechanism essential for cell survival (Levine and Kroemer, 2008). Dysfunctional autophagy, when defective or excessive, has been linked to human pathologies ranging from neurodegeneration and myopathies to cancer and inflammatory diseases (Levine and Deretic, 2007; Mizushima et al, 2008). The autophagy-associated clinical conditions can be due to increased or decreased autophagy levels, underscoring the need to understand both the induction of the autophagy pathway and its downregulation. Thus far, identifying the brakes in the system once it is set in motion by the upstream Tor-dependent signalling systems has eluded a proper definition. And yet, the autophagic process must be tightly regulated to support cell survival when needed but also to avoid cell death and injury through excessive autophagy.
A key signalling regulator of autophagy is the Akt/mTOR pathway. Inhibition of mTOR kinase by specific inhibitor, rapamycin or nutrient deprivation results in activation of autophagy (Mizushima et al, 2008). Once triggered, the morphologically detectable phase of autophagic execution can be divided in several stages, beginning with formation of isolation membrane and its elongation, followed by completion of an autophagosome and finally maturation through fusion with lysosomes and degradation of the lumenal content (Mizushima et al, 2008). More than 31 autophagy-related genes (ATG) have been identified in yeast with the core ATG subset being conserved in mammalian cells (Xie and Klionsky, 2007). Formation and elongation of an autophagic isolation membrane (phagophore) requires two unique protein conjugation systems, resulting in the formation of an Atg5–Atg12 conjugate, in a noncovalent complex with Atg16, and in the C-terminally lipid-conjugated LC3 (Atg8) (Xie and Klionsky, 2007; Yoshimori and Noda, 2008). These two systems cooperate in phagophore expansion allowing it to engulf cytosolic targets, to finally close and form a double-membrane organelle termed the autophagosome. Once an autophagosome is formed, the Atg5–Atg12/Atg16 complex is released, whereas a portion of LC3 remains trapped within the autophagosome and is degraded on autophagosome–lysosome fusion to form the lytic organelle called autolysosome in a process referred to as maturation or flux. The degradation of LC3 during flux could provide one level of feedback inhibition in the system.
In addition to the above conjugation systems, phosphatidylinositol 3-phosphate (PI3P) has been implicated in the autophagy pathway based on the pivotal role of the type III PI3Kinase hVps34, an enzyme that generates PI3P, and its autophagy-regulatory partner Beclin-1 (Atg6) (Liang et al, 1999; Levine and Kroemer, 2008; Yoshimori and Noda, 2008). Inhibition of hVps34 blocks formation of autophagosomes, whereas exogenous delivery of PI3P to cells stimulates autophagy (Kabeya et al, 2000; Petiot et al, 2000). The Beclin-1–hVps34 complex is additionally modified by Atg14 or Vps38 (UVRAG) to promote autophagy initiation and maturation stages, respectively (Itakura et al, 2008; Liang et al, 2008). Thus, much has been learned about the activation of the autophagic pathway. In sharp contrast, little is known about its intensity modulation and downregulation once hVps34 and Beclin-1 are activated (Wei et al, 2008). The location of PI3P formation and its enzymatic turnover in the context of autophagy in mammalian cells remain undefined. Furthermore, a role for a PI3P phosphatase has not been implicated in any of the autophagy systems, possibly given that Ymr1, the sole yeast myotubularin-like protein (Laporte et al, 1998) (myotubularins in mammalian cells act as PI3P phosphatases), has not been identified in screens for Atg proteins. Considering the apparent absence of a role for a PI3P turnover in yeast, in which nearly all known key autophagy factors have been pioneered and identified, it is possible that PI3P turnover might not be of significance for autophagy in mammalian systems and in general.
Myotubularins (MTM) are a family of lipid phosphatases (Figure 1A) that contain the consensus signature of the tyrosine and dual-specificity phosphatase, His-Cys-X2-Gly-X2-Arg. They specifically dephosphorylate PI3P and PI(3,5)P2 at the D3 position (Laporte et al, 2003; Clague and Lorenzo, 2005; Robinson and Dixon, 2006). Sixteen MTM family members (MTM1, MTMR1-15) have been identified in humans, but only nine of them seem to have catalytic activity (Alonso et al, 2004). PI3P synthesis takes place on membranes of diverse intracellular organelles; therefore, it is likely that different myotubularins control specific cognate intracellular pools of PI3P (Laporte et al, 2003; Clague and Lorenzo, 2005; Robinson and Dixon, 2006). However, the intracellular site of action and specific function of myotubularins remain largely unknown. Here, we tested the hypothesis that one or more myotubularins may control autophagy in mammalian cells. We screened all active myotubularin family members by siRNA knock-downs for their effects on autophagy. We found that Jumpy (also known as MTMR14) and, to a lesser extent MTMR6, affect autophagy. We show that Jumpy inhibits autophagy by acting at the autophagic isolation membrane stage, and that this function is associated with a specific form of human genetic disease called centronuclear myopathy (Tosch et al, 2006).
Figure 1.
Screening of active members of the myotubularin family identifies Jumpy as a negative regulator of autophagy. (A) Domains and members of catalytically active myotubularins. Asterisks, Jumpy mutations found in patients with centronuclear myopathy. C330S, R336Q and Y462C, Jumpy mutants used in this study. (B) RAW 264.7 cells transfected for 48 h with control (sc), MTMR6 or Jumpy siRNA, were pretreated for 30 min with 100 nM Bafilomycin A1 (BafA1), 10 μg/ml E64d and 10 μg/ml pepstatin, then incubated for 30 min in full or starvation media in the presence of BafA1, E64d and pepstatin. Cells were lysed and analysed by immunoblotting with anti-LC3 or anti-actin. Densitometric LC3-II/actin ratios are shown underneath the blot. Graph: LC3-II/actin ratio for Jumpy siRNA in full medium is equal to or exceeds LC3-II/actin ratio for control siRNA (sc) in starvation medium. (C, D) RAW 264.7 cells were transfected for 36 h with control (scramble) or Jumpy siRNA, transfected once more with corresponding siRNA and mRFP-GFP-LC3 DNA construct overnight and incubated for 2 h in full or starvation media. Cells were fixed and LC3 puncta analysed by confocal fluorescence microscopy. (C) Representative confocal images of RAW 264.7 cells in full or starvation media after transfection with control (scramble) or Jumpy siRNA (siJumpy). Red and yellow arrows indicate GFP−RFP+ and GFP+RFP+ puncta, respectively. (D) Quantitation of number of LC3 puncta per cell, total puncta per cell (GFP+RFP+ and GFP−RFP+ puncta) GFP+RFP+ puncta per cell and GFP−RFP+ puncta per cell. Data, mean±s.e.m. n=5 independent experiments, 30 cells per experiments. *P<0.05, **P<0.01, ***P<0.001 (t-test). Scale bars, 5 μm. (E, F) C2C12 myoblasts were transfected for 48 h with control (scramble) or Jumpy siRNA, followed by a second transfection overnight with corresponding siRNA and mRFP-GFP-LC3 DNA construct. Cells were fixed and LC3 puncta were counted by confocal fluorescence microscopy. (E) Representative confocal images of C2C12 cells in full media after transfection with control (scramble) or Jumpy siRNA (siJumpy). (F) Quantification of the number of LC3 puncta per cell. Data, mean±SEM for n=3 (independent experiments), 30 cells per experiments. *P <0.05 (t-test). Scale bars, 10 μm. (G, H) C2C12 cells were transfected for 48 h with control (sc) (lanes 1, 2, 5, 6) or Atg5 siRNA (lanes 3, 4, 6, 7) followed by a second transfection for 48 h with same siRNA and control (lanes 1, 3, 5, 7) or Jumpy siRNA (lanes 2, 4, 6, 8). Cells were incubated for 1 h with or without 100 nM Baf A1, 10 μg/ml E64d and 10 μg/ml pepstatin in full media, lysed and analysed by immunoblotting with anti-LC3 or anti-actin (G). (H) Densitometric LC3-II/actin ratios for samples treated with BafA1 and protease inhibitors from G (lanes 5–8). Inset shows Atg5 knock-down by immunoblotting.
