Abstract
Nonsense-mediated decay (NMD) is a eukaryotic quality control mechanism that degrades mRNAs carrying premature stop codons. In mammalian cells, NMD is triggered when UPF2 bound to UPF3 on a downstream exon junction complex interacts with UPF1 bound to a stalled ribosome. We report structural studies on the interaction between the C-terminal region of UPF2 and intact UPF1. Crystal structures, confirmed by EM and SAXS, show that the UPF1 CH-domain is docked onto its helicase domain in a fixed configuration. The C-terminal region of UPF2 is natively unfolded but binds through separated α-helical and β-hairpin elements to the UPF1 CH-domain. The α-helical region binds sixfold more weakly than the β-hairpin, whereas the combined elements bind 80-fold more tightly. Cellular assays show that NMD is severely affected by mutations disrupting the beta-hairpin binding, but not by those only affecting alpha-helix binding. We propose that the bipartite mode of UPF2 binding to UPF1 brings the ribosome and the EJC in close proximity by forming a tight complex after an initial weak encounter with either element.
Keywords: mRNA quality control, nonsense mediate decay (NMD), NMR, UPF1, UPF2, X-ray crystallography
Introduction
Among the different mechanisms evolved by eukaryotes to control the quality of mRNA, nonsense-mediated decay (NMD) is one of the most extensively studied (Behm-Ansmant et al, 2007; Doma and Parker, 2007; Isken and Maquat, 2007; Shyu et al, 2008). The NMD pathway involves recognition and targeting for degradation of transcripts containing premature termination codons (PTCs), which may result from DNA mutations, transcription errors or pre-mRNA-processing errors, notably splicing. Originally, it was thought that the major biological function of NMD was to protect cells from the potentially deleterious effects of truncated proteins (Behm-Ansmant et al, 2007). However, it is now clear that, in different organisms, 3–10% of transcriptome is naturally targeted by NMD (He et al, 2003; Rehwinkel et al, 2005; Behm-Ansmant and Izaurralde, 2006), and it may be a much more general mechanism for regulating transcriptome diversity arising from alternative or mis-splicing (Green et al, 2003; Isken and Maquat, 2007; Jaillon et al, 2008).
Nine NMD protein factors, SMG1–9, have been identified in higher eukaryotes, two of them very recently (Yamashita et al, 2009). The three UPF (UP-frameshift) proteins, UPF1 (SMG2), UPF2 (SMG3) and UPF3 (SMG4), constitute the conserved core of the NMD machinery and are found in almost all eukaryotes with a few possible exceptions among protists (Kadlec et al, 2006; Chen et al, 2008), suggesting that NMD has an ancient evolutionary origin. Despite the universal conservation of the three UPF proteins, significantly different mechanistic models have been proposed for NMD in different organisms (Conti and Izaurralde, 2005; Lejeune and Maquat, 2005). However, an evolutionarily consistent model for PTC recognition has recently begun to emerge (Amrani et al, 2004; Kertesz et al, 2006; Schwartz et al, 2006; Muhlemann et al, 2008; Brogna and Wen, 2009) on the basis of studies in the mammalian system (Buhler et al, 2006; Eberle et al, 2008; Ivanov et al, 2008; Silva et al, 2008; Singh et al, 2008). This model proposes that NMD is triggered by an inefficient termination event caused by the failure of PABP (and/or other 3′ UTR factors) to interact with the terminating ribosome (Brogna and Wen, 2009). In mammals, the exon junction complex (EJC) works within this model as an enhancer to increase NMD efficiency, but it is not absolutely required, as previously thought (Lejeune and Maquat, 2005). EJC is a multi-protein complex that is deposited by the splicing machinery on mRNA ∼24 nt upstream of the exon boundaries and marks the sites of intron excision (Kim et al, 2001; Le Hir et al, 2001). EJC is retained during subsequent mRNA maturation events, including nuclear export, but can recruit additional factors. Notably, in the nucleus, the EJC core factors, MAGOH–Y14, recruit UPF3b (Gehring et al, 2003; Chamieh et al, 2008) and subsequently UPF3b recruits UPF2 on export into the cytoplasm (Lykke-Andersen et al, 2000). The UPF3b–UPF2 interaction has been described at the atomic level and is mediated by the N-terminal RNP domain of UPF3b interacting with the third of the three MIF4G (middle domain of eIF4G) domains of UPF2 (Kadlec et al, 2004).
In mammalian cells, NMD is thought to occur during the first, ‘pioneer' round of translation (Ishigaki et al, 2001). The functional link between the ribosome stalled at a PTC and the EJC involves the recruitment of different factors in a complex and dynamic molecular architecture, beginning with the translation termination factors, eRF1 and eRF3 (eRF1-3) (Czaplinski et al, 1998; Ivanov et al, 2008). Subsequently UPF1 and SMG1 join eRF1-3 to form a transient complex called SURF (named after the component proteins) (Kashima et al, 2006). The downstream EJC makes contact with the SURF complex through the interaction of UPF2 (which is bound to UPF3b on the EJC), with UPF1 and SMG1 forming the so-called DECID complex (Kashima et al, 2006). At this stage, the conserved ternary core UPF complex is formed and SMG1 is stimulated to phosphorylate UPF1 on its C-terminal SQ-motifs (Kashima et al, 2006). Hyperphosphorylated UPF1 is recognized by SMG7 by a 14-3-3-like domain, also conserved in SMG5 and SMG6 (Fukuhara et al, 2005). SMG6 carries the endonuclease activity that initiates the degradation of nonsense mRNAs in metazoans, showing that NMD machinery contributes directly to their decay (Glavan et al, 2006; Huntzinger et al, 2008; Eberle et al, 2009). SMG7 promotes further destabilization of these transcripts in a DCP2- and XRN1-dependent manner (Unterholzner and Izaurralde, 2004). It has recently been shown that the decapping enzyme, DCP1, is recruited to the phospho-UPF1 through the proline-rich nuclear receptor co-regulatory protein 2 (PNRC2) (Cho et al, 2009). The interaction between the ribosome-associated SURF complex and the downstream EJC to form the DECID complex is primarily mediated through UPF2, which bridges between UPF1 on SURF and UPF3b on EJC. However, it has been reported that NMD can also occur either in a UPF2-independent process (Gehring et al, 2005) or in a UPF3b-independent manner (Chan et al, 2007; Tarpey et al, 2007).
UPF1 is a highly conserved ∼120 kDa protein that shows RNA-dependent ATPase and 5′-3′ RNA helicase activities in vitro (Cheng et al, 2007), both of which are required for NMD (Czaplinski et al, 1995; Weng et al, 1996; Bhattacharya et al, 2000). UPF1 has several additional cellular functions, including a function in maintaining genome stability (Azzalin and Lingner, 2006; Isken and Maquat, 2008), and a mouse knockout for UPF1 is embryonically lethal (Medghalchi et al, 2001). The crystal structure of the UPF1 superfamily 1 helicase domain (residues 295–914) has been determined (Cheng et al, 2007). UPF1 also has a unique highly conserved N-terminal cysteine–histidine-rich domain (CH-domain, residues 115–275) that binds three structural zinc atoms (Kadlec et al, 2006). The CH-domain contains the UPF2-binding site (Weng et al, 1996; Serin et al, 2001; Kadlec et al, 2006).
UPF2 is an ∼140 kDa perinuclear protein (hUPF2 comprises 1272 residues) characterized by three MIF4G domains (Mendell et al, 2000; Serin et al, 2001). UPF3b binds to the third MIF4G domain of UPF2 (Kadlec et al, 2004), whereas the function of the preceding N-terminal part of the protein is unknown. The UPF1-binding region of UPF2 is at the C-terminus of the protein and is separated from the third MIF4G domain by a conserved Glu/Asp-rich acidic region (He et al, 1997; Serin et al, 2001). The targeted knockout of UPF2, the only known function of which is in NMD, has severe effects on mouse haematopoietic stem cells, but milder effects on differentiated ones, suggesting an important role of NMD in proliferating cells (Weischenfeldt et al, 2008).
