Abstract
Of the approximately 250,000 species of flowering plants, nearly one in ten are members of the Compositae (Asteraceae), a diverse family found in almost every habitat on all continents except Antarctica. With an origin in the mid Eocene, the Compositae is also a relatively young family with remarkable diversifications during the last 40 My. Previous cytologic and systematic investigations suggested that paleopolyploidy may have occurred in at least one Compositae lineage, but a recent analysis of genomic data was equivocal. We tested for evidence of paleopolyploidy in the evolutionary history of the family using recently available expressed sequence tag (EST) data from the Compositae Genome Project. Combined with data available on GenBank, we analyzed nearly 1 million ESTs from 18 species representing seven genera and four tribes. Our analyses revealed at least three ancient whole-genome duplications in the Compositae—a paleopolyploidization shared by all analyzed taxa and placed near the origin of the family just prior to the rapid radiation of its tribes and independent genome duplications near the base of the tribes Mutisieae and Heliantheae. These results are consistent with previous research implicating paleopolyploidy in the evolution and diversification of the Heliantheae. Further, we observed parallel retention of duplicate genes from the basal Compositae genome duplication across all tribes, despite divergence times of 33–38 My among these lineages. This pattern of retention was also repeated for the paleologs from the Heliantheae duplication. Intriguingly, the categories of genes retained in duplicate were substantially different from those in Arabidopsis. In particular, we found that genes annotated to structural components or cellular organization Gene Ontology categories were significantly enriched among paleologs, whereas genes associated with transcription and other regulatory functions were significantly underrepresented. Our results suggest that paleopolyploidy can yield strikingly consistent signatures of gene retention in plant genomes despite extensive lineage radiations and recurrent genome duplications but that these patterns vary substantially among higher taxonomic categories.
Keywords: paleopolyploidy, whole-genome duplication, genome evolution, duplicate gene retention, Asteraceae, Compositae
Introduction
Polyploidy has long fascinated botanists because it is a prevalent process despite posing immediate and extensive challenges for an organism. A single genome duplication isolates an individual from its parental species and forces the nascent polyploid to overcome numerical inferiority and parental competition if it is to survive (Levin 1975). The concurrent duplication of all nuclear genes is accompanied by widespread changes in gene expression (Adams and Wendel 2005) and often chromosomal rearrangements (Levin 2002; Gaeta et al. 2007). Yet despite the potential for ecological and genomic havoc, polyploidy is remarkably frequent, especially among plants. By some accounts, 20–40% (Stebbins 1971) of extant flowering plant species are neopolyploids and as many as 70% are thought to have some polyploid ancestry (Masterson 1994; De Bodt et al. 2005; Cui et al. 2006).
Recent analyses of plant genomes have enhanced exploration of the history of polyploidy in plant evolution and support the botanical community's long-standing interest in polyploidy. Neopolyploidy is fairly easy to detect by changes in chromosome number, genome size, and nuclear gene copy numbers relative to putative progenitors. Some of these methods have been used for nearly a century to identify and study neopolyploidy to great effect (Stebbins 1971; Levin 2002), but methods for confidently assessing past polyploidy, or paleopolyploidy, were unavailable until recently. A primary obstacle to inferring paleopolyploidy is diploidization, a process of mutation, gene loss, and chromosomal rearrangement that commences immediately after a genome duplication and over time returns the polyploid to a diploid genetic system. Analyses of large genomic data sets can overcome the obscuring effects of diploidization to reveal signatures in plant genomes indicative of paleopolyploidy. Most striking are examples from studies of whole-genome sequences that infer paleopolyploidy through a combination of duplicate gene age distributions, gene family sizes, and identification of homoeologous regions. Such analyses have identified paleopolyploidy in the history of the Poaceae (Paterson et al. 2004; Yu et al. 2005) and indicate ancient duplications near the base of the Brassicaceae and the Rosids (Vision et al. 2000; Bowers et al. 2003; Schranz and Mitchell-Olds 2006; Jaillon et al. 2007; Tang et al. 2008). However, full-genome sequences are not available for many taxa and are not necessary for the identification of paleopolyploidy. Most of our knowledge of the phylogenetic distribution and prevalence of paleopolyploidy is based on analyses of the age distribution of duplicate genes from expressed sequence tag (EST) sequences. Bursts of gene duplication and/or an abrupt reduction in duplicate gene death create peaks in these age distributions and are used to infer paleopolyploid events. To date, analyses of ESTs have implicated paleopolyploidy in the history of over 15 plants, including Populus (Sterck et al. 2005) and Solanum (Schlueter et al. 2004; Blanc and Wolfe 2004b; Cui et al. 2006) plus the well-established paleopolyploid Arabidopsis (Vision et al. 2000; Blanc et al. 2003; Bowers et al. 2003; Blanc and Wolfe 2004b).
Despite the recent advancements in identifying paleopolyploidy in plants, we do not yet have assessments of paleopolyploidy from many lineages within a single family. Such studies are necessary for advancing research on paleopolyploidy because they place ancient duplication events in a phylogenetic context that permits evolutionary comparisons. However, collecting phylogenetically diverse genomic data for a large family of flowering plants is not trivial and until recently such data did not exist. One family for which substantial genomic data have recently become available is the Compositae (Asteraceae). With close to 25,000 species constituting approximately 10% of all angiosperms, the Compositae is the largest and one of the most diverse families of flowering plants (Funk et al. 2005; Stevens 2008). Members of the family occur in nearly every habitat on all continents except Antarctica and represent a full range of life histories from annuals to perennials and vines to trees. Evolutionary genomic analyses are now possible in the family because of the release of nearly 750,000 EST sequences for 18 species of Compositae by the Compositae Genome Project (CGP; http://compgenomics.ucdavis.edu) as well as nearly 17,000 ESTs for Gerbera (Laitinen et al. 2005). Combined with other ESTs available on GenBank, nearly 1 million EST sequences are now available for species across four tribes of the family, providing one of the most phylogenetically diverse collections of ESTs in any plant family.
