Abstract
Oligoadenylate synthetases (OASs) are interferon-inducible enzymes that participate in the first line of defense against a wide range of viral infection. Recent studies have determined that Oas1b, a member of the OAS gene family in the house mouse (Mus musculus), provides specific protection against flavivirus infection (e.g., West Nile virus, dengue fever virus, and yellow fever virus). We characterized the nucleotide sequence variation in coding and noncoding regions of the Oas1b gene for a large number of wild-derived strains of M. musculus and related species. Our sequence analyses determined that this gene is one of the most polymorphic genes ever described in any mammal. The level of variation in noncoding regions of Oas1b is an order of magnitude higher than the level reported for other regions of the mouse genome and is significantly different from the level of intraspecific variation expected under neutrality. Furthermore, a phylogenetic analysis of intronic sequences demonstrated that Oas1b alleles are ancient and that their divergence predates several speciation events, resulting in transspecific polymorphisms. The amino acid sequence of Oas1b is also extremely variable, with 1 out of 7 amino acid positions being polymorphic within M. musculus. Oas1b alleles are comparatively more divergent at synonymous positions than most autosomal genes and the ratio of nonsynonymous to synonymous substitution is remarkably high, suggesting that positive selection has been acting on Oas1b. The ancestry of Oas1b polymorphisms and the high level of amino acid polymorphisms strongly suggest that the allelic variation at Oas1b has been maintained in mouse populations by long-term balancing selection.
Keywords: balancing selection, Mus musculus, Oas1b, West Nile virus, flavivirus
Introduction
Organisms and their pathogens are engaged in a coevolutionary process resulting from their conflicting interests: hosts must retain their ability to resist and eventually clear infection, and pathogens must retain their infectivity and ability to evade host defense. Host defense genes are in the forefront of this process and bear the signature of the interactions between the host and its pathogens. Many defense genes have been shown to evolve rapidly at the protein level because of the action of positive selection. This pattern of evolution is caused either by a succession of selective sweeps of new resistance alleles (the “arms race” model of evolution) or by the persistence of dynamic polymorphisms due to selective fluctuations in allelic frequency (the “Red Queen” model) (for a review, see Woolhouse et al. 2002). The selective mechanism responsible for the maintenance of polymorphism in populations is generally referred to as balancing selection and can be caused by heterozygote advantage, frequency-dependent selection (Kojima 1971), or selection that varies in time and space (Hedrick et al. 1976). Although most balanced polymorphisms tend to be transient, in some very rare cases, balancing selection has been able to maintain alleles for very long periods of evolutionary time. At the major histocompatibility complex (MHC) in mammals (Hedrick and Thomson 1983; Figueroa et al. 1988) and at the plant resistance (R) genes (Bergelson et al. 2001), a remarkably large number of alleles have been maintained by balancing selection, and some of these polymorphisms are so ancient that they predate several speciation events, resulting in transspecific polymorphisms (Figueroa et al. 1988).
2′,5′-Oligoadenylate synthetases (OASs) are members of the interferon pathway, which plays an important antiviral role (for a review, see Hovanessian and Justesen 2007). Interferon upregulates the transcription of OAS genes and OAS proteins are converted to an enzymatically active form by a double-strand RNA-dependent process. From adenosine triphosphate molecules, activated OAS proteins synthesize ppp(A2′p)nA oligoadenylates (2-5A) that bind to the latent endoribonuclease RNase L leading to the dimerization and activation of RNase L, followed by the degradation of cellular and viral RNA. The OAS gene family has 4 members in humans (OAS1, OAS2, OAS3, and OASL) and 10 members in the house mouse, Mus musculus (Oas1a to Oas1h, Oas2, and Oas3) (Eskildsen et al. 2002; Mashimo et al. 2003). Some of the mouse Oas1 copies, such as Oas1b and Oas1c, have lost their synthetase activity (Rogozin et al. 2003). As these copies remain otherwise relatively conserved, it is likely that they have additional functions.
It was recently found that the Oas1b gene confers resistance against infection with West Nile virus and other flaviviruses (e.g., dengue fever virus, yellow fever virus, and Japanese encephalitis virus) (Mashimo et al. 2002; Perelygin et al. 2002). Resistant mice can be infected by flaviviruses but the virus titers in their tissues are 1,000–10,000 times lower than those of susceptible mice, and the spread of infection is much slower (Sabin 1952; Goodman and Koprowski 1962). The vast majority of laboratory strains are susceptible to flaviviral infection and develop severe encephalitis due to a nonsense mutation in exon 4 of Oas1b resulting in a truncated protein (Mashimo et al. 2002; Perelygin et al. 2002). In contrast, most wild mice are resistant (Sangster et al. 1998) and at least 2 functionally different resistance alleles exist in nature: the “major resistance” allele that confers resistance against the vast majority of flaviviruses and a “minor resistance” allele that protects efficiently against yellow fever virus yet has little effect on the outcome of infection with other flaviviruses (e.g., Murray Valley encephalitis virus [Sangster et al. 1993]). It was found that these 2 resistance alleles differ at 14 amino acid positions (i.e., 4.2%) (Perelygin et al. 2002). This level of amino acid divergence is extremely high and is comparable with the divergence usually observed between distantly related species (e.g., mouse and rat proteins differ on average by 5% at the amino acid level; Gibbs et al. 2004). Although these data suggest that balancing selection could be acting on this gene, this pattern could also result from the complex evolutionary history of the house mouse, a species in which hybridization between subspecies (Ferris et al. 1983; Orth et al. 1998; Bonhomme et al. 2007) and with closely related species is known to occur (Greene-Till et al. 2000; Orth et al. 2002).
