Abstract
The dangers of serious ADR (SADR) are well known by clinicians, pharmacologists, and the lay public. The ascertainment of the molecular mechanisms behind SADRs has made significant progress through genetics and gene expression measurements. But as the field of pharmacology adopts the same novel higher-density measurement modalities that have proven successful in other areas of biology, one wonders whether there can be more ways to benefit from the explosion of data created by these tools. The development of analytic tools and algorithms to interpret these biological data to create tools for medicine is central to the field of translational bioinformatics. After we introduce some of the types of SADR predictors we need, we will cover several publicly-available databases available for the study of SADRs, scaling from clinical to molecular measurements. We will then demonstrate recent examples of how bioinformatics methods coupled with data repositories can advance the science of SADRs.
INTRODUCTION
How a patient will respond to any drug-based therapy is not easily predicted, as it can vary from individual to individual. Some patients may not respond to a therapy, while others require only a small dose to achieve therapeutic effects. Side effects are commonly observed with drug-based therapies, however, adverse drug reactions (ADRs) can have dire consequences. ADRs, defined as unintended and undesired response after the use of drugs under normal dosage for therapeutic uses, are considered serious ADR (SADR) if they result in deaths, hospitalizations, significant or permanent disability, requiring intervention to prevent permanent and life-threatening situations. SADRs are a major clinical problem, accounting for an estimated over two million hospitalization annually, with over 100,000 deaths in the United States (US) alone (1). The rate of SADRs occurring in hospitalized patients in the US has been estimated to be 6–7% (1). Between 0.12–0.3% of US hospitalized patients had fatal SADRs (1). In effect, the fatal SADRs rank between the 4th and 6th leading causes of deaths in the US annually. Besides being a tremendous strain on the healthcare industry, SADRs also have an enormous impact on the pharmaceutical industry, accounting for most of the drugs withdrawn in the last decade (2). ADRs are also the top reason for drug discontinuation in patients. Moreover, a significant portion of investigational drugs fail because of toxicity of ADRs during clinical trials.
While susceptibility to ADRs may arise from both genetic and non-genetic factors, our current understanding of ADRs show that genetics play a pivotal role in drug responses and SADRs that is the primary focus of pharmacogenomics. However, despite significant progress in the last 50 years, our knowledge on the genetic factors contributing to ADRs and SADRs is still very limited. This is partly due to low-throughput technologies which prevented global genome-wide analyses. In the current era of sequenced genomes and high-throughput genomic technologies such as DNA microarrays and proteomics, the bottleneck has shifted away from one of molecular measurement technology and instead toward our ability to process, analyze, and use our large sets of data. This burgeoning field of translational bioinformatics has been concerned with developing informatics tools that facilitate the capture, storage, management, integration, dissemination, and utility of these large sets of biological data (3). The development of analytic tools and algorithms to interpret these biological data is central to the field of bioinformatics.
Given the rapid technological advancements of the past few years, this review will focus on existing knowledge-bases, resources, and recent methodological developments pertaining to the discovery of molecular factors associated with SADRs. Our goal in this review is to illustrate how methods in bioinformatics have been and will continue to be critical in translating the relevant genomic discoveries into clinical practice. We start this review addressing the kinds of molecular predictors we need related to ADRs, with a few case examples of gene expression profiles and gene variants predictive of adverse drug events and drug efficacy. After this introduction to the kinds of predictors we need to discover, we will then cover several of the most useful publicly-available repositories of data for the study of ADRs, scaling in resolution from clinical data to molecular measurements. We will then demonstrate several recent case-examples of how bioinformatics methods, when coupled with these data repositories, can advance the science of SADRs. Finally, we will end with the future challenges in this field.
MOLECULAR PREDICATORS FOR DRUG TOXICITY
Molecular predicators specifically for drug toxicity have traditionally revolved around gene variants. Thiopurine methyltransferase (TPMT), a phase II metabolizing enzyme, was first associated with the metabolism of 6-mercaptopurine and azathioprine back in 1980 (4). Lack of knowledge of the genotype of this gene can lead to 10-fold overdosing of these drugs, potentially leading to fatal hematopoietic toxicity (4).
Largely guided by an increasing understanding of drug pharmacology and the high degree of variation in drug metabolizing enzymes, genes encoding for drug metabolizing enzymes have been the most studied class in association to ADRs (5, 6). The highly polymorphic cytochrome P450 (CYP) enzyme system, a class of phase I metabolizing enzymes, are heavily studied because they are responsible for metabolizing approximately 67% of drugs (7). The Human Cytochrome P450 Allele Nomenclature Committee maintains a complete web-based list of all peer-reviewed alleles of CYP (8); as of this writing, it lists 200+ alleles from 29 CYP families.
Candidate gene sequencing approaches have uncovered many of the associations between genetic variants and SADRs. As an example, specific alleles in CYP2C9 have been linked to hemorrhages in patients taking warfarin (9). Polymorphisms in CYP2D6 have also been linked to many SADRs including tardive dyskinesia and bradycardia in patients taking antipsychotics and beta-blockers, respectively (10). Similarly, variations in dihydropyrimidine dehydrogenase and UDP-glucuronosyltransferase (encoded by UGT1A1) have also been shown to be linked to neurotoxicity (11) and neutropenia (12) found in patients taking 5-FU and irinotecan respectively.
While powerful in yielding specific polymorphisms predictive of drug toxicity, traditional candidate gene approaches have been limited by progress in biological knowledge. Strategies that have targeted only those genes known to participate in pharmacokinetics and pharmacodynamics otherwise ignore the many other genes that have yet to be biologically-linked to SADRs. Until recently, the ability to interrogate polymorphisms across the entire genome was hampered by both high costs and technology. Recently, technologies for genotyping have evolved while the costs have been driven down, so that genome-wide association studies (GWAS) to link variants to SADRs are possible (13). As of this writing, these tools can genotype nearly two million variants as a cost of only a few hundred dollars per individual.
The immediate application of these high-density genotyping tools in determining the underlying genetic factors in various disease states is apparent and has demonstrated utility (13, 14). Recently, several GWAS have been successfully applied toward the study of SADRs. These include hemorrhage associated with warfarin, hepatic toxicity with ximelagatran, and simvastatin-induced myopathy. A GWAS study of the most commonly used anticoagulant, warfarin, attempted to identify additional SNPs beyond those in VKORC1 and CYP2C9 that could explain the estimated remaining 50% of the variation in stable warfarin dosing (15). However, no other SNPs showed significant enough association in a subsequent validation set of patients (15). Ximelagatran, another anticoagulant, was slated to replace warfarin, but ultimately was not marketed due to hepatic toxicity observed in clinical trials. A GWAS explored why some patients taking ximelagatran exhibited elevated serum alanine aminotransferase (ALT) levels, a proxy for hepatic injury (15). Strong genetic associations between elevations in ALT and major histocompatibility locus alleles DRB*07 and DQA1*02 were identified (15), implicating antigen-presenting proteins in drug-induced liver toxicity. This finding was consistent with other genetic association studies of drug-induced liver toxicities (16, 17). Another recent GWAS including 85 patients with 80 mg simvastatin-induced myopathy and 90 control patients in a case-control design across 300,000 SNPs revealed a single strong association to the SLCO1B1 gene (18). This gene encodes an organic anion-transporter that is involved in statin uptake (19). Findings such as these clearly demonstrate a full reversal in strategy, from the older approach of developing hypotheses of gene variation based on biological findings and knowledge, instead to the development of hypotheses of biological processes based on primary gene variation findings.