Results
PI3P phosphatase screen identifies Jumpy and MTMR6 as autophagy regulators
One of the key assays to monitor autophagy is LC3 immunoblotting (Kabeya et al, 2000). On autophagy induction, LC3-I is C-terminally lipidated with phosphatidylethanolamine to form LC3-II. The LC3-I to LC3-II conversion can be conveniently monitored by electrophoretic mobility shift from a slower migrating form, LC3-I, to the faster migrating form LC3-II. LC3 conversion depends on PI3kinase activity (Kabeya et al, 2000). To test whether myotubularins are involved in autophagic processes, the enzymatically active members of the myotubularin family were knocked-down with siRNA in RAW 264.7 macrophages and baseline or induced LC3-II levels analysed by immunoblotting. RAW 264.7 macrophages were transfected for 48 h with control (scramble) siRNA or siRNA specific for each of the myotubularins, incubated for 2 h in full or starvation media and probed for LC3 conversion by quantifying LC3-II relative to the loading control. Among the eight catalytically active murine myotubularins (MTM1, MTMR1, 2, 3, 4, 6, 7 and Jumpy) (Figure 1A), only MTMR6, and Jumpy knock-downs changed LC3-II/loading control ratios (Supplementary Figure S1A–C). Knock-down of individual myotubularins were confirmed by RT–PCR or by immunoblotting (Supplementary Figure S2) except for MTMR7. Treatment with siRNA against Jumpy (Supplementary Figure S1B and D) and MTMR6 (Supplementary Figure S1C and F) increased LC3-II/GAPDH after 2 h starvation in comparison to control siRNA.
To discern whether LC3-II increase following Jumpy or MTMR6 knock-downs was due to autophagy stimulation or inhibition of LC3-II degradation, Bafilomycin A1 (BafA1) and lysosomal protease inhibitors were added during the 30-min treatment (Mizushima and Yoshimori, 2007). Jumpy and MTMR6 knock-downs increased LC3-II in starvation media in the presence of inhibitors in excess to cells transfected with control siRNA (Figure 1B). The effect of Jumpy knock-down on LC3-II was more pronounced than MTMR6. Of note is that increase in basal autophagy (full medium) as a result of Jumpy knock-down equaled or exceeded in magnitude LC3-II conversion caused by starvation (Figure 1B, graph). Thus, Jumpy is needed to prevent spurious activation of autophagy when it is not physiologically induced (e.g. by starvation). The effects of Jumpy siRNA on enhanced LC3-II levels were also pronounced in starvation media (Figure 1B, starvation lanes). These results indicate that Jumpy knock-down increases both basal and starvation-mediated autophagy. Importantly, the effect of Jumpy knock-down was stronger than or as strong as the effect of starvation (Figure 1B, graph), which is used as a key physiological inducer of autophagy applied as the gold standard in nearly all autophagic studies (Levine and Deretic, 2007; Levine and Kroemer, 2008; Mizushima et al, 2008). Next, we determined whether the raise in LC3-II level was accompanied by changes in autophagic flux. LC3-II levels were measured in the presence or absence of flux inhibitors. Jumpy siRNA increased autophagic flux both in full and starvation media (Supplementary Figure S3), indicating that Jumpy acts as a suppressor of the autophagy pathway.
Jumpy controls autophagosome formation and maturation in macrophages
As Jumpy siRNA showed the strongest effect on LC3-II levels, we focused on this PI3P phosphatase. To determine whether the increase in autophagosome formation caused by Jumpy-knock-down also led to maturation into autolysosomes, we used mRFP-GFP-LC3 probe developed to differentiate early from late autophagic organelles (Kimura et al, 2007). On autophagy induction, the lipidated LC3-II associates with autophagosomal membranes, resulting in the formation of punctate organelles that can be quantified by fluorescence microscopy (Kabeya et al, 2000). mRFP-GFP-LC3 allows distinction between early autophagic organelles (GFP+RFP+ puncta) and mature, acidified autolysosomes (GFP−RFP+ puncta) as the GFP signal is quenched in acidic compartments (Filimonenko et al, 2007; Kimura et al, 2007). RAW 264.7 macrophages knocked-down for Jumpy and transfected with mRFP-GFP-LC3 displayed increased total (GFP+RFP+ plus GFP−RFP+) LC3 puncta per cell in both full and starvation media (Figure 1C and D; Supplementary Figure S4). The number of GFP+RFP+ puncta per cell remained the same in full media and decreased in starvation media with Jumpy siRNA-treated cells in comparison to control (Figure 1C and D; Supplementary Figure S4). In contrast, Jumpy knock-down resulted in increased GFP−RFP+ puncta representing maturing autophagosomes (Figure 1C and D; Supplementary Figure S4). In conclusion, Jumpy knock-down increased the total number of autophagic organelles, indicating that reduced Jumpy expression stimulated autophagosome formation. Furthermore, Jumpy knock-down facilitated maturation of the newly formed autophagosomes resulting in increased autolysosome numbers instead of accumulation of autophagosomes. These data show that Jumpy affects formation of autophagosomes and their maturation into autolysosomes.
Jumpy inhibits autophagy in C2C12 cells
We next confirmed that the effects of Jumpy knock-downs on autophagy detected in macrophages applied to other cell types, by testing HeLa cells, mouse neuroblast Neuro-2A, MEF cells (Supplementary Figures S5 and S6A) and myoblast cell line C2C12 (Figure 1E–F; Supplementary Figure S6C). Muscle cells were of particular interest, as missense mutations in human populations within the Jumpy gene are associated with sporadic cases of centronuclear myopathy (Tosch et al, 2006). The mutations seen in patients result in a loss or decrease in Jumpy PI3P phosphatase activity (Tosch et al, 2006). Furthermore, overexpression of Jumpy results in cellular PI3P decrease (Tosch et al, 2006). Although Jumpy enzymatic activity is well defined (Tosch et al, 2006), its cellular function and the cell biological consequences of its loss are completely unknown. Therefore, we focused on testing further whether and how Jumpy regulated autophagy, by using mouse myoblast cell line C2C12. C2C12 myoblasts were transfected for 48 h with Jumpy siRNA and incubated in full or starvation media for 4 h in the presence or absence of BafA1. In both full and starvation media, Jumpy siRNA increased LC3-II levels compared with control samples. This was also observed in the presence of autophagic flux inhibitor, BafA1 (Supplementary Figure S6B and C), indicating an increase in autophagosome formation.
We next assessed the maturation state of the autophagosomes in C2C12 cells using the mRFP-GFP-LC3 construct (Kimura et al, 2007). C2C12 were transfected for 48 h with control or Jumpy siRNA, followed by a second transfection as above plus mRFP-GFP-LC3 and incubated for 4 h in full or starvation media. Although a knock-down of Jumpy did not change the number of GFP+RFP+ puncta (early autophagic organelles) per cell in comparison to control, Jumpy siRNA resulted in an increase in total and GFP−RFP+ (mature) LC3 puncta (Figure 1E and F; Supplementary Figure S6D and E). The results indicate that, as in RAW 264.7 macrophages, Jumpy inhibits basal and starvation-induced autophagy levels in C2C12 cells, and that its loss results in increased autophagy.