Using these substantial EST resources, we address a number of questions about paleopolyploidy and the evolution of the Compositae. A diversity of chromosome numbers occurs across the family ranging from n = 2 to n = 114 (Funk et al. 2005), and a paleopolyploidization has long been hypothesized at the base of the subfamily Heliantheae s. l. (Smith 1975; Robinson 1981). Consistent with an ancient genome duplication at the base of the Heliantheae s. l. are observations of multiple nuclear gene copy numbers in members of the subfamily (Yahara et al. 1989; Berry et al. 1995; Gentzbittel et al. 1995). Baldwin et al. (2002) provided further support for this hypothesis from a parsimony analysis of chromosome number evolution that strongly suggested a paleopolyploidization at the base of the subfamily. Contrary to these studies, Blanc and Wolfe (2004b) failed to find evidence of paleopolyploidy in their analysis of Helianthus duplicate gene age distributions. We revisit this question using larger EST data sets for more species than were available to Blanc and Wolfe (2004b), and perhaps more critically, we employ a variety of statistical analyses to identify significant features in age distributions consistent with paleopolyploidy in the Compositae.
We also use the broad genomic resources of the Compositae to examine the composition of genes retained in duplicate following paleopolyploidy. Recent analyses of eukaryotic genomes have shown that paleologs, genes retained in duplicate from paleopolyploidy (Chapman et al. 2006), are often biased with respect to function (Seoighe and Gehring 2004; Blanc and Wolfe 2004a; Maere et al. 2005; Blomme et al. 2006; Aury et al. 2006; Rensing et al. 2007; Scannell et al. 2007). We tested for evidence of biased paleolog retention following genome duplication in the Compositae and compared the results with previous analyses and models of gene retention. Past analyses of paleolog functional biases in plants, almost exclusively on Arabidopsis, have only addressed this question by evaluating the patterns of duplicate gene retention from a single species’ data. Our multispecies analysis goes beyond previous studies by testing whether patterns of biased paleolog retention are shared among species after tens of millions of years of divergence.
Materials and Methods
EST libraries for 18 Compositae species representing four tribes were downloaded from GenBank in February 2007 and assembled into unigenes (table 1). A 19th species, Helianthus paradoxus, was not included in the analyses because it is a homoploid hybrid species, and its parental genomes (Helianthus annuus and Helianthus petiolaris) were already represented. Prior to assembly, vector, and low-quality sequences were removed using Seqclean (http://compbio.dfci.harvard.edu/tgi/software/) with the UniVec database (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html). Contigs were assembled for each species using the program TGICL with default settings (http://compbio.dfci.harvard.edu/tgi/software/) (Quackenbush et al. 2000), and a unigene file containing assembled contigs and singletons was created.
Table 1.
EST Assembly and Gene Family Statistics for 18 Species of Compositae
| Species | ESTs | Unigenes | % Unigenes Duplicated | Gene Family Count | Duplications With Ks < 2 |
| Gerbera hybrida | 16,998 | 8,438 | 12.1a | 7,657 | 293 |
| Centaurea maculosa | 39,957 | 21,432 | 37.1 | 15,900 | 3,554 |
| Centaurea solstitialis | 40,406 | 23,267 | 37.9 | 17,062 | 3,954 |
| Carthamus tinctorius | 40,875 | 19,963 | 39.2 | 14,327 | 3,284 |
| Cichorium endivia | 30,170 | 19,480 | 31.0 | 15,269 | 2,508 |
| Cichorium intybus | 41,704 | 22,674 | 33.2 | 17,255 | 2,895 |
| Taraxacum officinale | 41,278 | 16,858 | 31.9 | 12,564 | 1,651 |
| Lactuca sativa | 80,735 | 27,907 | 32.4 | 21,018 | 3,013 |
| Lactuca serriola | 55,452 | 21,140 | 25.6 | 16,788 | 1,587 |
| Lactuca saligna | 30,689 | 12,448 | 30.6 | 9,439 | 1,091 |
| Lactuca perennis | 29,118 | 12,129 | 30.0 | 9,333 | 1,192 |
| Lactuca virosa | 30,056 | 12,732 | 27.8 | 9,924 | 998 |
| Helianthus annuus | 93,428 | 37,222 | 30.0 | 27,725 | 3,500 |
| Helianthus argophyllus | 35,702 | 17,845 | 27.5 | 14,586 | 2,018 |
| Helianthus ciliaris | 21,589 | 15,158 | 28.6 | 12,382 | 2,037 |
| Helianthus exilis | 33,958 | 20,101 | 33.4 | 15,490 | 2,973 |
| Helianthus petiolaris | 27,472 | 13,655 | 30.2 | 10,557 | 1,519 |
| Helianthus tuberosus | 40,361 | 22,013 | 39.2 | 16,030 | 4,077 |
Lower percentage of duplicate genes in Gerbera relative to other taxa is likely because this is a single tissue EST collection.
From these unigenes, duplicate gene pairs were identified and their divergence, in terms of substitutions per synonymous site per year (Ks), was calculated. Duplicate pairs were identified as sequences that demonstrated 40% sequence similarity over at least 300 bp from a discontiguous MegaBlast (Zhang et al. 2000; Ma et al. 2002). Reading frames for duplicate pairs were identified by comparison to available plant protein sequences. Each duplicated gene was searched against all plant proteins available on GenBank (Wheeler et al. 2007) using BlastX (Altschul et al. 1997). Best hit proteins were paired with each gene at a minimum cutoff of 30% sequence similarity over at least 150 sites. Genes that did not have a best hit protein at this level were removed before further analyses. To determine reading frame and generate estimated amino acid sequences, each gene was aligned against its best hit protein by Genewise 2.2.2 (Birney et al. 1996). Using the highest scoring Genewise DNA–protein alignments, custom Perl scripts were used to remove stop and “N” containing codons and produce estimated amino acid sequences for each gene. Amino acid sequences for each duplicate pair were then aligned using MUSCLE 3.6 (Edgar 2004). The aligned amino acids were subsequently used to align their corresponding DNA sequences using RevTrans 1.4 (Wernersson and Pedersen 2003). Ks values for each duplicate pair were calculated using the maximum likelihood method implemented in codeml of the PAML package (Yang 1997) under the F3×4 model (Goldman and Yang 1994).
