Abstract
Protein stability remains one of the main factors limiting the realization of the full potential of protein therapeutics. Poly(ethylene glycol) (PEG) conjugation to proteins has evolved into an important tool to overcome instability issues associated with proteins. The observed increase in thermodynamic stability of several proteins upon PEGylation has been hypothesized to arise from reduced protein structural dynamics, although experimental evidence for this hypothesis is currently missing. To test this hypothesis, the model protein α-chymotrypsin (α-CT) was covalently modified with PEGs with molecular weights (MW) of 700, 2000 and 5000 and the degree of modification was systematically varied. The procedure did not cause significant tertiary structure changes. Thermodynamic unfolding experiments revealed that PEGylation increased the thermal transition temperature (Tm) of α-CT by up to 6°C and the free energy of unfolding (ΔGU (25°C)) by up to 5 kcal/mol. The increase in stability was found to be independent of the PEG MW and it leveled off after an average of four PEG molecules were bound to α-CT. Fourier-transformed infrared (FTIR) H/D exchange experiments were conducted to characterize the conformational dynamics of the PEG-conjugates. It was found that the magnitude of thermodynamic stabilization correlates with a reduction in protein structural dynamics and was independent of the PEG MW. Thus, the initial hypothesis proved positive. Similar to the thermodynamic stabilization of proteins by covalent modification with glycans, poly(ethylene glycol) thermodynamically stabilizes α-CT by reducing protein structural dynamics. These results provide guidance for the future development of stable protein formulations.
Keywords: H/D exchange, PEGylation, protein engineering, protein thermodynamic stability, protein dynamics
Introduction
Advances in biotechnology have accelerated the discovery and development of many proteins as therapeutic agents. The increasing use of recombinant proteins by the pharmaceutical industry has highlighted the important issue of protein stability (Frokjaer and Otzen 2005). The inherent physical and chemical instabilities of proteins can result in protein conformational changes, denaturation, aggregation, precipitation, and/or adsorption to surfaces, events that cause loss of biopharmaceutical efficacy (Manning et al. 1989). Protein stability challenges can be encountered during purification, separation, storage, delivery, and administration. Several stabilizing strategies have been pursued to resolve these instability issues. These include the design of mutant proteins, the use of stabilizing excipients during formulation (e.g., sugars and polyols), and/or chemical modifications (e.g., by glycosylation or PEGylation) (Wang 1999; Wang 2000; Solá et al. 2006; Solá et al. 2007).
In recent years, several PEGylated protein products have been developed and approved by the FDA for therapeutic use in humans (Harris and Chess 2003). Approved PEGylated protein drugs include PEG-L-asparaginase (Oncaspar®), PEG-adenosine deaminase (Adagen®), PEG-interferon α2a (Pegasys®), PEG-interferon α2b (PEG-Intron®), PEG-granulocyte-colony stimulating factor (Neulasta®), and PEG-growth hormone receptor antagonist (Somavert®). Other PEG conjugated proteins are currently undergoing clinical trial, including PEG-arginine deiminase and PEG-uricase.
PEGylation refers to the attachment of one or several PEG molecules to the protein or peptide surface. PEGylation chemistry can involve several functionalities of the available amino acid side chains at the protein surface (i.e., NH2, COOH, SH, and OH). Historically, PEGylation has mostly been performed to improve protein half-life in the blood serum and to reduce immune responses. The prolonged circulating time is achieved by reducing glomerular filtration of the protein drug and by enhancing its proteolytic resistance (Abuchowski et al. 1977a; Abuchowski et al. 1977b). Reduced immunogenicity is a result of epitope shielding by the PEG molecules (Soares et al. 2002).
An additional advantage of PEGylated proteins is their increased in vitro thermodynamic stability. The increased stability is beneficial for product development and storage, where proteins can be submitted to several denaturing stresses (Cromwell et al. 2006; Perez et al. 2002). Increased melting temperatures (Tm) have been reported for several PEGylated proteins when compared to the unmodified protein. López-Cruz et al. reported an increase of 2°C in the Tm of Myceliophthora thermophila laccase upon PEG conjugation to 8 of its 14 available surface exposed lysine residues (López-Cruz et al. 2006). The Tm of recombinant human endostatin was increased from 47.6 to 62.6°C upon modification with a single PEG molecule of 5 kDa at the N-terminus (Nie et al. 2006). An increment of 2°C in the Tm was observed for human interferon-beta when it was modified with two PEG molecules of 40 kDa (Basu et al. 2006). It has been reported that after modification of α-chymotrypsin (α-CT) with PEG of 1.9 and 5 kDa the Tm is shifted to higher temperatures by as much as 5°C (Castellanos et al. 2005; Topchieva et al. 1995).
The molecular mechanism by which PEGylation causes increased thermodynamic stability remains uncertain. Several researchers have proposed that the protein structure becomes more rigid upon PEGylation, yet no experimental evidence has been provided to support this hypothesis (García-Arellano et al. 2002; López-Cruz et al. 2006; Soares et al. 2002; Yang et al. 1996). This is due to the lack of systematic studies regarding the effect of PEGylation parameters on the biophysical properties of proteins (i.e., structure, structural dynamics, and stability) which have to involve both, variation in the MW and number of PEG molecules attached to the protein. Understanding the mechanism by which PEGylation influences protein biophysical properties is vital to the development of PEG-based stabilizing strategies in the future.
In the present study, we employed a series of PEG-α-CT conjugates with varying PEGylation degree (1 to 9 PEG molecules bound to α-CT) and using PEG of different MW (700, 2000, and 5000 Da) as an experimental system to systematically study the correlation between changes in protein thermodynamic stability and structural dynamics.
