Abstract
We have found earlier that Tor1 binds to 5S rDNA chromatin but the functional significance has not been established. Here, we show that association with 5S rDNA chromatin is necessary for TOR complex 1 (TORC1) to regulate the synthesis of 5S ribosomal RNA and transfer RNAs (tRNAs) by RNA polymerase (Pol) III, as well as the phosphorylation and binding to Pol III-transcribed genes of the Pol III repressor Maf1. Interestingly, TORC1 does not bind to tRNA genes, suggesting that TORC1 modulates tRNA synthesis indirectly through Maf1 phosphorylation at the rDNA loci. We also find that Maf1 cytoplasmic localization is dependent on the SSD1-v allele. In W303 cells that carry the SSD1-d allele, Maf1 is constitutively nuclear but its nucleolar localization is inhibited by TORC1, indicating that TORC1 regulates nucleoplasm-to-nucleolus transport of Maf1. Finally, we show that TORC1 interacts with Maf1 in vivo and phosphorylates Maf1 in vitro, and regulates Maf1 nucleoplasm-to-nucleolus translocation. Together, these observations provide new insights into the chromatin-dependent mechanism by which TORC1 controls transcription by Pol III.
Keywords: Maf1, nucleolus, rDNA, RNA polymerase III, target of rapamycin (TOR)
Introduction
Cell growth is a process by which the cell increases its size and mass through the synthesis of proteins and other macromolecules. Ribosome biogenesis is crucial for growth by producing ribosomes, the machinery for protein synthesis. It accounts for the majority of nuclear transcription (up to 80%) in a eukaryotic cell (Warner, 1999; Moss and Stefanovsky, 2002). The cell tightly controls ribosome biogenesis in response to different growth and stress conditions. Deregulation of ribosome biogenesis is a prominent feature of many tumour cells: transformed cells overexpress the products of polymerase (Pol) I and III-transcribed genes; activation of Pol III-dependent transcription can promote oncogenic transformation (White, 2005; Johnson and Johnson, 2008). Ribosome biogenesis requires close coordination of all three RNA polymerases to produce individual components in an equal ratio (Warner, 1999; Moss and Stefanovsky, 2002). How the activity of all three RNA polymerases is controlled in a well-coordinated manner is a very interesting question. Recently, significant progress has been made that shows the cell has the intrinsic ability to balance the production of different ribosomal RNAs (rRNAs) (Laferte et al, 2006). However, the coordination of ribosome biogenesis is likely to be very complex and requires further understanding.
TOR (target of rapamycin) is an evolutionally conserved PI3K-related kinase and a central regulator of cell growth (Wullschleger et al, 2006; Tsang et al, 2007b) originally found in budding yeast (Heitman et al, 1991). TOR proteins form two functional complexes. In yeast, TOR complex 1 (TORC1) consists of Kog1, Lst8, and either Tor1 or Tor2, whereas TOR complex 2 (TORC2) consists of Avo1-3, Lst8 and Tor2 (Loewith et al, 2002). The anticancer drug rapamycin specifically inhibits TORC1 after forming a complex with its intracellular receptor FKBP12 (FK506-binding protein 12 kDa), and the subsequent binding of FKBP12-rapamycin to FRB domain of TOR (Zheng et al, 1995). Mutations of Tor1 at Ser1972 block FKBP12-rapamycin binding and confer rapamycin resistance (Zheng et al, 1995). TORC1 regulates a broad spectrum of growth-related processes including ribosome biogenesis, protein translation, nutrient import and autophagy (Wullschleger et al, 2006; Tsang et al, 2007b). TORC2 carries out a rapamycin-insensitive essential function of regulating actin cytoskeleton organization (Loewith et al, 2002).
TORC1 is a major regulator of transcription in yeast, including all ribosomal genes transcribed by three major RNA polymerases (Warner, 1999; Moss and Stefanovsky, 2002). Rapamycin and nutrient starvation cause rapid repression of ribosomal genes (Zaragoza et al, 1998; Powers and Walter, 1999). TOR is found in the cytoplasm as well as in the nucleus in mammals and yeast (Zhang et al, 2002; Drenan et al, 2004; Li et al, 2006), suggesting that TOR is likely to have nuclear functions such as gene regulation. Interestingly, TORC1 nuclear localization is essential for Pol I- but not Pol II-transcribed genes in yeast (Li et al, 2006). TORC1 in either cytoplasmic or nuclear form is sufficient to control Pol II gene transcription. TORC1 is further found to be associated with 35S promoter and 5S rDNA chromatins. This association is critically dependent on a helix-turn-helix (HTH) sequence, a classical DNA-binding motif. Interestingly, HTH deletion abolishes Tor1 association with rDNA chromatin but does not affect Tor1 localization in the nucleus, suggesting that this mutation specifically inhibits the ability of Tor1 to bind to rDNA chromatin. Together, these observations show that TORC1 association with rDNA chromatins is crucial for TORC1 to regulate 35S rDNA transcription. However, the significance of TORC1 association with rDNA chromatin in the regulation of Pol III-dependent genes, including 5S rDNA and tDNAs, remains to be established.
Maf1 is a key regulator of Pol III-dependent transcription (Geiduschek and Kassavetis, 2006; Willis and Moir, 2007) that was initially identified in yeast (Murawski et al, 1994). It was later found to be a negative regulator of Pol III-dependent transcription (Boguta et al, 1997; Pluta et al, 2001). Mammalian Maf1 has also been recently reported to have similar functions (Johnson et al, 2007; Goodfellow et al, 2008), indicating that Maf1 proteins are evolutionarily conserved. Maf1 is a major effector mediating diverse growth and stress signals to control the transcription of Pol III-dependent genes, including 5S rRNA tRNA genes (Upadhya et al, 2002). It does so by preventing Pol III occupancy on its targeted genes, likely as a result of Maf1 interaction with Pol III (Desai et al, 2005). Maf1 is a phosphoprotein present in the cytoplasm under nutrient-rich conditions (Moir et al, 2006; Oficjalska-Pham et al, 2006; Roberts et al, 2006). In response to TORC1 inhibition by starvation or rapamycin, Maf1 becomes dephosphorylated and accumulates in the nucleus, resulting in association with and repression of 5S rDNA and tDNA (tRNA gene). Although these observations clearly show that TORC1 regulates Maf1 phosphorylation, the detailed mechanisms such as how TORC1 acts on Maf1 remain obscure. Moreover, how Maf1 activity is controlled remains unresolved. Protein kinase A (PKA) has been shown to phosphorylate Maf1 N-terminus in vitro and regulate Maf1 nuclear localization (Moir et al, 2006). However, in a strain lacking Maf1 nuclear exportin, Maf1 is constitutively nuclear but Pol III transcription remains normally regulated by TORC1 (Towpik et al, 2008), indicating that there is an unknown mechanism by which TORC1 controls Maf1 activity.
In this study, we confirm that TORC1 is indeed associated with 5S rDNA and show that the ability of TORC1 to associate with rDNA chromatin is crucial for TORC1 to regulate Maf1 phosphorylation, and 5S rRNA and tRNA synthesis by Pol III. We further find that TORC1 is associated with Maf1 in vivo and phosphorylates Maf1 in vitro, and regulates Maf1 nucleoplasm-to-nucleolus relocalization. Together, these results show that TORC1 regulates Maf1 phosphorylation and subnuclear localization, and Pol III-dependent transcription in a chromatin-dependent mechanism.