Further cleaning of the data set was conducted to remove duplication events that could bias the results. All duplicate pairs containing identifiable transposable elements were removed from the analysis because duplication resulting from transposition may obscure a signal from paleopolyploidy. To reduce the possibility that identical genes are represented in the data set, but missed by the TGICL clustering due to alternative splicing, all Ks values from one member of a duplicate pair with Ks = 0 were removed. Further, to reduce the multiplicative effects of multicopy gene families on Ks values, we used simple hierarchical clustering to construct phylogenies for each gene family (Blanc and Wolfe 2004b), identified as single-linked clusters, and calculate the node Ks values. Node Ks values <2 were used in subsequent analyses.
To identify significant features in the age distribution, we employed three statistical tests. We used the bootstrapped K-S goodness of fit test of Cui et al. (2006) to assess if the overall age distributions deviated from a simulated null. Taxa that deviated significantly from the null were then analyzed with SiZer (Chaudhuri and Marron 1999) to identify significant features (α = 0.05) in our age distributions. SiZer uses changes in the first derivative of a range of kernel density estimates to find significant slope increases or decreases, and the combination may be used to identify peaks and their ranges (Chaudhuri and Marron 1999). We also used EMMIX to fit a mixture model of normal distributions to our data by maximum likelihood (Mclachlan et al. 1999). Peaks produced by paleopolyploidy are expected to be approximately Gaussian (Schlueter et al. 2004; Blanc and Wolfe 2004b), and this mixture model test identifies the number of normal distributions and their positions that could produce our observed age distributions. For our analyses, 1–10 normal distributions were fitted to the data with 1,000 random starts and 100 k-mean starts. The Bayesian information criterion was used to select the best model fit to the data.
Age distributions from lineages as phylogenetically diverse as the tribes of the Compositae are not directly comparable because of molecular evolutionary rate variation among nuclear genomes. To account for this rate heterogeneity, we corrected Ks values for each tribe using relative rate corrections based on Ks branch length ratios. A representative of each tribe was selected (Gerbera hybrida for Mutisieae, Centaurea solstitialis for Cardueae, Lactuca sativa for Cichorioideae, and H. annuus for Heliantheae) along with two outgroups, Solanum lycopersicon (unigenes constructed as above) and Arabidopsis thaliana (TAIR 7 cds), to calculate Ks branch lengths of orthologs across a constrained topology in PAML. Thirty-six putative orthologs with at least 300-bp alignment overlaps were identified among these taxa by reciprocal best Blast hits (supplementary table S1, Supplementary Material online). Using these orthologs, we calculated the Ks branch lengths for each gene in the Compositae ingroup across a constrained topology based on the majority rule consensus tree of maximum likelihood analyses of the 36 nuclear gene orthologs (supplementary fig. S1, Supplementary Material online). DNA sequences for each ortholog set were aligned in MUSCLE 3.6 (Edgar 2004), and maximum likelihood phylogenies were recovered using the default settings in PHYML 2.4.4 (Guindon and Gascuel 2003). The majority rule consensus tree for these 36 nuclear phylogenies was found using the CONSENSE program of PHYLIP 3.68 (Felsenstein 2008). Our consensus tree topology is supported by the supertree phylogeny of Funk et al. (2005) and many other analyses (Jansen and Palmer 1987, 1988; Jansen et al. 1991; Jansen and Kim 1996; Kim et al. 2005). Using this topology, the ratios of branch lengths for Centaurea, Lactuca, and Helianthus versus Gerbera were calculated for each gene. The mean ratio over all 36 orthologs for each lineage was applied as a relative rate correction to the Ks values for their respective taxa, and we used Tukey–Kramer analyses to identify statistically significant groups among our rate corrected data sets. We also applied the rate corrections to the Ks branch lengths in our 36 nuclear gene phylogenies and computed a mean rate corrected phylogeny.
Gene Ontology (GO) annotations of the Compositae ESTs were obtained through discontiguous MegaBlast searches against A. thaliana transcripts from TAIR (TAIR 7 released 25 April 2007) for the best hit with at least 100 bp and an e value of 1 × 10−10. To ensure that we had comprehensive coverage of the transcriptome, we pooled GO annotations for all species from the same tribe, excluding Gerbera because it was a relatively small, single tissue library. We tested for differences among GO annotations by chi-square tests with P values computed from 100,000 Monte Carlo simulations in R (R Development Core Team 2005). When chi-square tests were significant (P < 0.05), GO categories with residuals >|2| were implicated as major contributors to the significant chi-square statistic. Using this statistical framework, we tested for significant differences among each tribe's full GO annotations, paleologs versus nonpaleologs for each tribe, among each tribe's paleologs from a shared paleopolyploidization, and paleologs from a tribe-specific duplication versus paleologs from older genome duplications in the same lineage. Boundaries for each whole-genome duplication were defined by the mixture model results.