Materials and Methods
Chemicals
All chemicals were of the highest purity grade available from commercial sources. Bovine α-CT was purchased from Sigma-Aldrich (St. Louis, MO). Activated linear PEGs with MW of 2 and 5 kDa were purchased from Nektar Technologies (Huntsville, AL). Methyl-PEO12-NHS ester (685.71 g/mol, PEG700) and 2, 4, 6-trinitrobenzene sulfonic acid (TNBSA) were from Pierce (Rockford, IL).
PEGylation of α-CT
Chemical modification of the surface exposed lysine amino groups of α-CT with the activated PEG was carried out as previously reported (Castellanos et al. 2005). In brief, different PEG:α-CT molar ratios were employed during synthesis to obtain PEG-α-CT conjugates with varying degrees of modification. Un-reacted PEG and buffer salts were removed by dialyzing against deionized water at 4°C. The PEG-α-CT conjugates were subsequently lyophilized. The degree of protein modification was determined by titration of the free amino groups with TNBSA (Habeeb 1966).
Circular dichroism (CD) spectroscopy
Near-UV CD spectra were measured using an Olis DSM-10 UV-Vis CD spectrophotometer. For all measurements the protein concentration was adjusted to 0.6 mg/mL in 10 mM potassium phosphate buffer (pH 7.1 and 25°C). Spectra were recorded from 260 to 310 nm using 10 mm path length quartz cells. Each spectrum was obtained by averaging 6 scans at 2 nm resolution. Blank spectra were subtracted from the protein CD spectra.
Differential Scanning Calorimetry
All thermal denaturation experiments were performed using a CSC 6100 Nano II differential scanning calorimeter (Calorimetry Sciences Corp., Lindon, UT). Protein samples were prepared at a concentration of 1 mg/ml in 10 mM potassium phosphate buffer, pH 7.1. Heat capacity thermograms were determined at a 2°C/min scan rate from 20°C to 90°C. Baselines were run with the same buffer used for the samples. Analysis of heat capacity thermograms for α-CT was carried out as previously reported (Solá et al. 2006).
Kinetics of hydrogen/deuterium (H/D) exchange
Amide H/D exchange kinetics were measured with a ThermoNicolet Nexus 470 FTIR spectrometer and the exchange kinetics analyzed as previously reported (Solá and Griebenow 2006a; Zavodszky et al. 1998). α-CT and PEG-α-CT conjugates (4 mg) were dissolved in 10 mM potassium phosphate buffer, pH 7.1, and subsequently lyophilized. The lyophilized samples were dissolved in 200 μL of D2O and immediately transferred to a FTIR cell with CaF2 windows and a 25 μm Teflon spacer. Spectra (2000 – 1000 cm−1) were collected at 2 min intervals until 10 min, 5 min intervals until 30 min, and every 30 min until 360 min. For the first 30 min, 10 scans were averaged, and after 30 min, 56 scans, all at 2 cm−1 resolution. Spectra of buffer blanks were subtracted from all sample spectra. H/D exchange spectra were processed for quantitati ve analysis in the form of hydrogen exchange decay plots (X vs. time). The fraction of unexchanged backbone hydrogen atoms at time t was determined by the following equation:
where w(t) is the ratio of the baseline corrected absorbance of amide II (1550 cm−1) to amide I (1637.5 cm−1) at time t, w(0) is the amide II/amide I ratio of the undeuterated protein and w(∞) is the amide II/amide I ratio for the fully deuterated protein.
Results
PEGylation of α-CT and characterization of the PEG-α-CT conjugates
To study the effects of the PEGylation degree and of the PEG MW on protein stability, PEGs with three different MW were employed to synthesize PEG-α-CT conjugates with varying amount of bound PEG. PEG molecules were covalently linked to α-CT surface accessible lysine residues. The PEG employed was activated with a succimidyl ester group, which is displaced by amines via nucleophilic substitution. By controlling the PEG-to-α-CT molar ratio during the reaction, it was possible to prepare PEG-α-CT conjugates with up to 9 bound PEG molecules. α-CT has 14 surface reactive lysine residues. The degree of modification was determined by measuring the amount of unreacted amino groups using the trinitrobenzene sulfonic acid assay (Figure 1) (Habeeb 1966).
Figure 1.
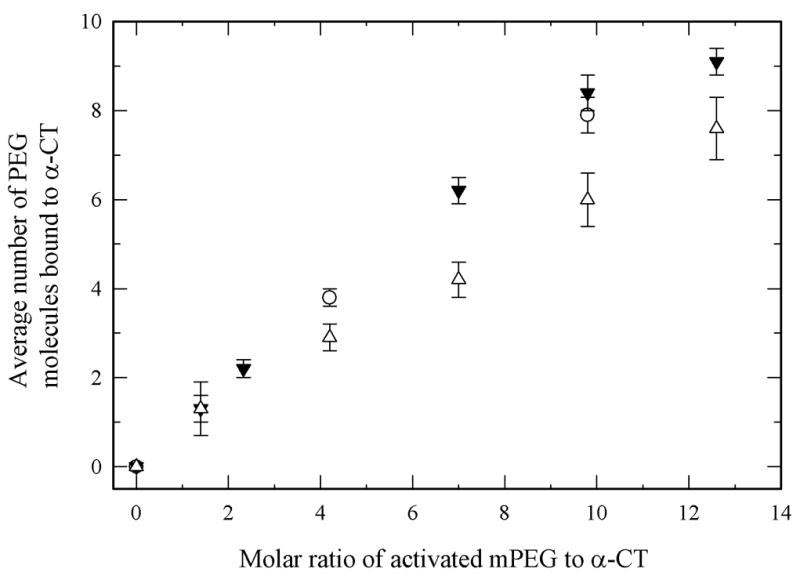
Degree of PEG-modification of α-CT accomplished during synthesis using various amounts of activated PEG with a molecular weight of 700 Da (open circles), 2000 Da (solid down triangles), and 5000 Da (open triangles).