Results
TORC1 association with 5S rDNA chromatin in a nutrient-dependent and rapamycin-sensitive manner
In addition to 35S promoter chromatin, we showed earlier that Tor1 is associated with 5S rDNA chromatin in a rapamycin-sensitive manner (Li et al, 2006) (Figure 1A and C), suggesting a similar role of TORC1 in Pol III regulation. We, therefore, further characterized TORC1 association with 5S rDNA and the functional significance. We found that Tor1 association with 5S rDNA was significantly reduced by starvation (Figure 1B). Little or no chromatin immunoprecipitation (ChIP) signal was detected in a tor1Δ strain, indicating the specificity of the ChIP result. Rapamycin-sensitive association of Tor1 with 5S rDNA and 35S promoter (35S-P) was further confirmed by real-time PCR (Figure 1D). In contrast, the repetitive CUP1 promoter, and intergenic spacer 1 (IGS1) and 35S rDNA coding region did not show any significant Tor1 binding.
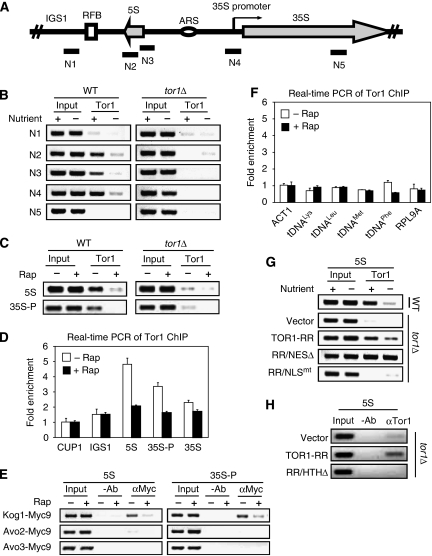
Figure 1.
TORC1 binds to 5S rDNA chromatin in a nutrient-dependent and rapamycin-sensitive manner. (A) Shown is the structure of a yeast rDNA repeat. N1-5 indicates the PCR primer sets used for ChIP assays. N4 covers 35S rDNA promoter region. (B) Tor1 associates with 5S rDNA and 35S promoter chromatin regions in a nutrient-dependent manner. Early log phase W303a wild-type (WT) and tor1Δ cells were starved for 30 min by nutrient depletion. ChIP assay was conducted using a Tor1-specific antibody. (C) Association of Tor1 with 5S rDNA and 35S promoter (35S-P) in a rapamycin-sensitive manner. Early log phase W303a WT and tor1Δ cells were treated without or with 100 nM rapamycin. ChIP assay was conducted with the PCR primer sets N2 and N4. (D) Real-time PCR quantification of Tor1 occupancy on various regions of rDNA sequences in the absence or the presence of rapamycin. ChIP samples were analysed by real-time PCR. Values are the average of three quantifications. Error bars represent the standard deviation. Fold enrichments were determined by comparing the ChIP signals of target regions with that of control (CUP1 promoter region). IGS1, 5S rDNA, 35S rDNA promoter (35S-P), 35S rDNA coding region (35S) were tested. (E) TORC1 but not TORC2 is associated with 5S rDNA and 35S promoter chromatin regions. Early log phase cells expressing Kog1-Myc9, Avo2-Myc9 or Avo3-Myc9 were treated without or with 100 nM rapamycin. ChIP assay was conducted by immunoprecipitation with a Myc-specific antibody (9E10) and the PCR primer set N2 and N4. (F) Real-time PCR quantification of Tor1 occupancy on various tDNA loci. ChIP samples were analysed by real-time PCR. Values are the average of three quantifications. Error bars represent the standard deviation. Fold enrichments were determined by comparing the ChIP signals of target regions with that of controls (ACT1 promoter region). A ribosome subunit gene (RPL9A) promoter was included as an unrelated control. (G) Nuclear localization controls TORC1 binding to 5S rDNA chromatin. Early log phase W303a WT and tor1Δ cells carrying a vector control or expressing Tor1-RR variants were starved for 30 min. The binding of WT Tor1, Tor1-RR, Tor1-RR/NLSmt and Tor1-RR/NESΔ to 5S rDNA was detected by ChIP assay with a Tor1-specific antibody and the PCR primer set N2. (H) HTH motif is required for TORC1 to bind to 5S rDNA. W303a tor1Δ cells carrying a vector control or expressing Tor1-RR or Tor1-RR/HTHΔ were cultured to early log phase and ChIP assay was conducted with a Tor1-specific antibody and the PCR primer set N2.
We also investigated which TOR complex is associated with 5S rDNA. We found that Kog1-Myc9 was associated with 5S rDNA in a rapamycin-sensitive manner (Figure 1E) similar to what was observed with 35S rDNA promoter. In contrast, Avo2-Myc9 and Avo3-Myc9 were not detected in these chromatin regions. Thus, TORC1 but not TORC2 is associated with 5S rDNA and 35S promoter in a nutrient-dependent and rapamycin-sensitive manner. As Pol III is responsible for the synthesis of both tRNAs and 5S rRNA, we investigated the association of Tor1 with tRNA genes (tDNA). To our surprise, we were unable to detect significant Tor1 association with tDNA over the background (ACT1, RPL9A), as judged by both conventional and real-time PCR (Figure 1F and data not shown). Unlike the rRNA genes, TORC1 does not seem to be associated with tDNA chromatin.
Association with 5S rDNA chromatin requires Tor1 nuclear localization and the HTH motif
TORC1 is normally localized in both the nucleus and cytoplasm. On nutrient starvation, however, TORC1 is rapidly excluded from the nucleus (Li et al, 2006). Tor1 nuclear import and export are mediated by the nuclear localization sequence (NLS) and nuclear export sequence (NES), respectively. In addition, Tor1's ability to bind to 35S promoter is dependent on an HTH motif. Mutations in these motifs specifically block Tor1 nucleocytoplasmic transport or association with 35S rDNA promoter (Li et al, 2006). When such a mutation is combined with a rapamycin-resistant (RR) mutation, it provides a valuable tool to delineate the underlying physiological function of Tor1 localization or association with rDNA. For example, Tor1-RR/NLSmt is excluded from the nucleus, preventing it from association from 35S promoter (Li et al, 2006). In contrast, Tor1-RR/NESΔ, concentrated in the nucleus, is capable of binding to 35S rDNA promoter in a starvation-insensitive manner. In addition, HTHΔ disrupts the ability of Tor1-RR to associate with 35S rDNA promoter. Interestingly, both Tor1-RR/NLSmt and Tor1-RR/NLSΔ retain the ability to regulate Pol II-dependent transcription, revealing distinct mechanisms for Tor1 to control different class of genes (Li et al, 2006).
To determine the requirement for TORC1 association with 5S rDNA chromatin, we studied Tor1-RR, Tor1-RR/NLSmt and Tor1-RR/NESΔ. We found that both endogenous wild-type Tor1 and Tor1-RR were associated with 5S rDNA chromatin in a nutrient-dependent manner (Figure 1G). Tor1-RR/NESΔ but not Tor1-RR/NLSmt was also found at 5S rDNA chromatin. Notably, the amount of Tor1-RR/NESΔ bound to 5S rDNA chromatin was considerably more than Tor1-RR, and this binding was resistant to nutrient starvation, indicating that Tor1 association with 5S rDNA chromatin is primarily regulated by Tor1 nuclear import. Tor1-RR/HTHΔ also failed to bind to 5S rDNA chromatin (Figure 1H). These observations show that TORC1 association with 5S rDNA chromatin is regulated by its nuclear localization and is dependent on the HTH motif. Strikingly, TORC1 shows the same requirement towards both 5S rDNA and 35S promoter chromatin, suggesting that TORC1 binding to both 5S and 35S rDNA chromatins is well coordinated.