Results
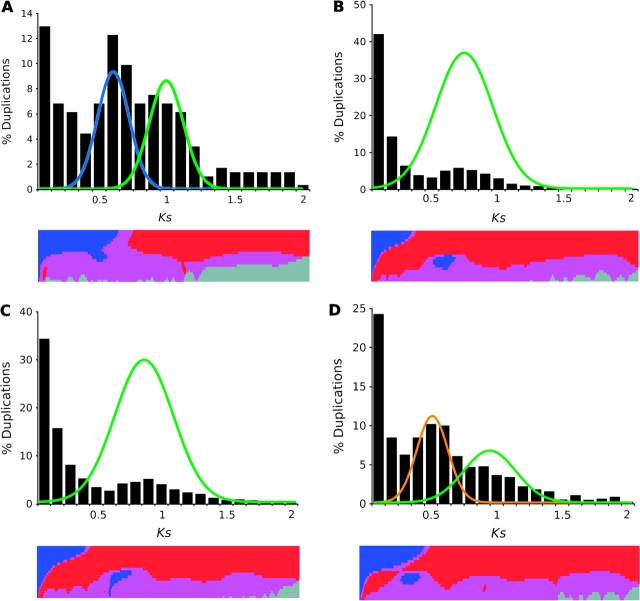
Our EST analyses provide robust evidence for several genome duplications in the history of the Compositae. The EST assemblies yielded a total of 344,462 unigenes for 18 species from seven genera and four tribes (table 1). On average, each species was represented by 19,136 unigenes distributed across 14,628 gene families with nearly 31% of the unigenes duplicated. Histograms of the age of gene duplication events, as inferred by Ks from our gene family phylogenies, demonstrated peaks consistent with paleopolyploidy in the ancestry of all 18 species (fig. 1 and supplementary fig. S2 [Supplementary Material online]). These Ks distributions also deviated significantly (P < 0.0001) from a null model of constant duplicate gene birth and death in a K-S goodness of fit test. Consistent with these results, SiZer analyses identified significant peaks (P < 0.05) in most species’ age distributions (table 2). These peaks were congruent for species within tribes, with members of the Heliantheae demonstrating a significant peak from Ks ∼ 0.35–0.65, the Cichorioideae from Ks ∼ 0.6–0.95, the Cardueae from Ks ∼ 0.5–0.85, and the Mutisieae from Ks ∼ 0.35–0.95 (fig. 1 and supplementary fig. S2 [Supplementary Material online]). Most importantly, the SiZer peaks overlap with peaks observed in histograms and provide strong support that the putative paleopolyploid signals are well distinguished from the background of small-scale gene duplications.
FIG. 1.—
Histograms of Compositae gene duplication ages with mixture model and SiZer analyses. (A) Gerbera hybrida, Mutisieae. (B) Carthamus tinctorius, Cardueae. (C) Cichorium intybus, Cichorioideae. (D) Helianthus argophyllus, Heliantheae. Plots of normal distributions were fitted from mixture model analyses; green lines represent the basal Compositae paleopolyploidization, whereas the blue and orange lines represent the independent Mutisieae and Heliantheae paleopolyploidizations, respectively. SiZer maps below histograms identify significant features at corresponding Ks values with blue areas indicating significantly increasing slopes, red indicating significantly decreasing slopes, purple representing no significant slope change, and gray indicating not enough data for the test.
Table 2.
Summary of Paleopolyploidy Analyses of ESTs for 18 Species of Compositae
| Tribe | Species | K-S Test | SiZer Range (Ks) | Number of Mixture Distributiona | Mixture Median (Ks) | % of Mixture Proportionb |
| 0.35–0.95 | 2 | 0.56 | 40.9 | |||
| Mutisieae | Gerbera hybrida | * | 0.95 | 25 | ||
| Cardueae | Centaurea maculosa | * | 0.52–0.87 | 1 | 0.69 | 33.5 |
| Cardueae | Centaurea solstitialis | * | — | 1 | 0.65 | 30.2 |
| Cardueae | Carthamus tinctorius | * | 0.39–0.82 | 1 | 0.67 | 31.6 |
| Cichorioideae | Cichorium endivia | * | 0.56–0.92 | 1 | 0.81 | 18c |
| Cichorioideae | Cichorium intybus | * | 0.55–0.92 | 1 | 0.81 | 35.5 |
| Cichorioideae | Taraxacum officinale | * | 0.62–0.97 | 1 | 0.86 | 34.2 |
| Cichorioideae | Lactuca sativa | * | 0.57–0.93 | 1 | 0.72 | 32.6 |
| Cichorioideae | Lactuca serriola | * | 0.56–0.96 | 1 | 0.77 | 32.8 |
| Cichorioideae | Lactuca saligna | * | 0.56–0.95 | 1 | 0.84 | 34.4 |
| Cichorioideae | Lactuca perennis | * | 0.74–0.93 | 1 | 0.81 | 35.3 |
| Cichorioideae | Lactuca virosa | * | 0.52–0.98 | 1 | 0.80 | 40 |
| Heliantheae | Helianthus annuus | * | 0.33–0.62 | 2 | 0.46 | 21 |
| 0.90 | 15.3 | |||||
| Heliantheae | Helianthus argophyllus | * | 0.28–0.63 | 2 | 0.45 | 35.4 |
| 0.89 | 26.9 | |||||
| Heliantheae | Helianthus ciliaris | * | 0.26–0.63 | 2 | 0.48 | 34.4 |
| 0.91 | 29 | |||||
| Heliantheae | Helianthus exilis | * | 0.28–0.55 | 2 | 0.44 | 35 |
| 0.90 | 25.6 | |||||
| Heliantheae | Helianthus petiolaris | * | 0.30–0.65 | 2 | 0.45 | 28.1 |
| 0.91 | 27.5 | |||||
| Heliantheae | Heilanthus tuberosus | * | 0.29–0.53 | 2 | 0.44 | 30.4 |
| 0.83 | 23 |
The number of normal distributions fitted by the mixture model to the significant SiZer ranges or histogram peaks.
The percentage of total duplications contained in each mixture model component.
Reduced percentage of duplications relative to other Cichorioideae due to the presence of a large number of introgressed genes early in the age distribution. See M. S. Barker and L. H. Rieseberg (in preparation) for a more complete explanation.
*P < 0.0001.
Maximum likelihood mixture model analyses of the Ks distributions revealed a further level of complexity. Members of the Cardueae and Cichorioideae each demonstrated a single normal distribution consistent with paleopolyploidy, whereas all members of the Mutisieae and Heliantheae contained two distributions, as is apparent from some species’ histograms (fig. 1 and supplementary fig. S2 [Supplementary Material online]). The single distribution observed in the Cardueae and Cichorioideae comprised an average of 32% of the duplications in these taxa, with a peak center of Ks = 0.67 for the Cardueae and Ks = 0.81 for the Cichorioideae (table 2). These results agree with histograms of duplicate gene ages for these taxa that demonstrate only a single peak and suggest a solitary paleopolyploidization. The two distributions observed in members of the Heliantheae and Mutisieae are each similar in scale to peaks from single genome duplications (table 2), suggesting that each of these peaks are the products of separate duplication events. In the Heliantheae, the mixture model places the centers of the successive duplications at Ks = 0.45 and Ks = 0.89. These two distributions correspond to two peaks in the Helianthus histograms that are difficult to distinguish visually because their tails overlap. Only the first of these two peaks is identified by SiZer, most likely because the prominent peak produced by the most recent paleopolyploidization obscures the positive slope of the older peak's left tail. Supporting this interpretation are significant declines near Ks = 0.95–1.05 in the Helianthus SiZer maps (fig. 1 and supplementary fig. S2 [Supplementary Material online]) that correspond to the right tails of the mixture model distributions (table 2 and supplementary table S2 [Supplementary Material online]). Similarly, concurrent genome duplications with centers at Ks = 0.56 and Ks = 0.95 are inferred for the Mutisieae with the two mixture model distributions aligning with two peaks in the Gerbera histogram.