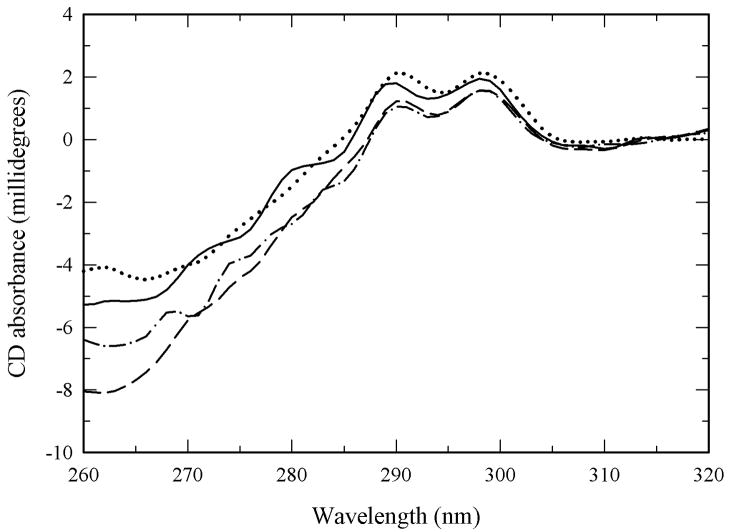
To assess the effect of PEGylation on the protein tertiary structure, the PEG-α-CT conjugates were characterized by near-UV CD spectroscopy. Previous work showed that perturbations of α-CT tertiary structure cause a significant decrease in the spectral intensity in the near-UV CD spectra. The near-UV CD spectrum of non-modified α-CT displays two maxima at 288 and 297 nm, which completely disappear upon thermal unfolding (Castellanos et al. 2005). No major spectral changes are observed as a result of PEGylation, which rules out large perturbations to the tertiary structure of the bioconjugates. It was found that neither the molecular weight of the PEG nor the degree of PEGylation adversely impacted structural integrity of α-CT (Figure 2). These results agree with previously published work which revealed no significant changes in secondary and tertiary structure of α-CT after PEGylation (Castellanos et al. 2005; Castillo et al. 2006).
Figure 2.
Near UV-CD spectra of α-CT (solid line) and PEG-α-CT conjugates: (PEG700)8-α-CT (dashed line), (PEG2000)6-α-CT (dash-dot), and (PEG5000)8-α-CT (dotted line).
Effect of PEGylation on protein thermodynamic stability
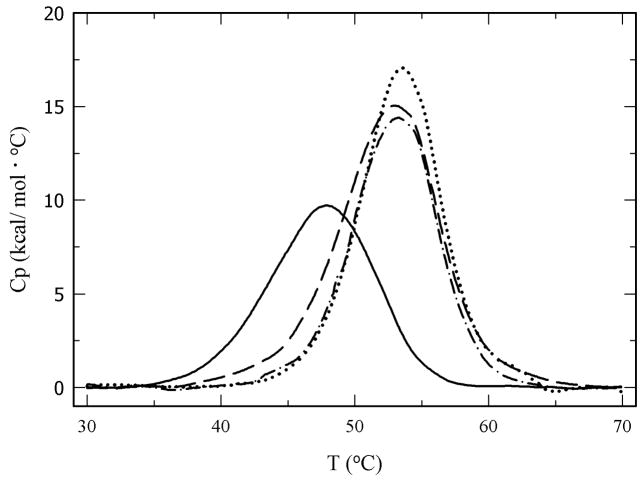
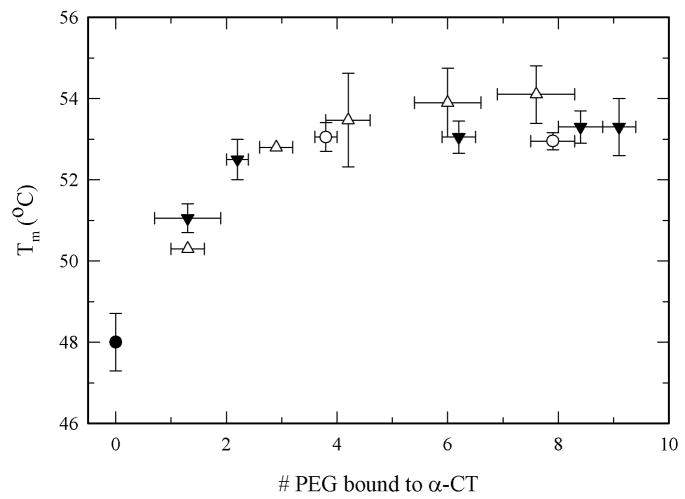
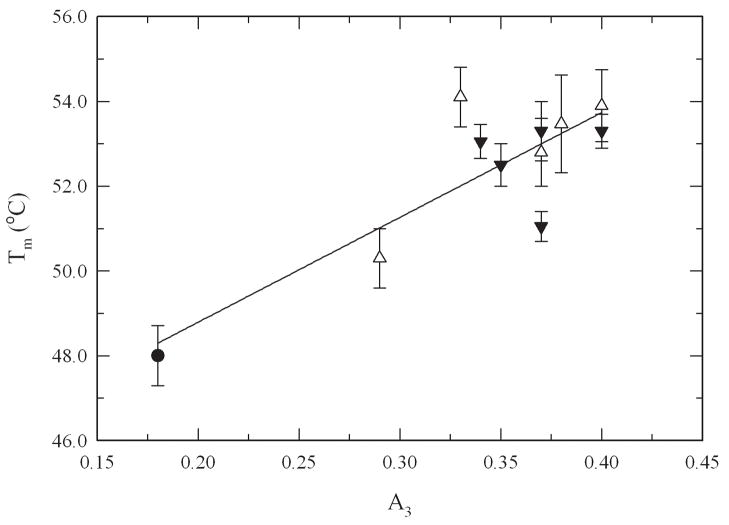
To determine the role of the extent of PEGylation and PEG MW on protein thermodynamic stability, thermal unfolding experiments using differential scanning calorimetry (DSC) were carried out. Figure 3 shows the thermograms for unmodified α-CT and for three PEG-α-CT conjugates with PEGs of 700, 2000, and 5000 Da. Unmodified α-CT displays a two-state thermally induced unfolding transition with the denaturation midpoint (Tm) at 48.0°C. Modification of α-CT with PEG resulted in a shift in the endothermic denaturation transition to higher temperatures. This resulted in increased Tm values for the PEG-α-CT conjugates at increasing PEG molar content until a plateau was reached (Figure 4). The effect of the PEGylation degree on the Tm levels off at around 53.0°C after the 4th PEG molecule is bound to the protein. A maximum increase of 6°C in the Tm was achieved. Analysis of the denaturation endotherms permitted the calculation