We assessed the variation of the Oas1b gene and we analyzed its molecular evolution in the house mouse and related species. Coding and noncoding regions of Oas1b were sequenced in a sample of wild-derived strains representative of the entire genetic diversity of this species. The Oas1b gene is extremely polymorphic within M. musculus and even within its subspecies, Mus musculus musculus and Mus musculus domesticus. Polymorphisms at Oas1b are very ancient and predate several speciation events. Our data demonstrate that the Oas1b gene has evolved under balancing selection for more than 2.8 Myr and constitute one of the very few cases of old transspecific polymorphisms maintained by long-term balancing selection in a mammal.
Materials and Methods
Sampling
The house mouse M. musculus is a complex species that originated in India less than 1 MYA (Boursot et al. 1996). From Northern India, this species gradually colonized the periphery of the Euro-Asiatic continent where 3 well-defined subspecies are found (M. m. musculus from Eastern Europe to Northern China; M. m. domesticus in Western Europe, North Africa, and the Middle East; and Mus musculus castaneus in South East Asia). These 3 subspecies still hybridize in natural conditions so that a clear-cut hybrid zone exists in Europe, whereas the subspecies seem to intermix to a greater extent in Asia. Mice from Japan are hybrids between M. m. musculus and M. m. castaneus and are sometimes referred to as “Mus musculus molossinus” (Yonekawa et al. 1986). We obtained DNA samples from 34 wild-derived strains of house mice, 23 from the “Conservatoire de la souris sauvage” (Université Montpellier II, Montpellier, France), and 11 from the Jackson laboratory (Bar Harbor, ME). These strains have been maintained in captivity for many generations so that they are homozygous at most loci, making the determination of haplotypes trivial. This sample includes strains from each of the 3 major subspecies, M. m. musculus (11 strains), M. m. domesticus (12 strains), and M. m. castaneus (4 strains); strains with undefined subspecific status from Iran, India, and Pakistan (4 strains); and hybrid strains from Japan (5 strains). In addition, we obtained DNA from other species within the genus Mus to determine the phylogenetic context of Oas1b evolution. These species include the 4 European species of mice, Mus spretus (3 strains), Mus macedonicus (3 strains), Mus spicilegus (4 strains), and Mus cypriacus (1 strain), which collectively diverged from the house mouse ∼1.4 MYA, the Indian species Mus famulus (2.8 MYA), and the Southeast Asian species Mus caroli (3.2 MYA). As an outgroup, we used the species Coelomys pahari that diverged from the genus Mus 6.5 MYA.
Molecular Analysis
Each of the 6 exons of Oas1b was amplified by polymerase chain reaction (PCR). We also amplified partial fragments from introns 2, 3, 4, and 5, from a fragment containing the 3′ untranslated region (3′ UTR), and from a noncoding region located 10 kb downstream of the Oas1b gene. The products of amplification were purified and sent for sequencing to the company Macrogen (Seoul, Korea). To assess the variation of the Oas1b gene, we compared it with the level of variation in 2 neutral regions of the mouse genome. We selected 2 autosomal segments located at least 1 Mb from any known gene, 1 on chromosome 12 (from nucleotides 64,811,904 to 64,813,152 on the February 2006 assembly of the mouse genome at http://genome.ucsc.edu) and 1 on chromosome 19 (from nucleotides 19,726,706 to 19,727,769). These fragments were amplified by PCR in each wild-derived strain and sequenced. The list of primers used for amplification is available on request from the corresponding author. DNA sequences have been deposited at the EMBL database under GenBank accession numbers AM887890–AM887932.
Data Analysis
Sequences were aligned and manipulated using the Bio-Edit platform (Hall 1999). Distances, including synonymous (ds) and nonsynonymous (dn) distances, were calculated using the Nei and Gojobori (1986) method as implemented in the MEGA3 software package (Kumar et al. 2004). Phylogenetic analyses were performed using the Neighbor-Joining method (as implemented in MEGA3) and the maximum likelihood method (using PHYML [Guindon et al. 2005]). The nucleotide diversity was estimated using the parameter π and Watterson's θ, and Tajima's D (Tajima 1989) was calculated to assess the effect of selection on polymorphisms. We also tested for selection using the Hudson–Kreitman–Aguade test (known as the HKA test [Hudson et al. 1987]). The HKA test is based on the assumption that, in the absence of selection, silent site polymorphisms and divergence are expected to be the same across all loci. Basically, the test compares the ratio of polymorphism with divergence between a gene of interest (Oas1b in our case) and a neutral region of the genome (the neutral regions on chromosomes 12 and 19, see the Molecular Analysis). If the difference between the 2 ratios is statistically significant from the null hypothesis using a goodness of fit test, we can then reject the hypothesis of neutrality. We also used the McDonald and Kreitman (1991) test (known as the MK test): if variation at a locus is neutral, then the rate of substitution between species and the amount of variation within species are a function of the mutation rate. Therefore, under neutrality, the ratio of nonsynonymous to synonymous fixed differences between species should be the same as the ratio of nonsynonymous to synonymous polymorphisms within species. If the 2 ratios differ significantly, we can reject the hypothesis of neutrality. The DnaSP program (Rozas et al. 2003) was used to perform the HKA and MK tests. We tested the possibility that gene conversion between alleles and/or between paralogous copies of Oas1b affected the sequence of Oas1b alleles using the method of Sawyer (1989) implemented in the GENECONV program.