The mixture model also identified a number of smaller distributions in many species that were not recovered in other analyses (supplementary table S2, Supplementary Material online). All species contained one to three distributions in their duplication-rich initial peaks (i.e., Ks = 0–0.1) that likely represent a mixture of tandem and other small-scale duplications in addition to alleles, segmental duplications, or neopolyploidy. One ancillary peak, a distribution observed from Ks ∼ 1.2–2, was particularly robust. Despite duplicate gene loss and the large error in estimating Ks beyond saturation, we observed this distribution in all 18 species surveyed. Taking into account the antiquity of this age range, the feature may correspond to an ancient polyploidy shared by all asteroids or possibly all eudicots, as has been proposed from other analyses of plant genomes (Vision et al. 2000; Bowers et al. 2003; De Bodt et al. 2005; Cui et al. 2006; Jaillon et al. 2007).
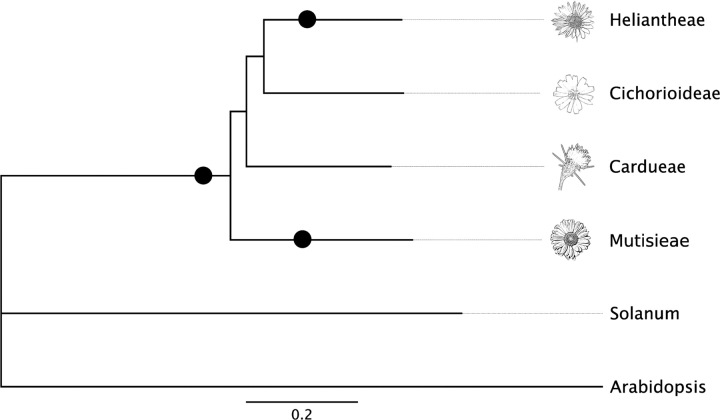
Considering the phylogenetic distribution of our sampled taxa (fig. 2), the simplest explanation for the observed duplication events is a shared paleopolyploidization at or near the base of the Compositae with independent duplications in the ancestry of the Heliantheae and Mutisieae. Our analyses of the overlap and phylogenetic placement of paleopolyploidizations support this hypothesis. To account for rate heterogeneity and make a valid comparison of Ks values across the Compositae, we calculated a rate correction based on the ratio of derived Ks branch lengths relative to the basal branch Gerbera for 36 ortholog phylogenies across a well-established topology (Jansen and Palmer 1987, 1988; Jansen et al. 1991; Jansen and Kim 1996; Funk et al. 2005; Kim et al. 2005) and used the mean ratios to correct all Ks values (table 3). This step revealed a substantial amount of nuclear rate heterogeneity across the Compositae with a nearly 30% difference in background molecular evolutionary rate between the fastest (Heliantheae) and slowest (Cardueae) lineages. When this correction was applied to the Ks distributions, two categories emerged across all taxa (table 3); a common paleopolyploid peak near Ks = 0.75 in the Cardueae, Cichorideae, and Heliantheae and a paleopolyploidization restricted to the Heliantheae at Ks = 0.37. Tukey–Kramer analyses indicate that the means of these two categories are significantly different. Although the Mutisieae is not included in the rate correction, the Ks = 0.95 Gerbera peak is probably the same as the Ks = 0.75 peak in other tribes, especially considering that the derived taxa do not show evidence of an additional, older duplication. Thus, we interpret these results as evidence of a single paleopolyploidization near the base of the Compositae with independent duplications in the Heliantheae and Mutisieae.
FIG. 2.—
Phylogeny of Compositae tribes and outgroups displaying observed paleopolyploid events and the rapid radiation of tribes. Branch lengths are mean rate corrected Ks values from 36 nuclear orthologs (table S1, Supplementary Material online). Black dots indicate inferred paleopolyploidizations in the evolution of the Compositae. Topology is a consensus phylogeny of the 36 nuclear orthologs (supplementary fig. S1, Supplementary Material online).
Table 3.
Nuclear Gene Relative Rate Corrections for Three Tribes of the Compositae
| Rate Corrected Paleopolyploidizations (Ks)b | |||
| Tribe | Relative Rate (% Ks)a | Tribe | Family |
| Cardueae | −12 | — | 0.75 ± 0.012** |
| Cichorioideae | +9 | — | 0.74 ± 0.018** |
| Heliantheae | +22 | 0.37 ± 0.007* | 0.74 ± 0.012** |
Percent difference relative to the Mutisieae (Gerbera), based on 36 nuclear gene phylogenies (supplementary table S1, Supplementary Material online).
Rate corrections applied to paleopolyploid peak means, as inferred from mixture model ranges, recovers two statistically significant classes indicated with superscripts (P < 0.001).