To show that Jumpy siRNA-induced LC3-II increase required autophagy machinery, Atg5, protein essential for LC3-II formation (Mizushima et al, 2001), was knocked-down and LC3-II level measured after 48 h transfection with control (sc) and/or Atg5 and Jumpy siRNA (Figure 1). Atg5 knock-down inhibited LC3-II increase observed with Jumpy siRNA treatment (Figure 1) showing that Jumpy knock-down enhances autophagy in Atg5-dependent manner. As expected with MEF Atg7−/−, no LC3-II was observed in cells transfected with control or Jumpy siRNA (Supplementary Figure S5).
Jumpy controls autophagic degradation
To test whether the increase of autophagy associated with Jumpy knock-down resulted in a functional autophagic process, we examined effects on long-lived proteins, a canonical substrate for autophagic turnover. Transfection with Jumpy siRNA resulted in increased rate of proteolysis in C2C12 cells observed both in full and starvation media compared with control siRNA (Figure 2A).
Figure 2.
Jumpy blocks autophagic degradation. (A) Proteolysis of long-lived proteins in C2C12 myoblasts. C2C12 cells were transfected with control (scramble) or Jumpy siRNA (siJumpy), labelled overnight in media containing [3H] leucine, washed, incubated for 2 h in complete media (containing cold leucine) and incubated for 4 h in full or starvation media. Leucine release was calculated from radioactivity in the tricarboxylic acid-soluble form relative to total cell radioactivity. Results shown represent mean±s.e.m. for combined data from three independent experiments. (B) Quantitation of p62 protein levels. C2C12 cells were transfected for 48 h with control (sc) or Jumpy siRNA and p62 levels analysed by immunoblotting, quantitated by densitometry and represented as a percentage of control. Data are mean±s.e.m. (n=4 independent experiments). (C) C2C12 cells were transfected for 48 h with control (sc) or Jumpy siRNA, incubated for 4 h with or without 100 nM Bafilomycin A1 in full media, lysed, probed for endogenous p62 and actin by immunoblotting and percentage of p62 were quantitated (mean±s.e.m., n=3). (D) C2C12 cells were transfected for 48 h with control (sc) (lanes 1, 2) or Beclin siRNA (lanes 3, 4) followed by a second transfection for 48 h with same siRNA and control (lanes 1, 3) or Jumpy siRNA (lanes 2, 4). Cells were lysed, probed for p62, Beclin and actin by immunoblotting and percentage of p62 were quantitated (mean±s.e.m., n=5). (E–M) C2C12 cells were transfected for 48 h with GFP, GFP-Jumpy (Jumpy), GFP-MTM1 (MTM1) or GFP-MTMR2 (MTMR2), fixed and immunostained with anti-p62 antibody (red). p62 puncta were quantitated by confocal fluorescence microscopy. Representative confocal images of GFP (E), GFP-Jumpy (F), GFP-MTM1 (G), GFP-MTMR2 (H) transfected cells, immunostained for p62 (I), (J), (K) and (L), respectively. Scale bars, 5 μm. White lines represent outline of the cells. Quantitation of p62 puncta per cell (M). Bars, SEM (n=3 independent experiments, with an average of 30 cells per experiments). (N, O) C2C12 cells were transfected for 48 h with GFP, GFP-Jumpy (Jumpy), GFP-MTM1 (MTM1) or GFP-MTMR2 (MTMR2), lysed, analysed for p62, GFP and actin by immunoblotting (N) and percentage of p62 were quantitated (mean±s.e.m., n=3) (O). (P, Q) C2C12 cells were transfected for 48 h with GFP or GFP-Jumpy (Jumpy), incubated with or without 100 nM Bafilomycin A1 (BafA1) for 4 h, lysed, analysed for p62, GFP and actin by immunoblotting (P) and percentage of p62 were quantitated (mean±s.e.m., n=4) (Q). *P<0.05, ***P<0.001, †ns (t-test).
A common adapter for autophagic substrates, p62 (Bjorkoy et al, 2005), is degraded along with the substrates it delivers to autophagosomes (Bjorkoy et al, 2005; Filimonenko et al, 2007; Komatsu et al, 2007). We tested whether Jumpy affected p62 levels. After 48 h transfection with Jumpy siRNA, p62 protein levels were decreased by 20±2% compared with control samples (Figure 2B). This decrease was rescued after 4 h treatment with BafA1, indicating that the degradation was lysosomal in nature, consistent with p62 proteolysis in autolysosomes (Figure 2C). When we knocked-down Beclin-1, a key autophagy regulator (Liang et al, 1999) acting in complexes with hVps34, this completely erased the incremental increase in p62 degradation observed in cells treated with Jumpy siRNA (Figure 2D), indicating that Jumpy knock-down resulted in elevated p62 degradation through autophagy. These data along with the increased LC3-I to LC3-II conversion detected by immunoblotting, elevated LC3 puncta formation, increased autophagic flux and increased stable protein degradation show that Jumpy negatively controls autophagy.
Jumpy but not other myotubularins inhibits p62 degradation
We next tested, in an experimental set up converse to siRNA knock-down studies, whether Jumpy overexpression inhibited autophagy. As p62-positive protein aggregates increase with decreased autophagy (Filimonenko et al, 2007; Komatsu et al, 2007), we monitored the number of p62 puncta per cell. C2C12 myoblasts were transfected with GFP or GFP-Jumpy expression constructs and immunostained for endogenous p62. Cells transfected with Jumpy showed an eight-fold increase in p62 aggregates compared with controls (Figure 2E, F, I, J and M). To establish a specificity for Jumpy, C2C12 cells were also transfected with GFP-MTM1 or GFP-MTMR2. Overexpression of MTM1 and MTMR2 led to fewer p62 aggregates than Jumpy overexpression (Figure 2G, H, K, L and M), indicating that Jumpy specifically affects p62 degradation relative to other myotubularins tested. In contrast to MTM1- and MTMR2-transfected cells used as comparison controls, p62 did not appear in Lamp1-positive compartments in Jumpy-transfected even after addition of BafA1 (Supplementary Figure S7), showing that the p62 aggregates induced by Jumpy overexpression were not delivered to lysosomes. Accumulation of p62 was also observed by immunoblotting in lysates from cells overexpressing Jumpy (Figure 2N and O). Again, MTM1 and MTMR2 showed no effect like Jumpy in increasing levels of p62, supporting the immunofluorescence findings. To verify that accumulation of p62 was due to inhibition of autophagy, cells transfected with GFP or GFP-Jumpy were treated with BafA1 for 4 h. In contrast to control samples, BafA1 did not increase p62 levels in GFP-Jumpy-transfected cells (Figure 2P and Q), indicating that in cells overexpressing Jumpy autophagic degradation of p62 was already inhibited. The p62 accumulation observed in Jumpy-transfected cells correlated with an increase in large tdTomato-LC3 puncta (Supplementary Figure S8) similar to the large LC3 puncta accumulating in Atg14-depleted cells (Zhong et al, 2009). Collectively, our data unequivocally show that the PI3P phosphatase Jumpy inhibits autophagy.