Here, we characterize the interaction between human UPF1 and UPF2 by a variety of techniques, including X-ray crystallography, electron microscopy (EM), NMR, small-angle X-ray scattering (SAXS), isothermal calorimetry and in vitro and in vivo mutagenesis. We present crystal structures of the combined CH- and helicase domains (residues 115–914) of UPF1 in complex with the C-terminal region of human UPF2 (residues 1105–1198), providing the first information on both the relative arrangement of the two UPF1 domains and the structural basis for the interaction between UPF1 and UPF2. We show that the free C-terminal region of UPF2 is unstructured but co-folds on binding to UPF1, with an α-helical element binding on one side of the CH-domain and a β-hairpin element on the other. This mode of interaction of UPF2 with UPF1 is a good example of ‘clamp-type fuzzy complex' (Tompa and Fuxreiter, 2008) in which an intrinsically disordered protein region partially folds on binding to a partner protein (Dyson and Wright, 2002). Possible rationales for this mode of UPF1–UPF2 interaction will be discussed in the light of the current understanding of the mechanism of NMD.
Results
Overview of the X-ray and NMR structural work
The UPF1–UPF2 complex was obtained by the co-expression of the two proteins in Escherichia coli or by a reconstitution using UPF2 purified under denaturing conditions. We determined several different structures of the UPF2–UPF1 complex, including two structures with the CH-domain alone (data not shown because of a relatively low resolution, see methods) and two with the combined CH- and helicase domains of UPF1 (residues 115–914) (Table I). The most complete picture of the complex emerges from a monoclinic (P21) crystal form of the complex containing both domains of UPF1 with UPF2(1105–1198) at 2.9 Å resolution. In this structure, both the helical and β-hairpin segments of UPF2 have good and unambiguous electron density (Figure 1A, Supplementary Figure S1), although the linker between the two is only poorly defined. A second orthorhombic (I222) crystal form of the same complex diffracting to the higher resolution of 2.5 Å shows a relatively poor definition of the CH-domain (probably because of mobility through a lack of strong crystal contacts) and a very weak density for only the UPF2 β-hairpin region; indeed crystal contacts preclude binding of the helical segment. We have also determined the structure of a slightly extended construct of the CH-domain of UPF1 alone (residues 115–287) at the considerable higher resolution of 1.5 Å resolution compared with the original structure (Kadlec et al, 2006) (data not shown). This CH-domain construct is better expressed and has a properly configured C-terminal region, as in the full-length UPF1 structures. It was thus used in subsequent solution work, notably in NMR studies.
Table 1.
Data collection and refinement statistics of UPF1(115–914)–UPF2(1105–1198) complex
| UPF1(115–914)–UPF2(1105–1198) orthorhombic form | UPF1(115–914)–UPF2(1105–1198) monoclinic form | |
|---|---|---|
| Beamline (ESRF) | ID29 | ID14-4 |
| Wavelength (Å) | 0.976 | 0.940 |
| Space group | I222 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 64.9, 129.1, 311.3 | 92.4, 97.3, 124.6 |
| α, β, γ (deg) | 90.0, 90.0, 90.0 | 90.0, 102.4, 90.0 |
| Resolution (Å) | 30–2.5 (2.5–2.6)a | 30–2.85 (2.85–2.90)a |
| Rmerge | 11.1 (76.7) | 8.6 (75.8) |
| I/σI | 11.2 (1.9) | 11.1 (2.1) |
| Completeness (%) | 98.4 (98.0) | 97.3 (97.1) |
| Redundancy | 3.71 (3.78) | 3.80 (3.88) |
| Refinement | ||
| Resolution (Å) | 46.5–2.50 (2.50–2.57)a | 30–2.85 (2.85–2.92)a |
| Total no. of reflections/free | 42869/2266 | 46534/2463 |
| Rwork | 0.196 (0.289) | 0.200 (0.332) |
| Rfree | 0.240 (0.309) | 0.248 (0.372) |
| Number of atoms | ||
| Protein atoms | 6168 (+3 Zn) | 13085 (+6 Zn) |
| Water molecules | 254 | — |
| Sulphate ions | 8 | 23 |
| Average B-factors (Å2) | 45.9 | 65.6 |
| RMSD values | ||
| Bond lengths (Å) | 0.012 | 0.011 |
| Bond angles (deg) | 1.39 | 1.23 |
| Ramachandran plotb | ||
| Favoured (%) | 96.0 | 96.2 |
| Allowed (%) | 99.9 | 99.8 |
aValues in parentheses are for highest-resolution shell.
bMolprobity http://molprobity.biochem.duke.edu/.
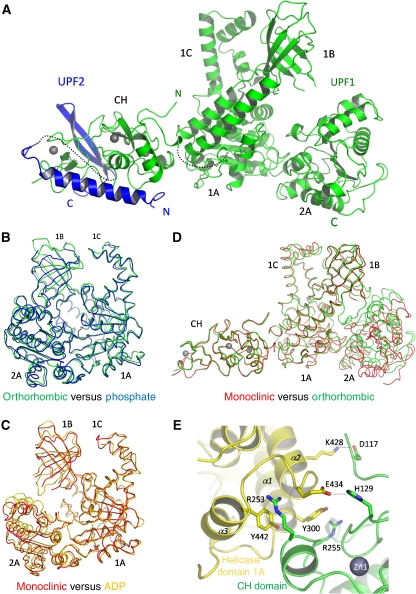
Figure 1.
Structure of the UPF1(115–914)–UPF2(1105–1198) complex. (A) Ribbon diagram of the complete structure, with UPF1 in green and UPF2 in blue. The missing links between the UPF1 CH- and helicase domains and between the N and C-terminal parts of UPF2 are represented as dotted lines. (B) Superposition of the closed form of the helicase domain (orthorhombic crystal, green) with the previously described helicase domain in the phosphate-bound form (PDB ID 2gk7, blue). The RMSD between the two structures is 1.03 Å for 591 aligned Cα atoms. The RMSD values between the orthorhombic form and the AMPPNP (PDB ID 2gjk) and ADP (PDB ID 2gk6) forms are, respectively, 1.81 and 2.00 Å. (C) Superposition of the open form of the helicase domain (monoclinic crystal, red) with the previously described helicase domain in the ADP-bound form (PDB ID 2gk6, gold). The RMSD between the two structures is 1.35 Å for 584 aligned Cα atoms. The RMSD values between the monoclinic form and the phosphate and AMPPNP forms are, respectively, 2.48 and 3.07 Å. (D) Superposition of UPF1 from the monoclinic (red) and orthorhombic (green) crystal forms showing that the relative orientations of the CH and helicase domains are the same in each case, although the helicase conformation is different. (E) The principal interacting residues from the CH- (green) and helicase (yellow) domains of UPF1 are represented as sticks. The same interactions are found in both monoclinic and orthorhombic crystal forms. These residues are well conserved (Supplementary Figure S2).
In parallel, we measured two-dimensional NMR spectra on various complexes using 15N and 15N/13C-labelled proteins to derive structural and dynamic information under solution conditions. Backbone signals could be assigned for the CH-domain of UPF1 alone, allowing the mapping of the UPF2-binding site. This was carried out both in the context of the complex with the full C-terminal region of UPF2 and also by titrating synthetic peptides of the separate alpha-helical and beta-hairpin motifs to the 15N-labelled UPF1 CH-domain. These NMR chemical shift perturbation data give strong additional evidence for two separate binding sites of UPF2 on the CH-domain of UPF1, as well as the presence of residual disordered regions in bound UPF2.