of the enthalpy (ΔHm) and heat capacity changes (ΔCp) associated with the unfolding event (Table I). These values allowed the calculation of the Gibbs free energy of unfolding at 25°C (ΔGU(25°C)) by using the Gibbs-Helmholtz equation (ΔGU(T)= ΔHm(1 − T/Tm) − ΔCp[(Tm − T) + Tln(T/Tm)]) (Pace 1990; Solá et al. 2006). Similar to the Tm, the free energy of protein unfolding (ΔGU(25°C)) increased for the PEGylated proteins. Stabilization of up to 5 kcal/mol was achieved for the most modified PEG-protein conjugates. Interestingly, it was observed that the MW of the PEG did not influence the magnitude of thermodynamic stabilization of the protein. Similar to the stabilization effects of protein glycosylation, stabilization by PEGylation was solely dependent on the amount of PEG bound to the protein.
Figure 3.
Thermograms for α-CT and PEG-α-CT conjugates obtained by differential scanning calorimetry: (PEG700)8-α-CT (dashed line), (PEG2000)6-α-CT (dash-dot), and (PEG5000)8-α-CT (dotted line).
Figure 4.
Effect of PEG-size and PEGylation degree on the Tm of α-CT and PEG-α-CT conjugates: α-CT (solid circle), PEG 700 Da (open circles), PEG 2000 (solid triangles) and PEG 5000 Da (open triangles).
Table I.
Thermodynamic analysis of the differential scanning calorimetry data.
| Sample/Degree of PEGylationa | Tm (°C) | ΔHm (kcal/mol) | ΔCp (kcal/mol K) | ΔGu(25 °C) (kcal/mol) |
|---|---|---|---|---|
| α-CT | 48.0 ± 0.8 | 120 ± 18 | 3.2 ± 0.2 | 6 ± 1 |
| PEG700-α-CT | ||||
| 3.8 ± 0.2 | 53.1 ± 0.4 | 128 ± 18 | 1.9 ± 0.5 | 9 ± 1 |
| 7.9 ± 0.4 | 53.0 ± 0.2 | 141 ± 4 | 2.6 ± 0.6 | 8.9 ± 0.8 |
| PEG2000-α-CT | ||||
| 1.3 ± 0.6 | 51.1 ± 0.4 | 144 ± 4 | 1 ± 1 | 10.3 ± 0.8 |
| 2.2 ± 0.2 | 52.5 ± 0.8 | 149 ± 8 | 1.5 ± 0.5 | 10.7 ± 0.5 |
| 6.2 ± 0.3 | 53.1 ± 0.8 | 126 ± 2 | 0.13 ± 0.05 | 10.0 ± 0.5 |
| 8.4 ± 0.4 | 53.3 ± 0.5 | 123 ± 4 | 1.8 ± 0.5 | 8.3 ± 0.4 |
| 9.1 ± 0.3 | 53.3 ± 0.5 | 118 ± 5 | 1.2 ± 0.7 | 9 ± 1 |
| PEG5000-α-CT | ||||
| 1.3 ± 0.3 | 50.3 ± 0.7 | 128 ± 2 | 4 ± 1 | 5.9 ± 0.9 |
| 2.9 ± 0.3 | 52.8 ± 0.8 | 127 ± 8 | 3 ± 2 | 7.1 ± 0.2 |
| 4.2 ± 0.4 | 53 ± 1 | 113 ± 25 | 4 ± 2 | 6.1 ± 0.8 |
| 6.0 ± 0.6 | 53.9 ± 0.8 | 119 ± 22 | 0.2 ± 0.7 | 9.1 ± 0.1 |
| 7.6 ± 0.7 | 54.1 ± 0.7 | 120 ± 22 | −0.3 ± 2 | 11 ± 2 |
In mol of PEG/mol of α-chymotrypsin.