Jumpy localizes to autophagic isolation membranes and autophagosomes
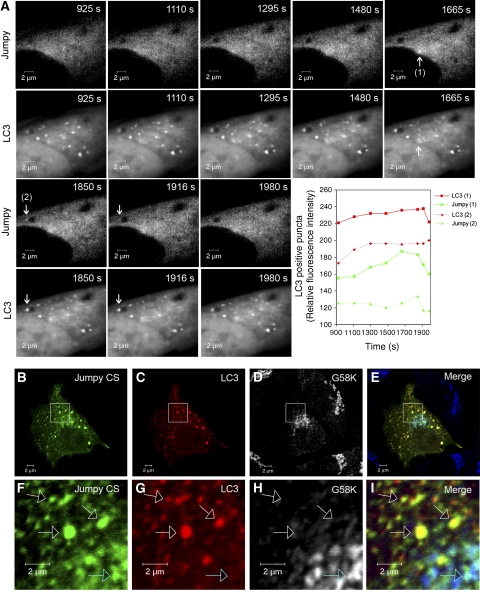
Next, we asked whether Jumpy acted directly on autophagic membranes, by examining the localization of Jumpy relative to autophagic organelles in C2C12 cells. As GFP-Jumpy WT was found mainly diffuse in the cytosol of C2C12 myoblasts (Figure 2F), we considered the possibility that Jumpy recruitment to autophagic membranes might be transient. C2C12 myoblasts were transfected with GFP-Jumpy WT and Cherry-LC3, autophagy was induced by starvation, and GFP-Jumpy WT dynamics relative to Cherry-LC3 autophagosomes was monitored by live confocal fluorescence microscopy. We detected association of GFP-Jumpy WT with Cherry-LC3 puncta (Figure 3A; Supplementary Movie S1), suggesting that Jumpy is physically present, albeit transiently, on autophagic organelles. The mean duration of Jumpy association with autophagosomes was 7.5 min (±2.5 min; range) (Figure 3A, graph), during which a cycle of adsorption and desorbtion of GFP-Jumpy WT was completed.
Figure 3.
Jumpy localizes to autophagosomes. (A) Transient association of Jumpy WT with LC3+ organelles. Time lapse sequence of C2C12 myoblasts transfected for 24 h with GFP-Jumpy WT and Cherry-LC3 and analysed by live confocal microscopy in a 5LIVE Zeiss microscope. EBSS was added to the cells and z-stacks collected at 3-min intervals for a total of 45 min. The collected images were processed to generate a maximum projection (collapsing a 3D image into an x–y projection) for each time point. Arrows indicate colocalization of GFP-Jumpy WT with LC3. Graph: Plots of relative fluorescence intensity (GFP fluorescence pixel intensity) over time of GFP-Jumpy WT colocalizing with Cherry-LC3+-positive puncta indicated by arrows. (B–I) Association of Jumpy CS mutant with autophagic organelles. C2C12 cells were transfected for 24 h with GFP-Jumpy C330S (Jumpy CS) and tdTomato-LC3 (LC3), fixed and immunostained with anti-G58K (Alexa 648-labelled secondary antibody). Jumpy CS (green), LC3 (red) and G58K (white in D, H or blue in E, I). Boxed areas (B–E) are shown at higher magnification in the corresponding panel below (F–I). Scale bars, 2 μm. White arrows indicate colocalization between Jumpy CS, LC3 but not G58K, blue arrow indicates colocalization between Jumpy CS and G58K but not LC3.
It has been reported that the catalytically inactive Jumpy variant C330S (Jumpy CS, expected to act as a substrate-locked mutant), in contrast to the diffuse cytosolic localization of Jumpy WT localizes to distinct intracellular compartments, that is, Golgi and undefined peripheral punctate structures (Tosch et al, 2006). To characterize the nature of the peripheral Jumpy CS profiles (Figure 3B) and given our functional findings, we tested whether they might be autophagosomal in nature. We first ascertained that GFP-Jumpy CS is not a substrate for autophagy (Supplementary Figure S6F). Next, we examined Jumpy CS localization relative to LC3. C2C12 cells were transfected with GFP-Jumpy CS and tdTomato-LC3 and immunostained for Golgi marker G58K. As anticipated (Tosch et al, 2006), GFP-Jumpy CS partially colocalized with the Golgi marker G58K but was also present on peripheral structures as noted earlier (Tosch et al, 2006) (Figure 3B–I; Supplementary Table S1). Importantly, we found that the peripheral Jumpy CS-positive G58K-negative structures were positive for LC3 (Figure 3B–I; Supplementary Table S1). A similar colocalization between tdTomato-LC3 and Jumpy CS was observed in RAW 264.7 macrophages (Supplementary Figure S9A–C). The localization of Jumpy CS to LC3-positive organelles was not due to Jumpy CS being an autophagic substrate (e.g. as a protein aggregate) as GFP-Jumpy CS was not a substrate for autophagy (Supplementary Figure S6F).
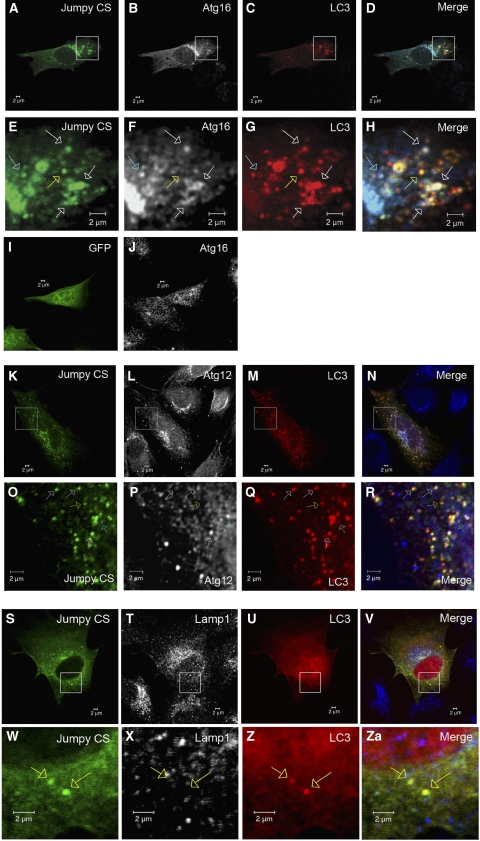
To determine how early Jumpy acts in autophagic organelle development, we analysed Jumpy CS localization relative to autophagic isolation membrane (phagophore) markers, Atg16 and Atg12. Atg16 forms a multimeric complex with Atg5–Atg12 and is found exclusively on phagophores in mammalian cells or the preautophagosomal structures in (PAS) yeast (Mizushima et al, 2003; Fujita et al, 2008). C2C12 cells were transfected with GFP-Jumpy CS and tdTomato-LC3 and immunostained for endogenous Atg16. A portion of Jumpy CS-positive structures were positive for both LC3 and Atg16, consistent with the phagophore stage (Figure 4A–H; Supplementary Table S1). Another subset of profiles was only positive for Jumpy CS and LC3 (i.e. were Atg16 negative), indicating that Jumpy CS was also present on complete autophagosomes. Of further note is that in cells transfected with Jumpy CS, Atg16, predominantly present in the cytosol, redistributed to the perinuclear region profiles that were LC3 negative (Figure 4A, B, I and J).
Figure 4.