Regulation of Pol III-dependent transcription requires TORC1 nuclear localization and chromatin association
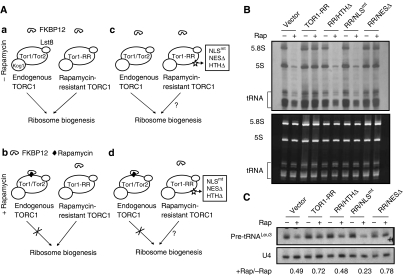
TORC1 proteins in the nucleus and cytoplasm have distinct functions in the regulation of Pol I and Pol II genes: TORC1 nuclear localization and association with 35S promoter chromatin are required for the regulation of Pol I- but not Pol II-transcribed genes (Li et al, 2006). We wondered whether the control of Pol III-transcribed genes has similar requirements and addressed this question with different Tor1-RR variants with distinct localizations. Tor1-RR contains the Ser1972 → Ile mutation that prevents the binding of FKBP12-rapamycin to Tor1, conferring dominant rapamycin-resistance (Zheng et al, 1995). On adding rapamycin, endogenous TORC1 is knocked down, whereas TORC1 consisting of a Tor1-RR variant is not affected (Figure 2A), allowing specific analysis of these variants for their regulatory functions. In the absence of rapamycin, endogenous TORC1 (containing either Tor1 or Tor2) and the TORC1-containing Tor1-RR are capable of promoting ribosome biogenesis (Figure 2A a). In the presence of rapamycin, however, endogenous TORC1, but not the Tor1-RR-containing TORC1, is chemically knocked down by rapamycin (Figure 2A b). In the absence of rapamycin treatment, cells expressing Tor1-RR variants have normal ribosome biogenesis because of the presence of functional endogenous TORC1 (Figure 2A c) (Li et al, 2006). When treated with rapamycin, endogenous TORC1 is inhibited, whereas TORC1 containing a Tor1-RR variant (NESΔ, NLSmt or HTHΔ) is not affected (Figure 2A d), allowing us to specifically examine the effect of different mutations (NESΔ, NLSmt or HTHΔ) on ribosome biogenesis (Figure 2A d). It is worth noting that all three Tor1-RR variants still retain the ability to regulate Pol II-transcribed genes and Gln3 phoshorylation, indicating that they can form an otherwise functional TORC1 complex necessary for Gln3 regulation (Li et al, 2006).
Figure 2.
Transcription of Pol III-dependent genes requires TORC1 nuclear localization and its ability to associate with chromatin. (A) A strategy to study the regulation of ribosome biogenesis by TORC1. See main text for detailed description. (B) Synthesis of rRNAs and tRNAs in yeast cells expressing Tor1-RR variants. Early log phase W303a tor1Δ cells carrying vector control or expressing Tor1-RR variants were treated without or with rapamycin for 30 min, and then metabolically labelled with [5, 6-3H]-Uracil. Newly synthesized rRNAs and tRNAs were detected by autoradiography (upper panel). Total RNAs were stained by ethidium bromide (lower panel). (C) Transcription of pre-tRNALeu3 in yeast cells expressing Tor1-RR variants. Early log phase W303a tor1Δ cells carrying vector control or expressing Tor1-RR variants were treated without or with rapamycin for 30 min. The level of pre-tRNALeu3 was determined by northern blot with U4 small nuclear RNA as a loading control. The numbers represent the ratio of transcript levels after and before rapamycin treatment.
To analyse the functions of different Tor1-RR variants, we metabolically labelled yeast cells with 3H-Uracil to monitor rRNA synthesis. In the absence of rapamycin, 5.8S rRNA (transcribed by Pol I) and 5S rRNA (transcribed by Pol III) were actively synthesized because of the presence of the endogenous TORC1 (Figure 2B). On rapamycin treatment, 5S and 5.8S rRNA synthesis was strongly inhibited in the control cells, or cells expressing Tor1-RR/NLSmt or Tor1-RR/HTHΔ. In contrast, 5S and 5.8S rRNA synthesis remained relatively normal in cells expressing Tor1-RR and Tor1-RR/NESΔ. The synthesis of tRNAs showed essentially the same behaviour as 5S and 5.8S rRNAs. To verify this result, we performed northern blot to detect labile precursor tRNA species (pre-tRNALeu3), which is another method to monitor tRNA synthesis (Upadhya et al, 2002; Oficjalska-Pham et al, 2006). Indeed, tRNA synthesis showed the similar response to the TOR1-RR variants (Figure 2C). These results indicate that nuclear localization and association with rDNA chromatin are required for Tor1 to control Pol III-dependent transcription. It is interesting to note that unlike 5S rRNA, tRNA synthesis is only moderately affected by rapamycin (Figure 2), which is consistent with earlier observations (Upadhya et al, 2002; Oficjalska-Pham et al, 2006).
Maf1 phoshorylation and cytoplasmic localization depend on TORC1 nuclear localization and chromatin association
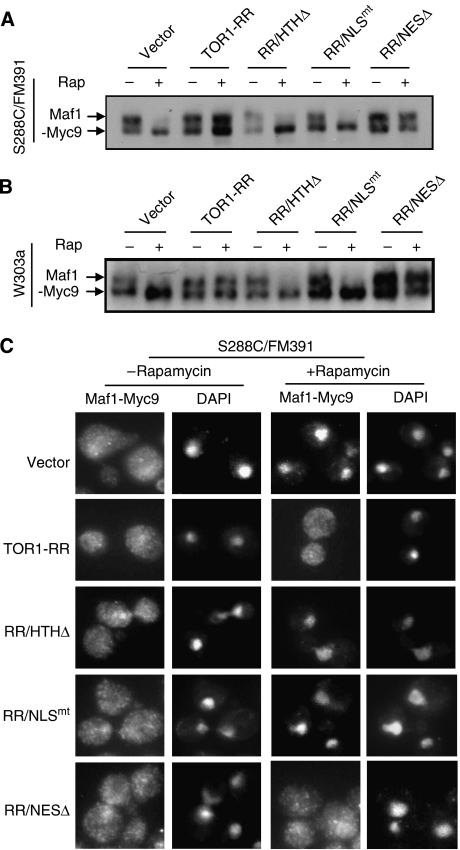
Maf1 negatively regulates Pol III-dependent transcription in response to nutrient limitation or rapamycin treatment. Maf1 is a phosphoprotein whose phosphorylation is regulated by TORC1 (Moir et al, 2006; Oficjalska-Pham et al, 2006; Roberts et al, 2006). We investigated where Maf1 phosphorylation occurs using different Tor1-RR variants. In agreement with earlier studies (Moir et al, 2006; Oficjalska-Pham et al, 2006; Roberts et al, 2006), rapamycin inhibited Maf1 phosphorylation in the control cells of both S288C/FM391 and W303a strains (Figure 3A and B). In cells expressing Tor1-RR or Tor1-RR/NESΔ, however, Maf1 remained phosphorylated under the same condition. In contrast, rapamycin inhibited Maf1 phosphorylation in cell expressing Tor1-RR/NLSmt and Tor1-RR/HTHΔ, indicating that Maf1 phosphorylation critically depends on TORC1 nuclear localization and chromatin association.
Figure 3.