Structure of the combined CH- and helicase domains of UPF1
Previous structures of the UPF1 helicase core (residues 295–914) showed that it comprises two RecA-like sub-domains (denoted 1A and 2A) with two unique insertions into domain 1A, denoted 1B (a β barrel domain) and 1C (a helical domain) (Cheng et al, 2007). Three states of the enzyme were described, closed forms with either AMPPNP (PDB code 2gjk) or phosphate bound (PDB code 2gk7) and a more open form with ADP bound (PDB code 2gk6). The differences arise mainly because of rigid body motions of the four sub-domains. The orthorhombic and monoclinic crystal forms that we have determined of the UPF1(115–914)–UPF2(1105–1198) complex also show different relative arrangements of the sub-domains (Figure 1B–D). In the high-resolution orthorhombic form, the helicase is in a very well-ordered closed configuration, with only a narrow cleft between the two RecA-like domains (Figure 1B). This conformation shows the highest similarity with the previously described phosphate-bound form (Cheng et al, 2007), with a RMSD value of 1.03 Å for 591 aligned Cα atoms (Figure 1B). Indeed, we observe a tightly bound sulphate at the position of the phosphate/gamma phosphate of AMPPNP. In the monoclinic crystal form, parts of UPF1 are less well ordered, especially the β-barrel domain 1B. Domains 1A and 2A are in a more open conformation that resembles most closely the ADP-bound form with an RMSD value of 1.35 Å for 584 aligned Cα atoms (Figure 1C). The individual domains 1A and 2A do not differ significantly in structure in all crystal forms to date. However, in our orthorhombic form, some of the flexible loops in domains 1B and 1C are better defined than in the previously determined structure (Figure 1B). The different conformations observed for the helicase domain in our two crystal forms may be because of the difference in crystallization conditions and crystal packing, but highlight the intrinsic flexibility of the helicase quaternary structure.
Despite the difference in the orientations of the helicase sub-domains, the two crystal forms show the same orientation and interface of the CH-domain with respect to the helicase domain (Figure 1D). One end of the elongated CH-domain (which contains both N- and C-terminal elements of the domain) packs against the external surface of the helicase sub-domain 1A (notably N-terminal helices α1, α2 and α3), with the rest of the CH-domain extending away from the helicase. Residues 280–287, forming the linker region between the two domains, are poorly ordered. The domain interface involves specific hydrogen bonds, as well as van der Waals contacts (Figure 1E). Arg253 and His129 side chains (CH domain) form, respectively, a hydrogen bond with the main chain carboxyl of Val437 and a salt bridge with Glu434 (helicase). In addition, Asp298 (helicase) forms a hydrogen bond with the main chain amino group of Gln256 (CH domain). Finally Asp117 (CH-domain) forms a salt bridge with Lys428 (helicase). Helicase Tyr300, which stacks on Arg255, and Tyr442, which stacks on Arg253, are also crucial elements in the interface. The total buried area of this interface is 1163.1 Å2 (565.7 Å2 for the helicase domain and 597.4 Å2 for the CH-domain), as determined by the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). Although relatively modest, the fact that the same interface is observed in two distinct crystal forms and involves largely conserved residues (Supplementary Figure S2) suggests that the observed rigid orientation of the CH- and helicase domains is biologically significant and not a crystal-packing artefact. Furthermore, as the helicase itself is in a different state (closed or open) in the two different crystal forms, this shows that the rigid attachment of the CH-domain to domain 1A is compatible with different relative configurations of domain 2A. However, it cannot be ruled out that the ability to undergo functional conformational changes between these configurations may be affected by the presence of the CH-domain.
To determine whether the observed domain configuration of UPF1 depended on the presence of bound UPF2, we attempted to crystallize UPF1 in the absence of UPF2, but this was unsuccessful. Instead we carried out EM and SAXS studies on UPF1(115–914), with and without bound UPF2(1105–1198). Negatively stained EM images were of sufficient quality to allow a three-dimensional reconstruction of each sample. The reconstructions obtained for unbound and UPF2-bound UPF1 were very similar to each other and to the crystal structure of the UPF1–UPF2 complex, with, in each case, the CH-domain bound to the side of the helicase domain (Supplementary Figure S3).
To confirm that this is also valid for the proteins in solution, we analysed free and UPF2-bound UPF1(115–914) by SAXS and compared the measured scattering data with the theoretical scattering curve calculated from the crystal structure models. In both cases, the calculated curves show a satisfactory fit to the measured ones (Supplementary Figure S4A,B). In the case of UPF1(115–914) alone, the best fit was obtained with the crystal structure of the helicase in the closed conformation (χ=0.87), whereas for the UPF1–UPF2 complex, the best fit was with the helicase in the open conformation (χ=0.95). For both UPF1 and the UPF1–UPF2 complex, the measured radius of gyration is very close to that calculated from the respective crystal structures (see below) (Supplementary Figure S4D). The increased radius of gyration of the complex compared with the free UPF1 and the change in form of the distance distribution (Supplementary Figure S4C), which shows that the complex clearly has more mass at large distances from the centre of the mass, are fully consistent with the crystallographically observed binding of UPF2 on the CH-domain at the periphery of UPF1. Furthermore, the model-independent ab initio envelopes calculated from the data show an elongated shape that accommodates the crystal structures well, with the CH-domain protruding away from the helicase domain, and in the case of the UPF2 complex, with more volume associated with the CH-domain (Supplementary Figure S5). Minor discrepancies in these comparisons are likely to occur for two reasons. First, the crystal structures lack some loops in UPF1 and in the flexible linker of UPF2 connecting the two UPF1-binding elements; second, in solution, helicase probably fluctuates between open and closed conformations with perhaps a broader amplitude than that sampled by the crystal structures. As a final control, we calculated the scattering curve from an atomic model in which the CH-domain is displaced towards the cleft formed by the 1A and 2A domains of UPF1 (both for free and UPF2-bound UPF1). This severely deteriorated the fits to the experimental data (respectively, χ=2.82 and χ=5.42 for free and UPF2-bound UPF1, data not shown).
Taken together, the crystallographic, EM and SAXS results indicate that the UPF1 CH-domain is bound to the helicase domain in a fixed, distal orientation, pointing away from the ATPase active site, irrespective of the presence or not of bound UPF2(1105–1207). This conclusion is further reinforced by an ATPase assay conducted on free and UPF2-bound UPF1, which shows that UPF2(1105–1207) does not alter UPF1 RNA-dependent ATPase activity (Supplementary Figure S6).
Structure of UPF2 in the UPF1(115–914)–UPF2(1105–1198) complex
The crystal structure of the UPF1(115–914)–UPF2(1105–1198) complex in the monoclinic form allows the identification of two UPF1-interacting regions of UPF2, separated by a flexible linker, in agreement with what was originally proposed for yeast UPF2 (He et al, 1996). The N-terminal part of the UPF2 (residues 1108–1128) fragment forms a long, slightly curved, amphipathic α-helix (Figure 2A). The C-terminal part folds into a β-hairpin (residues 1167–1189) comprising strand βA, strand βB and an intervening loop, followed by a short α-helix (residues 1193–1198) (Figure 2B). Structures of the CH-domain alone with UPF2(1167–1207) show that this α-helix extends to at least residue 1203 (data not shown). Between the two, residues 1129–1166 form an extended linker, the first part of which (residues 1129–1139) is observed with weak electron density wrapping around the CH-domain, whereas the following glycine-rich peptide is not visible at all.