Jumpy localizes to autophagic isolation membranes. (A–H) C2C12 cells were transfected for 24 h with GFP-Jumpy C330S (Jumpy CS) and tdTomato-LC3 (LC3), fixed and immunostained with anti-Atg16 (Alexa 648-labelled secondary antibody). Jumpy CS (green), LC3 (red) and Atg16 (white in B and F or blue in D and H). Boxed areas (A–D) are shown at higher magnification in the corresponding panel below (E–H). White arrows indicate colocalization among Jumpy CS, LC3 and Atg16, yellow arrow indicates colocalization between Jumpy CS and LC3 but not Atg16, blue arrow indicates colocalization between Jumpy CS and Atg16 but not LC3. (I, J) C2C12 cells were transfected for 24 h with GFP (I), fixed and immunostained with anti-Atg16 (J). (K–R) C2C12 cells were transfected for 24 h with GFP-Jumpy C330S (Jumpy CS) and tdTomato-LC3 (LC3), fixed and immunostained with anti-Atg12(M) antibody. Jumpy CS (green), LC3 (red) and Atg12 (white in L, P or blue in N, R). Boxed areas (K–N) are shown at higher magnification in the corresponding panel below (O–R). White arrows indicate colocalization among Jumpy CS, LC3 and Atg12, yellow arrow indicates colocalization between Jumpy CS and LC3 but not Atg12, blue arrow indicates colocalization between Jumpy CS and Atg12 but not LC3. (S–ZA) C2C12 cells were transfected for 24 h with Jumpy CS and Cherry-LC3 (LC3), fixed and immunostained with anti-lamp1. Jumpy CS (green), LC3 (red) and Lamp1 (white in T, X or blue in V, ZA). Boxed areas (S–V) are shown at higher magnification in the corresponding panel below (W–ZA). Yellow arrows indicate colocalization between Jumpy CS and LC3 but not Lamp1. Scale bars, 2 μm.
To firm up the notion that Jumpy CS associates with autophagic isolation membranes, we next analysed Jumpy CS localization relative to endogenous Atg12. A portion of Jumpy CS-positive structures were positive for both tdTomato-LC3 and Atg12 (Figure 4K–R; Supplementary Table S1), whereas another subset of profiles were only positive for Jumpy CS and LC3 (i.e. were Atg12 negative) (Figure 4K–R), confirming that Jumpy CS is present on both autophagic isolation membranes (phagophores) and autophagosomes. A similar colocalization was observed among B10-tagged Jumpy CS, GFP-LC3 and Atg12 (Supplementary Figure S10A), validating that Jumpy CS localizes to isolation membranes irrespective of its tag. Furthermore, those Jumpy CS-positive structures that were positive for tdTomato-LC3 did not colocalize with Lamp1 (Figure 4S–ZA; Supplementary Table S1), showing that Jumpy CS is not present on autolysosomes. Furthermore, GFP-Jumpy CS did not colocalize with the endosomal PI3P-binding protein, EEA1 (Supplementary Figure S10B and Table S1), indicating that Jumpy CS associates specifically with autophagic membranes. Collectively, these data show that Jumpy has a function in autophagy by associating with early autophagic organelles, acting as early as at the phagophore stage.
Jumpy regulates recruitment of the PI3P-binding autophagy factor WIPI-1 (Atg18) to autophagic membranes
WIPI-1 is the mammalian orthologue of the yeast Atg18, the only thus far characterized autophagic PI3P-binding protein that participates in the formation of autophagosomes (Proikas-Cezanne et al, 2007; Xie and Klionsky, 2007). WIPI-1 partially colocalizes with LC3 positive membranes but not with mature autophagosomes (Proikas-Cezanne et al, 2004, 2007). As Jumpy is present on Atg16-positive membranes and the relationship between Atg16 and Atg18 is not known, we first examined intracellular distribution of WIPI-1 relative to Atg16. We found that WIPI-1 structures colocalized with Atg16 (Figure 5A–C). Thus, WIPI-1 puncta represents autophagic isolation membranes. As WIPI-1 is recruited to autophagic membranes in a PI3P-dependent manner (Proikas-Cezanne et al, 2007) and both WIPI-1 and Jumpy were present on autophagic isolation membranes, we hypothesized that Jumpy levels might regulate WIPI-1 puncta formation. We monitored GFP-WIPI-1 distribution in C2C12 myoblast cells by measuring the number of WIPI-1 puncta per cell. Starvation induced a 10-fold increase in the number of WIPI-1 puncta (Figure 5D, E and I). The WIPI-1 puncta formation was inhibited by wortmannin (Figure 5F and I) confirming that the formation of WIPI-1 puncta required PI3P synthesis and that they were not the result of nonspecific GFP-WIPI-1 protein aggregation. Next, the effect of Jumpy knock-down was examined. C2C12 cells were transfected with control or Jumpy siRNA, followed by second transfection with GFP-WIPI-1, and autophagy was induced by starvation. Jumpy siRNA resulted in a five-fold increase in the number of GFP-WIPI-1 puncta per cell in full media (Figure 5D, G and I) and a 50% increase in starvation media (Figure 5E, H and I). These data show that Jumpy prevents PI3P-dependent WIPI-1 recruitment to autophagic membranes.
Figure 5.
Jumpy siRNA increases recruitment of PI3P-binding Atg factor WIPI-1 (Atg18) to autophagic membranes. (A–C) C2C12 were transfected for 48 h with Jumpy siRNA, followed by a second transfection overnight with Jumpy siRNA and GFP-WIPI-1, incubated for 4 h in starvation media, fixed and immunostained with anti-Atg16 antibody (red). GFP-WIPI1-transfected cell (A), endogenous Atg16 immunostaining (B), colocalization of GFP-WIPI-1 and Atg16 is indicated in yellow in merge image (C). Boxed areas are shown at higher magnification as inset. Scale bars, 10 and 5 μm in insets. (D–I) C2C12 cells were transfected for 48 h with control (scramble) (D, E) or Jumpy siRNA (F–H), followed by a second transfection overnight with corresponding siRNA and GFP-WIPI-1. Cells were incubated for 4 h in full (D, G) or starvation media (E, F, H) in presence (F) or absence (E–H) of 100 nM wortmannin, fixed and analysed by confocal fluorescence microscopy. Quantitation of WIPI-1 puncta per cell (I). Bars, s.e.m. (n=3 independent experiments, with an average of 30 cells per experiment). *P<0.05, **P<0.01 (t-test).
Jumpy increases Atg9 association with autophagic organelles
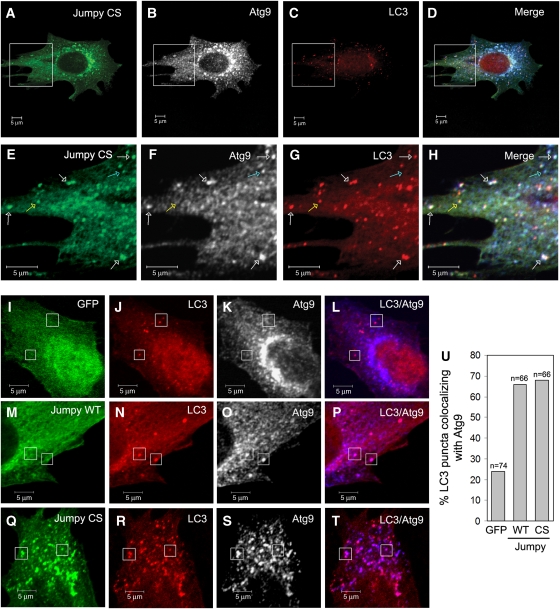
In yeast, Atg18 has a function in the retrieval of Atg9 from PAS (Xie and Klionsky, 2007). Atg9, the only known mammalian transmembrane Atg protein, is required for autophagosome formation and is present on PAS in yeast and on LC3-positive compartments in mammalian cells (Noda et al, 2000; Young et al, 2006). The Atg9 cycling is believed to be essential for autophagosome formation (Xie and Klionsky, 2007). As Jumpy impaired WIPI-1 (Atg18) recruitment to autophagic membranes, we next asked whether Jumpy affected Atg9 distribution vis-a-vis autophagic organelles. First, we examined the localization of Jumpy CS relative to exogenously expressed Atg9. C2C12 cells were transfected with GFP-Jumpy CS, tdTomato-LC3 and HA-Atg9 and immunostained for HA. We found more than 50% colocalization between Jumpy CS and Atg9 with a portion of Jumpy CS- and Atg9-positive structures colocalizing with LC3 (Figure 6A–H). Next, we measured the percentage of LC3 puncta colocalizing with Atg9 in cells transfected with GFP, GFP-Jumpy CS or GFP-Jumpy WT. Both Jumpy WT and Jumpy CS increased by three-fold the percentage of LC3 puncta that were Atg9 positive compared with control GFP-transfected cells (Figure 6I–U). This shows that Jumpy causes Atg9 accumulation on autophagic organelles most likely by blocking its PI3P and Atg18-dependent cycling.