TORC1 regulates Maf1 phoshorylation and nucleocytoplasmic transport. (A) Maf1 phosphorylation requires TORC1 nuclear localization and chromatin association in S288C/FM391 cells. Exponentially growing S288C/FM391 cells carrying a vector control only or expressing Tor1-RR variants were treated without or with 100 nM rapamycin for 30 min. Phosphorylation of Maf1-Myc9 was determined by electrophoretic mobility (arrows) by western blot with a Myc-specific antibody. (B) Maf1 phosphorylation requires TORC1 nuclear localization and chromatin association in W303a cells. Exponentially growing W303a cells carrying a vector control only expressing Tor1-RR variants were treated without or with 100 nM rapamycin for 30 min. Phosphorylation of Maf1-Myc9 was determined by electrophoretic mobility (arrows) by western blot with a Myc-specific antibody. (C) Maf1 is predominantly cytoplasmic and rapamycin treatment causes Maf1 nuclear accumulation in S288C/FM391 cells. Exponentially growing S288C/FM391 cells carrying a vector control only or expressing Tor1-RR variants were treated without or with 100 nM rapamycin for 30 min. Maf1-Myc9 localization was determined by indirect immunofluorescence (IF) with a Myc-specific antibody. The nucleus was stained by DAPI.
Maf1 is normally localized in the cytoplasm and rapamycin causes Maf1 to enter the nucleus, which has been proposed to control Maf1 activity (Moir et al, 2006; Oficjalska-Pham et al, 2006; Roberts et al, 2006). To further characterize the role of TORC1 regulation, we investigated Maf1 localization in cells expressing different Tor1-RR variants. In agreement with earlier findings, exponentially growing S288C/FM391 cells showed Maf1 distribution in the cytoplasm (Figure 3C). Interestingly, Maf1 was also clearly detectable in the nucleus. Rapamycin treatment caused Maf1 to enrich in the nucleus (Figure 3C). The same phenomenon was observed in cells expressing Tor1-RR/NLSmt and Tor1-RR/HTHΔ. In contrast, Maf1 remained throughout the entire cell in the presence of rapamycin in cells expressing Tor1-RR or Tor1-RR/NESΔ. Thus, regulation of Maf1 cytoplasmic retention requires TORC1 nuclear localization and rDNA chromatin association.
It has been reported that artificially forced enrichment of Maf1 in the nucleus is insufficient to inhibit Pol III transcription of tRNA (Moir et al, 2006; Willis and Moir, 2007). In S288C/FM391 cells lacking the Maf1 nuclear exportin Msn5, Maf1 is enriched in the nucleus but Maf1 phosphorylation and Pol III-dependent transcription remain fully regulated by nutrient availability (Towpik et al, 2008). These observations suggest that there is a novel regulatory mechanism for Maf1 within the nucleus. Interestingly, we found that in W303a cells, Maf1 was constitutively nuclear (Supplementary Figure 1). However, Maf1 remained phosphorylated in a rapamycin-sensitive manner (Figure 3B). In addition, Pol III-dependent transcription in these cells was regulated in a rapamycin-sensitive manner (Figure 2B and C). Maf1 nuclear regulation occurs in both S288C/FM391 and W303, two most popular laboratory strains, indicating that the nuclear mechanism is a common regulation and is not strain specific. Thus, W303 strain provides an excellent model to investigate nuclear regulation of Maf1.
One key difference between S288C/FM391 and W303 is that S288C/FM391 contains an allele of SSD1 called SSD1-v, whereas W303 strain carries an SSD1-d allele, based on their ability to suppress the lethality of sit4Δ mutation (Sutton et al, 1991). Interestingly, overexpression of SSD1 suppresses RNA Pol III mutations (Stettler et al, 1993), raising the possibility that Ssd1 is involved in Maf1 regulation. To test this hypothesis, we cloned SSD1-v from S288C/FM391 into a centromere plasmid and introduced it into W303a. Indeed, SSD1-v suppressed the rapamycin hypersensitive phenotype of the W303a tor1Δ strain (Supplementary Figure 2A), confirming an earlier observation (Reinke et al, 2004). In W303a cells carrying the SSD1 plasmid, there was a significant increase in Maf1 proteins in the cytoplasm (Supplementary Figure 2B), suggesting that Ssd1 is involved in the regulation of Maf1 cytoplasmic localization.
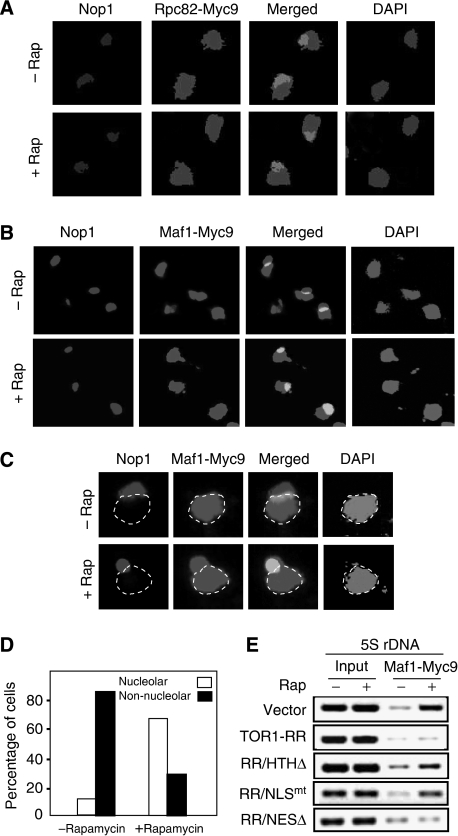
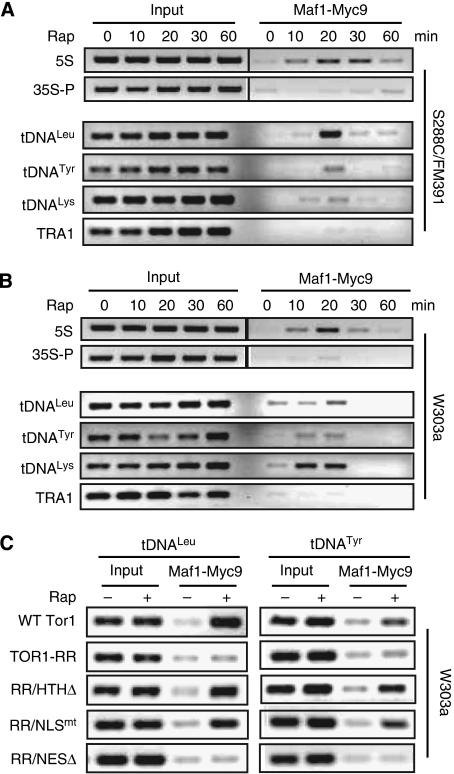
TORC1 controls Maf1 nucleolar localization and association with Pol III-dependent genes
The nucleolus is a subnuclear compartment in which 5S rDNA is transcribed by Pol III. We found that Pol III-specific subunit Rpc82 was distributed throughout the entire nucleus, which did not change significantly in response to rapamycin treatment (Figure 4A; see Supplementary Figure 3A for colour images). Thus, a simple mechanism for TORC1 to regulate Pol III activity and Maf1 within the nucleus is to control Maf1's accessibility to the nucleolus. Indeed, we therefore investigated Maf1 localization in W303a cells and found that Maf1 in exponential W303a cells was localized in the nucleus but excluded from the nucleolus (Figure 4B–D; see Supplementary Figure 3B and C for colour images). After rapamycin treatment, however, Maf1 rapidly entered the nucleolus as indicated by co-staining with Nop1, a nucleolar marker. Thus, TORC1 normally keeps Maf1 outside the nucleolus. We further studied Maf1 association with Pol III-transcribed genes by ChIP assay. In exponentially growing cells, Maf1 was poorly associated with 5S rDNA and tDNA (Figure 5A and B, 0 min). On rapamycin treatment, however, Maf1 occupancy at these chromatin regions increased significantly. Interestingly, Maf1 association with 5S rDNA and tDNA chromatins peaked at around 20 min of rapamycin treatment and then tapered (Figure 5), indicating that Maf1 only transiently associates with Pol III-transcribed genes. The peak Maf1 occupancy at Pol III-transcribed genes occurred earlier in W303a than S288C/FM391 cells. As Maf1 is already localized in the nucleus in W303a cells, a logic explanation for this phenomenon is that it takes less time for Maf1 to travel to the nucleolus in this genetic background. The presence of Tor1-RR or Tor1-RR/NESΔ, but not Tor1-RR/HTHΔ or Tor1-RR/NLSmt, prevented the rapamycin-induced increase of Maf1 association with Pol III-transcribed genes (Figures 4E and 5C), suggesting that TORC1 regulates Maf1 occupancy on Pol III-dependent genes within the nucleus and at rDNA chromatin.