Results
The coding sequence of Oas1b was obtained for 34 wild-derived strains of the house mouse (fig. 1). Fifty-five amino acid variants (at 51 positions) were detected within the species M. musculus. This number is extremely large considering that this gene is only 377 amino acid long, that is, about 1 out of 7 amino acid positions is variable. As expected, we did not observe a single case of heterozygosity and the determination of linkage between polymorphisms was trivial. The lack of apparent heterozygosity strongly suggests that our PCR primers amplified only Oas1b and not other paralogous copies. The premature stop codon responsible for the susceptible phenotype of the laboratory strains was found in 3 wild-derived strains (SKIVE, SF, and DOT), and another stop codon, also in exon 4, was found in strain MBS. Single base pair deletions resulting in frameshifts were detected in exons 1 and 4 of strains CTP and 22MO, respectively, and an in-frame deletion of 39 nt resulting in a 13 amino acids deletion was found in exon 5 of the BZO strain. Out of the 55 variable amino acids, 24 polymorphisms are shared between at least 2 subspecies. These shared amino acid polymorphisms result from the same mutations at the DNA level indicating that they are not the product of convergent evolution. In contrast, a single amino acid difference is fixed in one of the M. musculus subspecies (at amino acid position 45, in M. m. musculus, including M. m. molossinus). This pattern of variation extends to interspecific comparisons: there is not a single fixed amino acid that differentiates M. musculus from other species. In contrast, 5 polymorphisms are shared between M. musculus and either M. spicilegus or M. spretus, and this number is presumed to be an underestimate due to the limited availability of strains of species other than M. musculus. The lack of fixed differences between taxa and the abundance of shared polymorphisms suggests that many polymorphisms are older than the origin of M. musculus and at least 5 of them are older than the split between M. musculus and other European species (M. spretus, M. spicilegus, M. macedonicus, and M. cypriacus), which occurred ∼1.4 MYA.
FIG. 1.—
Variable amino acids at the Oas1b gene. The phenotypes associated with each allele are designated as follows: (s) susceptible strains carrying the premature stop codon in exon 4 (Mashimo et al. 2002; Perelygin et al. 2002), (ps) strains predicted to be susceptible based on their amino acid sequences, (r) resistant strains similar to the functionally characterized resistant allele (Sabin 1952; Goodman and Koprowski 1962; Mashimo et al. 2002; Perelygin et al. 2002), (pr) strains predicted to be resistant based on their amino acid sequence, and (mr) strains carrying an allele similar to the minor resistance allele.
To confirm the ancestry of Oas1b alleles, the synonymous divergence (ds) between M. m. musculus and M. m. domesticus alleles was compared with the divergence of 183 autosomal genes for which a M. m. domesticus (from the C57BL/6J strain) and a M. m. musculus (from the CZECHII/EiJ strain) sequence are available in databases (fig 2A). On average, Oas1b alleles are much more divergent at synonymous sites than most autosomal genes and the ds between some Oas1b alleles can be as high as 4%. Only a handful of genes show such high ds, including several MHC genes. Figure 2A also shows that the distribution of intersubspecific divergences is almost indistinguishable from the distribution of interspecific divergences. On average, M. m. musculus and M. m. domesticus Oas1b alleles differ by 2.85% at synonymous sites, whereas they differ on average by 2.22% from European species. This clearly implies that allelic variation at the Oas1b gene is as old or older than the split between M. musculus and its European relatives.
FIG. 2.—
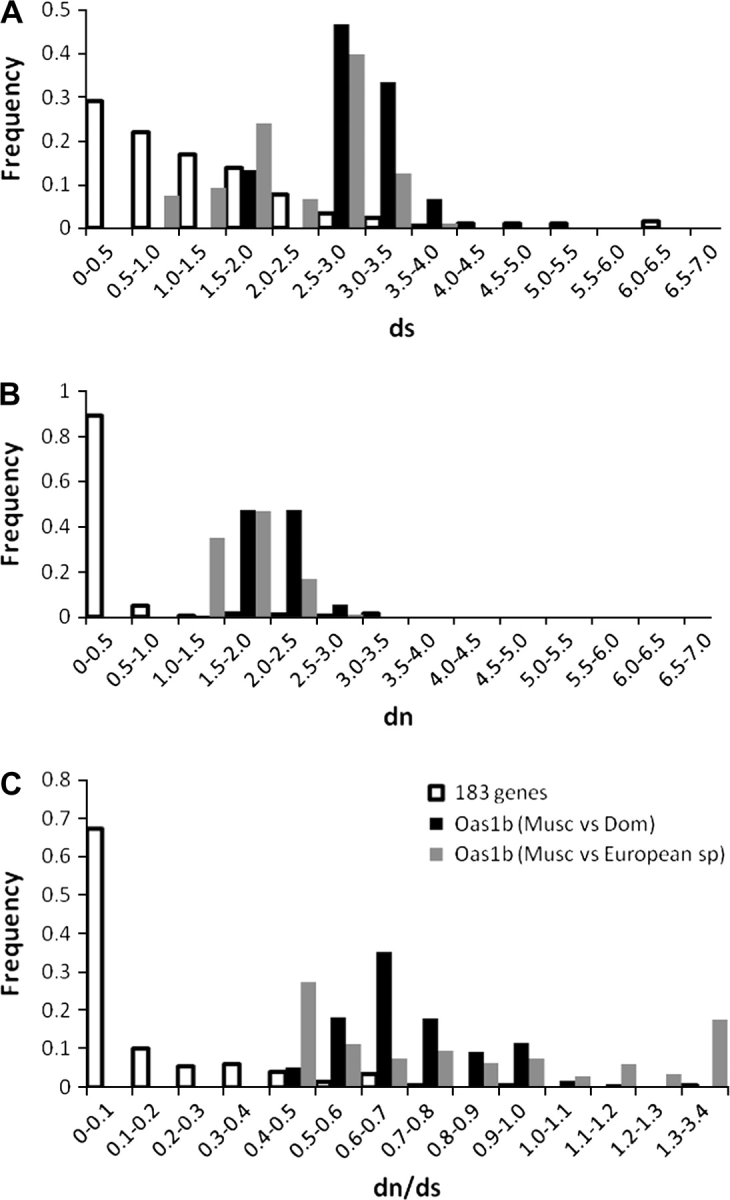
Comparison of the synonymous divergence (ds, panel A), the nonsynonymous divergence (dn, panel B), and their ratio (dn/ds, panel C) between Oas1b alleles and between 183 autosomal genes for which a Mus musculus musculus and Mus musculus domesticus alleles are available in the database. The dn/ds distribution is based on 153 genes because the ds value for 30 genes is 0, making dn/ds infinite.