MOLECULAR PREDICATORS FOR DRUG EFFICACY
The study of drug efficacy is just as important as the study of drug toxicity in the SADRs; indeed, the failure of efficacy of a drug in a patient with a lethal illness may carry severe consequences. Drug efficacy in cancer therapy is of particular importance, as SADRs often develop as a result of cytotoxic chemotherapeutic regimens. For instance, cancer patients treated with cisplatin can suffer from SADRs affecting hearing, the nervous system and kidneys, but such toxicities may be judged to be an acceptable risk if the therapeutic efficacy against an otherwise fatal disease is high. If SADRs are an element of the risk-benefit equation for chemotherapeutic use, then accurate predictors for efficacy are crucial.
Two prime examples of successful single-target based predicators of drug efficacy are imatinib mesylate (Gleevec) and trastuzumab (Herceptin). The approval of imatinib mesylate for chronic myelogenous leukemia (CML) marked the fastest anticancer drug to be approved by the FDA, after only three months of review (20). Almost all patients with CML had a chromosomal translocation event between chromosome 9 and 22 (known as Philadelphia chromosome) which activates the tyrosine kinase fusion oncoprotein Bcr/Abl (21). Imatinib mesylate acts by inhibiting Bcr/Abl activation and has revolutionized CML therapy by dramatically increasing CML survival rates. On the other hand, only 11% of the breast cancer patients receiving trastuzumab, a monoclonal antibody targeted to human epidermal growth receptor 2 (HER2), achieved tumor regression in the initial clinical trials (22). However, by targeting the treatment to only patients with HER2 overexpression, based on immunohistochemistry or cytogenetic assays, a much higher rate (34–50%) of patients demonstrate tumor regression (23). Trastuzumab is also the first example of a FDA approved drug with a companion FDA approved immunohistochemistry diagnostic test, HercepTest (Dako, Carpinteria, CA), designed to identify the patients most likely to benefit from the drug. This is because HER2 overexpression is only observed in ~25% of patients with breast cancer. Both trastuzumab and imatinib mesylate have acquired additional indications: trastuzumab as part of various combination therapies for breast cancer and imatinib for kit (CD117)-positive gastrointestinal stromal tumors (GIST), showing broad utility of single target drugs.
The single-target drugs, such as imatinib mesylate and trastuzumab, are just two examples of several that demonstrate efficacy in a subset of a larger patient population. Over the last seven years, molecular predicators based on genomic data, particularly gene expression signatures, have led the way for improved selection of these patients. Successful chemosensitivity predications, based on gene expression signatures to determine which subsets of patients would respond best to specific chemotherapeutic drugs, have demonstrated their utility in cancer therapies such as breast cancer and childhood leukemia. A review of the fortyone studies which utilized gene expression signature to predict drug chemosensitivity was recently published (24). One of the earliest examples of these predications was a 92-gene signature to predict docetaxel response in breast cancer patient (25). Gene expression signatures have been successfully combined with drug sensitivity data on cancer cell lines to predict how patients respond (26). In one study, bioinformatics methods were used to link drug sensitivity data from the National Cancer Institute (NCI)-60 cell lines were used to identify genes that best discriminate responses to etoposide, adriamycin, cyclophosphamide and 5-fluroururacil (5-FU) (27). Additionally, the patient gene expression signatures were compared against other gene expression signatures generated from known pathway activations, thereby allowing drugs targeting matching pathways to be prime therapeutic candidates (27).
Even GWAS have been performed to investigate drug efficacy. Studies examining responses to anti-Tumor Necrosis Factor therapy in patients with Rheumatoid Arthritis (28), interferon-beta therapy in Multiple Sclerosis patients (29), ilperidone (30), and thiazide diuretic (31) are enhancing our understanding of why certain individuals fail to respond to certain drugs. A recent GWAS yielded gene variants associated with successful smoking cessation, and selection of cessation medications (32). It is becoming clearer that patient stratification based on drug efficacy and toxicity patterns is needed to prevent SADRs.
For the most up-to-date list of GWAS, the National Institute of Health (NIH) maintains a list of published high density GWAS, with the phenotype studied, p-values of significant SNP allele associations, along with the sizes of the patients for both the discovery and replication studies.
CLINICAL REPOSITORIES AVAILABLE TO STUDY SADR
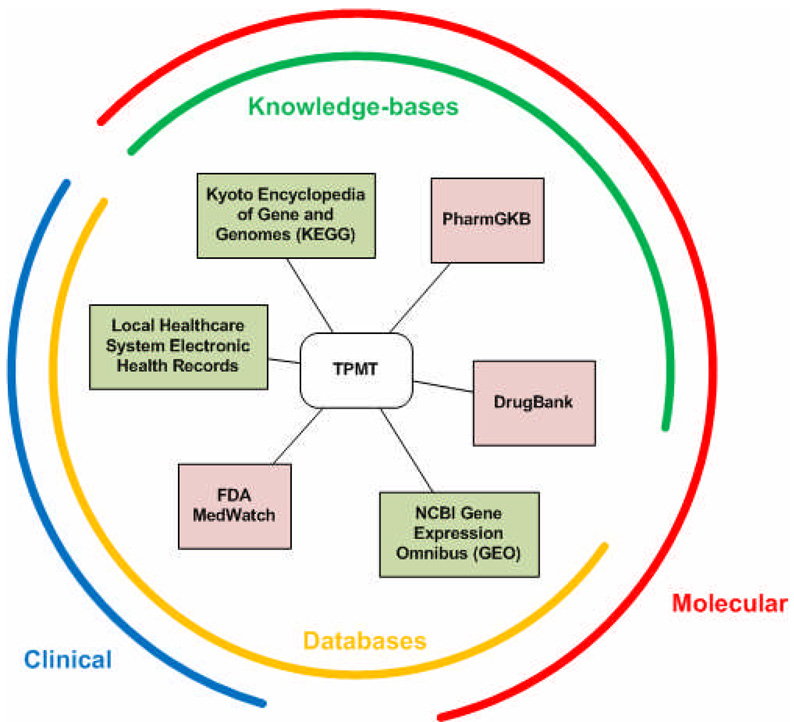
While molecular data has been shared across communities of investigators for more than two decades, the number of repositories holding various kinds of biological and molecular measurements has continued to grow exponentially. In 2008, the annual Molecular Biology Database Collection issue of Nucleic Acids Research grew by 10% to include more than a thousand databases (33). Studies into the nature of SADRs have been performed on both clinical and molecular fronts, with many of the data and results deposited into various types of databases, some of which are publicly-accessible. At the same time, many publicly-available databases could be used in the study of SADRs, even though they are not primarily labeled as SADR repositories. Here, we will illustrate some example clinical and molecular repositories and their applicability to the study of SADRs (Figure 1).