Figure 4.
Maf1 is normally excluded from the nucleolus and inhibition of TORC1 promotes Maf1 nucleolar localization and 5S rDNA association. (A) Rpc82 is localized in both nucleus and nucleolus, which is not affected by rapamycin treatment. Exponentially growing W303a cells expressing Rpc82-Myc9 were treated without or with 100 nM rapamycin for 30 min. Rpc82-Myc9 localization was determined by IF with a Myc-specific antibody. The nucleolus was stained with a Nop1 antibody and the nucleus was stained by DAPI. (B) Maf1 is normally absent from the nucleolus and rapamycin causes Maf1 nucleolar localization. Exponentially growing W303a cells expressing Maf1-Myc9 were treated without or with 100 nM rapamycin for 30 min. Maf1-Myc9 localization was determined by IF with a Myc-specific antibody. The nucleolus was stained with a Nop1 antibody and the nucleus was stained by DAPI. (C) Shown are enlarged images of representative cells from Figure 4B. The dotted line indicates the nucleoplasm. (D) Quantification of cells with different Maf1 localization (white bar, cells with Maf1 in the nucleolus; black bar, cells with Maf1 excluded from the nucleolus) (N⩾200). (E) Inhibition of Maf1 association with 5S rDNA chromatin is dependent on TORC1 nuclear localization and rDNA chromatin binding. W303a cells carrying a vector control or expressing Tor1-RR variants were treated without and with 100 nM rapamycin and ChIP assay was performed with a Myc-specific antibody.
Figure 5.
Inhibition of TORC1 causes transient Maf1 association with Pol III-transcribed genes. (A) Rapamycin causes Maf1 to transiently associate with 5S rDNA and tDNA chromatins in S288C/FM391 cells. Exponentially growing cells expressing Maf1-Myc9 was treated without or with 100 nM rapamycin for different times. Maf1 association with 5S rDNA (upper panel) and tDNAs (lower panel) was determined by ChIP. 35S rDNA promoter and TRA1 were used as controls. (B) Rapamycin causes Maf1 to transiently associate with 5S rDNA and tDNAs in W303a cells. Exponentially growing cells expressing Maf1-Myc9 was treated without or with 100 nM rapamycin for different times. Maf1 association with 5S rDNA (upper panel) and tDNAs (lower panel) was determined by ChIP. 35S promoter (35S-P) and TRA1 were used as controls. (C) TORC1 association with chromatin is antagonistic to Maf1 binding. Early log phase W303a cells carrying a vector control or expressing Tor1-RR variants were treated with 100 nM rapamycin for 20 min. The binding of Maf1-Myc9 to tDNALeu and tDNATyr was detected by ChIP assay.
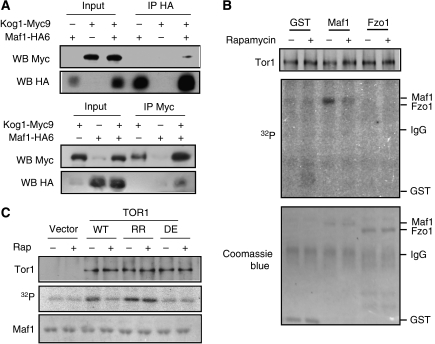
TORC1 interacts with Maf1 in vivo and phosphorylates Maf1 in vitro
Although TORC1 is well known to regulate Maf1 phosphorylation, precisely where TORC1 is placed in the Maf1 regulatory pathway is not clear. We asked whether TORC1 is capable of interacting with Maf1. When immunoprecipitated with an HA-specific antibody, Kog1-Myc9 was detected only in the presence of Maf1-HA6 (Figure 6A, upper panel). Conversely, Maf1-HA6 was found in the anti-Myc immunocomplex only in the presence of Kog1-Myc9 (Figure 6A, lower panel). These results show that TORC1 specifically interacts with Maf1, raising the possibility that Maf1 is a TORC1 substrate. We, therefore, tested whether TOR kinase phosphorylates Maf1 in vitro. Indeed, immunoprecipitated Tor1 phosphorylated bacterially produced GST-Maf1 but not GST or GST-Fzo1 (Figure 6B). Phosphorylation of Maf1 is due to Tor1 kinase as wild-type Tor1 and Tor1-RR, but not the kinase-dead Tor1(DE) phosphorylated Maf1 (Figure 6C). Tor1(DE) contains the D2294E mutation in the catalytic domain (Zheng et al, 1995). Importantly, rapamycin inhibited the Maf1 phosphorylation activity of Tor1 but not Tor1-RR (Figure 6B and C). Together, these results provide the first evidence that TORC1 is closely involved in Maf1 regulation by phosphorylating Maf1 directly or through an associated kinase within the protein complex.
Figure 6.
TORC1 interacts with Maf1 in vivo and phosphorylates Maf1 in vitro. (A) TORC1 specifically interacts with Maf1. Maf1-HA was immunoprecipitated from early log phase cells. The presence of Kog1-Myc9 in the immunocomplex was detected by western blot (WB) with a Myc-specific antibody (upper panel). A reverse co-IP was shown in the lower panel. (B) Tor1 phosphorylates GST-Maf1 in vitro. W303a WT cells were treated without and with 200 nM rapamycin for 60 min and Tor1 was immunoprecipitated by a Tor1-specific antibody. Kinase assay was performed by incubation of Tor1 and bacterially expressed GST, GST-Maf1 and GST-Fzo1 (AA 1–343) with [γ-32P]-ATP. The immunoprecipitated Tor1 was analysed by western blot (upper panel). Protein phosphorylation was detected by autoradiography (middle panel). The recombinant protein input was stained by Coomassie blue (lower panel). (C) Phosphorylation of Maf1 is due to intrinsic Tor1 protein kinase activity. Yeast cells carrying a vector control or expressing Tor1 WT, Tor1-RR (rapamycin-resistant) or Tor1(DE) (kinase dead) were treated without and with 200 nM rapamycin. Tor1 was immunoprecipitated with a Tor1-specific antibody and incubated with bacterially expressed recombinant GST-Maf1 in the presence of [γ-32P]-ATP. The immunoprecipitated Tor1 was analysed by western blot (upper panel). GST-Maf1 phosphorylation was analysed by autoradiography (middle panel). Total GST-Maf1 was stained with Coomassie blue (lower panel).