The persistence of ancestral polymorphisms at Oas1b was further examined by performing a phylogenetic analysis on intronic sequences. Figure 3A shows a phylogenetic tree built using intron 5 sequences. This tree differs drastically from the species tree and shows strong evidence for transspecific polymorphisms. Two clades supported by high bootstrap values and by several indels are apparent on the tree. Each of these clades contains M. m. domesticus sequences and sequences from other species. M. m. domesticus sequences of clade 1 are more closely related to M. macedonicus, M. spicilegus, and M. spretus sequences than they are to M. m. domesticus sequences belonging to clade 2. Similarly, M. m. domesticus alleles in clade 2 are more closely related to M. spicilegus strain ZRU and to M. famulus than to clade 1. This clearly demonstrates that M. m. domesticus contains 2 deeply divergent allelic lineages at the Oas1b locus. These 2 lineages have separated before the split between M. musculus and M. famulus, which took place ∼2.8 MYA. These lineages have also been maintained in M. spicilegus because sequences of this species are found in both clades.
FIG. 3.—

(A) Maximum likelihood phylogeny of Oas1b alleles based on intron 5 sequences. Arrows represent phylogenetically informative insertions (+) and deletions (−). For instance, the arrow labeled +5 on the branch leading to clade 1 indicates that all sequences in this clade share a 5-bp insertion. Bootstrap values higher than 75% are indicated. The tree was built using the HKY model of substitution but other models and methods produced nearly identical trees. Mus musculus musculus and Mus musculus molossinus strains are in blue, Mus musculus domesticus in orange, Mus musculus castaneus in purple, and European species (Mus spretus, Mus spicilegus, Mus macedonicus, and Mus cypriacus) in green. KTK is Mus caroli, and FAM is Mus famulus. The origin of the strains is shown on figure 1. (B) Maximum likelihood phylogeny of Oas1b alleles based on introns 2, 3, and 4 and on the 3′ UTR.
Phylogenetic analyses were also performed on introns 2, 3, and 4 and on the 3′ UTR (fig. 3B). The phylogenetic trees for each of these sections of Oas1b are drastically different. Trees based on intron 4 and on the 3′ UTR are similar to the intron 5 tree and show that M. m. domesticus alleles, and to a lesser extent M. m. musculus alleles, fall into 2 distinct groups, supported by high bootstrap values. These groups are separated by long branches suggestive of their ancestry, although the phylogenetic analysis does not provide strong statistical support for transspecific polymorphism in intron 4 and in the 3′ UTR. The 2 divergent M. m. domesticus clades differ by 2.35% (±0.52) and 2.70% (±0.61), for intron 4 and the 3′ UTR, respectively, whereas they differ by 1.80% (±0.52) and 2.70% (±0.62) from M. famulus, again suggesting that ancient alleles were maintained in M. musculus since before its split from M. famulus. The composition of the domesticus groups is the same for intron 5 and the 3′ UTR but is different from intron 4 tree. Strains DIK, PERC and PERA belong to the same clade as strains DGA and DJO for intron 5 and the 3′ UTR, but these strains are closer to strains SF, SKIVE, and DOT on the intron 4 tree. This suggests that recombination is affecting the evolutionary history of Oas1b alleles by shuffling intronic sequences, although it appears the homogenizing effect of recombination has not been sufficient to mask the persistence of ancestral polymorphisms at Oas1b. Trees built using introns 2 and 3 show a radically different pattern. The tree based on intron 3 sequences also shows some level of mixing between the subspecies of M. musculus and M. spicilegus and M. macedonicus, but on this tree the M. famulus sequence falls clearly outside the diversity of M. musculus alleles. In addition, the divergences between M. musculus sequences are much smaller than the one observed for introns 4 and 5 and for the 3′ UTR. The most divergent groups of M. m. domesticus sequences on the intron 3 tree differ by 0.68% (±0.19), which is almost 5 times smaller than the divergence between groups for intron 5 (3.29 ± 0.66%). On the trees based on intron 2, M. m. domesticus and M. m. musculus sequences form monophyletic groups of closely related sequences with no evidence of transspecific polymorphism, and the topology of this tree is very similar to the species tree.