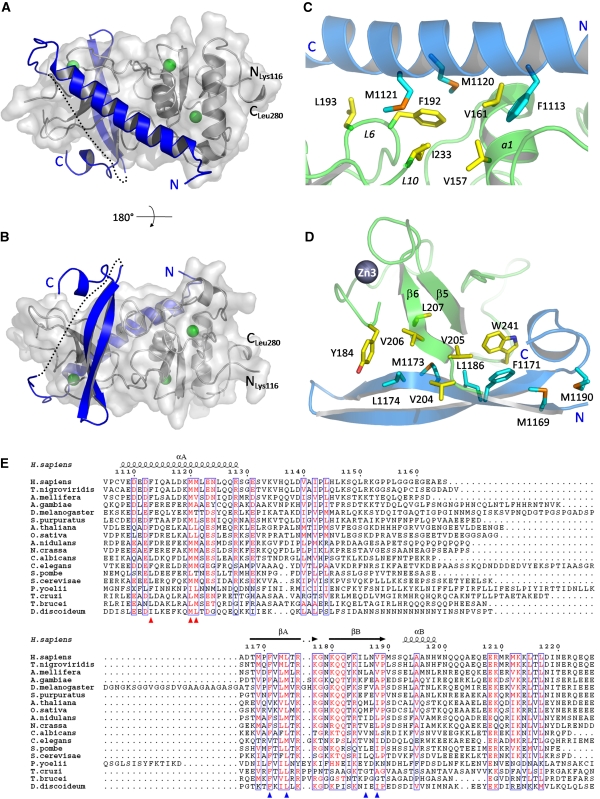
Figure 2.
UPF2 binds on two opposite surfaces of the UPF1 CH domain. (A, B) UPF2 (blue) is represented as ribbons and UPF1 (grey) as ribbons and a transparent surface. UPF1 zinc atoms are shown in green. The UPF2 missing linker is represented as a dotted line. The two views differ by a rotation of 180 degrees around the horizontal axis. (C) The principal residues of the UPF2 N-terminal helix (cyan) and the UPF1 CH-domain (yellow), which form the hydrophobic interface between the two molecules, are represented as sticks. (D) The main interacting residues of UPF2 C-terminal β-hairpin (cyan) and UPF1 CH domain (yellow) are represented as sticks. Met 1169 and Met 1190 do not interact directly with UPF1 binding but form part of a small hydrophobic core important for the stability of the bound form of UPF2. (E) Sequence alignment of the UPF1-binding domain of representative UPF2 proteins from yeast to human. Residues with similarity >70% are displayed in red. The secondary structure of the UPF1-bound human UPF2 is indicated as α (alpha-helix) and β (beta-strand). Red and blue triangles indicate the main UPF1-interacting residues belonging to the N-terminal helical region and the C-terminal β-hairpin, respectively. The alignment was generated with ClustalX (Thompson et al, 2002) and showed using ESPript (http://espript.ibcp.fr/ESPript/ESPript/).
The two UPF2 regions bind apparently independently on two opposite faces of the UPF1 CH-domain, and in both cases, the interaction between the two proteins has a strong hydrophobic component. The binding site for the UPF2 α-helix is formed by the UPF1 residues, Val157, Val161, Phe192, Leu193 and Ile233 and the aliphatic part of Arg236 belonging to loops L6 and L10 and helix α1, which create a hydrophobic surface contacting UPF2 residues, Phe1113, Ile1114, Leu1117, Met1120, Met1121 and Leu1125 (Figure 2C and Supplementary Figure S7A). In particular, Phe1113 contacts Val157 and Val161; Met1120 and Met1121 both interact with Phe192 and also with, respectively, Val161 and Ile233; and Leu1125 and Arg1128 contact Leu193. In addition, there are hydrogen-bond interactions between Asp1110 and Ser152, as well as between Asn1124 and Asn190. The total buried surface area on helix binding is 1826.8 Å2 (978.8 Å2 for UPF2 and 848.0 Å2 for UPF1), as determined by PISA.
On the other side of the CH-domain, the UPF2 β-hairpin is inserted between loops L10 and L7, which form another hydrophobic surface involving residues Leu176, Tyr184, Phe196, Trp241, Leu242 and the region 204-VVVL-207 (Figure 2D and Supplementary Figure S7B). The main interactions involve UPF2 residues from strand βA, including Leu1174, which contacts UPF1 residues Tyr184, Val204 and Val206, and Phe1171, which interacts with Val205. Met1173 is buried in the centre of the interface, notably contacting Phe196. Residues Leu1186, Val1188, Pro1189 and Leu1194, belonging to the UPF2 strand βB, contact Trp241 of UPF1 loop L10. In addition, UPF2 Arg1176 contacts UPF1 Tyr184 (Supplementary Figure S7B) (see below for in vitro mutations of these residues). The β-hairpin of UPF2 is also stabilized by many intra-molecular interactions, including a small hydrophobic core comprising Met1169 and Phe1171 from the N-terminal end of strand βA, and Val1188, Met1190, Leu1194 and Ala1195 from the C-terminal end of strand βB. In addition, there are at least 10 inter-strand hydrogen bonds. The loop of the β-hairpin, containing highly conserved Gly1178 (residues 1177–1181) is poorly ordered and apparently flexible. The total buried surface area on hairpin binding is 1666 Å2 (833.4 Å2 for UPF2 and 832.6 Å2 for UPF1), as calculated by PISA. However, this calculation almost certainly underestimates the solvent exclusion effect due to hairpin binding, as it assumes the hairpin structure is present in the unbound state of UPF2. NMR spectra of the unbound peptide (residues 1167–1207) indicate that it is unstructured in solution (see below). Thus, folding of the extended peptide 1167–1207 into the hairpin structure, notably forming the mini-hydrophobic core, in itself buries an additional 1275 Å2 of the solvent accessible surface (649 Å2 for residues 1167–1179, 626 Å2 for residues 1180–1198).
Alignments show that the UPF2 residues involved in interacting with UPF1 are highly conserved in type throughout evolution, notably in positions of key hydrophobic residues and the glycine-containing hairpin loop (Figure 2E). However, the flexible linker between the two binding regions is highly divergent in sequence and length. This is discussed further below.
The superposition of the UPF1 CH-domain structure in the presence (monoclinic form) and in the absence of UPF2 (original structure, PDB code 2iyk) shows that, whereas the helicase proximal part of the domain (comprising the N-terminal region 118–190 and the C-terminal region 240–270) does not change significantly, the distal region rotates slightly to accommodate binding of the UPF2 β-hairpin, which would otherwise clash with residues 198–204 (loop L7) (Figure 3). This loop is better defined in the structure of the complex and is only partially ordered in the high-resolution structure of the CH-domain. NMR 15N relaxation data of the CH-domain alone indicate dynamics of the sub-nanosecond time scales of this loop (Supplementary Figure S8). The L7 loop movement allows the parallel β-strand addition of UPF1 residues 203-SVV-205 to residues 1172-VML-1174 of the UPF2 strand βA, with the formation of three hydrogen bonds between the respective main chains, an important mediator of the UPF1–UPF2 interaction. In addition, there is a significant rotamer change of Phe196 on UPF2 binding. In contrast, there are no significant conformational changes at the binding site of the α-helical part of UPF2. The conserved loop L9 (residues 219–224) is poorly ordered in all structures, although it is in the vicinity of the long linker connecting the two interacting regions of UPF2.
Figure 3.
Changes in the UPF1 CH domain on UPF2 binding. Superposition of the UPF1 CH domain in the UPF2 complex (green) and alone (blue, PDB code 2iyk). UPF2 is shown in red. The distal region, notably loop L7, rotates to allow UPF2 binding, notably β-strand addition, whereas the helicase proximal end, including the N- and C-terminal parts, is unchanged.
Verification of the interaction model in solution by NMR
The CH-domain of UPF1(115–287) gives a good 1H, 15N correlation spectrum. Using an 15N/13C doubly labelled sample, 73% of the backbone signals could be assigned (see Supplementary data). To obtain spectra of the UPF1 (115–287)–UPF2(1105–1207) complex, with only one or other component labelled, we reconstituted complexes with, respectively, labelled or unlabelled UPF1 and unlabelled or labelled UPF2 after a purification of the UPF2 under denaturating conditions (see Materials and methods). We first confirmed that the refolding protocol does not affect the structural integrity of the UPF1–UPF2 complex. The 1H,15N HSQC spectra of fully 15N-labelled UPF1(115–287)–UPF2(1105–1207), obtained by co-expression and without refolding, superimpose very well with the corresponding spectra of 15N-UPF1(115–287)–UPF2(1105–1207) and UPF1(115–287)–15N-UPF2(1105–1207), which were recorded on samples that had been obtained by refolding (Supplementary Figure S9A). The line widths of the amide proton signals are consistent with the formation of a monomeric stoichiometric complex in solution (data not shown).