Effects of PEGylation on structural dynamics
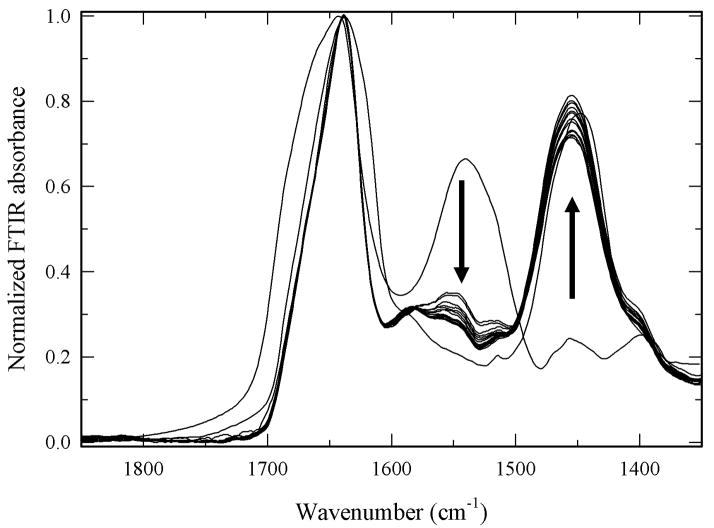
Hydrogen/deuterium exchange is a versatile method for obtaining information on the structural dynamics of macromolecular systems (Ferraro and Robertson 2004; Hvidt and Nielsen 1966; Privalov and Tsalkova 1979). Measuring the time course of H/D exchange with FTIR allows to asses changes in global protein structural dynamics (Li et al. 2002; Zavodszky et al. 1998). H/D exchange experiments were carried out following the protocol previously reported (Solá and Griebenow 2006a). Figure 5 shows the spectroscopic results from a typical FTIR H/D exchange experiment for α-CT including both, the spectra of the undeuterated and completely deuterated protein. H/D exchange kinetics were determined by following the decrease in the absorbance of the amide II band (N–H, 1500–1600 cm−1) relative to the amide I band (C=O, 1600–1700 cm−1) (Hvidt and Nielsen 1966). The exchange data were plotted in the form of decay plots, where the fraction of unexchanged amide hydrogen atoms (X) decreases over time due to the H/D exchange process (for details see Materials and Methods section). Quantitative analysis of the decay plots was carried out by fitting a two-exponential decay model of the following form:
where A1, A2, and A3 are the fractions of the fast, slow, non-exchanging amide protons and kHX,1 and kHX,2 are the apparent exchange rate constants for the fast and slow amide protons. Under experimental conditions, A3 refers to the unexchanged fraction of amide protons, which is an indicator of structural dynamics; reduction of protein structural dynamics results in an increase in unexchanged amide protons (Castillo et al. 2008).
Figure 5.
Measurement of amide H/D exchange by FTIR spectroscopy. Results from a typical H/D exchange experiment for α-CT (pD 7.1 at 25°C). The arrows highlight both the decreasing amide II band (N–H; 1550 cm−1) and the increasing amide II′ band (N-D; 1450 cm−1).
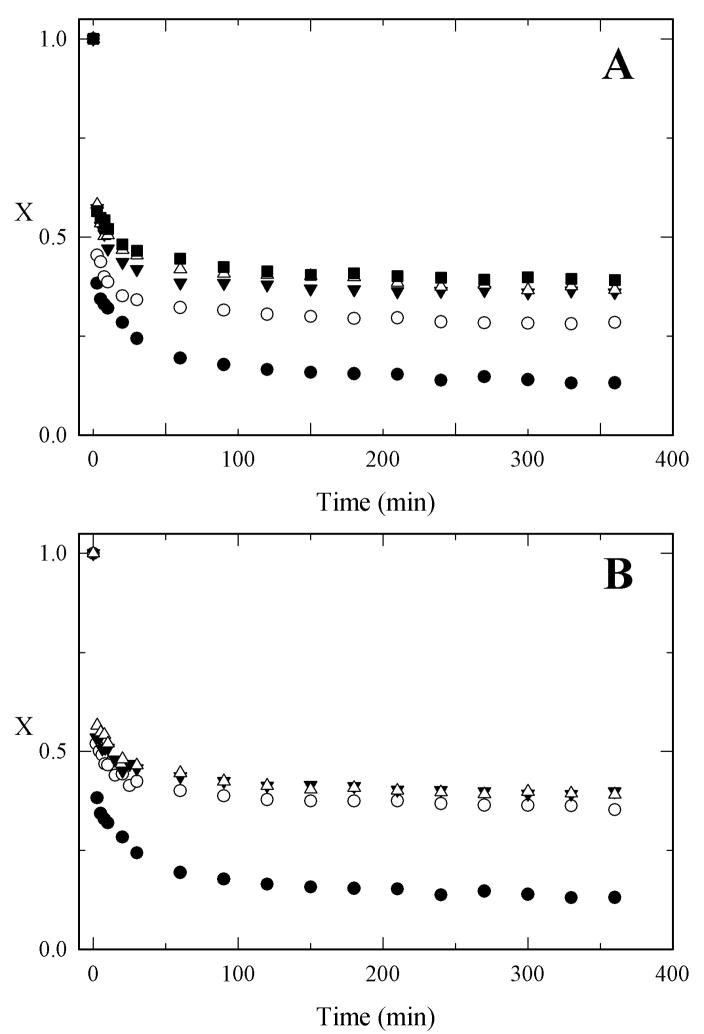
In figure 6A, the H/D exchange decay plots for the PEG5000-α-CT conjugates with varying amount of bound PEG are presented. As the amount of PEG per mol of enzyme is increased, both the rate constant and the fraction of fast exchanging amide hydrogens are reduced. Conversely, there is an increase in the fraction of non-exchanging amide hydrogens (A3) (Figure 6, Table II). This observation is indicative of a more rigid protein core as a result of PEGylation. The structural dynamics of the protein become reduced as the PEGylation degree increases until it levels off after the 4th PEG molecule is attached. The effect of the PEG MW on H/D exchange rates is presented in figure 6B for PEG-α-CT conjugates with comparable degrees of PEGylation (50% of modified lysines). Similar to the effects on thermodynamic stability, the MW of the PEG chain is not a significant factor in reducing the protein structural. Therefore, the reduction in protein structural dynamics only depended on the degree of modification, not the molecular weight of the PEG.