Figure 1.
A well-known pharmacogene, TPMT, appears in a variety of publicly-accessible information sources. Knowledge-bases, such as KEGG, PharmGKB, and DrugBank, hold existing knowledge on TPMT, while databases, such as clinical electronic health records, MedWatch, and GEO, hold observed measurements or characteristics of TPMT. Both knowledge and data on TPMT can span from the clinical (or whole organism) realm to the molecular realm. Information sources in pink are pharmacology-specific, while green sources are general use, and not specific for pharmacology.
The focus of the clinical side of SADR studies lies primarily with detection and prevention. For many countries, pharmacovigilance is a key strategy toward detecting and preventing SADRs. Naturally, effective pharmacovigilance requires cooperation of the pharmaceutical and biotechnology industry, regulatory agencies, and academic institutions. Clinical trials are typically the first place where ADRs arise. One such example is the identification of a UGT1A1 variant responsible for susceptibility to tranilast-induced hyperbilirubinemia, found during a phase III clinical trial testing the efficacy of tranilast in reducing the re-stenosis rate percutaneous trans-luminal coronary angioplasty (34). Clinical trials are great for identifying SADRs, however, due to the limited number of participants, short duration of the trials, and characteristics of the study participants that may not reflect the patients who are later prescribed the drug (e.g. children, ethnicities), clinical trials often fail to detect SADRs.
Many SADRs are detected through post-marketing spontaneous reporting systems (SRS) established by many countries following the discovery of the teratogenic effects of thalidomide. SRS are valuable tools -- they allow the early detection of rare, new SADRs in a cost effective manner. In fact, SRS are responsible for majority of the drug withdraws from the marketplace, though others argue these systems are still insufficient to find current SADRs (35). Global drug safety monitoring efforts spearheaded by the World Health Organization (WHO) include VigiBase, a database containing drug safety information from 82 nations with more than seven million reports (36). Similarly, many countries have also set up their own SRS, such as the United Kingdom’s Yellow Card System, Japan’s Pharmaceuticals and Medical Devices Agency (PMDA), The United States Food and Drug Administration’s MedWatch (also known as Adverse Event Reporting System[AERS]) and the Canadian Vilgilance Online Database (CVOD) which contains ADR events dating back to 1965 (36). For example, MedWatch lists 608 reported adverse effects for Vioxx from a recent quarter, while the CVOD breaks down its reported adverse effects for Vioxx by 511 submitted by pharmacists and 168 submitted by consumers. While the nomenclature for drugs is not universally standardized within or between these systems, newer vocabularies such as the National Library of Medicine’s RxNorm could be used to resolve across these systems (37).
Large and small health care institutions are moving toward electronic computerized systems as a way to streamline the entire health care process. This has opened the door for use of electronic health records (EHRs) to identify ADRs. Indeed several studies have demonstrated the time reduction advantage of mining electronic medical records over traditional manual chart reviews in identifying ADRs. These studies employed a combination of methods to detect ADRs, including rule-based triggers, drug-drug interactions, drug allergy information, natural language processing, and Naranjo algorithm, a questionnaire which estimates the probability of an ADR (38–40). Moreover, all of them revealed that a significant portion (~25%) of ADRs is preventable. Together, these clinical studies into ADRs have shown that clinical improvements can be made to avoid SADRs. While patient-specific clinical records are typically not publicly-available and require institutional review board approval for study, the widespread adoption of EHRs essentially means this type of data has at least been locally-available in many locales. Toward the goal of improving sensitivity by pooling resources across locales, the FDA recently launched a more active drug safety surveillance system called Sentinel Initiative (41). In cooperation with Centers for Medicare and Medicaid Services, the Veterans Administration, the Department of Defense, and both public and private organizations, the FDA will have access to 25+ million electronic medical records so that active monitoring of SADRs can be performed. It is clear that a global, collaborative effort will be required to prevent SADRs. One recent example of this collaboration is the establishment of The Predictive Safety Testing Consortium (PSTC) between the public and private sectors. Early results led to the approval of seven new biomarkers to monitor renal toxicity.
MOLECULAR KNOWLEDGE-BASES AND REPOSITORIES FOR STUDYING SADR
The focus of the molecular arm of SADR studies lies primarily with determining the underlying molecular mechanisms in SADRs. There are many molecular repositories and databases currently available for the bioinformatics-enabled SADR researcher (Table 1). Indeed, of the more than 1000 molecular databases listed earlier (33), 29 of them have the word “drug” in them, up from 3 four years ago. Here, we will first cover the popular knowledge-bases containing known genes related to drug use and effects, then cover the repositories of molecular data which can be exploited to discover new genes associated with drugs.
Table 1.
Translational bioinformatics resources for SADR studies
| Pharmacogenomic knowledge-bases | |
| DrugBank – GenoBrowse | http://www.drugbank.ca/genobrowse |
| FDA Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels | http://www.fda.gov/cder/genomics/genomic_biomarkers_table.htm |
| PharmGKB | http://www.pharmgkb.org/ |
| Chemical Effects in Biological Systems (CEBS) | http://cebs.niehs.nih.gov |
| Genetic databases | |
| Catalog of Published Genome-Wide Association Studies | http://genome.gov/gwastudies/ |
| NIH Genetic Association Database (GAD) | http://geneticassociationdb.nih.gov/ |
| NCBI Database of Genotype and Phenotype (dbGAP) | http://www.ncbi.nlm.nih.gov/gap/ |
| Molecular databases | |
| NCBI Gene Expression Omnibus (GEO) | http://www.ncbi.nlm.nih.gov/geo/ |
| EBI ArrayExpress | http://www.ebi.ac.uk/microarray-as/ae/ |
| Connectivity Map | http://www.broad.mit.edu/cmap/ |
| Pathway and interaction databases | |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | http://www.genome.ad.jp/kegg/ |
| Comparative Toxicogenomics Database (CTD) | http://ctd.mdibl.org/ |
| Chemical databases | |
| Chemical Entities of Biological Interest (CHEBI) | http://www.ebi.ac.uk/chebi/ |
| NCBI PubChem | http://pubchem.ncbi.nlm.nih.gov/ |
| Developmental Therapeutics Program (DTP) | http://dtp.nci.nih.gov/ |
One of the most comprehensive repositories of known genes affecting pharmacokinetics and pharmacodynamics is the Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB) (42). PharmGKB curators continually scan the pharmacogenomics literature for the most up-to-date information related to drugs, disease, and genes. For example, a search in PharmGKB for "CYP2D6” returns almost 500 hits with links to various publications, drugs, pathways, phenotypes, and diseases, as of this writing.