Figure 6.
Jumpy colocalizes with and regulates Atg9 distribution to autophagic organelles. (A–T) C2C12 cells were transfected for 24 h with GFP, GFP-Jumpy WT or GFP-Jumpy C330S (Jumpy CS) and Tomato-LC3 (LC3) and HA-Atg9, fixed and immunostained with anti-HA. GFP (I), Jumpy WT (M), Jumpy CS (green) (A, E, Q), LC3 (red) (C, G, J, N, R) and Atg9 (white in B, F, K, O, S or blue in D, H, I, P, T). Boxed areas (A–D) are shown at higher magnification in the corresponding panel below (E–H). White arrows indicate colocalization between Jumpy CS, LC3 and Atg9, blue arrow indicates colocalization between Jumpy CS and Atg9 but not LC3 and yellow arrow indicates colocalization between Jumpy CS and LC3 but not Atg9. White boxes (I–T) show LC3 puncta location. Scale bars, 5 μm. (U) Percentage of LC3 puncta colocalizing with Atg9. n, number of puncta counted.
Jumpy variant associated with centronuclear myopathy is defective in downregulating autophagy
Two Jumpy missense variants have been described in patients with centronuclear myopathy (Tosch et al, 2006) (i) a missense mutation, R336Q (RQ), within the catalytic site that abrogates Jumpy phosphatase activity and (ii) the missense Y462C (YC) mutation outside of the phosphatase domain, which has a mild effect (80% of wild-type phosphatase activity) (Tosch et al, 2006). We tested these naturally occurring Jumpy variants for their effects on autophagy. We first measured p62 puncta accumulation in C2C12. Both Jumpy WT and Jumpy YC increased the number of p62 puncta compared with control (YFP) (Figure 7A, B, D, F, G, I and K). In contrast, Jumpy RQ did not promote accumulation of p62 puncta and showed a similar number of puncta as seen in controls (YFP) (Figure 7A, E, F, J and K). These data suggest that a loss of Jumpy PI3P phosphatase activity associated with the R336Q Jumpy allele leads to increased autophagy.
Figure 7.
Jumpy R336Q mutant associated with centronuclear myopathy is defective in inhibiting autophagy. (A–K) C2C12 cells were transfected for 48 h with YFP, YFP-Jumpy WT (Jumpy WT), YFP-Jumpy C330S (Jumpy CS), YFP-Jumpy Y462C (Jumpy YC) or YFP-Jumpy R336Q (Jumpy RQ), fixed and immunostained with anti-p62 antibody (red). p62 puncta were quantitated by confocal fluorescence microscopy. Representative confocal images of YFP (A), YFP-Jumpy WT (B), YFP-Jumpy C330S (C), YFP-Jumpy Y462C (D) and YFP-Jumpy R336Q (E) transfected cells, immunostained for p62 (F), (G), (H), (I) and (J), respectively. Scale bars, 5 μm. Quantitation of p62 puncta per cell (K). Bars, s.e.m. (n=3 independent experiments, 30 cells per experiments). *P<0.05 (t-test), ns: nonsignificant. (L) HeLa cells were transfected for 24 h with YFP, YFP-Jumpy WT (Jumpy WT) or YFP-Jumpy R336Q (Jumpy RQ), incubated with or without 50 ng/μl rapamycin (BafA1 (100 nM) was present in both control and rapamycin treated cells) for 2 h, lysed and analysed for LC3, YFP and actin by immunoblotting. Densitometric LC3-II/actin ratios are shown underneath the blot.
Strikingly, overexpression of Jumpy CS promoted accumulation of p62 puncta and large tdTomato-LC3 puncta as in the case of Jumpy WT overexpression (Figure 7B, C, G, H and K; Supplementary Figure S8). One explanation for this finding is that Jumpy CS acts as a substrate trap and thus blocks PI3P accessibility to its effectors which results in inhibition of autophagy ostensibly similar to the effects of Jumpy WT. Consistent with this possibility, similar mutations in the active site of tyrosine phosphatases result in catalytically inactive phosphatases with substrate-trapping properties (Garton et al, 1996; Flint et al, 1997). This also agrees with the findings that Jumpy CS, in contrast to Jumpy WT, YC and RQ, is mostly found on membranes and not in the cytosol (Figure 7A–E) and that Jumpy CS as Jumpy WT increase number of large LC3 puncta and promote Atg9 accumulation on autophagic organelles (Supplementary Figure S8; Figure 6I–U), supporting the idea that this mutant remains associated with membrane sites in which unimpeded access to specific PI3P patches is needed to promote the PI3P-dependent stages of autophagy.
The effects of Jumpy WT and RQ on LC3 lipidation were compared next. Cells were transfected with YFP-Jumpy WT or YFP-Jumpy RQ construct, autophagy induced in the presence of BafA1 to prevent autophagic degradation of LC3-II, and levels of LC3-II assessed by immunoblotting. Overexpression of Jumpy WT inhibited LC3-II conversion associated with autophagy induction in HeLa (Figure 7L) or C2C12 cells (Supplementary Figure S11), in keeping with the role of Jumpy phosphatase activity in inhibition of early stages of autophagy. In contrast, overexpression of the inactive Jumpy RQ mutant did not decrease LC3-II conversion but instead increased LC3-II levels, suggesting that Jumpy RQ acts as a dominant negative mutant (Figure 7L; Supplementary Figure S11). These experiments show that the Jumpy RQ variant associated with centronuclear myopathy is unable to downregulate autophagy.
Discussion
Our study establishes that the PI3P phosphatase Jumpy negatively regulates autophagy. This represents a paradigm shift from the notion that autophagy initiation in mammalian cells is controlled exclusively by regulating PI 3-kinase activity, a concept that now has to account for the role of a specific PI3P phosphatase. Jumpy acts on discrete domains of nascent autophagic membranes, and orchestrates succession of Atg factors controlling autophagy initiation. At a macro-level, Jumpy acts as a ‘brake' in the autophagy pathway. This brake is applied at the PI3P-dependent initiation stage of the execution phase of autophagy. A model emerges in which it is the balance between the PI3P production (regulated by hVp34) and the PI3P hydrolysis (regulated by Jumpy) that determines induction and baseline levels of autophagy, rather than the forward reaction alone, which has been thus far the nearly exclusive view of the process. In addition, our findings link a human myopathy to altered regulation of autophagy by Jumpy, highlighting the significance of autophagosomal PI3P regulation by a phosphatase in a physiologically and medically relevant context.