The placement of genome duplications in relation to lineage divergences also supports our interpretation of the data. We placed paleopolyploidizations onto a rate corrected phylogeny that was generated by recalculating the Ks branch lengths of the 36 ortholog trees using the relative rate corrections. From these trees, we estimated the mean divergences among our taxa using the corrected branch lengths. This yielded a mean divergence of Ks = 0.62 for the Mutisieae and all other tribes, the earliest tribal divergence in our phylogeny (fig. 2). In support of our interpretation that a genome duplication occurred near the base of the Compositae, our estimate of the divergence of the Mutisieae is more recent than the basal paleopolyploidization observed among all sampled Compositae taxa. This divergence date is also consistent with an independent duplication at Ks = 0.56 early in the evolution of the Mutisieae. Similarly, the divergence of the Cichorideae and the Heliantheae, at Ks = 0.50, supports our inference of an independent Heliantheae paleopolyploidization at Ks = 0.37. Additionally, our summary phylogeny of 36 nuclear orthologs is consistent with other studies of Compositae phylogeny that have demonstrated a rapid origin of extant tribes (Kim et al. 2005) with our results suggesting that all tribes originated during a relatively narrow window of divergence (Ks = 0.50–0.62).
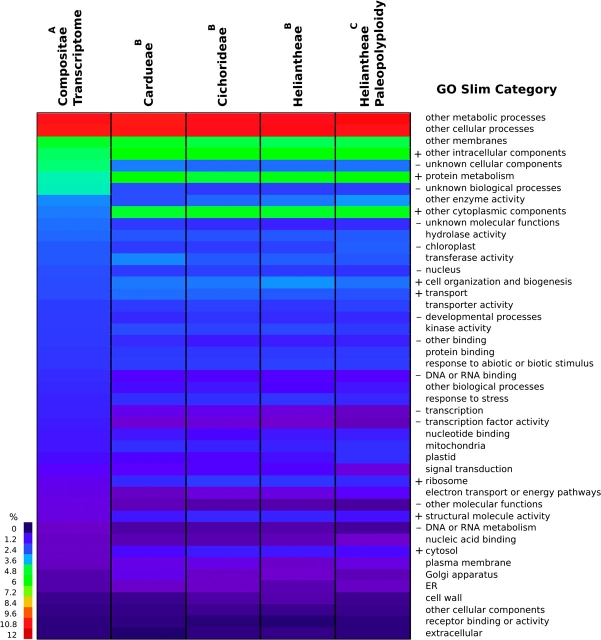
GO annotations provide another line of support for our interpretation of a shared paleopolyploidization near the base of the Compositae (fig. 3; GO figure and supplementary table S3 [Supplementary Material online] GO annotations as Microsoft Excel file). Consistent with a common paleopolyploidization, the pooled GO annotations for paleologs from the genome duplication at the base of the family were not significantly different (χ2 = 71.6, P = 0.90) among the Cardueae, Cichorideae, and Heliantheae. In contrast, the paleologs from the basal Heliantheae duplication were slightly different from the genes retained in duplicate from the older, family-wide duplication (χ2 = 60.5, P = 0.049) with higher retention of plastid-targeted genes from the more recent, tribal duplication event. Total GO annotations were also not significantly different among each of these tribes (χ2 = 90.7, P = 0.42), as would be expected if the ESTs provided thorough coverage of a common Compositae transcriptome. However, the paleologs from the basal Compositae genome duplication were significantly different (P < 0.00001) from each of their tribes’ nonpaleolog fractions. In general, genes associated with structural or cellular organization GO slim categories such as ribosomes, cytosol, structural molecular activity, cytoplasmic and cellular components, and cell organization and biogenesis were overrepresented in duplicate from the basal family paleopolyploidization, whereas genes were underrepresented for regulatory or developmental categories such as transcription and transcription factors, binding, molecular functions, and DNA or RNA metabolism.
FIG. 3.—
GO annotations of Compositae whole transcriptome and paleologs. The leftmost column displays the pooled Compositae transcriptome of 18 species, whereas the remaining columns represent paleologs retained in each tribe from the basal Compositae genome duplication and the basal Heliantheae genome duplication. Colors represent percent of transcriptome a particular GO category composes. Superscripts indicate significantly different groups as determined by chi-square tests (P < 0.05). GO categories that are significantly enriched or reduced among paleologs relative to nonpaleologs in at least three comparisons are indicated with ± signs.
Discussion
Although a previous study (Blanc and Wolfe 2004b) failed to identify paleopolyploidy in their analyses of Compositae ESTs, our research uncovered strong evidence of past genome duplications. This discrepancy is most likely the result of differences in data analyses rather than data quality or quantity. Like our research, Blanc and Wolfe (2004b) used CGP data and the number of ESTs in their analyses is on par with our smallest data sets. However, advances in the statistical analyses of duplicate gene age distributions—particularly the combined use of K-S goodness of fit tests, SiZer, mixture models, and relative rate corrections—permit the identification and phylogenetic placement of genome duplications not possible from a simple visual inspection of histograms. Our analysis of six Helianthus EST data sets also provides independent validation of a paleopolyploidization at the base of the Heliantheae s. l. that was previously suggested by studies of cytology, phylogeny, and gene copy number (Smith 1975; Robinson 1981; Yahara et al. 1989; Berry et al. 1995; Gentzbittel et al. 1995; Baldwin et al. 2002). Significantly, this observation supports both our statistical approach and the interpretation that peaks in age distributions of duplicate genes, in this study and others, are indeed ancient whole-genome duplications.
Considering the size and rapid diversification of the Compositae, the potential importance of paleopolyploidy in initiating major angiosperm radiations cannot be overlooked. Although the Compositae is the largest family of flowering plants and represents nearly 10% of angiosperm diversity, the family is thought to be relatively young and likely evolved no more than 50 Ma (Funk et al. 2005). Driving some of this diversification was the Oligocene radiation of all Compositae tribes during a relatively short time frame of approximately 12 My, as estimated from analyses of chloroplast sequences (Kim et al. 2005). Our nuclear genome analyses also support a rapid tribal radiation, with the divergence of all tribes from the Mutisieae to Heliantheae diversifying during a mean Ks range of 0.12 in our 36 nuclear gene phylogenies (fig. 2). This is comparable to the amount of synonymous site divergence observed between orthologs of distantly related species of the same or sister genera in the Compositae (Barker MS, unpublished data), underscoring the brisk rate of this radiation. Intriguingly, the basal Compositae paleopolyploidization occurs near Ks = 0.75 in our nuclear gene phylogeny, a synonymous site divergence of only 0.13 from the initiation of tribal radiations. A similar pattern is observed for the Heliantheae, a lineage that accounts for 25% of the Compositae and nearly 2.5% of all angiosperms. Phylogenetic analyses (Baldwin et al. 2002) suggest that the Heliantheae paleopolyploidization is not likely shared by its small sister lineage the Arthroismeae, a pattern consistent with a polyploid-induced radiation in the Heliantheae. Though paleopolyploidy preceded these two major radiations, we cannot establish a causal relationship between paleopolyploidy and diversification of the Compositae with our present data. However, paleopolyploidy preceding bursts of diversification appears to be a recurring theme in genomic analyses (Taylor et al. 2001; Paterson et al. 2004; Aury et al. 2006; Scannell et al. 2006), and with additional data, the Compositae provides an opportunity to statistically test this apparent relationship.