Discussion
RNA Pol III is responsible for the synthesis of 5S rRNA and tRNAs, essential components for ribosomal biogenesis and protein synthesis. Elevated Pol III products are a hallmark of cancer cells (White, 2005). It was recently shown that overexpression of Pol III products promotes oncogenic transformation (Johnson et al, 2008; Marshall et al, 2008). Given that both TORC1 and Pol III have a key function in cancer growth, it is important to understand the detailed mechanism by which TORC1 regulates Pol III-dependent transcription. Here, we report a nuclear role of TORC1 in the regulation of Pol III activity. TORC1 nuclear localization and association with rDNA chromatin are regulated by nutrient availability and crucial for Pol III-dependent gene transcription (Figure 2B and C). In contrast to Pol II-transcribed genes, the cytoplasmic form of TORC1 is unable to regulate the expression of Pol III-transcribed genes. These results suggest that TORC1 mainly regulates Pol III activity in the nucleus and at rDNA chromatin.
Kinases are traditionally thought to regulate gene expression by controlling the cytoplasm-to-nucleus transport of specific transcription factors. However, evidences are emerging that kinases are associated with target genes to generate rapid response to environmental changes (Edmunds and Mahadevan, 2006). TORC1 association with 5S rDNA provides such a platform for rapid Pol III regulation. To understand the mechanism of TORC1 regulation of Pol III, we investigated how TORC1 controls Maf1, a key Pol III repressor. We found that Tor1-RR variants defective in nuclear localization or rDNA association fail to regulate Maf1 phosphorylation (Figures 3A and B, 2B and C), suggesting that TORC1 regulates Maf1 phosphorylation at the chromatin. Furthermore, we found that TORC1 interacts with Maf1 in vivo and Tor1 phosphorylates recombinant Maf1 in vitro (Figure 6). Together, these results provide the first evidence that TORC1 is closely involved in Maf1 phosphorylation, either directly or through an associated kinase. It was shown earlier that Maf1 dephosphorylation requires several PP2A enzymes, including Pph3, Pph21 and Pph22 (Oficjalska-Pham et al, 2006). Many TORC1 substrates (e.g. S6K1) are dephosphorylated by PP2A, and TORC1 is known to regulate certain PP2A enzymes (Duvel and Broach, 2004). Thus, it is likely that TORC1 regulates reversible Maf1 phosphorylation–dephosphorylation to generate rapid responses.
Maf1 is localized primarily in the cytoplasm in the S288C/FM391 strain. Starvation or rapamycin promotes Maf1 relocalization from the cytoplasm to the nucleus. Maf1 nucleocytoplasmic transport is regulated by both TORC1 and PKA (Moir et al, 2006). Interestingly, Maf1 is predominantly localized in the nucleus of W303 cells that carry SSD1-d instead of SSD1-v in S288C strain (Supplementary Figure 1). Our data show that Maf1 cytoplasmic localization requires SSD1-v allele: introducing SSD1-v into W303 cells is sufficient to render Maf1 cytoplasmic localization. However, nuclear accumulation of Maf1 per se is not sufficient to repress transcription by Pol III (Willis and Moir, 2007). TORC1 still regulates Maf1 phosphorylation (Figure 3B) and Pol III-dependent transcription (Figure 2B and C), which is in agreement with a recent report that Maf1 phosphorylation and Pol III transcription are normal in S288C/FM391 cells lacking Maf1 nuclear exportin (Towpik et al, 2008). Here, we show that Maf1 is excluded from the nucleolus and is absent from Pol III-transcribed genes under normal growth. On TORC1 inhibition, however, Maf1 rapidly enters the nucleolus and associates with Pol III-transcribed chromatins (Figure 5A and B). These observations provide a mechanistic explanation for TORC1 to control Maf1 activity within the nucleus.
HTHΔ mutation affects the ability of Tor1 to bind to both 5S and 35S rDNA. As Maf1 is only detected at 5S rDNA, it is logical to propose that TORC1 complex phosphorylates Maf1 at 5S rDNA chromatin, causing Maf1 to dissociate from this chromatin region and be excluded from the nucleolus. This also explains why Maf1 level is normally low at 5S rDNA. As the HTHΔ also affects TORC1 binding to 35S rDNA promoter, we cannot completely rule out the possibility that TORC1 complex also phosphorylates Maf1 at 35S rDNA promoter. As we and others have not detected Maf1 at 35S rDNA promoter, this scenario seems to be highly unlikely. Although TORC1 is not detectable at tDNA chromatins, its control of Maf1 binding to tDNA and tRNA synthesis remains HTH dependent. This observation suggests that TORC1-dependent phosphorylation at rDNA chromatin is sufficient to control Maf1 activity towards both 5S rDNA and tDNA, likely through controlling Maf1 availability in the nucleolus as it has been reported that tDNAs are also localized in the nucleolus (Thompson et al, 2003).
Coordinated regulation of Pol I and Pol III is necessary for the balanced production of 5S and 35S rRNAs. In response to rapamycin treatment, transcription of these genes decreases in a synchronous manner (Zaragoza et al, 1998), indicating that TORC1 is key to such transcriptional coordination. TORC1 concomitantly binds to 5S and 35S promoter chromatins in a nutrient-dependent and rapamycin-sensitive manner, which may provide a simple yet efficient mechanism to co-regulate Pol I- and Pol III-dependent genes. This possible mechanism is particularly well suited for acute responses to environmental changes. rRNAs, which are transcribed by two different major RNA polymerases, have to be stoichiometrically maintained. Therefore, multiple levels of control are likely involved to balance different ribosomal components (Chedin et al, 2007). A recent study shows that up-regulation of 35S rRNA synthesis in the CARA yeast strain (producing Rrn3-A43 fusion) leads to a concomitant increase of other ribosomal components (Laferte et al, 2006). Thus, cells have the ability to balance the production of ribosome components, which is TORC1 independent.
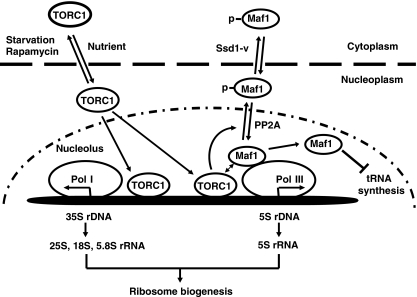
In conclusion, we propose the following model for TORC1 to regulate Pol I and Pol III-dependent transcription (Figure 7). Under favourable growth conditions, TORC1 is localized in the nucleus and is associated with both 5S rDNA and 35S promoter chromatins to regulate the synthesis of rRNAs. On starvation, TORC1 dissociates from rDNAs and exits from the nucleus, resulting in transcriptional repression. At 5S rDNA chromatin, TORC1 regulates Maf1 by phosphorylation, preventing Maf1 from accumulation in the nucleolus and association with Pol III-transcribed genes. In certain yeast genetic background, TORC1-regulated phosphorylation also promotes Maf1 nuclear export, which further reduces the accessibility of Maf1 to Pol III-transcribed genes. Thus far, we have not been able to detect TORC1 association with tDNAs. Phosphorylation of Maf1 at rDNA chromatins may be sufficient to control Maf1 nucleolar localization, which in turn controls the transcription of tRNA genes. It is interesting to note that rapamycin dramatically inhibits rDNA transcription but only has a moderate effect on tRNA synthesis. One explanation is that TORC1 association with rDNA chromatin generates more robust regulation. Further investigation in this area should provide more detailed mechanisms by which TORC1 controls different Pol III-transcribed genes.
Figure 7.