Despite its critical regulatory role, the mechanistic details of PI3P function during initiation of autophagy are surprisingly lacking (Yoshimori and Noda, 2008). Both in yeast and mammalian cells, PI3P production is necessary for autophagy initiation, the Atg16-directed recruitment of the Atg5–Atg12 complex to PAS or autophagic isolation membranes (Fujita et al, 2008) and for the linked conversion of LC3-I into LC3-II (Kabeya et al, 2000; Xie and Klionsky, 2007). More recently, PI3P-enriched compartments, derived from endoplasmic reticulum, have been proposed to possibly have a function in autophagosome biogenesis (Axe et al, 2008). However, how PI3P is connected to these molecular events is far from being understood. In yeast, PI3P is enriched on inner surfaces of isolation membranes and autophagosomes (Obara et al, 2008) and its production is required for the proper recruitment to autophagic membranes of Atg18, the only thus far characterized PI3P effector among the Atg factors (Reggiori et al, 2004). Atg18 is implicated in the so-called process of retrieval of Atg9 from PAS to peripheral compartments (Reggiori et al, 2004). The purpose of this Atg9 cycling during autophagy remains unclear (Young et al, 2006; Suzuki and Ohsumi, 2007), although it clearly has a function in autophagosome formation (Xie and Klionsky, 2007). In resting mammalian cells, Atg9 is in an equilibrium between the trans-Golgi network and late endosomes, the locations from where it promptly redistributes to LC3+ autophagic organelles, ranging from nascent phagophores to early degradative but still LC3-positive autophagic vesicles (Young et al, 2006). Our data now indicate that overexpression of Jumpy wild type or its CS mutant, freezes autophagic organelles in an Atg9+ and LC3+ dual-positive state. Thus, one can infer that Jumpy regulates specific PI3P domains on early autophagic membranes, at the site and time when key molecular decisions are made to initiate the process of autophagosome formation.
The function of WIPI-1, the mammalian orthologue of yeast Atg18, while presently unknown holds promise in deciphering the precise role of PI3P in autophagy initiation. WIPI-1 is recruited to autophagic membranes after autophagy induction showing partial colocalization with LC3 and Atg16 (observed in this study) and accumulation on cup-shaped membranes (Proikas-Cezanne et al, 2004). As with yeast Atg18, the association of WIPI-1 to autophagic membranes requires PI3P (Proikas-Cezanne et al, 2007). As Jumpy associates with autophagic organelles, in particular phagophores, and regulates WIPI-1 and Atg9 trafficking, we conclude that Jumpy inhibits autophagy by dephosphorylating a pool of PI3P directly involved in WIPI-1 (Atg18) recruitment to autophagic membranes. One of the candidate processes affected by Jumpy through WIPI-1 is the cycling of Atg9, but additional roles in positioning and organization of phagophores cannot be excluded.
Although autophagic machinery is conserved from yeast to humans, Jumpy is not found in yeast (Tosch et al, 2006). Noteworthy, yeast encodes a myotubularin-like protein, Ymr1 (Laporte et al, 1998), although its role in autophagy has not been examined. It remains possible that Ymr1 has a function in autophagy, perhaps observable only under certain physiological conditions similarly to one of the myotubularins identified here as affecting autophagy: the mammalian myotubularins affecting autophagy include Jumpy characterized here, the yet to be examined in full MTMR6, and potentially MTMR7. MTMR7 did not show any effects in the siRNA screen shown in Supplementary Figure S1; it turned out, however, that the mRNA entry for MTMR7 used in the initial screen was based on unstable mRNA species, and when a more stable mRNA species was targeted with siRNAs, MTMR7 mRNA knock-down could be documented and showed some effects on LC3-II (Supplementary Figure S13). The extra layers of regulation in mammalian cells may be necessary to accommodate the additional functions of autophagy in mammals including development of metazoan-specific tissues (Cecconi and Levine, 2008) such as muscles. Of note, autophagosomes are noticeably smaller in skeletal muscles than in other tissues (Mizushima et al, 2004), thus, another role of myotubularins could be the regulation of autophagosomal size, which may be tissue specific and thus perhaps idiosyncratic to multicellular eukaryotes.
Autophagy has been implicated in several types of myopathies, such as Danon disease, Pompe disease and X-linked myopathy with excessive autophagy (Kundu and Thompson, 2008; Levine and Kroemer, 2008; Mizushima et al, 2008). Recently, autophagy has been reported to have an important function during denervation- or fasting-induced skeletal muscle wasting and atrophy (Mammucari et al, 2007; Zhao et al, 2007). Given the general importance of autophagy in muscle physiology and pathology, and the unanticipated link uncovered here between autophagy and Jumpy as a myopathy risk locus, we propose that excessive or improperly regulated autophagy contributes to the pathogenesis of centronuclear myopathy. Centronuclear myopathies are congenital disorders characterized by muscle atrophy and by a peculiar positioning of the nuclei in the center of the skeletal muscle fibres (Pierson et al, 2005). Although, several genes involved in the disease, including dynamin 2, amphiphysin 2, Jumpy and MTM1, have been identified, their cellular functions in relationship to muscle physiology are not understood (Blondeau et al, 2000; Taylor et al, 2000; Bitoun et al, 2005; Tosch et al, 2006; Nicot et al, 2007). Here, we have shown that Jumpy R336Q, a naturally occurring mutation in centronuclear myopathy (Tosch et al, 2006), disables Jumpy in suppressing autophagy. This suggests that autophagy dysregulation may be an important pathogenesis determinant in centronuclear myopathy. Missense mutations in the MTM1 gene, which encodes another prominent member of the myotubularin family, is the most common cause leading to centronuclear myopathy (Laporte et al, 2003). MTM1 protein is found on ruffles at the plasma membrane (Blondeau et al, 2000) and on endocytic compartments (Cao et al, 2007). Although, our screen in macrophages did not identify MTM1 as an autophagy regulator, we cannot exclude the possibility that in muscle cells MTM1 contributes to the regulation of the autophagy pathway.
In summary, our study shows a role for the PI3P phosphatase Jumpy in regulating the early stage of autophagy, Furthermore, we have uncovered a link between a disease-associated Jumpy mutation and autophagy. These findings represent advance in our understanding of the regulation autophagy at the level of PI3P on autophagic membranes during initiation, which now no longer involves only the forward reaction mediated by the hVPS34–Beclin-1 complex but also depends on the Jumpy-dependent turnover of PI3P pools that are relevant for autophagy.
Materials and methods
Cell cultures
C2C12 muscle myoblast, and murine RAW264.7 macrophage cell lines were from ATCC and were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS) and L-glutamine (complete media). HeLa cells were maintained in DMEM, 10% FBS.
Chemicals, antibodies and plasmid constructs
All chemicals were purchased from Sigma-Aldrich except for BafA1 and rapamycin (LC laboratories). Generation of EYFP-Jumpy-Wild-Type, -C330S, -R336Q and -Y462C, EGFP-Jumpy-Wild-Type, -C330S, B10-Jumpy CS, EGFP-MTM1, EGFP-MTMR2 (Tosch et al, 2006) and GFP-WIPI-1 (Proikas-Cezanne et al, 2007) plasmid constructs are described elsewhere. tdTomato-LC3 and Cherry-LC3 were from T. Johansen (Bjorkoy et al, 2005; Pankiv et al, 2007). mRFP-GFP-LC3 and HA-mAtg9 were from T. Yoshimori (Kimura et al, 2007) and S. Tooze (Young et al, 2006), respectively. The following antibodies were used in this study: LC3 (Sigma or from T Ueno), Atg12 (M) and Atg16 from N. Mizushima (Mizushima et al, 2001, 2003), Atg12 (C) (Cell signaling), Beclin-1 (Santa Cruz), Lamp1 (Clone 1D4B, RDI /Fitzgerald), G58K (Sigma), Atg5 (Novus), p62 (Progen), GFP and Actin (Abcam). The specificity of Atg16, Atg12 (M) and Atg12 (C) antibody staining were validated in C2C12 with Atg16 and Atg12 staining colocalizing with GFP-LC3 under starvation conditions (Supplementary Figure S12).