DrugBank is a unique knowledgebase of drugs, including both FDA approved ones as well as experimental ones, drug actions, and their molecular targets (43). A DrugCard entry stores all associated information aggregated from various sources, such as GenBank and Chemical Entities of Biological Interest (ChEBI), pertaining to a specific drug. With 4,700+ drugs, 100+ data fields from 20+ databases, as of this writing, DrugBank effectively bridges the various nomenclature and identifiers found in diverse drug databases into cohesive DrugCard entries (43). The recently added GenoBrowse feature in DrugBank summarizes the specific genes and SNP alleles along with literature references responsible for various ADRs and functional effects. As of this writing, GenoBrowse lists adverse effects and drug function information on ~60 drugs. For instance, five distinct alleles in four different genes have been linked to ADRs in patients taking 5-FU.
A smaller knowledge-base called the “Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels” is run by the Food and Drug Administration (FDA) itself, with a summary webpage on all approved drug label changes related to genomic biomarkers. These include recommendations for physicians prescribing specific drugs to pay particular attention to subsets of patient populations. These drug labeling changes are slowly altering clinical care.
Other knowledge-bases exist linking chemicals with genes and proteins; while these knowledge-bases include facts on more than just drugs, they may hold utility for bioinformatics-based SADR studies. The Chemical Effects in Biological Systems (CEBS) Knowledge Base is a systems toxicogenomic repository founded on systems biology principles that store toxicological information from gene expression, protein, clinical chemistry and histopathology, and metabolic experiments (44). For instance, clinical and pathology data along with proteomic and gene expression experiments studying the toxic effects of acetaminophen on liver is stored within CEBS. The Comparative Toxicogenomics Database (CTD) is another curated knowledge-base that captures the relationships between chemicals, genes and diseases with over 4,000 chemicals, 14,000 genes and 3,000 diseases (45). For example, sirolimus has 75 known interacting genes, associations with 236 diseases, and 162 pathways associations. Along this line, the Kyoto Encyclopedia of Genes and Genomes (KEGG) is a foundational knowledge-base that stores curated pathway information (molecular interactions and cellular processes) from published literature (46), giving network context of how genes, diseases and metabolites are inter-related.
Gene expression microarrays have enabled the measurement of RNA expression levels across the genomes of several organisms for more than a decade (47, 48). With this track record, it is no surprise that whole-genome molecular profiling studies on drug toxicity and responsiveness using gene expression microarrays outnumber those using high-density SNP measurements. This is primarily because toxicity effects in cell lines and model organisms, such as rat and mouse, are much more readily available than samples from human patients. By far the largest repository of gene expression experiments, the Gene Expression Omnibus (GEO) maintained by the National Library of Medicine, contains data from many such drug-related experiments (49). The largest single toxicogenomics dataset in GEO is one comprised of 5,288 microarrays (50). The study was performed by Iconix Biosciences to explore the liver drug responses across 1,695 rats treated with various doses of 344 compounds. The European Bioinformatics Institute (EBI) also stores similar gene expression experiments in the ArrayExpress database. As of this writing, there are 29 toxicogenomic experiments in ArrayExpress.
One large-scale gene expression study, known as the Connectivity Map, consists of 164 chemicals tested mostly on the MCF7 breast-cancer cell line (51). Pattern-matching algorithms can be used on these gene expression signatures to find hidden similarities in effect across these chemicals. Data from the Connectivity Map has been deposited into GEO. But beyond this single experiment, data from many other gene expression studies are also present in GEO. We have estimated that gene expression data from at least 213 drugs is available in GEO, tested across many doses and tissues, and contained in many separate experiments (52). As we will show later in this review, these public repositories can also serve as resources for larger meta-analyses. Public databases on the bioactivities of small molecules have primarily been the result of government-based initiatives. The NCI Developmental Therapeutics Program (DTP), established by Congress in 1955 as the Cancer Chemotherapy National Service Center, provides a rich repository of cancer cell lines and measurements made on those cell lines, including the NCI-60 panel (53). The DTP has an emphasis on cancer, transplantable animal and human tumors, small molecules, and compound screening services. Toward that end, the DTP has made public the data from screening thousands of compounds in those cancer cell lines and animal models, providing critical preclinical and research tools that has resulted in discovery of 40 chemotherapeutic drugs, including cetuximab (Erbitux), a monoclonal antibody that inhibits epidermal growth factors (EGFR) approved to treat colorectal (54) and head and neck cancers (55). The DTP compound susceptibility measurements have been joined with gene expression measurements leading predictors of chemotherapeutic efficacy (26, 56, 57).
The Molecular Libraries Initiatives (MLI) is a generalization of the methods of the DTP, established by NIH in 2003 as one of the five Roadmap Initiatives designed to expedite the translation of research discoveries to bedside. The major goal of the MLI is to acquire, screen 500,000 small molecules in high-throughput bioassays, and release the screening results in a database called PubChem (58). Today, PubChem contains information on 18 million chemical compounds as studied across nearly a thousand bioassays, including those from the DTP. While PubChem is not specific to the study of SADRs, there are bioassay measurements relevant to SADRs, including, for example, growth inhibition assays that gives clues into drug chemosensitivities in various cells or organisms.
The public availability of these knowledge-bases and databases directly and indirectly related to SADRs is slowly breaking down the barriers in studying SADRs and invites creative integration of data between disparate experimental modalities. Bioinformatics already plays a central role in aggregating and storing these increasingly large sets of data and knowledge from both the clinical and molecular domains. More importantly, the development of analytic tools and algorithms required to analyze and interpret these results and knowledge will be pivotal in translating research findings into the clinic.