Figure 6.
H/D exchange decay plots for α-CT and PEG-α-CT conjugates. (A) Effect of PEGylation degree on H/D exchange rates of α-CT (solid circle), (PEG5000)1.3-α-CT (open circles), (PEG5000)2.9-α-CT (solid down triangles), (PEG5000)4.2-α-CT (open triangles), and (PEG5000)6.0-α-CT (solid squares). (B) Effect of the PEG size on H/D exchange rates for α-CT (solid circles), (PEG700)8-α-CT (open circles), (PEG2000)6-α-CT (solid down triangles), and (PEG5000)8-α-CT (open triangles).
Table II.
Kinetic parameters derived from amide H/D exchange kinetic analysis for α-CT and PEG-α-CT conjugates.
| Sample/Degree of PEGylation | A1a | kHX,1 (min−1) | A2a | kHX,2 (min−1) | A3b |
|---|---|---|---|---|---|
| α-CT | 0.64 | 1.66 | 0.14 | 0.040 | 0.22 |
| PEG700-α-CT | |||||
| 3.8 ± 0.2 | 0.66 | 1.35 | 0.12 | 0.013 | 0.22 |
| 7.9 ± 0.4 | 0.49 | 1.56 | 0.14 | 0.034 | 0.37 |
| PEG2000-α-CT | |||||
| 1.3 ± 0.6 | 0.53 | 1.61 | 0.11 | 0.025 | 0.37 |
| 2.2 ± 0.2 | 0.56 | 0.83 | 0.09 | 0.025 | 0.35 |
| 4.2 ± 0.3 | 0.55 | 0.66 | 0.12 | 0.008 | 0.33 |
| 8.4 ± 0.4 | 0.47 | 1.96 | 0.13 | 0.030 | 0.40 |
| 9.1 ± 0.3 | 0.51 | 2.04 | 0.12 | 0.029 | 0.37 |
| PEG5000-α-CT | |||||
| 1.3 ± 0.3 | 0.57 | 1.14 | 0.14 | 0.033 | 0.29 |
| 2.9 ± 0.3 | 0.45 | 0.97 | 0.19 | 0.046 | 0.37 |
| 4.2 ± 0.4 | 0.48 | 0.77 | 0.14 | 0.018 | 0.38 |
| 6.0 ± 0.6 | 0.44 | 1.31 | 0.16 | 0.025 | 0.40 |
| 7.6 ± 0.7 | 0.50 | 0.84 | 0.16 | 0.023 | 0.33 |
Ai is the fraction of amide protons that exchange with a rate constant kHX,i.
A3 is the unexchanged fraction of amide protons.
Discussion
In recent years, protein PEGylation has become a widely employed technology to improve the in vivo lifetime of protein and peptide drugs. Accordingly, the number of PEG-protein conjugates approved by the FDA and the amount of research articles on the topic are steadily increasing (Veronese and Pasut 2005). A benefit of protein PEGylation is the increase in protein thermodynamic stability which can result in a more robust protein product that can better withstand the denaturing conditions encountered in product formulation and has a longer shelf-life.
The primary goal of this work was to determine the mechanism by which PEGylation increases the thermodynamic stability of proteins in order to advance the understanding of protein biophysics and protein stabilization. Our results show that the magnitude of stabilization increased proportionally with the degree of modification before leveling off (Figure 4). There is no significant difference between the stabilization effects of PEG of different MW (Table 1). This is important since PEG polymers of less than 1 kDa can be synthesized with uniform molecular weight, thus avoiding polydispersity issues. The in vivo implications of this finding, however, will have still to be established.
To account for the observed stabilization effect of PEGylation, several researchers proposed that chemical modification causes reduced protein structural dynamics. By measuring H/D exchange for the different PEG-α-CT conjugates, we demonstrate that after PEGylation the kinetics of H/D exchange and extent of hydrogen exchange are indeed reduced (Figure 6). This observation demonstrates a reduction in protein structural dynamics and a more rigid protein core. The increase in protein thermodynamic stability correlates with this reduction in protein structural dynamics.
A group of authors has suggested that the reduction of protein dynamics upon PEG modification might be a consequence of the amphiphilic nature of PEG. Water molecules solvate the hydrophilic regions of bound PEG, while the hydrophobic regions interact with hydrophobic clusters on the protein surface. The net effect of this is the removal of water molecules from the surface of the protein to produce a more rigid protein structure (García-Arellano et al. 2002; Longo and Combes 1999; Solá and Griebenow 2006b). Computer models suggest that the PEG chains fold upon themselves and form loosely folded domains on the surface of proteins, possibly excluding water from the protein surface (Manjula et al. 2003). Using small-angle X-ray scattering, Svergun et al solved the first solution structure for a PEG-protein conjugate (Svergun et al. 2008). This low resolution structure of PEGylated hemoglobin shows that part of the PEG polymer sits on the surface of the protein and the other part extends away of the protein. Their structure also revealed a more compact and rigid protein and partial dehydration of the protein surface.