By comparing NMR spectra comprising different regions of UPF2, we defined the interaction surface of the two proteins. Substantial chemical shift changes comparing the 1H,15N HSQC spectra of 15N-labelled UPF1(115–287) free and bound to UPF2(1105–1207) indicate a large binding interface (Figure 4A). However, spectra of 15N-UPF1(115–287), with either bound UPF2(1105–1227) or UPF2(1105–1207), superimpose very well (Supplementary Figure S9B), showing that the last 20 residues at the C terminus of UPF2 are not necessary for UPF1 binding. This is further confirmed by comparing the 1H,15N HSQC spectra of 15N-UPF2(1105–1227) or 15N-UPF2(1105–1207) bound to unlabelled UPF1(115–287). The additional NMR signals corresponding to the last 20 residues of the larger UPF2 construct have chemical shifts that are consistent with an unstructured peptide chain (Supplementary Figure S9C). Indeed, these extreme C-terminal residues can also be readily proteolysed without affecting the stability of the complex (see truncation analysis below).
Figure 4.
Interaction of UPF2 with the CH-domain of UPF1 studied using NMR. Overlay of 1H, 15N HSQC spectra of the 15N-labelled UPF1 CH domain (residues 115–287), free (green) and in complex with unlabelled UPF2 comprising (A) the complete bipartite binding motif (residues 1105–1207), (B) the helical motif (residues 1105–1129) and (C) the β-hairpin motif (residues 1167–1207) (blue). The corresponding complexes are indicated schematically. Assigned chemical-shift perturbations are mapped onto the surface of UPF1 and are coloured in red. Cyan residues could not be analysed because of a lack of NMR signal.
To further characterize the bipartite binding region in UPF2, which interacts with UPF1, NMR spectra were recorded using two synthetic peptides, corresponding to the helical (residues 1105–1129) and β-hairpin (residues 1167–1207) motifs of UPF2, and to 15N-labelled UPF1 (115–287). As expected, both peptides bind to different regions in UPF1 (Figure 4B and C). The chemical shift perturbations on binding of the two peptides mapped onto the three-dimensional structure of UPF1 agree very well with the crystal structure. On binding of the UPF2 helix peptide, residues Leu193, Ile233, Val161 and Val157 in UPF1 show large chemical-shift perturbations, consistent with the interactions seen in the crystal structure (Figure 4B). For the UPF2 β-hairpin peptide, UPF1 residues Val206, Val205 and Trp241 show strong chemical-shift perturbations that cluster on the corresponding binding region seen in the crystal structure (Figure 4C). Finally, the NMR spectrum of the free 15N-UPF2(1105–1207) shows that the protein is present in an almost completely unfolded state in solution and folding is observed on addition of UPF1(115–287) (Supplementary Figure S9D). NMR measurements on the β-hairpin peptide show that it also unfolded in solution, whereas the helix peptide shows some NOEs that indicate a fractional population of helical conformation (data not shown).
We used isothermal titration calorimetry to investigate the thermodynamics of binding of UPF2(1105–1207) and of each of the two UPF2 elements separately to UPF1(115–287) (Supplementary Figure S10). The low dissociation constant, Kd, of 0.2 μM for the combined helical and β-hairpin elements in UPF2(1105–1207) is consistent with the high stability of the UPF1–UPF2 complex observed throughout purification. The separate helical and β-hairpin elements show different affinities for UPF1. α-helix (1105–1129) shows the weaker binding with a Kd of 92 μM, whereas β-hairpin (1167–1207) has an ∼sixfold higher affinity with a Kd of 16 μM (Supplementary Figure S10). These values are fully consistent with the corresponding NMR titration results (data not shown). Interestingly, different relative thermodynamic contributions are observed for the binding of the two elements. Binding of the α-helix alone is enthalpy driven (consistent with a major contribution of hydrogen bond formation), whereas for β-hairpin, the interaction is entropy driven (consistent with a major contribution of water release due to the hydrophobic effect) (Supplementary Figure S10D).
Altogether, these data confirm that UPF2 used a disordered bipartite motif that couples UPF1 binding to folding of the two interacting elements, with the β-hairpin element having the stronger interaction.
In vitro mutational analysis
To analyse the role of the different UPF2 C-terminal elements or particular residues in the formation of the complex with UPF1, we cloned variant His-tagged UPF2 constructs, co-expressed them with untagged UPF1(115–914) and tested their ability to retain UPF1 during Ni2+ resin purification. We first defined by deletion analysis the UPF2 region required for complex formation. It was found that the region 1105–1198 was minimal for a stable stoichiometric complex, having the same behaviour as longer constructs encompassing this region, the largest tested being 1090–1237 (data not shown). This confirms that region 1199–1237 is dispensable, even though the crystallographic structure shows that the C-terminal α-helix is extended until residue 1203 and there are some universally conserved features in the sequence alignment up to 1220 (Figure 2E). Consistent with the ITC results, a construct comprising residues 1151–1207, in which the N-terminal helix is deleted, is able to partially retain UPF1 during co-purification, whereas just the N-terminal helical region (residues 1105–1151) has an affinity too weak to retain UPF1 (data not shown). Internal deletion of the glycine-rich region of the linker (residues 1153–1164) does not affect UPF2 binding (data not shown).
We then analysed the importance of the principal UPF1-interacting residues of UPF2 by cloning different His-tagged UPF2(1105–1227) single-residue mutants and testing them as described above (Figure 5A). In almost all cases, mutations were chosen to introduce charged side chains in otherwise hydrophobic residues involved in interactions with UPF1, as experience shows that alanine mutations are usually insufficient to significantly affect protein–protein interactions. The mutation of conserved Phe1113 (F1113E) in the N-terminal alpha-helix almost completely abolishes UPF1 binding, whereas mutations of Met1120E and Met1121E show a weaker effect. Mutations of β-hairpin hydrophobic residues Phe1171E, Met1173E and Leu1174E have very severe effects on the UPF1–UPF2 interaction, completely impairing UPF1 retention by UPF2. Mutation of Arg1176 (R1176E) also has an effect on complex formation. The Arg1176 side chain interacts with Tyr184 of UPF1, which, when mutated, also disrupts complex formation (Kadlec et al, 2006). Met1169 and Phe1171 in the β-hairpin, Leu1194 and Ala1195 in the C-terminal small alpha-helix, and Met1190 in between these two secondary structure elements form hydrophobic interactions that maintain the C-terminal alpha helix packed on the β-hairpin. Thus, even though it does not directly contact UPF1, the mutation Met1169E impairs complex formation, probably by disrupting the mini-hydrophobic core of UPF2 and thus preventing correct folding on UPF1 binding.
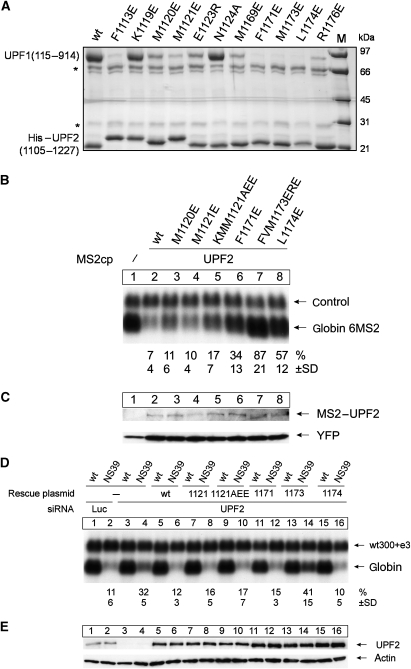
Figure 5.