For both M. m. domesticus and M. m. musculus, the nucleotidic diversity in introns 4 and 5 and in the 3′ UTR is 5–10 times higher than the nucleotidic diversity in introns 2 and 3 and in 2 putatively neutral regions of the mouse genome (table 1). This level of polymorphism is among the highest reported for any genomic region of the house mouse and is clearly outside the range reported by Zhang et al. (2005) for 44 genomic segments (∼7 single nucleotide polymorphisms/kb within M. m. domesticus). The values of Tajima's D were negative for introns 2, 3, and 4 and for the 2 neutral regions. In introns 5 and in the 3′ UTR, Tajima's D takes positive values, although we did not detect any significant deviation from neutrality. We tested if the excess of polymorphisms at the 3′ end of Oas1b was significantly different from the level of intraspecific variation expected under neutrality. To this end, an HKA test was performed using the 2 noncoding fragments on chromosomes 12 and 19 as neutral regions of reference (see Materials and Methods). We were able to reject the null hypothesis of neutrality for introns 4 and 5 and the 3′ UTR (bold in table 1), although not all comparisons yielded significant deviations from neutrality. In contrast, we cannot reject the neutrality hypothesis for introns 2 and 3 and for the region located 10 kb downstream of Oas1b.
Table 1.
Summary Statistics for Different Noncoding Regions of Oas1b and for 2 Neutral Regions of the Mouse Genome (on chromosomes 12 and 19; see text)
| Regions | La | Nb | Sc | π (%)d | θ (%)e | Df | HKA Chromosome 12g | HKA Chromosome 19h | ||
| χ2 | P Value | χ2 | P Value | |||||||
| Intron 2 | 1525 | |||||||||
| Mus musculus musculus | 13 | 23 | 0.309 | 0.486 | −1.05 | 0.248 | 0.618 | 0.593 | 0.441 | |
| Mus musculus domesticus | 12 | 11 | 0.144 | 0.238 | −1.78 | 1.808 | 0.179 | 0.134 | 0.715 | |
| Intron 3 | 725 | |||||||||
| M. m. musculus | 11 | 6 | 0.152 | 0.285 | −1.61 | 0.176 | 0.675 | 0.919 | 0.338 | |
| M. m. domesticus | 10 | 17 | 0.625 | 0.837 | −0.31 | 1.672 | 0.196 | 0.088 | 0.767 | |
| Intron 4 | 710 | |||||||||
| M. m. musculus | 13 | 33 | 1.356 | 1.649 | −0.31 | 2.656 | 0.103 | 4.568 | 0.033 | |
| M. m. domesticus | 11 | 27 | 1.603 | 1.436 | −0.59 | 7.360 | 0.007 | 5.777 | 0.016 | |
| Intron 5 | 690 | |||||||||
| M. m. musculus | 13 | 41 | 1.305 | 1.978 | −0.79 | 1.851 | 0.174 | 3.461 | 0.063 | |
| M. m. domesticus | 11 | 26 | 1.556 | 1.319 | 1.62 | 5.271 | 0.022 | 2.817 | 0.093 | |
| 3′ UTR | 667 | |||||||||
| M. m. musculus | 8 | 23 | 1.395 | 1.438 | 1.14 | 2.147 | 0.143 | 4.920 | 0.027 | |
| M. m. domesticus | 11 | 21 | 1.444 | 1.162 | 1.37 | 6.025 | 0.014 | 2.963 | 0.085 | |
| 3′ fragment | 645 | |||||||||
| M. m. musculus | 9 | 7 | 0.267 | 0.399 | −1.03 | 0.023 | 0.879 | 0.264 | 0.608 | |
| M. m. domesticus | 5 | 4 | 0.372 | 0.298 | 1.70 | 1.121 | 0.289 | 0.204 | 0.651 | |
| Chromosome 12 | 817 | |||||||||
| M. m. musculus | 10 | 13 | 0.435 | 0.277 | −0.99 | 0.109 | 0.742 | |||
| M. m. domesticus | 9 | 1 | 0.032 | 0.053 | −1.14 | 0.518 | 0.475 | |||
| Chromosome 19 | 1093 | |||||||||
| M. m. musculus | 11 | 8 | 0.150 | 0.257 | −1.46 | 0.109 | 0.742 | |||
| M. m. domesticus | 10 | 8 | 0.176 | 0.266 | −1.46 | 2.833 | 0.092 | |||
NOTE.—P values <0.05 are indicated in bold.
Length of the fragment in base pairs.
Number of strain sequenced.
Number of segregating sites.
Nucleotidic diversity.
Watterson's estimator of nucleotidic diversity.
Tajima's D.
Results of the HKA test using the region on chromosome 12 as neutral region of reference.
Results of the HKA test using the region on chromosome 19 as neutral region of reference.
We also examined the possibility that gene conversion (Perelygin et al. 2006) could have affected the divergence of Oas1b alleles. Using GENECONV, we were unable to detect a single significant instance of gene conversion between Oas1b and any other member of the Oas1 gene family. Therefore, the large divergences between Oas1b intronic sequences result from the persistence of alleles over a long period of evolutionary time and not from the effect of gene conversion between paralogous copies. This analysis clearly indicates that Oas1b polymorphisms are ancient and strongly suggests that balancing selection has acted on Oas1b polymorphisms. It further suggests that the sites under balancing selection are located at the C-terminus of the Oas1b protein because the strongest evidence for transspecific polymorphism is found for introns 4 and 5 and for the 3′ UTR.