A working model for TORC1-dependent regulation of transcription by Pol I and Pol III. TORC1 is localized in the nucleus and is associated with both 5S and 35S rDNA chromatins, promoting their transcription by Pol I and Pol III, respectively, in a nutrient-dependent and rapamycin-sensitive manner. The concomitant TORC1 association/dissociation with 5S rDNA and 35S promoter provides a simple yet efficient mechanism to coordinate the synthesis of ribosomal RNAs in response to environmental changes. TORC1 at 5S rDNA and possibly also 35S rDNA promoter regulates the Maf1 phosphorylation, and subsequently Pol III-transcribed genes, including 5S rRNA and tRNA genes.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study are: W303a (MATa ura3-1 leu2-3, -112 his3-11, -15 trp1-1 ade2-1 can1-100); S288C/FM391 (MATa hisD1 leu2D0 met15D0 ura3D0); SZy1007(W303a tor1∷KanMX); SZy1701(W303a MAF1∷ MYC9-TRP); SZy2009 (S288C/FM391 maf1∷KanMX); SZy1565(S288C/FM391 trp1∷ URA3); SZy1707(SZy1565 AVO2∷MYC9-TRP1); SZy1709 (SZy1565 AVO3∷MYC9-TRP1); BLY03 (bar1 ura3 leu2 trp1 ade2 his3D200 gal1 can1 YCG1:KAN); SZy1705 (BLY03 KOG1∷MYC9-TRP). TOR1-RR and TOR1-RR variants together with TOR1 native promoter were cloned into the centromere plasmid pRS315. Wild-type TOR1 (pYDF72), kinase dead TOR1 (pYDF71-DE) and TOR1-RR (pYDF71) were cloned under GAL1 promoter. MAF1-MYC9 and MAF1-HA6 were constructed by cloning MAF1-MYC9 and MAF1-HA6 together with MAF1's own promoter into centromere plasmid pRS416. GST-MAF1 and GST-FZO1 were made by inserting MAF1 or FZO1(AA1-343) between SalI and EcoRI restriction site of pGEX-4T-1.
Cell extracts and western blot
Yeast cells were cultured in synthetic complete medium to early log phase (OD600=0.4). Rapamycin or drug vehicle (methanol) was added to final concentration of 100 nM to cell cultures for various times. Cells were collected and lysed in lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 2 mM PMSF, Roche protease complete inhibitor cocktail and phosSTOP tablet) with glass beads beating at 4°C. Crude lysates were cleared by centrifugation and the supernatants were collected for western blot analysis.
RNA preparation, northern blot
Total yeast RNA was extracted by hot phenol method as described before (Bertram et al, 2002). A measure of 25 μg total RNA was separated on 10% poly acrylamide gel containing 6 M Urea. RNA was then transferred to nylon membrane and detected by hybridization with [γ-32P]-ATP-labelled pre- tRNALeu3 probe (5′-CCAAACAACCACTTATTTGTTGA-3′) (Li et al, 2000) or the small nuclear RNA U4 probe (5′-CACCGAATTGACCATGAGGAGACGGTCTGG-3′) as control. Quantification of northern blot signal was performed by Bio-Rad Gel-dot system. Picture of autoradiograph film was taken by camera associated with Bio-Rad Gel-dot, and further analysed by Bio-Rad Quality One-4.6.3 software. The signal was measured by subtracting a nearby area. An adjusted value of pre-tRNALeu3 was obtained by dividing the value of pre-tRNALeu3 by that of U4. Remaining signal after rapamycin treatment was quantified by dividing the adjusted value of rapamycin-treated sample by the adjusted value of non-treated sample.
Metabolic labelling
For metabolic labelling (Udem and Warner, 1972), yeast cells in SD-Leu plus one third of the normal uracil were grown to OD600=0.4, and treated with 100 nM rapamycin for 30 min. Cells were then collected and washed with SD-Ura, resuspended in 0.5 ml SD-Ura plus 15 μCi/ml [5, 6-3H]-Uracil and incubated at 30°C for 5 min. Total RNA was extracted and separated on 10% poly acrylamide gel containing 6 M urea, and analysed by ethidium bromide staining and 3H autoradiography.
Immunofluorescence
Indirect immunofluorescence (IF) staining was carried out as described before (Bertram et al, 2000). Briefly, yeast cells were fixed by adding formaldehyde to cell cultures to a final concentration of 3.7%. Cells were collected and washed with phosphate buffer (0.5 mM MgCl2, 40 mM KH2PO4-K2HPO4 pH 6.5), resuspended in potassium buffer containing 1.2 M sorbitol, and treated with 50 μg/ml zymolyase. Maf1-Myc9 and Rpc82-Myc9 were stained by incubation with a Myc-specific polyclonal antibody A14 at 1:200 and the nucleolus was stained with a Nop1 monoclonal antibody at 1:1000. The nucleus was stained with 50 ng/ml DAPI in VectaShield mounting medium.
ChIP assays
Early log phase yeast cells were treated with the drug carrier methanol or 100 nM rapamycin and fixed with 1% formaldehyde for 30 min. The ChIP assay was performed as described before (Tsang et al, 2003, 2007a). For immunoprecipitation of Tor1-associated DNA, 0.5 mg total protein extracts were incubated with 5 μl of Tor1 antibody or control IgG for overnight at 4°C with rotation. For immunoprecipitation of Maf1-Myc9-associated DNA, 1 mg total protein extracts were incubated with 10 μl of mouse monoclonal Myc antibody (9E10). Protein G-Sepharose beads were used to recover the antibody-antigen–DNA complexes in all experiments. The total input DNA was prepared in the same way except that the antibody immunoprecipitation steps were omitted. The primer pairs used to detect protein binding along rDNA sequences were described earlier (Li et al, 2006). The primer pairs for detecting protein binding at tDNA loci are:
tDNALeu, AGACGAGCAGCTTATCCCATAATGA and GTGACGCCTGGTCGGTAAAAAGAT;
tDNATyr, TTCACTCTGAACCATCTTGGAAGGA and CAAGTCTGGGAAGTGAATGGAGACA;
tDNALys, GTTTACCGTCCATTAACATGTGCATT and AATCTTGGAAAAATAGTGAACCGGG.
TRA1 locus was used as a negative control (Roberts et al, 2006). Quantification of ChIP results was performed using Quantity One software (Bio-Rad).
Real-time PCR analysis
Quantitative real-time PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) kit in 96-well plate. Each PCR contained 1 μl DNA template and 5 pmol of primers in 25 μl reaction volume. The primer pairs used for detecting Tor1 association at different loci were reported earlier: IGS1, 5S, 35S rDNA promoter, 35S coding region, ACT1 and CUP1 (Li et al, 2006); tDNALys and tDNAPhe (Roberts et al, 2006) and tDNALeu (Oficjalska-Pham et al, 2006). Real-time PCR (95°C for 10 min followed by 95°C for 15 s and 60°C for 1 min for 40 cycles) was performed using an ABI PRISM 7900 (Applied Biosystems) according to the manufacturer's recommendations. Quantification was determined by comparative threshold cycle (CT) method as described (Harismendy et al, 2003). The optimum CT value was determined by SDS 2.2 software (Applied Biosystems) and verified by amplification curve analysis. Fold differences were calculated based on the ChIP occupancy of a sample gene relative to that of the corresponding control gene.