BIOINFORMATICS SUCCESS STORIES FOR THE STUDY OF ADRS
Recent success studies from several groups highlight the elegant use disparate datasets toward the study of SADRs enabled by bioinformatics methodologies. In each of these cases, two or more knowledge-bases and databases were linked because of a commonality across those sets. Drawing on the knowledge stored in DrugBank, which contains information on both approved and experimental drugs and their targets, Yildirim and colleagues constructed a drug-target network and used network properties to describe drugs and how they relate to other datasets (59). By clustering drugs based on Anatomical Therapeutic Chemical (ATC) classification along with their targets, the drug-target network confirmed that most drugs are ‘follow-on’ drugs, that is, drugs targeting proteins already targeted by another drug. In comparing the drug target proteins with essentiality proteins, or proteins encoded by genes whose orthologs in model organisms are found to be essential, drug target proteins show different topographical signatures, have less gene expression coexpression, and high tissue specificity. Moreover, an additional comparison of drug target proteins to disease proteins previously implicated in Mendelian disorders indicated that most drugs are palliative (targeting proteins not causal for disease) rather than etiology-based (proteins causing disease). This last finding is not surprising, as most drugs address the clinical symptoms, rather than the underlying disease pathogenesis. Coupled with lack of complete understanding of mechanistic underpinnings of most drugs which often lead to off-target effects, it is not unexpected that ADRs are common and SADRs unanticipated.
As an example of the study of drug toxicity, Huang and colleagues sought to determine the genetic variants associated with cytotoxicity from various drug treatments (60, 61). They took advantage of the lymphoblastoid cell lines which have been used to maintain individuals’ DNA needed for the Human Genome Project and subsequent HapMap projects. Huang applied various drug treatments to these cell lines and then measured cytotoxicity. Genome-wide gene expression profiling was also performed on the same cell lines. Since the SNP genotypes had already been determined for these cell lines as part of the HapMap project, Huang was able to borrow from those measurements to identify SNPs responsible for cytotoxicity and drug sensitivity to cisplatin (61), carboplatin (60) and daunorubicin (62). While acknowledging that lymphoblastoid cell lines are not necessarily the best cells in which to probe for drug toxicity, this research methodology is an example of how making new measurements from the same resources on which publicly-available data was already measured can enable research findings greater than the “sum of the parts”.
The activities of chemical compounds on the NCI-60 cancer cell lines are stored in a repository with the intent to serve as early research resource for drug development leads. Two studies utilizing data from these screenings exemplify how the data can be extended to make novel discoveries. First, while the NCI-60 cell lines are comprised of cells from various cancers, including leukemias, melanomas, breast, ovarian, renal, prostate, colon, lung and central nervous system (CNS) cancers, other cancer cell lines such as those from bladder cancers are not represented. Lee and colleagues sought to take advantage of existing NCI-60 data with application toward those unrepresented cancers in a methodology called “coexpression extrapolation” or COXEN (63). This was done by first identifying a common molecular dataset or experimental modality between the NCI-60 and the bladder cancer cells, which in this case were gene expression measurements. Then, chemosensitive or resistant gene expression signatures were linked to the drug activities measured in NCI-60 cells. Based on coexpression patterns between the bladder and NCI-60 cells, a multivariate algorithm was used to extrapolate and predict drug activity on the bladder cells (63). In this fashion, the authors performed in silico screening and identified a novel drug candidate for bladder cancer cell.
Fliri and colleagues had a different goal in mind, trying to find pre-clinical markers that could be predictive of post-marketing side effects (64). Fliri first constructed biological activity spectra (biospectra) from in vitro protein binding assays of prescription drugs (65). Using hierarchical clustering methods, Fliri found regions of similarity between biospectra, structure similarity, and side effect profiles from drug information labels, across the same drugs. This research methodology shows how coded drug label information and molecular measurements can be linked if these pieces of data apply to the same set of pharmaceuticals.
In the last case example, Campillos and colleagues exploited the side effect information from prescription drug labels to identify novel molecular activities for existing drugs (66). Similarity in side effects was classified based on the Unified Medical Language System (UMLS). The UMLS is a 20+ year old initiative from the National Library of Medicine to build a large comprehensive standardized vocabulary for the “language of biomedicine and health” (67) over 100 biomedical vocabularies. UMLS contains over 1 million concepts in biomedicine (e.g. concept C0206131 “adipocytes”), unified across lexical variation and terminology (e.g. “adipocyte”, “mature fat cell”), language (e.g. “lipozyten” in German), and original coding system (e.g. M0026722 used by the librarians in MeSH, 24826007 used by the pathologists in SNOMED-CT) (68). Campillos used UMLS to represent side effects, and a weighting scheme to account for the rareness and interdependence of side effects (66). As similarity in side effects correlated with shared target(s) between drugs, Campillos reasoned that side effect similarity could be used to predict novel targets between any two “unexpected” drug pairs. One such example was fluoxetine which was predicted to target Dopamine Receptor D3 (DRD3), because of its shared side effects with rabeprazole. By combining side effect similarity with chemical similarity, 13 out of 20 novel target predications were experimentally validated (66), thereby identifying novel off-target effects which could be used to driven novel indications for these drugs.
These success studies share a several unifying themes related to bioinformatics. First, publicly-accessible repositories or databases is crucial in advancing existing knowledge. All of the studies mentioned used resources freely available to the public, such as the data from NCI-60 or the HapMap genotypes. Second, novel data abstraction or representation of existing information, such as biospectra or side effect similarity, can reveal novel relationships. Third, unified vocabularies, such as those found in ATC or UMLS, are not just essential for data interpretation and analyses, but also enable novel lines of questioning. Finally, profound insights can be gained from creative integration of data from different experimental modalities.
NEWER TYPES OF MOLECULAR MEASUREMENTS
Much of our existing knowledge of genetic factors involved in SADRs centers around single nucleotide polymorphisms; however, polymorphisms of other kinds have also been identified and point to additional new knowledge that will need to be uncovered for a better mechanistic understanding of SADRs. For instance, copy number variants (CNVs) and deletions have also been shown to contribute to SADRs. Some individuals with extra copies of specific variants of CYP2D6, responsible for metabolizing numerous drugs, including opioids such as codeine, can suffer from toxic effects. This has resulted in label changes mandated by the FDA to inform consumers of genetic risk factors‥ The glutathione transferases GSTM1 and GSTT1, responsible for metabolizing anticancer drugs such as cisplatin and 5-FU, have been found to be deleted in many ethnicities (70). Copy number changes can be surveyed across the genome using the same technologies currently used to survey single nucleotide polymorphisms.
Besides nucleotide base changes, epigenetic modifications can also result in clinical and molecular phenotypes. For instance, the high CYP1B1 expression in the prostate cancer cells compared to normal cells has been linked to hypomethylation in the promoter and enhancer regions of CYP1B1 (71). Hypomethylation allows transcription factors and enhancers access to promoter / enhancer regions and increase gene expression. Moreover, methylation differences may contribute to variable drug response. For example, the CYP24 methylation pattern is different in tumor-derived endothelial cells, compared with normal endothelial cells (72). The methylation, or silencing, of CYP24 in tumor cells resulted in reduced responsiveness of CYP24 to a pharmaceutical.