Of the myriad mechanisms by which paleopolyploidy may have promoted diversification of the Compositae, the creation of novel genes via sub- and neofunctionalization is likely among the most prominent. Sub- and neofunctionalization are well-established consequences of gene duplication (Blanc and Wolfe 2004a; Adams and Wendel 2005; Aury et al. 2006; Sémon and Wolfe 2007) that may facilitate diversification through the evolution of novel phenotypes (Benderoth et al. 2006) or differential functional resolution (Causier et al. 2005). In the Compositae, there is evidence that paleopolyploidy has yielded duplicate genes associated with the evolution of the family's characteristic composite inflorescence. Chapman et al. (2008) found that the CYCLOIDEA (CYC) gene family, a family of transcription factors associated with branching and floral symmetry, has experienced a significant expansion in the Compositae. Ten members of the CYC family were recovered in Helianthus, much more than the one to five copies found in all other investigated plants. They proposed a paleopolyploid origin for some copies, and our analyses clarify their interpretation of the evolutionary history of the gene family. Based on our observation of two genome duplications in the history of Helianthus, it is likely that the 40–45 Ma duplication shared across all branches of the CYC phylogeny (Chapman et al. 2008) is derived from the basal Compositae paleopolyploidization, whereas other duplications, such as the 26–31 Ma CYC2 duplications, are probably products of the basal Heliantheae paleopolyploidization. These duplications have likely been significant in the evolution of the Compositae because Chapman et al. (2008) observed that some CYC2 copies have experienced positive selection and have their expression subfunctionalized among the disk and ray florets of the composite inflorescence. Similarly, Broholm et al. (2008) have reported numerous copies of CYC2 in Gerbera and through a series of genetic experiments demonstrated that the various copies control the specification of flower types in the inflorescence. These results suggest that some of the unique characteristics of the Compositae inflorescence are a by-product of gene and genome duplications. Given the unique floral architecture and diversity of the Compositae, both Chapman et al. (2008) and Broholm et al. (2008) suggest that the evolution of floral novelties may have played a role in the family's various radiations, and our data intimate paleopolyploidy in the evolution of this floral diversity.
Despite substantial diversification and divergence since the paleopolyploidization near the base of the Compositae, the genes retained in duplicate from the event are strikingly similar across the three tribes we examined. Our analyses demonstrate that although 33–38 My have passed since the divergence of the tribes (Kim et al. 2005), the lineages have retained the same distribution of GO categories among paleologs. Moreover, the profile of paleologs from both genome duplications in the Heliantheae are nearly identical, with only a difference in the amount of plastid-targeted genes distinguishing the more recent paleopolyploidization (fig. 3 and table S3). Parallel paleolog retention has also been observed from the single paleopolyploidization in yeast (Scannell et al. 2007) and between independent genome duplications in Arabidopsis and Cleome (Schranz and Mitchell-Olds 2006). Further, analyses in Arabidopsis have demonstrated that genes retained in duplicate from an older paleopolyploidization are likely to be retained as paleologs in subsequent genome duplications (Seoighe and Gehring 2004; Maere et al. 2005; Chapman et al. 2006).
Although the paleologs within the Compositae are largely consistent across lineages and duplications, they are not consistent with the pattern of duplicate genes retained from other ancient genome duplications in flowering plants. Paleologs in Arabidopsis (Seoighe and Gehring 2004; Blanc and Wolfe 2004a; Maere et al. 2005) and Cleome (Schranz and Mitchell-Olds 2006) are enriched for genes associated with transcription and signaling. These observations lead some authors to link paleopolyploidy with the evolution of regulatory complexity (Blanc and Wolfe 2004a; De Bodt et al. 2005; Maere et al. 2005), a perspective that is further supported by models of duplicate gene retention that are based upon the biased retention of regulatory genes (Freeling and Thomas 2006; Birchler and Veitia 2007). In contrast, Compositae paleologs are significantly enriched for genes associated with structural components or cellular organization, and regulatory and developmental genes such as transcription factors are significantly underrepresented. Analyses outside of flowering plants have also found other patterns of paleolog retention. For example, paleologs in yeast (Scannell et al. 2007) and Physcomitrella (Rensing et al. 2007) are enriched for genes associated with the GO categories ribosomal proteins, kinases, or metabolism, whereas paleologs in Paramecium (Aury et al. 2006) demonstrate no functional biases but instead have preferential retention of genes involved in macromolecular complexes.
These data suggest that mutation rates and/or patterns of intrinsic selection on different gene categories––while consistent within lineages––vary substantially among higher taxonomic categories. Considering the ecological and morphological diversity of the Compositae, it is difficult to imagine that extrinsic selection has played a large role in such uniform paleolog retention within this group. Possibly, different categories of genes differ consistently in mutation or subfunctionalization rates, leading to parallel patterns of retention. Alternatively, duplicates from some gene categories may be favored by phylogenetically conserved selection. Additional information, such as gene expression and genomic data for other tribes and related families, would provide further insight into the forces that determine the fates of duplicate genes in the Compositae and other flowering plants. Our finding of repeated taxonomic-specific patterns of duplicate gene retention demonstrates that further genomic comparisons within and among plant lineages should be fruitful for elucidating the forces that govern the evolution of gene families and genomic novelty.