Mutational analysis of the UPF2–UPF1 interface. (A) In vitro binding of UPF2 mutants. His-tagged UPF2(1105–1207) mutants were co-expressed with UPF1(115–914) and loaded on Ni2+ resin. The resin was washed with 10 CV of buffer containing 50 mM imidazole, 2 CV of buffer containing 100 mM imidazole and the proteins eluted with buffer containing 500 mM imidazole and analysed on a 10–16% SDS–PAGE. Contaminants are indicated with asterisks. The small UPF2 fragments run slightly differently because of charge variations among the mutants. (B) In vivo mutations and NMD tethering assay. Northern blot analysis of RNA from HeLa cells that were transfected with vectors for the 6MS2 reporter (6MS2) and the control (ctrl), together with MS2 (cp, lane 1), MS2-tagged UPF2 (lane 2) or the indicated mutants of UPF2 (lanes 3–8). (C) Cytoplasmic extracts from cells used in (A) were analysed with an MS2-specific antibody to visualize MS2-UPF2, or with a GFP-specific antibody to visualize the co-transfected GFP. (D) Northern blot analysis of RNA from HeLa cells transfected with Luciferase siRNA (negative control, lanes 1–2) or with a UPF2-targeting siRNA (lanes 3–16). The NMD reporter plasmids β-globin wt or NS39 and a transfection efficiency control (Gehring et al, 2003) were transfected, together with a plasmid expressing the indicated siRNA-insensitive mutants of UPF2 (lanes 5–16). The numbers indicate changes in mRNA abundance±s.d. determined by the analysis of five independent experiments. (E) Immunoblot analysis of the UPF2 expression in the lysate from cells used in (D) with a UPF2-specific antibody; actin served as control for comparable loading.
A mutational analysis of the putative UPF2-interacting residues of UPF1 was carried out before the UPF1–UPF2 complex structure was known (Kadlec et al, 2006). The results are fully consistent with the structural data now available. UPF1 mutations V161E/R162E, F192E and Y125E, in residues now shown to be directly interacting with the α-helical region of UPF2, strongly reduced UPF2 binding. On the opposite side of the UPF1 CH-domain, another set of mutations, E182R/Y184D, V204D and V206E, were shown to have a stronger effect, as observed for the mutation of their UPF2 counterpart. These residues are now observed to be directly contacting the β-hairpin region of UPF2.
In vivo mutational analysis
Experiments in vitro show that point mutations within UPF2 can almost completely disrupt the interaction with UPF1 by altering the hydrophobic contacts established by the two proteins; this effect is qualitatively similar whether the residues come from either of the two UPF2 elements mediating the interaction. The equivalent UPF2 mutants, in the context of the full-length protein, were used in UPF2 tethering or RNAi knockdown rescue assays (Gehring et al, 2003, 2005), to analyze their importance for in vivo NMD efficiency. In the tethering assay, mutations of residues Phe1171 and Leu1174, located in the β-hairpin region, decrease NMD efficiency between 30 and 50%, reaching ∼80% for the triple mutant, FVM1173ERE. However, mutations located in the N-terminal α-helix (M1120E and M1121E) do not reduce NMD activity significantly, except for a slight decrease in the case of the triple mutant, KMM1121AEE (Figure 5B). In the rescue assay, in which native UPF2 is siRNA depleted and the function is rescued by a transfection of wild-type or mutant UPF2 that are RNAi insensitive, we observe very little effect on NMD efficiency compared with wild-type for the α-helix mutants and a significant effect among β-hairpin mutants only for the triple mutant, FVM1173ERE, which also has the biggest loss of function in the tethering assay (Figure 5D). For both assays, the expression level of UPF2 mutants was checked by western blot (Figure 5C and E). The results of the two assays thus show the same trend, but with the tethering assay being more sensitive to UPF2 mutations. This can be explained by the fact that, in the tethering assay, activity is completely dependent on the tethered UPF2, there being no other upstream factors present (for example, UPF3b and EJC). On the contrary, for the rescue assay, the presence of residual wild-type UPF2 could reduce sensitivity; moreover, other factors, notably UPF3b, could provide additional bridging interactions. We also note that in the rescue assay, the FVM1173ERE mutant seems to have a dominant-negative function, perhaps resulting from the defective UPF2 interacting with UPF3b on the EJC, thus preventing the residual native UPF2 from binding and triggering NMD.
In conclusion, our in vitro binding studies and in vivo functional studies show that the β-hairpin region has the dominant function in making a functional UPF2–UPF1 interaction and that the complete disruption of this interaction severely impairs NMD. However, NMD is apparently tolerant to milder disruptions of the interaction of the C-terminal domain of UPF2 with UPF1, suggesting that alternative interactions, either involving other regions of UPF1 and UPF2 or other NMD factors, such as SMG-1, help stabilize the triggering complex (Yamashita et al, 2009).
Discussion
UPF1 and UPF2 are essential proteins for NMD. The interaction of EJC-associated UPF2 with UPF1 triggers UPF1 phosphorylation by SMG-1, initiating a series of downstream events that finally lead to the degradation of PTC-containing mRNAs. It was previously established that the C-terminal region of UPF2 is important for binding to the CH-domain of UPF1, but the structural details of this interaction have been hitherto unknown. Here, we show that the interaction is mediated by two distinct α-helical and β-hairpin elements of UPF2 that bind on opposite surfaces of the UPF1 CH-domain, separated by a flexible linker. Sequence comparisons show that this mode of binding is probably conserved throughout evolution, as the key feature of the C-terminal region of UPF2 are well conserved (Figure 2E). Complementarily, the interacting residues of the UPF1 CH-domain are also highly conserved (Kadlec et al, 2006). As previously noted, the only exceptions to this conservation are certain protists that seem to be mutated in the UPF2-binding regions of UPF1, for example, Encephalitozoon cuniculi (Kadlec et al, 2006). Interestingly, a putative UPF2 homologue exists in E. cuniculi (NP_584637), which lacks the entire C-terminal region beyond the third MIF4G domain.
NMR measurements on UPF2(1105–1207) show that the unbound C-terminal region of UPF2 is intrinsically disordered, with the secondary structures co-folding only on UPF1 binding. Indeed, there is a growing literature about the mediation of protein–protein interactions by intrinsically disordered proteins or intrinsically disordered regions (IDR) of proteins (Dyson and Wright, 2002, 2005; Meszaros et al, 2007; Tompa and Fuxreiter, 2008). These interactions have several characteristic features, most of which are shown by the UPF1–UPF2 complex, which distinguish them from more classical protein–protein interactions between globular regions of proteins (Meszaros et al, 2007). These include a large interaction surface (comparable with classical interfaces), but with the IDR having a much higher proportion of buried residues; thus, large interfaces can be achieved with shorter polypeptide lengths. Second, analyses have shown that IDRs interact with a higher proportion of hydrophobic residues (Meszaros et al, 2007), as has been pointed out above for UPF2. It is interesting to note that the mode of binding of UPF2 to UPF1 has surprising similarities to that of a peptide from SNARE protein SNAP-25 binding to botulinum neurotoxin A, which also shows helical and beta elements interacting in a bipartite manner on opposite sides of the toxin, but with the difference that the extended intervening peptide is ordered in this case (Breidenbach and Brunger, 2004). The similarity even extends to the presence of two close methionines on the α-helical segment of SNAP-25 making hydrophobic interactions, as in UPF2 (compare Figure 2C with Figure 3A in (Breidenbach and Brunger, 2004)). Consistent with previous analyses (Meszaros et al, 2007), the disordered linker (residues 1130–1167) between the α-helical and β-hairpin of UPF2 is highly divergent in sequence and length compared with the interacting regions (Figure 2E). However, the first part of this linker (1130–1145) has certain conserved features and is partially visible in the electron density in the vicinity of residues 219–224 of UPF1 loop L9, suggesting that both these regions may be involved in additional interactions at some stage during complex assembly.