Previous investigations have correlated reduced protein dynamics with an increase in protein thermodynamic stability. Solá and Griebenow reported increased thermodynamic stability of α-CT upon chemical glycosylation (Solá and Griebenow 2006a). Therein, the increased thermodynamic stability by glycosylation correlated with a reduction in the protein dynamics. Reduced protein structural dynamics have been proposed as the main factor accounting for the high thermostability of proteins of extremophiles. For example, adenylate kinase from the thermophilic archaeon Sulfolobus acidocaldarius posses a more rigid and compact structure than its homologue from porcine muscle cytosol (Bonisch et al. 1996). Measuring H/D exchange with FTIR, Petsko showed that the more stable thermophilic enzyme, 3-isopropylmalate dehydrogenase (IPMDH) from Thermus thermophilus HB8, has reduced conformational flexibility when compared to its homologous IPMDH from Escherichia coli (Zavodszky et al. 1998). Stabilization of proteins by ligand binding also correlates with a loss in structural dynamics. For example, the Tm of bovine serum albumin increased from 59.0°C to 79.8°C when bound to anilinonaphthalene sulphonate, which was attributed to a reduction in protein dynamics (Celej et al. 2003). On the other hand, a chemical modification that increased protein dynamics, as measured by H/D exchange, lead to the destabilization of cytochrome c of around 4 kcal/mol (de Jongh et al. 1995).
In this report we show that the increase in protein thermodynamic stability was accompanied by a reduction of protein structural dynamics. PEGylation is currently a well established technique for improving the pharmacokinetics and pharmacodynamics of protein drugs because of its in vivo stabilization effects. It can also be used to improve protein products shelf-life, a current limitation of protein pharmaceuticals (Wang 1999; Wang 2000). Understanding the mechanism by which proteins become stabilized by chemical modifications will greatly benefit biopharmaceutical products since protein drugs can potentially be modified with infinitely diverse chemical structures, whose limit is solely dictated by the chemist inventiveness.
Figure 7.
Correlation between α-CT’s structural dynamics and its thermodynamic stability. α-CT (solid circle), PEG 2000 (solid down triangles) and PEG 5000 Da (open triangles). The linear regression was performed with the program Sigma Plot (R2 = 0.71; p < 0.01).
Acknowledgments
This publication was made possible by grant number S06 GM08102 from the National Institute for General Medical Sciences (NIGMS) at the National Institutes of Health (NIH) through the Support of Competitive Research (SCORE) Program. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS. JARM was supported by a fellowship from the NIH Research Initiative for Scientific Enhancement (RISE) Program (contract number R25 GM061151) and by the Fellowship Program of the Puerto Rico Development Company (PRIDCO).
References
- Abuchowski A, McCoy JR, Palczuk NC, van Es T, Davis FF. Effect of covalent attachment of polyethylene glycol on immunogenicity and circulating life of bovine liver catalase. J Biol Chem. 1977a;252(11):3582–3586. [PubMed] [Google Scholar]
- Abuchowski A, van Es T, Palczuk NC, Davis FF. Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J Biol Chem. 1977b;252(11):3578–3581. [PubMed] [Google Scholar]
- Basu A, Yang K, Wang ML, Liu S, Chintala R, Palm T, Zhao H, Peng P, Wu DC, Zhang ZF, et al. Structure-function engineering of interferon-beta-1b for improving stability, solubility, potency, immunogenicity, and pharmacokinetic properties by site-selective mono-PEGylation. Bioconjugate Chem. 2006;17(3):618–630. doi: 10.1021/bc050322y. [DOI] [PubMed] [Google Scholar]
- Bonisch H, Backmann J, Kath T, Naumann D, Schafer G. Adenylate kinase from Sulfolobus acidocaldarius: Expression in Escherichia coli and characterization by Fourier Transform Infrared Spectroscopy. Arch Biochem Biophys. 1996;333(1):75–84. doi: 10.1006/abbi.1996.0366. [DOI] [PubMed] [Google Scholar]
- Castellanos IJ, Al-Azzam W, Griebenow K. Effect of the covalent modification with poly(ethylene glycol) on alpha-chymotrypsin stability upon encapsulation in poly(lactic-co-glycolic) microspheres. J Pharm Sci. 2005;94(2):327–340. doi: 10.1002/jps.20243. [DOI] [PubMed] [Google Scholar]
- Castillo B, Mendez J, Al-Azzam W, Barletta G, Griebenow K. On the relationship between the activity and structure of PEG-α-chymotrypsin conjugates in organic solvents. Biotech Bioeng. 2006;94(3):565–574. doi: 10.1002/bit.20863. [DOI] [PubMed] [Google Scholar]
- Castillo B, Solá RJ, Ferrer A, Barletta G, Griebenow K. Effect of PEG modification on subtilisin Carlsberg activity, enantioselectivity, and structural dynamics in 1,4-dioxane. Biotech Bioeng. 2008;99(1):9–17. doi: 10.1002/bit.21510. [DOI] [PubMed] [Google Scholar]