The high level of divergence of Oas1b alleles is also apparent at nonsynonymous sites (fig. 2B). The average divergence at nonsynonymous sites (dn) between M. m. musculus and M. m. domesticus is 2.00%. Very few genes show such a high dn and in our sample of 183 genes, only 5 genes (all involved in host–pathogens interactions) had a dn greater than 2.00%. This high level of dn is consistent with the exceptionally large number of amino acid polymorphisms at Oas1b. We then calculated the dn/ds ratio for all genes in our data set. A high value for the ratio dn/ds indicates that selection favors nonsynonymous substitutions, that is, amino acid changes, and is one of the predictions of balancing selection (Garrigan and Hedrick 2003). The dn/ds ratio is much higher for the Oas1b gene than it is for the vast majority of genes (fig. 2C). In general, the dn/ds ratio is very low (<0.1 and [Gibbs et al. 2004]) because most genes are evolving under strong purifying selection. In contrast, the dn/ds ratio is always higher than 0.4 for Oas1b and a number of pairwise comparisons are higher than 1. The Oas1b dn/ds ratio was also calculated between M. musculus and the other species in our sample (fig. 4), and values of dn/ds as high as 3.0 were obtained for some comparisons. Although it is plausible that an undetermined fraction of nonsynonymous changes resulted from a recent relaxation of selection on Oas1b (suggested by the presence of inactivating mutations in several strains) and caused an overestimation of dn, values of dn/ds higher than 1 are not consistent with neutral evolution. Instead, the high values of dn/ds suggest that selection in favor of nonsynonymous mutations is indeed acting on Oas1b and confirm the action of balancing selection on Oas1b polymorphisms.
FIG. 4.—
Relationship between synonymous divergence (ds) and the ratio dn/ds for Oas1b alleles in the genus Mus. The plot is best fitted to a power regression with a strong R2 value (0.67).
The balancing selection hypothesis further predicts an excess of nonsynonymous polymorphisms within species. In M. m. domesticus and M. m. musculus, 65% and 82% of all polymorphisms are nonsynonymous, respectively (table 2). In contrast, the number of fixed nonsynonymous and synonymous differences between M. m. domesticus and either M. famulus or M. caroli are basically identical. For M. m. musculus, the difference is even more striking as the number of fixed synonymous differences is higher than the number of fixed nonsynonymous differences, for both comparisons. The MK test shows that the ratio of nonsynonymous to synonymous fixed difference differs significantly from the ratio of nonsynonymous to synonymous polymorphisms in M. m. musculus but not in M. m. domesticus, most likely because of the conservative nature of the MK test (table 2). Although the excess of nonsynonymous polymorphisms in both M. m. musculus and M. m. domesticus is consistent with the action of balancing selection at Oas1b, the result of the MK test needs to be interpreted with caution because some nonsynonymous changes could reflect a recent relaxation of selection on Oas1b.
Table 2.
MK Neutrality Tests
| Mus famulus | Mus caroli | |||||
| Synonymousa | Nonsynonymousb | P Value | Synonymous | Nonsynonymous | P Value | |
| Mus musculus musculus | ||||||
| Polymorphic changes | 4 | 18 | 0.0095 | 4 | 18 | 0.0059 |
| Fixed differences | 15 | 12 | 21 | 16 | ||
| Mus musculus domesticus | ||||||
| Polymorphic changes | 14 | 26 | 0.5529 | 14 | 26 | 0.3065 |
| Fixed differences | 8 | 9 | 13 | 13 | ||
NOTE.—Tests were performed with either M. famulus or M. caroli as outgroups.
Synonymous (silent) mutations.
Nonsynonymous (replacement) mutations.
Discussion
Our analysis revealed an extremely high level of variation at the 3′ end of the Oas1b gene and provides strong evidence for transspecific polymorphism in the genus Mus. The nucleotide diversity in several introns of Oas1b is an order of magnitude higher than the diversity in putatively neutral regions of the mouse genome (the regions on chromosomes 12 and 19 and the 44 regions analyzed in Zhang et al. [2005]). The phylogenetic analysis of Oas1b introns further indicates that the allelic lineages found in M. musculus are older than the split between M. musculus and M. famulus, which occurred 2.8 MYA. Thus, this polymorphism is the second oldest reported in mice, after the genes of the MHC (Edwards et al. 1997). Although the presence of such divergent allelic lineages within a species could result from incomplete lineage sorting, this is very unlikely because the maintenance of alleles over 2.8 Myr would require an unrealistically large long-term effective population size. Assuming a conservative generation time of 2 generations/year, a long-term population size of more than one million individuals would be necessary to maintain the level of variation observed at the 3′ end of Oas1b (using θ = 4Neμ, where θ is the nucleotidic diversity, μ the mutation rate per site per generation, and Ne the effective population size). This value of Ne is 3–10 times higher than previous estimates (Eyre-Walker et al. 2002) and it is extremely unlikely that such a large effective population size was maintained for more than 2.8 Myr, considering that house mice went through several episodes of speciation that are typically associated with population bottlenecks. It is also unlikely that a recent hybridization event could account for the presence of divergent lineages in M. musculus. Although M. m. domesticus occasionally interbreed with M. spretus under natural conditions (Greene-Till et al. 2000; Orth et al. 2002), the molecular divergence between these 2 species is much smaller than the divergence between Oas1b allelic lineages and experimental crosses between M. musculus and its Asian relatives fail to produce viable birth. Instead, our data strongly suggest that Oas1b polymorphisms have been maintained in populations for the last ∼3 Myr by the action of balancing selection. We provide several lines of evidence that strongly support this hypothesis, including an extremely high nucleotidic diversity in introns 4 and 5 and in the 3′ UTR, a significant deviation from neutrality as revealed by the HKA test, a high dn/ds ratio and an excess of nonsynonymous polymorphisms. Balancing selection has been shown to act on a small number of loci in mammals (Hedrick and Thomson 1983; Figueroa et al. 1988; Shyue et al. 1995; Bamshad et al. 2002; Verrelli et al. 2002; Newman et al. 2006; Baysal et al. 2007) but rarely results in transspecific polymorphisms, probably because hosts eventually evolve resistance alleles that become fixed (Hedrick 2004). To our knowledge, there are only 2 well-documented cases of long-term (i.e., transspecific) balanced polymorphisms at host defense genes in mammal, the MHC (Figueroa et al. 1988; Edwards et al. 1997; Gutierrez-Espeleta et al. 2001) and the TRIM5α gene in Old World monkeys (Newman et al. 2006), making Oas1b only the third known case.