A recently discovered gene regulatory mechanism involving microRNAs (miRNA) has also been connected to SADRs. First discovered in 1993, ,miRNA are small noncoding RNA that typically bind to the 3’ untranslated region (UTR) of mRNAs and target them for degradation or translational repression (73). Tsuchiya and colleagues recently showed how one miRNA, miR-27b, regulates the expression of CYP1B1, which metabolizes polycyclic aromatic hydrocarbons and 17beta-estradiol (74). As CYP1B1 is found to be highly expressed in cancer tissues, this study essentially links miRNAs to drug chemosensitivity. Moreover, a separate study found that another miRNA, miR-24, binds to the 3’ UTR of dihydrofolate reductase (DHFR) and regulates its level. A SNP (miRSNP) in the 3’UTR of DHFR in the presumed binding site of miR-24 results in overexpression of DHFR and increased resistance to methotrexate, which is metabolized by DHFR (75).
The nascent field of pharmacoproteomics refers to the study of how proteins change in response to drug treatments, and has already contributed toward our understanding of SADRs. In particular, two-dimensional gel electrophoresis (2DE) has proved useful in understanding the underlying mechanisms in SADRs in several drugs. One such example is the study of the toxic liver effects of methapyrilene, an antihistamine and sleep aid that was eventually withdrawn from the market. The liver toxicity was found to be due to the protein adducts formed by methapyrilene, specifically in mitochondrion of rat liver being used to model this effect (76). Additionally, the toxicity effects of cyclosporine A, an immunosuppressant, in rat kidney (77) and brain (78) were also discovered with the aid of 2DE. Protein arrays, which are conceptually similar to high density SNP and gene expression measurements, are just emerging as potent tools to aid new studies in drug toxicity.
Together, these studies across the scale of molecular measurements, from genetic copy number variation, miRNA, epigenetics, and proteomics, show that SADRs are the result of variations and defects across many genomic levels and point to the increasing complexities associated with elucidating underlying mechanisms responsible for SADRs. As more of these data become available, we will undoubtedly see new calls for public-disclosure of these data from funding agencies and journals, similar to the calls that led to the current availability of gene expression, polymorphism, and DNA data. The first creative applications of bioinformatics methods to these datasets, especially when “mixed” with previously collected data, are likely to be of high impact.
PRESPECTIVES AND CHALLENGES
Understanding the molecular determinants of drug toxicity and efficacy of a few drugs has already transformed clinical care, particularly in cancer therapies. The development of bioinformatics tools is foundational and necessary to study and analyze the ever-increasing data and knowledge stored in databases and knowledge-bases for studying SADRs. More importantly, recent studies employing bioinformatics methodologies are paving the way toward elucidating molecular underpinnings of SADRs. In the near future, bioinformatics will be integral in continuing the revolution to bridge the gap between molecular and clinical domains. Existing capabilities limit the focus toward post-market surveillance, however, a more proactive pre-market review of drug toxcitiy and efficicay is becoming a possibility.
There has not been a more opportune time to realize the potential of pharmacogenomics. We have now at our disposal many genomic technologies to leverage toward the identification of genetic risk factors involved in SADRs, and to translate findings into the clinic. However, there are still many challenges ahead. Determining the molecular risk factors in SADRs, whether genetic or not, lies fundamentally with finding the patients who suffer from SADRs first. Because SADRs are rare events, the identification of patients who suffer from SADRs and the collection of their biospecimens is not a trivial problem. The problem of finding these patients is compounded by the lack of objective, standardized diagnosis of SADRs for all drugs. As example, drug-induced liver toxicity has no established diagnosis criteria, and instead relies on diagnosis by exclusion. In addition, patients who suffer from SADRs comprise only a small percentage of the overall patients that use specific drugs, and SADRs sometimes affect only specific populations (e.g. ethnic, elderly).
Finding these patients requires a global effort that spans the pharmaceutical and biotechnology industry, regulatory agencies, and educational institutions to collaborate in order to identify the appropriate patients. Towards this end, both regulatory agencies and academic institutions have taken initiatives to better identify SADR patients, collect and store biospecimens from these patients, and compile family histories and pertinent information (e.g. diet, geographic location) to enable more fruitful SADR studies.
Specifically, several consortiums have been established to identify genetic factors in SADRS. Examples of these include the recently formed Serious Adverse Events Consortium, the International Warfarin Consortium(79), and the European Consortium, EUDRAGENE(80), established to study six SADRs (Table 2). Often overlooked, these consortiums are essential first step toward identifying the drugs causing SADRs and the affected patients. Some of these consortia have committed to releasing data to the public within a defined time-frame, and this kind of data will fuel bioinformatics-enabled discoveries on the nature of SADRs.
Table 2.
Pharmacovigilance and genetic SADR consortiums
| Spontaneous Reporting Systems | |
Vigibase
|
http://www.umcproducts.com/DynPage.aspx?id=4910&mn=1107 |
Adverse Event Reporting System (AERS)
|
http://www.fda.gov/cder/aers/default.htm |
Canada Vigilance Online Database
|
http://www.hc-sc.gc.ca/dhpmps/medeff/databasdon/index-eng.php |
| SADR consortiums | |
Serious Adverse Event Consortium
|
http://www.saeconsortium.org/ |
International Warfarin Consortium
|
http://www.pharmgkb.org/views/project.jsp?pId=56 |
European collaboration for studying genetic basis of adverse drug reations (EUDRAGENE)
|
http//www.eudragene.org |
Canadian Genotype-specific Approaches to Therapy in Childhood program (GATC)
|
http://www.genomebc.ca/research_tech/research_projects/health/gatc.htm |
United States Drug Induced Liver Injury Network (DILIN)
|
http://dilin.dcri.duke.edu/ |
Pharmacogenetics of antimicrobial drug-induced liver injury (DILIGEN)
|
http://www.diligen.org/ |
Patient stratification is another key problem. Stratifying patients based on drug efficacy and toxicity may present an economical challenge to the pharmaceutical and biotechnology industries as the costs associated with drug development may not be offset by sales to only a subset of the ideal patient population.
Many current efforts are focused on finding SNPs associated with SADRs, with a strong push towards GWAS using high density SNP measurements to study drug response variability. There have only been a few successful studies thus far. It is too early to know if these GWAS are sensitive enough to detect all the genes involved, or whether an SADR represents a single phenotype or a variety of molecular phenotypes that are impossible to distinguish clinically. One major challenge that will need to be addressed is the lack of reproducibility between these high density genetic association studies. Moreover, variations beyond SNPs, such as CNVs, microRNAs, and epigenetics, have already been shown to be important in SADRS and will also need to be examined on a genome-wide scale. Additional measurement technologies and tools might also be required to identify the genes and underlying mechanisms important in ADRs, such as protein-protein interactions.