Co-immunoprecipitation
BLY03 cells expressing endogenous Kog1-Myc9 or plasmid-borne Maf1-HA6 or both were cultured to early log phase and lysed in IP buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.003% Triton X-100, 2 mM PMSF, Roche Complete protease inhibitor cocktail and phosSTOP tablet). Cell lysate was collected by centrifugation and 1 mg/0.5 ml total protein was incubated with either HA (12CA5) or MYC (9E10) antibody for 1 h with gentle rotation at 4°C. Protein G beads were added into total protein after washing with IP buffer and further incubated for 2 h with rotation. Beads were then washed with IP buffer extensively by spin down and boiled in 2.5 × SDS sample buffer. Kog1-Myc9 and Maf1-HA6 were detected by western blot.
GST fusion protein expression and purification
GST, GST-MAF1 and GST-FZO1 (AA1-343) plasmid was transformed into JM109 and protein expression was induced by 0.1 mM IPTG at 37°C for 3 h. Cells were collected and lysed in GST-binding buffer (25 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA) plus 0.5% Triton X-100, Roche Complete protease inhibitor cocktail, 2 mM PMSF for 1 min. Cells were immediately frozen at −80°C for 30 min then thawed at 37°C and subject to sonication. Lysate was collected and incubated with pre-washed glutathione beads for binding at 4°C for 2 h. To elute recombinant protein, beads were washed extensively with GST-binding buffer, and incubated with elution buffer (10 mM reduced glutathione, 50 mM Tris–HCl pH 8.0, 5% glycerol) at room temperature for 15 min.
In vitro kinase assay
W303a wild-type or W303a tor1Δ cells containing vector, TOR1 wild-type (pYDF72), TOR1-RR (pYDF71), and kinase dead TOR1-DE (pYDF71-DE) were cultured to early log phase and induced by changing to YP-galactose medium for 2 h. Cells were then treated without and with 200 nM of rapamycin for 1 h before lysed in IP buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.003% Triton X-100, 2 mM PMSF, Roche complete protease inhibitor cocktail and PhosSTOP tablet). TORC1 from each sample was immunoprecipitated with a specific Tor1 antibody and incubated with 100 μM ATP, 50 μCi [γ-32P]-ATP and 0.5 μg recombinant GST, Maf1 or Fzo1(AA1-343) in 50 μl kinase buffer (1 × PBS, 20% glycerol, 0.003% Triton X-100, 4 mM MgCl2, 10 mM DTT, protease inhibitor) for 30 min. Kinase assay was stopped by heating at 100°C for 5 min in 2.5 × SDS sample buffer. Samples were then subject to SDS–PAGE and stained with coomassie blue. Phosphorylation was detected by 32P autoradiography. Immunoprecipitation of Tor1 was analysed by western blot using a Tor1-specific antibody.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Yu-Ju Chen for contribution during early phase of this project, Jun-Hung Cho for constructing the GST plasmids, Dr Olivier Lefebvre for technical advice, and other colleagues in the Pol I and Pol III field for helpful discussions. This work was supported by NIH grants R01-CA099004 and R01-CA123391.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bertram PG, Choi J, Carvalho J, Ai WD, Zeng CB, Chan TF, Zheng XFS (2000) Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J Biol Chem 275: 35727–35733 [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Chan T-F, Ai W, Zheng XFS (2002) Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol 22: 1246–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M, Czerska K, Zoladek T (1997) Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene 185: 291–296 [DOI] [PubMed] [Google Scholar]
- Chedin S, Laferte A, Hoang T, Lafontaine D, Riva M, Carles C (2007) Is ribosome synthesis controlled by pol I transcription? Cell Cycle 6: 11–15 [DOI] [PubMed] [Google Scholar]
- Desai N, Lee J, Upadhya R, Chu Y, Moir RD, Willis IM (2005) Two steps in Maf1-dependent repression of transcription by RNA polymerase III 10.1074/jbc.M412375200. J Biol Chem 280: 6455–6462 [DOI] [PubMed] [Google Scholar]
- Drenan RM, Liu X, Bertram PG, Zheng XFS (2004) FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the golgi apparatus. J Biol Chem 279: 772–778 [DOI] [PubMed] [Google Scholar]
- Duvel K, Broach J (2004) The role of phosphatases in TOR signaling in yeast. Curr Top Microbiol Immunol 279: 19–38 [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC (2006) Cell signaling: protein kinases seek close encounters with active genes. Science 313: 449–451 [DOI] [PubMed] [Google Scholar]
- Geiduschek E, Kassavetis G (2006) Transcription: adjusting to adversity by regulating RNA polymerase. Curr Biol 16: R849–R851 [DOI] [PubMed] [Google Scholar]
- Goodfellow S, Graham E, Kantidakis T, Marshall L, Coppins B, Oficjalska-Pham D, Gerard M, Lefebvre O, White R (2008) Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol 378: 481–491 [DOI] [PubMed] [Google Scholar]
- Harismendy O, Gendrel CG, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O (2003) Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J 22: 4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Johnson DL, Johnson SAS (2008) Cell biology: RNA metabolism and oncogenesis. Science 320: 461–462 [DOI] [PubMed] [Google Scholar]
- Johnson S, Zhang C, Fromm J, Willis I, Johnson D (2007) Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell 26: 367–379 [DOI] [PubMed] [Google Scholar]
- Johnson SAS, Dubeau L, Johnson DL (2008) Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem 283: 19184–19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S (2006) The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components 10.1101/gad.386106. Genes Dev 20: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tsang C, Watkins M, Bertram P, Zheng X (2006) Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Li Y, Moir RD, Sethy_Coraci IK, Warner JR, Willis IM (2000) Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol 20: 3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo J, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth N, White R (2008) Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 133: 78–89 [DOI] [PubMed] [Google Scholar]
- Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, Willis IM (2006) Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. PNAS 103: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T, Stefanovsky VY (2002) At the center of eukaryotic life. Cell 109: 545–548 [DOI] [PubMed] [Google Scholar]
- Murawski M, Szczesniak B, Zoladek T, Hopper A, Martin N, Boguta M (1994) maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim Pol 41: 441–448 [PubMed] [Google Scholar]
- Oficjalska-Pham D, Harismendy O, Smagowicz W, Gonzalez de Pered A, Boguta M, Sentenac A, Lefebvre O (2006) General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell 22: 623–632 [DOI] [PubMed] [Google Scholar]
- Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M (2001) Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol 21: 5031–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P (1999) Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell 10: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J III, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T (2004) TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 279: 14752–14762 [DOI] [PubMed] [Google Scholar]
- Roberts D, Wilson B, Huff J, Stewart A, Cairns B (2006) Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell 22: 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P (1993) A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet 239: 169–176 [DOI] [PubMed] [Google Scholar]
- Sutton A, Immanuel D, Arndt KT (1991) The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol 11: 2133–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR (2003) Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towpik J, Graczyk D, Gajda A, Lefebvre O, Boguta M (2008) Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1 10.1074/jbc.M709157200. J Biol Chem 283: 17168–17174 [DOI] [PubMed] [Google Scholar]
- Tsang C, Li H, Zheng X (2007a) Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J 26: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C, Qi H, Liu L, Zheng X (2007b) Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today 12: 112–124 [DOI] [PubMed] [Google Scholar]
- Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XFS (2003) Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J 22: 6045–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S, Warner J (1972) Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol 65: 227–242 [DOI] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis I (2002) Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- White R (2005) RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Willis I, Moir R (2007) Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci 32: 51–53 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Zaragoza D, Ghavidel A, Heitman J, Schultz MC (1998) Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol 18: 4463–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shu L, Hosoi H, Murti KG, Houghton PJ (2002) Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem 277: 28127–23381 [DOI] [PubMed] [Google Scholar]
- Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL (1995) TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82: 121–130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File