It has been proposed that protein–protein interactions mediated by IDRs allow very specific recognition (because of the large interaction surface) but only moderate interaction strength (because of the entropy cost of ordering on binding), thus being suitable for processes requiring transient interactions. Our NMR, ITC and crystallographic results show that the α-helical and β-hairpin regions of UPF2 can bind independently to the UPF1 CH-domain, with estimated dissociation constants (Kd) of, respectively, 92 and 16 μM, whereas the two combined elements in the UPF2 C-terminal region have a Kd of 0.2 μM. What then might be the biological rationale for using an IDR-mediated mode of binding in the particular case of the UPF1–UPF2 interaction? Two points are relevant for discussion here. First the particular situation that arises in NMD whereby ribosome-bound UPF1 needs to recognize and bind tightly to EJC-bound UPF2, and second, possible competition between UPF1 binding to release factors and to UPF2.
With regard to the first point, the recognition and binding of UPF1 and UPF2 is not simply a problem of two freely diffusing proteins, but one in which each component is part of a large complex bound to the same mRNA. Productive interaction of these two relatively slowly diffusing complexes may be topologically constrained by the local environment. We suggest that recognition is more efficiently achieved by UPF2 using a long, flexible fishing line, which can rapidly explore a large volume, equipped with two separate hooks to ‘catch' UPF1. Either UPF2-binding element could make the initial encounter with UPF1, followed rapidly by binding of the other to form a tight complex. The concomitant folding of the secondary structure elements would also be equivalent to reeling in the fishing line and hence aid in bringing the stalled ribosome and EJC in closer proximity, perhaps allowing additional interactions to take place. In a model system, this ‘fly-casting' mechanism has been shown to speed up molecular recognition (Shoemaker et al, 2000).
With regard to competition for UPF1 binding with the release factors eRF1–3, it has recently been shown that, in mammalian cells, the CH-domain of UPF1 is the major region of interaction with the GTPase domain of both eRF3 and UPF2 (Ivanov et al, 2008), although this has been contested in yeast (Takahashi et al, 2008). It is not yet clear whether the UPF2- and eRF3-binding sites on the CH-domain overlap, but given that UPF2 potentially binds to a significant proportion of the surface, this seems possible. Again, the bipartite nature of UPF2 binding to UPF1 may have a special role here, in allowing UPF1 to simultaneously bind eRF3 and one or other elements of UPF2 in an initial interaction. This would subsequently be reinforced with both elements interacting within the DECID complex or after phosphorylation of UPF1, with a concomitant disassociation from eRF3 (Kashima et al, 2006). If the helical region of UPF2 competes for the eRF3-binding site of UPF1, this could explain its lesser importance in the NMD assays.
In a series of in vitro experiments, the effect of the C-terminal half of UPF2 (denoted UPF2S), alone or with full-length UPF3b, on the RNA-binding, RNA-dependent ATPase and helicase activities of UPF1 was determined (Chamieh et al, 2008). UPF2S (residues 770–1204) contains consecutively the third MIF4G domain (the UPF3b-binding site), the acidic region (1025–1094) and the C-terminal UPF1-binding site, and thus can bridge UPF1 to UPF3b, forming a ternary complex. Comparative measurements were determined using two UPF1 constructs, UPF1-L (115–914, the same construct as used here) and UPF1-ΔCH (295–914), that is, without the UPF2-binding CH-domain. It was observed initially that removal of the CH-domain doubles the helicase activity of UPF1-L and triples the ATPase activity without markedly affecting the RNA-binding ability. Thus, it was proposed that the CH-domain has a cis-inhibitory regulatory effect on the biochemical activities of UPF1-L, although this effect is moderate. Adding UPF2S to UPF1-L significantly reduces the RNA-binding ability of UPF1, slightly enhances the ATPase activity but does not affect the helicase activity. Adding UPF2S with UPF3b to UPF1-L (i.e., forming the ternary complex) again significantly reduces the RNA-binding ability of UPF1, further enhances the ATPase activity (nearly to the level obtained by removal of the CH domain) and increases the helicase activity (although not to the level obtained by removal of the CH domain). Thus, it was proposed that UPF1–2–3 ternary complex formation largely reverses the inhibitory effect of the CH-domain, thus perhaps triggering a remodelling function of UPF1.
Our results are generally consistent with this model, although it should be borne in mind that we have worked only with the C-terminal extremity of UPF2, lacking the UPF3b-binding site and the acidic region. The only discrepancy is that our combined crystallographic results, EM reconstructions and SAXS studies suggest that whether or not UPF1 has bound UPF2 (or partially bound, as in one of the crystal forms), the CH-domain is found docked on the side of the helicase 1A domain, orientated away from the helicase-active site. From our results, it is difficult to determine how binding of the C-terminal extremity of UPF2 to the CH-domain could influence the interaction of the CH-domain with the helicase, owing to the fact that, as shown in Figure 1D, any UPF2-induced conformational changes are restricted to the helicase distal part of the CH domain. Furthermore, we explicitly show that UPF2 binding does not affect the RNA-dependent ATPase activity of UPF1 (Supplementary Figure S6). Our SAXS results also seem to exclude the possibility that the CH-domain detaches part of the time in solution and thus, by virtue of its flexible linkage with the helicase, sterically interferes with the helicase function. Furthermore, our two different crystal forms of the UPF2–UPF1 complex show the helicase in different states (phosphate bound or ADP bound), suggesting that the fixed docking of the CH-domain on helicase domain 1A is compatible with different configurations of domain 2A. However, it cannot be ruled out that the ability to undergo necessary functional conformational changes, required for ATPase and helicase activities, may be affected by the presence of the bound CH domain.
Our observations are fully consistent with the results of Chamieh et al (2008) in that UPF2S binding to UPF1 does not change the ATPase activity or helicase activity significantly. The major negative effect of UPF2S binding (with or without UPF3b in addition) is on the RNA-binding ability of UPF1, which, we suggest, could be because of the acidic region (38 Glu/Asp between 1025–1094, i.e. 54%) having an inhibitory effect on RNA binding by electrostatic competition. The enhancement of UPF1 ATPase and helicase activity in the ternary UPF3b–UPF2S–UPF1 complex is then most likely because of an additional interaction between UPF3b and UPF1. Indeed, such an interaction has been implicated in a number of recent papers (Ohnishi et al, 2003; Ivanov et al, 2008; Takahashi et al, 2008) consistent with the fact that UPF2-independent NMD has been reported (Gehring et al, 2005) and is also discussed above in connection with our in vivo results showing that NMD is relatively tolerant to a mild disruption of the UPF1–UPF2 interaction.
Materials and methods
For full methods see Supplementary data.
Protein expression, purification and crystallization
UPF1(115–914) and His-tagged UPF2(1105–1198) were cloned, respectively, into pCDF-Duet1 and pProExHTb expression vectors and co-expressed in E.coli BL21Star(DE3) grown at 20°C after induction. The complex was purified on Ni2+ resin before and after His-tag removal using TEV protease. The last purification step was size exclusion chromatography in a buffer containing 20 mM Tris (pH 7), 150 mM NaCl and 4 mM DTT (buffer A). The protein was concentrated to 15 mg/ml and AMPPCP was added before crystallization to a final concentration of 5 mM. Crystallization trials were carried out with a Cartesian robot and yielded three different crystal forms. Cubic form: crystals appeared within ∼1 day in 1.2–1.6 M Na–K phosphate (pH 6.5–7.5), both in the presence and absence of AMPPCP. Orthorhombic form: Crystals appeared within 3–4 days in 1.6 M ammonium sulphate, 100 mM MES pH 6 and 10% dioxane. Monoclinic form: Crystals grow within ∼3–4 days in 1.5–1.6 M ammonium sulphate, 100 mM MES pH 6.3–6.5 and 2% v/v glycerol.