Technological advances must be coupled by development of novel algorithms and analytical tools to evaluate these high-throughput screens in order to yield clinically relevant results. Though there is often caution from investigators in the sharing of data, having unfettered data is the most efficient way for bioinformaticians to develop better quantitative and analytical methods, from which all benefit. In addition, bioinformatics-enabled findings are not the end goal for SADR studies, but instead a means to an end. Accurate, cost effective and rapid turnaround clinical tests must still be developed to enable broad use.
Our existing knowledge of pharmacogenomics has come a long way in the five decades since genetics was first suggested to be involved in drug response variability, but our available data has greatly surpassed our existing knowledge. Measurements drive analytical tools, tools drive data, and data enable novel questions. These questions might not be asked by traditional chemists or drug-discovery engineers, but instead by computationally-enabled scientists. Continued open-mindedness by academics and industries towards this new group of investigators, towards open sharing of data, and towards pooling of rare resources, will move our field of pharmacology towards the prevention of SADRs and make a more individualized, personalized medicine become a reality.
Acknowledgements
The work was supported by grants from the Lucile Packard Foundation for Children's Health, National Institute of General Medical Sciences (R01 GM079719), National Library of Medicine (T15 LM007033), Howard Hughes Medical Institute, and the Pharmaceutical Research and Manufacturers of America Foundation.
Footnotes
Conflict of Interest Statement
None
References
- 1.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. Jama. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 2.Giacomini KM, Krauss RM, Roden DM, Eichelbaum M, Hayden MR, Nakamura Y. When good drugs go bad. Nature. 2007;46:975–977. doi: 10.1038/446975a. [DOI] [PubMed] [Google Scholar]
- 3.Butte AJ. Translational bioinformatics: coming of age. J Am Med Inform Assoc. 2008;15:709–714. doi: 10.1197/jamia.M2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. American journal of human genetics. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 5.Evans WE, Johnson JA. Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annual review of genomics and human genetics. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. [DOI] [PubMed] [Google Scholar]
- 6.Pirmohamed M, Park BK. Genetic susceptibility to adverse drug reactions. Trends in pharmacological sciences. 2001;22:298–305. doi: 10.1016/s0165-6147(00)01717-x. [DOI] [PubMed] [Google Scholar]
- 7.Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, et al. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug metabolism and disposition: the biological fate of chemicals. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 8.Sim SC, Ingelman-Sundberg M. The human cytochrome P450 Allele Nomenclature Committee Web site: submission criteria, procedures, and objectives. Methods in molecular biology (Clifton, NJ. 2006;320:183–191. doi: 10.1385/1-59259-998-2:183. [DOI] [PubMed] [Google Scholar]
- 9.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. Jama. 2002;287:1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 10.Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–1671. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, McLeod HL, McMurrough J, Gonzalez FJ, Fernandez-Salguero P. Molecular basis of the human dihydropyrimidine dehydrogenase deficiency and 5-fluorouracil toxicity. The Journal of clinical investigation. 1996;98:610–615. doi: 10.1172/JCI118830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. The Journal of clinical investigation. 1998;101:847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature reviews. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 14.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma SK, Balamurugan A, Saha PK, Pandey RM, Mehra NK. Evaluation of clinical and immunogenetic risk factors for the development of hepatotoxicity during antituberculosis treatment. American journal of respiratory and critical care medicine. 2002;166:916–919. doi: 10.1164/rccm.2108091. [DOI] [PubMed] [Google Scholar]
- 17.O'Donohue J, Oien KA, Donaldson P, Underhill J, Clare M, MacSween RN, et al. Co-amoxiclav jaundice: clinical and histological features and HLA class II association. Gut. 2000;47:717–720. doi: 10.1136/gut.47.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. The New England journal of medicine. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 19.Konig J, Seithel A, Gradhand U, Fromm MF. Pharmacogenomics of human OATP transporters. Naunyn-Schmiedeberg's archives of pharmacology. 2006;372:432–443. doi: 10.1007/s00210-006-0040-y. [DOI] [PubMed] [Google Scholar]
- 20.FDA approves Gleevec for leukemia treatment. FDA consumer. 2001;35:6. [PubMed] [Google Scholar]
- 21.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 22.Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz CC, Dantis L, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 23.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 24.Minna JD, Girard L, Xie Y. Tumor mRNA expression profiles predict responses to chemotherapy. J Clin Oncol. 2007;25:4329–4336. doi: 10.1200/JCO.2007.12.3968. [DOI] [PubMed] [Google Scholar]
- 25.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 26.Butte AJ, Tamayo P, Slonim D, Golub TR, Kohane IS. Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc Natl Acad Sci U S A. 2000;97:12182–12186. doi: 10.1073/pnas.220392197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, et al. Genomic signatures to guide the use of chemotherapeutics. Nature medicine. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Batliwalla F, Li W, Lee A, Roubenoff R, Beckman E, et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Molecular medicine (Cambridge, Mass. 2008;14:575–581. doi: 10.2119/2008-00056.Liu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byun E, Caillier SJ, Montalban X, Villoslada P, Fernandez O, Brassat D, et al. Genome-wide pharmacogenomic analysis of the response to interferon beta therapy in multiple sclerosis. Archives of neurology. 2008;65:337–344. doi: 10.1001/archneurol.2008.47. [DOI] [PubMed] [Google Scholar]
- 30.Lavedan C, Licamele L, Volpi S, Hamilton J, Heaton C, Mack K, et al. Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Molecular psychiatry. 2008 doi: 10.1038/mp.2008.56. [DOI] [PubMed] [Google Scholar]
- 31.Turner ST, Bailey KR, Fridley BL, Chapman AB, Schwartz GL, Chai HS, et al. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Archives of general psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galperin MY. The Molecular Biology Database Collection: 2008 update. Nucleic Acids Res. 2008;36:D2–D4. doi: 10.1093/nar/gkm1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danoff TM, Campbell DA, McCarthy LC, Lewis KF, Repasch MH, Saunders AM, et al. A Gilbert's syndrome UGT1A1 variant confers susceptibility to tranilast-induced hyperbilirubinemia. The pharmacogenomics journal. 2004;4:49–53. doi: 10.1038/sj.tpj.6500221. [DOI] [PubMed] [Google Scholar]
- 35.Lenzer J. FDA is incapable of protecting US "against another Vioxx". BMJ. 2004;329:1253. doi: 10.1136/bmj.329.7477.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammond IW, Gibbs TG, Seifert HA, Rich DS. Database size and power to detect safety signals in pharmacovigilance. Expert opinion on drug safety. 2007;6:713–721. doi: 10.1517/14740338.6.6.713. [DOI] [PubMed] [Google Scholar]