There are 3 main categories of mechanisms that can maintain allelic diversity for long periods of time: frequency-dependent selection, heterozygote advantage, and selection that varies in time and space. At this point, it is not possible to determine with certainty the mechanism responsible for the long-term maintenance of Oas1b alleles and more data about Oas1b‘s allelic diversity in natural populations are needed. However, our data point to some type of frequency-dependent mechanism. Under this model, the resistance allele that protects against the most common pathogenic genotypes will also be the most common one in the host population. In this situation, rare flaviviral genotypes that evade host defense will be advantaged and increase in frequency. Because hosts carrying rare alleles have in turn the highest fitness, the frequency of the rare allele will increase and the frequency of the common allele will decrease leading to a dynamic polymorphism. The dn/ds ratios were plotted against their ds values, which is a proxy of the divergence time between alleles (fig. 4). Less divergent (younger) alleles have higher dn/ds than older ones. This suggests that balancing selection has favored rare alleles over more common ones, assuming that young alleles are, in general, at lower frequency in populations than older alleles. A similar relationship is also expected if nonsynonymous sites become saturated faster than synonymous sites. However, this seems unlikely because most values of dn are lower than 3% and saturation is not expected within this range.
Whatever the mechanism involved in the maintenance of Oas1b alleles, a functional difference between the Oas1b alleles under balancing selection is implicit. Balancing selection also implies some type of interactions, direct or indirect between Oas1b and flaviviruses. The 2 alleles that have been functionally characterized (the major and minor resistance alleles) differ in the specificity of their response to flaviviruses, the minor resistance allele protecting against a smaller diversity of flaviviral genotypes than the major resistance allele. The amino acid polymorphisms responsible for these functional differences have yet to be mapped to specific regions of Oas1b as these 2 alleles differ at a number of sites spread across the entire gene. Although Oas1b retained a number of functional features found in other OAS, it has lost its synthetase activity and seems to protect against flaviviruses by a still unknown RNase L–independent pathway (Scherbik et al. 2006). As the double-strand RNA-binding motifs of Oas1b are located at the N-terminus of the protein, balancing selection is probably not caused by the interactions between flaviviral RNA and the Oas1b protein. The C-terminus of the Oas1b protein contains a domain with strong sequence similarity to an extracellular domain of interleukin receptors 3 and 5 (Ferguson W, Boissinot S, unpublished observation). This domain encompasses the CFK motif responsible for Oas1b tetramerization and plays an important role in protein–protein binding in the interleukin receptor family. This domain is in the immediate vicinity of a number of polymorphic sites including one of the polymorphisms shared between the different subspecies of M. musculus and between M. musculus and M. spicilegus. As this region has the potential to mediate protein–protein interactions, it is plausible that interactions between the Oas1b protein and a flaviviral protein, or a host protein which is itself under balancing selection, are responsible for the pattern of balancing selection at Oas1b, although this hypothesis needs to be tested experimentally.
Acknowledgments
The work was conducted in part with equipment from the Core Facilities for Imaging, Cellular and Molecular Biology at Queens College. We thank Else Fjerdingstad, George Jackman, and Peter Novick for their helpful comments on the manuscript. We thank Francois Bonhomme and the “Conservatoire de la souris sauvage” for providing DNA samples. This research was supported by National Institutes of Health grant AI058968-01A1 and PSC-CUNY grant 66253-0035 to S.B.