Crystallographic data collection and structure determination
Details are given in Table I. Data collection was carried out at 100 K at the European Synchrotron Radiation Facility, with crystals cryoprotected with 20–30% glycerol. All data were integrated with XDS (Kabsch, 1993) and analysed with CCP4i. Molecular replacement was carried out with PHASER (McCoy, 2007), model building with COOT (Emsley and Cowtan, 2004) and refinement using REFMAC5 (Murshudov et al, 1997). Orthorhombic crystals (I222) of the UPF1(115–914)–UPF2(1105–1198) complex diffracted to a 2.5 Å resolution. The structure was solved by molecular replacement using the phosphate-bound form of the UPF1 helicase domain (pdb entry 2GK7) and the structure of the CH-domain (pdb entry 2IYK). Monoclinic crystals (P21) of the UPF1(115–914)–UPF2(1105–1198) complex diffracted to a 2.9 Å resolution, with two complexes in the asymmetric unit. The structure was solved by molecular replacement using the UPF1 helicase domains, 1A and 2A, separately and the CH-domain as search models. Refinement was carried out using tight NCS and TLS. Numerous sulphates are found to be bound to the helicase domain in both crystal forms.
SAXS
X-ray scattering data were collected at the Bio-SAXS beamline (ID14-EH3) at the European Synchrotron Radiation Facility. For both UPF1 and the UPF1–UPF2 complex, data were collected at three different concentrations (∼2, 5 and 10 mg/ml). From the corrected scattering curves, the pair-distribution functions were computed using GNOM (Svergun, 1992). The program, DAMMIN (Svergun, 1999), was used to generate the low-resolution ab initio shapes, which were superimposed and averaged using DAMAVER (Volkov and Svergun, 2003).
NMR data collection and assignment
NMR spectra were recorded at 300 K on a Bruker DRX600 spectrometer equipped with a cryogenic probe. For backbone 1H, 15N and 13C assignment of UPF1, standard triple-resonances experiments were recorded (Sattler et al, 1999). Spectra were processed using NMRPipe (Delaglio et al, 1995) and analysed with NMRVIEW (Johnson, 2004). Longitudinal (T1) and transverse (T2) 15N relaxation and {1H}–15N heteronuclear NOE experiments of UPF1 were recorded as described (Farrow et al, 1994). Chemical-shift perturbations were calculated as CSP=(5*Δδ(1H)2+(Δδ (15N))2)1/2. Further details are provided in Supplementary data.
His-tag pull-down assays
Mutagenesis of UPF2 was carried out using a Quick-Change site-directed mutagenesis kit and confirmed by sequencing. His-tag-fused UPF2(1105–1227) mutants and UPF1(115–914) were cloned and co-expressed as described above. The soluble part of cell lysate was loaded on the Ni2+ resin and washed with 10 CV of buffer A containing 50 mM imidazole and 2 CV containing 100 mM imidazole; the elution was carried out with a 500 mM imidazole buffer and analysed on a 10–16% SDS–PAGE stained with Coomassie blue.
In vivo NMD assays
Mutagenesis of UPF2 was carried out using a Quick-Change site-directed mutagenesis kit and confirmed by sequencing. The β-globin 6MS2 plasmid construct and transfection control have been described previously (Gehring et al, 2003, 2005). HeLa cells were grown in DMEM and transfected by calcium phosphate precipitation in 6-well dishes with 1.0 μg of an MS2–UPF2 fusion construct, with 0.5 μg of the control plasmid, 2 μg of the 6MS2 reporter vector and 0.2 μg of a GFP expression plasmid. Immunoblot analysis was carried out using 20 μg of cytoplasmic extracts for SDS–polyacrylamide gel electrophoresis. Total cytoplasmic RNA was analysed by northern blotting as described by Gehring et al (2003). Signals were quantified in an FLA-3000 fluorescent image analyser (Raytest). Percentages±s.d. were calculated from three independent experiments. siRNA transfection and UPF2 complementation with a siRNA-insensitive UPF2 expression plasmid (1.2 μg/well) were carried out as previously described by Gehring et al (2005).
For EM, isothermal titration calorimetry, ATPase assay, protein production and isotope labelling for NMR see Supplementary data.
PDB depositions
Atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 2wjv for the monoclinic form of the UPF1–UPF2 complex and 2wjy for the orthorhombic form (in this form, UPF2 is very poorly ordered and not included in the model).
Supplementary Material
Supplementary data
Acknowledgments
We thank the ESRF for providing access to synchrotron beamlines, Drs Thibaut Crépin and Andrew McCarthy for their help with crystallographic data collection and Dr Adam Round for his assistance with the small-angle scattering analysis. The technical platforms of the Partnership for Structural Biology (PSB) were extensively used, notably the robotic crystallization facility. AM is supported by a PhD fellowship (Ref. SFRH/BD/22323/2005) from the Portuguese Foundation for Science and Technology (FCT). We thank Joel Sussman and Peter Tompa for discussions about intrinsically disordered proteins.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J (2006) The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle 5: 1496–1498 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Izaurralde E (2006) Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev 20: 391–398 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E (2007) mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett 581: 2845–2853 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT (2004) Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 432: 925–929 [DOI] [PubMed] [Google Scholar]
- Brogna S, Wen J (2009) Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol 16: 107–113 [DOI] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O (2006) EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93 [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA II, Wilkinson MF (2007) An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Su LH, Sun CH (2008) Incomplete nonsense-mediated mRNA decay in Giardia lamblia. Int J Parasitol 38: 1305–1317 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H (2007) Structural and functional insights into the human Upf1 helicase core. EMBO J 26: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK (2009) Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 17: 316–325 [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1: 610–623 [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX Pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R (2007) RNA quality control in eukaryotes. Cell 131: 660–668 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2002) Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 12: 54–60 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O (2008) Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003 [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE (2005) Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE (2003) Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E (2006) Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, Rio DC, Brenner SE (2003) Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19: i118–i121 [DOI] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A (1996) Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA 2: 153–170 [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A (1997) Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE (2008) The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, Le Mouel A, Lepere G, Schachter V, Betermier M, Cohen J, Wincker P, Sperling L, Duret L, Meyer E (2008) Translational control of intron splicing in eukaryotes. Nature 451: 359–362 [DOI] [PubMed] [Google Scholar]
- Johnson BA (2004) Using NMRView to visualize and analyze the NMR spectra of Macromolecules. Methods Mol Biol 278: 313–352 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kadlec J, Guilligay D, Ravelli RB, Cusack S (2006) Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12: 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S (2004) The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11: 330–337 [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D (2006) Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34: 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G (2001) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293: 1832–1836 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE (2005) Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol 17: 309–315 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131 [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10: 99–105 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol 20: 8944–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B, Tompa P, Simon I, Dosztanyi Z (2007) Molecular principles of the interactions of disordered proteins. J Mol Biol 372: 549–561 [DOI] [PubMed] [Google Scholar]
- Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R (2008) Recognition and elimination of nonsense mRNA. Biochim Biophys Acta 1779: 538–549 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc 34: 93–158 [Google Scholar]
- Schwartz AM, Komarova TV, Skulachev MV, Zvereva AS, Dorokhov Iu L, Atabekov JG (2006) Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry (Mosc) 71: 1377–1384 [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA 97: 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A (2008) Messenger RNA regulation: to translate or to degrade. EMBO J 27: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L (2008) Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameters in indirect transform methods using perpetual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Svergun DI (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76: 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino S, Kontani K, Nishina H, Katada T (2008) Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14: 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J et al. (2007) Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39: 1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics, Chapter 2, Unit 2.3 [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M (2008) Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci 33: 2–8 [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT (2008) NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22: 1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW (1996) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, Kurata R, Hirano H, Anderson P, Ohno S (2009) SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data