- 37.Parrish F, Do N, Bouhaddou O, Warnekar P. Implementation of RxNorm as a terminology mediation standard for exchanging pharmacy medication between federal agencies. AMIA Annu Symp Proc. 2006:1057. [PMC free article] [PubMed] [Google Scholar]
- 38.Seger AC, Jha AK, Bates DW. Adverse drug event detection in a community hospital utilising computerised medication and laboratory data. Drug Saf. 2007;30:817–824. doi: 10.2165/00002018-200730090-00007. [DOI] [PubMed] [Google Scholar]
- 39.Gurwitz JH, Field TS, Judge J, Rochon P, Harrold LR, Cadoret C, et al. The incidence of adverse drug events in two large academic long-term care facilities. The American journal of medicine. 2005;118:251–258. doi: 10.1016/j.amjmed.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Gandhi TK, Weingart SN, Borus J, Seger AC, Peterson J, Burdick E, et al. Adverse drug events in ambulatory care. The New England journal of medicine. 2003;348:1556–1564. doi: 10.1056/NEJMsa020703. [DOI] [PubMed] [Google Scholar]
- 41.Kuehn BM. FDA turns to electronic "sentinel" to flag prescription drug safety problems. Jama. 2008;300:156–157. doi: 10.1001/jama.300.2.156. [DOI] [PubMed] [Google Scholar]
- 42.Hernandez-Boussard T, Whirl-Carrillo M, Hebert JM, Gong L, Owen R, Gong M, et al. The pharmacogenetics and pharmacogenomics knowledge base: accentuating the knowledge. Nucleic acids research. 2008;36:D913–D918. doi: 10.1093/nar/gkm1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters M, Stasiewicz S, Merrick BA, Tomer K, Bushel P, Paules R, et al. CEBS--Chemical Effects in Biological Systems: a public data repository integrating study design and toxicity data with microarray and proteomics data. Nucleic acids research. 2008;36:D892–D900. doi: 10.1093/nar/gkm755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattingly CJ, Rosenstein MC, Davis AP, Colby GT, Forrest JN, Jr, Boyer JL. The comparative toxicogenomics database: a cross-species resource for building chemical-gene interaction networks. Toxicol Sci. 2006;92:587–595. doi: 10.1093/toxsci/kfl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic acids research. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 48.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 49.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic acids research. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Natsoulis G, Pearson CI, Gollub J, B PE, Ferng J, Nair R, et al. The liver pharmacological and xenobiotic gene response repertoire. Molecular systems biology. 2008;4:175. doi: 10.1038/msb.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science (New York, NY. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 52.Lin YA, Chiang A, Lin R, Yao P, Chen R, Butte AJ. Methodologies for extracting functional pharmacogenomic experiments from international repository; AMIA Annual Symposium proceedings / AMIA Symposium; 2007. pp. 463–467. [PMC free article] [PubMed] [Google Scholar]
- 53.Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, et al. An information-intensive approach to the molecular pharmacology of cancer. Science (New York, NY. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 54.New treatments for colorectal cancer. FDA consumer. 2004;38:17. [PubMed] [Google Scholar]
- 55.Cetuximab approved by FDA for treatment of head and neck squamous cell cancer. Cancer biology & therapy. 2006;5:340–342. [PubMed] [Google Scholar]
- 56.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 57.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2008 doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nature biotechnology. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 60.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Molecular cancer therapeutics. 2008;7:3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. American journal of human genetics. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, Badner JA, et al. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer research. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Analysis of drug-induced effect patterns to link structure and side effects of medicines. Nature chemical biology. 2005;1:389–397. doi: 10.1038/nchembio747. [DOI] [PubMed] [Google Scholar]
- 65.Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biological spectra analysis: Linking biological activity profiles to molecular structure. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:261–266. doi: 10.1073/pnas.0407790101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science (New York, NY. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 67.Medicine, N.L.o. About the UMLS Resources. http://www.nlm.nih.gov/research/umls/about_umls.html.
- 68.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geisler SA, Olshan AF. GSTM1, GSTT1, and the risk of squamous cell carcinoma of the head and neck: a mini-HuGE review. American journal of epidemiology. 2001;154:95–105. doi: 10.1093/aje/154.2.95. [DOI] [PubMed] [Google Scholar]
- 71.Tokizane T, Shiina H, Igawa M, Enokida H, Urakami S, Kawakami T, et al. Cytochrome P450 1B1 is overexpressed and regulated by hypomethylation in prostate cancer. Clin Cancer Res. 2005;11:5793–5801. doi: 10.1158/1078-0432.CCR-04-2545. [DOI] [PubMed] [Google Scholar]
- 72.Chung I, Karpf AR, Muindi JR, Conroy JM, Nowak NJ, Johnson CS, et al. Epigenetic silencing of CYP24 in tumor-derived endothelial cells contributes to selective growth inhibition by calcitriol. The Journal of biological chemistry. 2007;282:8704–8714. doi: 10.1074/jbc.M608894200. [DOI] [PubMed] [Google Scholar]
- 73.Ruvkun G. The perfect storm of tiny RNAs. Nature medicine. 2008;14:1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 74.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer research. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 75.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(135):13–18. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lijinsky W, Reuber MD, Blackwell BN. Liver tumors induced in rats by oral administration of the antihistaminic methapyrilene hydrochloride. Science (New York, NY. 1980;209:817–819. doi: 10.1126/science.7403848. [DOI] [PubMed] [Google Scholar]
- 77.Steiner S, Aicher L, Raymackers J, Meheus L, Esquer-Blasco R, Anderson NL, et al. Cyclosporine A decreases the protein level of the calcium-binding protein calbindin-D 28kDa in rat kidney. Biochemical pharmacology. 1996;51:253–258. doi: 10.1016/0006-2952(95)02131-0. [DOI] [PubMed] [Google Scholar]
- 78.Varela MC, Arce A, Greiner B, Schwald M, Aicher L, Wahl D, et al. Cyclosporine A-induced decrease in calbindin-D 28 kDa in rat kidney but not in cerebral cortex and cerebellum. Biochemical pharmacology. 1998;55:2043–2046. doi: 10.1016/s0006-2952(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 79.Owen RP, Altman RB, Klein TE. PharmGKB and the International Warfarin Pharmacogenetics Consortium: the changing role for pharmacogenomic databases and single-drug pharmacogenetics. Human mutation. 2008;29:456–460. doi: 10.1002/humu.20731. [DOI] [PubMed] [Google Scholar]
- 80.Molokhia M, McKeigue P. EUDRAGENE: European collaboration to establish a case-control DNA collection for studying the genetic basis of adverse drug reactions. Pharmacogenomics. 2006;7:633–638. doi: 10.2217/14622416.7.4.633. [DOI] [PubMed] [Google Scholar]