References
- Bamshad MJ, Mummidi S, Gonzalez E, et al. (11 co-authors) A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci USA. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Lawrence EC, Ferrell RE. Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA. BMC Biol. 2007;5:12. doi: 10.1186/1741-7007-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Bonhomme F, Rivals E, Orth A, Grant GR, Jeffreys AJ, Bois PR. Species-wide distribution of highly polymorphic minisatellite markers suggests past and present genetic exchanges among house mouse subspecies. Genome Biol. 2007;8:R80. doi: 10.1186/gb-2007-8-5-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot P, Din W, Anand R, Darviche D, Dod B, Von Deimling F, Talwar GP, Bonhomme F. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J Evol Biol. 1996;9:391–415. [Google Scholar]
- Edwards SV, Chesnut K, Satta Y, Wakeland EK. Ancestral polymorphism of Mhc class II genes in mice: implications for balancing selection and the mammalian molecular clock. Genetics. 1997;146:655–668. doi: 10.1093/genetics/146.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen S, Hartmann R, Kjeldgaard N, Justesen J. Gene structure of the murine 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2002;59:1212–1222. doi: 10.1007/s00018-002-8499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD, Smith NGC, Gaffney D. Quantifying the slightly deleterious mutation model of molecular evolution. Mol Biol Evol. 2002;19:2142–2149. doi: 10.1093/oxfordjournals.molbev.a004039. [DOI] [PubMed] [Google Scholar]
- Ferris SD, Sage RD, Huang CM, Nielsen JT, Ritte U, Wilson AC. Flow of mitochondrial DNA across a species boundary. Proc Natl Acad Sci USA. 1983;80:2290–2294. doi: 10.1073/pnas.80.8.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa F, Gunther E, Klein J. MHC polymorphism pre-dating speciation. Nature. 1988;335:265–267. doi: 10.1038/335265a0. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Hedrick PW. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution Int J Org Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, et al. (228 co-authors) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Goodman GT, Koprowski H. Study of the mechanism of innate resistance to virus infection. J Cell Comp Physiol. 1962;59:333–373. doi: 10.1002/jcp.1030590313. [DOI] [PubMed] [Google Scholar]
- Greene-Till R, Zhao Y, Hardies SC. Gene flow of unique sequences between Mus musculus domesticus and Mus spretus. Mamm Genome. 2000;11:225–230. doi: 10.1007/s003350010041. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Espeleta GA, Hedrick PW, Kalinowski ST, Garrigan D, Boyce WM. Is the decline of desert bighorn sheep from infectious disease the result of low MHC variation? Heredity. 2001;86:439–450. doi: 10.1046/j.1365-2540.2001.00853.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hedrick P. Estimation of relative fitnesses from relative risk data and the predicted future of haemoglobin alleles S and C. J Evol Biol. 2004;17:221–224. doi: 10.1046/j.1420-9101.2003.00635.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Ginevan M, Ewing E. Genetic polymorphism in heterogeneous environment. Annu Rev Ecol Syst. 1976;7:1–32. [Google Scholar]
- Hedrick PW, Thomson G. Evidence for balancing selection at HLA. Genetics. 1983;104:449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian AG, Justesen J. The human 2′-5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie. 2007;89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K. Is there a constant fitness for a given genotype? No! Evolution. 1971;25:281–285. doi: 10.1111/j.1558-5646.1971.tb01881.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Glaser P, Lucas M, Simon-Chazottes D, Ceccaldi PE, Montagutelli X, Despres P, Guenet JL. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics. 2003;82:537–552. doi: 10.1016/s0888-7543(03)00176-9. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Lucas M, Simon-Chazottes D, Frenkiel MP, Montagutelli X, Ceccaldi PE, Deubel V, Guenet JL, Despres P. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc Natl Acad Sci USA. 2002;99:11311–11316. doi: 10.1073/pnas.172195399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Newman RM, Hall L, Connole M, et al. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci USA. 2006;103:19134–19139. doi: 10.1073/pnas.0605838103. (11 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth A, Adama T, Din W, Bonhomme F. Natural hybridization between two subspecies of the house mouse, Mus musculus domesticus and Mus musculus castaneus, near Lake Casitas, California. Genome. 1998;41:104–110. doi: 10.1139/g97-109. [DOI] [PubMed] [Google Scholar]
- Orth A, Belkhir K, Britton-Davidian J, Boursot P, Benazzou T, Bonhomme F. Natural hybridization between two sympatric species of mice Mus musculus domesticus L. and Mus spretus Lataste. C R Biol. 2002;325:89–97. doi: 10.1016/s1631-0691(02)01413-0. [DOI] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci USA. 2002;99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Zharkikh AA, Scherbik SV, Brinton MA. The mammalian 2′-5′ oligoadenylate synthetase gene family: evidence for concerted evolution of paralogous Oas1 genes in Rodentia and Artiodactyla. J Mol Evol. 2006;63:562–576. doi: 10.1007/s00239-006-0073-3. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Aravind L, Koonin EV. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J Mol Biol. 2003;326:1449–1461. doi: 10.1016/s0022-2836(03)00055-x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sabin AB. Genetic, hormonal and age factors in natural resistance to certain viruses. Ann N Y Acad Sci. 1952;6:936–944. doi: 10.1111/j.1749-6632.1952.tb39968.x. [DOI] [PubMed] [Google Scholar]
- Sangster MY, Heliams DB, MacKenzie JS, Shellam GR. Genetic studies of flavivirus resistance in inbred strains derived from wild mice: evidence for a new resistance allele at the flavivirus resistance locus (Flv) J Virol. 1993;67:340–347. doi: 10.1128/jvi.67.1.340-347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster MY, Mackenzie JS, Shellam GR. Genetically determined resistance to flavivirus infection in wild Mus musculus domesticus and other taxonomic groups in the genus Mus. Arch Virol. 1998;143:697–715. doi: 10.1007/s007050050324. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. RNase L plays a role in the antiviral response to West Nile virus. J Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyue SK, Hewett-Emmett D, Sperling HG, Hunt DM, Bowmaker JK, Mollon JD, Li WH. Adaptive evolution of color vision genes in higher primates. Science. 1995;269:1265–1267. doi: 10.1126/science.7652574. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, McDonald JH, Argyropoulos G, Destro-Bisol G, Froment A, Drousiotou A, Lefranc G, Helal AN, Loiselet J, Tishkoff SA. Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am J Hum Genet. 2002;71:1112–1128. doi: 10.1086/344345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- Yonekawa H, Gotoh O, Tagashira Y, Matsushima Y, Shi LI, Cho WS, Miyashita N, Moriwaki K. A hybrid origin of Japanese mice “Mus musculus molossinus”. Curr Top Microbiol Immunol. 1986;127:62–67. [PubMed] [Google Scholar]
- Zhang J, Hunter KW, Gandolph M, Rowe WL, Finney RP, Kelley JM, Edmonson M, Buetow KH. A high-resolution multistrain haplotype analysis of laboratory mouse genome reveals three distinctive genetic variation patterns. Genome Res. 2005;15:241–249. doi: 10.1101/gr.2901705. [DOI] [PMC free article] [PubMed] [Google Scholar]