Transient transfection by nucleoporation
RAW 264.7, C2C12, Neuro2A, MEF cells, and HeLa cells were harvested at day 2 of culture and re-suspended in the appropriate electroporation buffer (Amaxa Biosystems, MD). A measure of 5–10 μg plasmid DNA and/or 1.5 μg siRNA were mixed with 0.1 ml of cell suspension, transferred to an electroporation cuvette, and nucleofected with Amaxa Nucleofector apparatus (Amaxa Biosystems, MD). Jumpy, Atg5 and Beclin-1 knock-downs were achieved by using siGENOME SMARTpool reagent (Dharmacon) specific for Mus musculus for RAW 264.7, C2C12, Neuro2A and MEF cells transfection or for Homo Sapiens for HeLa transfection (Dharmacon). All effects of myotubularins, Jumpy, Atg5 or Beclin siRNAs were compared with siCONTROL Nontargeting siRNA pool (Dharmacon), which is labelled as scramble or sc in figures.
Fluorescence confocal microscopy and Immunoblotting
Procedures for fluorescence confocal microscopy, immunofluorescence and western immunoblotting were described elsewhere (Vergne et al, 2005; Delgado et al, 2008). Co-localization of GFP-Jumpy CS with different cellular markers was defined using the Zeiss LSM 510 co-localization co-efficient program (0 being no colocalization and 1 total colocalization). Only pixels above cytosolic fluorescence were evaluated using LSM 510 co-localization and crosshair functions.
p62 analysis
p62 protein levels were determined by immunoblotting, measured using NIH Image J software, and expressed as a percentage of control (scramble siRNA, GFP or YFP-transfected cells). p62 accumulation was analysed by immunofluorescence microscopy and quantitated by counting the number of endogenous p62 puncta (⩾I μm) per cell using Zeiss LSM Image Browser software (Filimonenko et al, 2007).
Proteolysis of long-lived proteins
C2C12 myoblast cells were transfected with siRNA and seeded in 12-well plates (8 × 104 cells per well). Four hours later, cells were labelled overnight in media containing 1 μCi/ml 3Hleucine, washed to remove unincorporated label, and pulsed for 2 h in full media containing cold leucine to allow degradation of short-lived proteins. Finally, cells were incubated in full or starvation media for 4 h. Trichloroacetic acid (TCA)-precipitable radioactivity of the cells monolayers and the TCA-soluble radioactivity in the media were determined. Leucine release (a measure of proteolysis of stable proteins) was calculated as a ratio between TCA-soluble supernatant and total cell-associated radioactivity.
Live confocal fluorescence microscopy
Recruitment of GFP-Jumpy WT was analysed by live confocal microscopy using a 5LIVE Zeiss microscope. EBSS was added to the cells and z-stacks collected at 3-min intervals for a total of 45 min. The collected images were processed to generate a maximum projection (collapsing a 3D image into an x–y projection) for each time point, as described earlier for 4D live confocal imaging (Chua and Deretic, 2004; Kyei et al, 2006; Roberts et al, 2006). Jumpy association was quantified as relative fluorescence units (RFUs) of GFP-Jumpy on LC3+-puncta substracted for RFU of GFP-Jumpy in cytosol and normalized to RFUc–RFUp at time corresponding to 925 s.
Supplementary Material
Supplementary Materials
Supplementary Movie S1
Review Process File
Acknowledgments
This work was supported by grants AI45148 from the National Institutes of Health to VD, INSERM, CNRS, Collège de France, and grants from AssociationFrançaise contre les myopathies, Agence Nationale de la Recherche and Foundation pour la recherche Médicale to JL, and grant SFB773 from the Deutsche Forschungsgemeinschaft to TP-C. This project was supported in part by the Dedicated Health Research Funds from the University of New Mexico School of Medicine to IV.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T (2004) Protein tyrosine phosphatases in the human genome. Cell 117: 699–711 [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun M, Maugenre S, Jeannet PY, Lacene E, Ferrer X, Laforet P, Martin JJ, Laporte J, Lochmuller H, Beggs AH, Fardeau M, Eymard B, Romero NB, Guicheney P (2005) Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet 37: 1207–1209 [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau F, Laporte J, Bodin S, Superti-Furga G, Payrastre B, Mandel JL (2000) Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum Mol Genet 9: 2223–2229 [DOI] [PubMed] [Google Scholar]
- Cao C, Laporte J, Backer JM, Wandinger-Ness A, Stein MP (2007) Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic 8: 1052–1067 [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua J, Deretic V (2004) Mycobacterium tuberculosis reprograms waves of phosphatidylinositol 3-phosphate on phagosomal organelles. J Biol Chem 279: 36982–36992 [DOI] [PubMed] [Google Scholar]
- Clague MJ, Lorenzo O (2005) The myotubularin family of lipid phosphatases. Traffic 6: 1063–1069 [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V (2008) Toll-like receptors control autophagy. EMBO J 27: 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 179: 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK (1997) Development of ‘substrate-trapping' mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA 94: 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16 L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton AJ, Flint AJ, Tonks NK (1996) Identification of p130(cas) as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol Cell Biol 16: 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19: 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A et al. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Kundu M, Thompson CB (2008) Autophagy: basic principles and relevance to disease. Annu Rev Pathol 3: 427–455 [DOI] [PubMed] [Google Scholar]
- Kyei GB, Vergne I, Chua J, Roberts E, Harris J, Junutula JR, Deretic V (2006) Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. EMBO J 25: 5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Bedez F, Bolino A, Mandel JL (2003) Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet 12 (Spec No 2): R285–R292 [DOI] [PubMed] [Google Scholar]
- Laporte J, Blondeau F, Buj-Bello A, Tentler D, Kretz C, Dahl N, Mandel JL (1998) Characterization of the myotubularin dual specificity phosphatase gene family from yeast to human. Hum Mol Genet 7: 1703–1712 [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V (2007) Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 7: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6: 458–471 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116 (Pt 9): 1679–1688 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3: 542–545 [DOI] [PubMed] [Google Scholar]
- Nicot AS, Toussaint A, Tosch V, Kretz C, Wallgren-Pettersson C, Iwarsson E, Kingston H, Garnier JM, Biancalana V, Oldfors A, Mandel JL, Laporte J (2007) Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet 39: 1134–1139 [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 148: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Noda T, Niimi K, Ohsumi Y (2008) Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells 13: 537–547 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P (2000) Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 275: 992–998 [DOI] [PubMed] [Google Scholar]
- Pierson CR, Tomczak K, Agrawal P, Moghadaszadeh B, Beggs AH (2005) X-linked myotubular and centronuclear myopathies. J Neuropathol Exp Neurol 64: 555–564 [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A (2007) Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett 581: 3396–3404 [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A (2004) WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 23: 9314–9325 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ (2004) The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 6: 79–90 [DOI] [PubMed] [Google Scholar]
- Roberts EA, Chua J, Kyei GB, Deretic V (2006) Higher order Rab programming in phagolysosome biogenesis. J Cell Biol 174: 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson FL, Dixon JE (2006) Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol 16: 403–412 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 581: 2156–2161 [DOI] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE (2000) Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA 97: 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosch V, Rohde HM, Tronchere H, Zanoteli E, Monroy N, Kretz C, Dondaine N, Payrastre B, Mandel JL, Laporte J (2006) A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum Mol Genet 15: 3098–3106 [DOI] [PubMed] [Google Scholar]
- Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V (2005) Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci USA 102: 4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Sinha S, Levine B (2008) Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 4: 949–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Yoshimori T, Noda T (2008) Toward unraveling membrane biogenesis in mammalian autophagy. Curr Opin Cell Biol 20: 401–407 [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119(Pt 18): 3888–3900 [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6: 472–483 [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z (2009) Distinct regulation of autophagic activity by Atg14 L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 11: 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Supplementary Movie S1
Review Process File