Supplementary Material
Supplementary table S1–S3 and figures S1–S2 are available at Molecular Biology and Evolution online (http://mbe.oxfordjournals.org/). Unigene files for all species available at http://msbarker.com
Acknowledgments
We thank K. Dlugosch, E. Baack, H. Dempewolf, and K. Adams for comments and discussion. This research was supported by the National Science Foundation Plant Genome Program (No. 0421630) to L.H.R., R.W.M., and S.J.K., as well as National Institutes of Health Training Grant 2 T32 GM007757 to M.S.B. We thank S. Tang, Z. Lai, R. Kesseli, D. Still, and J. Boore and the Joint Genomics Institute sequencing team for contributions to the EST sequencing component of the CGP.
References
- Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005;21:539–543. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J, Jaillon O, Duret L, et al. (43 co-authors) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Baldwin BG, Wessa BL, Panero JL. Nuclear rDNA evidence for major lineages of Helenioid Heliantheae (Compositae) Syst Bot. 2002;27:161–198. [Google Scholar]
- Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J. Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci USA. 2006;103:9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry ST, Leon AJ, Hanfrey CC, Challis P, Burkholz A, Barnes SR, Rufener GK, Lee M, Caligari PDS. Molecular marker analysis of Helianthus annuus L. 2. Construction of an RFLP linkage map for cultivated sunflower. Theor Appl Genet. 1995;91:195–199. doi: 10.1007/BF00220877. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Thompson J, Gibson T. PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 1996;24:2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004a;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004b;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, Bodt SD, Simillion C, Maere S, Peer YVD. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, Ingram R, Xue Y, Schwarz-Sommer Z, Davies B. Evolution in action: following function in duplicated floral homeotic genes. Curr Biol. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Chapman BA, Bowers JE, Feltus FA, Paterson AH. Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc Natl Acad Sci USA. 2006;103:2730–2735. doi: 10.1073/pnas.0507782103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M, Leebens-Mack J, Burke J. Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol Biol Evol. 2008;25:1260–1273. doi: 10.1093/molbev/msn001. [DOI] [PubMed] [Google Scholar]
- Chaudhuri P, Marron JS. SiZer for exploration of structures in curves. J Am Stat Assoc. 1999;94:807–823. [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. (13 co-authors) Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package). Version 3.68. Distributed by the author. Seattle (WA): Department of Genome Sciences, University of Washington; 2008. Available from: http://evolution.gs.washington.edu/phylip. [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Funk V, Bayer R, Keeley S, et al. (12 co-authors) Everywhere but Antarctica: using a supertree to understand the diversity and distribution of the Compositae. Biol Skr. 2005;55:343–374. [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzbittel L, Vear F, Zhang Y, Bervillé A, Nicolas P. Development of a consensus linkage RFLP map of cultivated sunflower (Helianthus annuus L.) Theor Appl Genet. 1995;90:1079–1086. doi: 10.1007/BF00222925. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J, Noel B, et al. (56 co-authors) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Kim K-J. Implications of chloroplast DNA for the classification and phylogeny of the Asteraceae. In: Hind DJN, Beentje HJ, editors. Compositae Systematics: proceedings of the International Compositae Conference; Kew; 1994. 1996. Vol. 1. London: Royal Botanic Gardens, Kew. p. 317–339. [Google Scholar]
- Jansen RK, Michaels HJ, Palmer JD. Phylogeny and character evolution in Asteraceae based on chloroplast DNA restriction site mapping. Syst Bot. 1991;16:98–115. [Google Scholar]
- Jansen RK, Palmer JD. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae) Proc Natl Acad Sci USA. 1987;84:5818–5822. doi: 10.1073/pnas.84.16.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Palmer JD. Phylogenetic implications of chloroplast DNA restriction site variation in the Mutisieae (Asteraceae) Am J Bot. 1988;75:753–766. doi: 10.1002/j.1537-2197.1988.tb13496.x. [DOI] [PubMed] [Google Scholar]
- Kim K, Choi K, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae) Mol Biol Evol. 2005;22:1783–1792. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- Laitinen RAE, Immanen J, Auvinen P, et al. (11 co-authors) Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae) Genome Res. 2005;15:475–486. doi: 10.1101/gr.3043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. Minority cytotype exclusion in local plant populations. Taxon. 1975;24:35–43. [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- Maere S, Bodt SD, Raes J, Casneuf T, Montagu MV, Kuiper M, Peer YVD. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- Mclachlan G, Peel D, Basford K, Adams P. The EMMIX software for the fitting of mixtures of normal and t-components. J Stat Softw. 1999;4:2. [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing, reference index version 2.x.x. Vienna (Austria): R Foundation for Statistical Computing; 2005. ISBN 3-900051-07-0. Available from: http://www.R-project.org. [Google Scholar]
- Rensing S, Ick J, Fawcett J, Lang D, Zimmer A, Peer YVD, Reski R. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol Biol. 2007;7:130. doi: 10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H. A revision of the tribal and subtribal limits of the Heliantheae (Asteraceae) Smithson Contrib Bot. 1981;51:1–102. [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC. Mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–876. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17:505–512. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Smith E. The chromosome numbers of North American Coreopsis with phyletic interpretations. Bot Gaz. 1975;136:78–86. [Google Scholar]
- Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold; 1971. [Google Scholar]
- Sterck L, Rombauts S, Jansson S, Sterky F, Rouze P, Peer YVD. EST data suggest that poplar is an ancient polyploid. New Phytol. 2005;167:165–170. doi: 10.1111/j.1469-8137.2005.01378.x. [DOI] [PubMed] [Google Scholar]
- Stevens PF. Angiosperm phylogeny website. Version 9, June 2008 [Internet] 2008. Available from: http://www.mobot.org/MOBOT/research/APweb/ [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Peer YVD, Meyer A. Genome duplication, divergent resolution and speciation. Trends Genet. 2001;17:299–301. doi: 10.1016/s0168-9525(01)02318-6. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, et al. (30 co-authors) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara T, Kawahara T, Crawford DJ, Ito M, Watanabe K. Extensive gene duplications in diploid Eupatorium (Asteraceae) Am J Bot. 1989;76:1247–1253. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, et al. (117 co-authors) The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.