- Celej MS, Montich GG, Fidelio GD. Protein stability induced by ligand binding correlates with changes in protein flexibility. Protein Sci. 2003;12(7):1496–1506. doi: 10.1110/ps.0240003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell MEM, Hilario E, Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8(3):E572–E579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJongh HHJ, Goormaghtigh E, Ruysschaert JM. Tertiary Stability of Native And Methionine-80 Modified Cytochrome-C Detected By Proton Deuterium-Exchange Using Online Fourier-Transform Infrared Spectroscopy. Biochemistry. 1995;34(1):172–179. doi: 10.1021/bi00001a021. [DOI] [PubMed] [Google Scholar]
- Ferraro DM, Robertson AD. EX1 hydrogen exchange and protein folding. Biochemistry. 2004;43(3):587–594. doi: 10.1021/bi035943y. [DOI] [PubMed] [Google Scholar]
- Frokjaer S, Otzen DE. Protein drug stability: A formulation challenge. Nat Rev Drug Discovery. 2005;4(4):298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- García-Arellano H, Valderrama B, Saab-Rincon G, Vázquez-Duhalt R. High temperature biocatalysis by chemically modified cytochrome c. Bioconjugate Chem. 2002;13(6):1336–1344. doi: 10.1021/bc025561p. [DOI] [PubMed] [Google Scholar]
- Habeeb AFSA. Determination of free amino groups in protein by trinitrobenzene sulfonic acid. Anal Biochem. 1966;14:328–336. doi: 10.1016/0003-2697(66)90275-2. [DOI] [PubMed] [Google Scholar]
- Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discovery. 2003;2(3):214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Prot Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- Li J, Cheng X, Lee JC. Structure and Dynamics of the Modular Halves of Escherichia coli Cyclic AMP Receptor Protein. Biochemistry. 2002;41(50):14771–14778. doi: 10.1021/bi026383q. [DOI] [PubMed] [Google Scholar]
- Longo MA, Combes D. Thermostability of modified enzymes: a detailed study. J Chem Technol Biotechnol. 1999;74(1):25–32. [Google Scholar]
- López-Cruz JI, Viniegra-Gonzalez G, Hernandez-Arana A. Thermostability of native and pegylated Myceliophthora thermophila laccase in aqueous and mixed solvents. Bioconjugate Chem. 2006;17(4):1093–1098. doi: 10.1021/bc0503465. [DOI] [PubMed] [Google Scholar]
- Manjula BN, Tsai S, Upadhya R, Perumalsamy K, Smith PK, Malavalli A, Vandegriff K, Winslow RM, Intaglietta M, Prabhakaran M, et al. Site-specific PEGylation of hemoglobin at cys-93(beta): Correlation between the colligative properties of the PEGylated protein and the length of the conjugated PEG chain. Bioconjugate Chem. 2003;14(2):464–472. doi: 10.1021/bc0200733. [DOI] [PubMed] [Google Scholar]
- Manning MC, Patel K, Borchardt RT. Stability of Protein Pharmaceuticals. Pharm Res. 1989;6(11):903. doi: 10.1023/a:1015929109894. [DOI] [PubMed] [Google Scholar]
- Nie YJ, Zhang X, Wang XC, Chen JH. Preparation and stability of N-terminal mono-PEGylated recombinant human endostatin. Bioconjugate Chem. 2006;17(4):995–999. doi: 10.1021/bc050355d. [DOI] [PubMed] [Google Scholar]
- Pace CN. Measuring and increasing protein stability. Trends Biotechnol. 1990;8(4):93–8. doi: 10.1016/0167-7799(90)90146-o. [DOI] [PubMed] [Google Scholar]
- Perez C, Castellanos IJ, Costantino HR, Al-Azzam W, Griebenow K. Recent trends in stabilizing protein structure upon encapsulation and release from bioerodible polymers. J Pharm Pharmacol. 2002;54(3):301–313. doi: 10.1211/0022357021778448. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Tsalkova TN. Micro- and macro-stabilities of globular proteins. Nature. 1979;280(23):693–696. doi: 10.1038/280693a0. [DOI] [PubMed] [Google Scholar]
- Soares AL, Guirmaraes GM, Polakiewicz B, Pitombo RND, Abrahao-Neto J. Effects of polyethylene glycol attachment on physicochemical and biological stability of E. coli L-asparaginase. Int J Pharm. 2002;237(1–2):163–170. doi: 10.1016/s0378-5173(02)00046-7. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Al-Azzam W, Griebenow K. Engineering of protein thermodynamic, kinetic, and colloidal stability: Chemical glycosylation with monofunctionally activated glycans. Biotech Bioeng. 2006;94(6):1072–1079. doi: 10.1002/bit.20933. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Griebenow K. Chemical glycosylation: New insights on the interrelation between protein structural mobility, thermodynamic stability, and catalysis. FEBS Lett. 2006a;580:1685–1690. doi: 10.1016/j.febslet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Griebenow K. Influence of modulated structural dynamics on the kinetics of alpha-chymotrypsin catalysis - Insights through chemical glycosylation, molecular dynamics and domain motion analysis. FEBS J. 2006b;273(23):5303–5319. doi: 10.1111/j.1742-4658.2006.05524.x. [DOI] [PubMed] [Google Scholar]
- Solá RJ, Rodríguez-Martínez JA, Griebenow K. Modulation of protein biophysical properties by chemical glycosylation: biochemical insights and biomedical implications. Cell Mol Life Sci. 2007;64(16):2133–2152. doi: 10.1007/s00018-007-6551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Ekstrom F, Vandegriff KD, Malavalli A, Baker DA, Nilsson C, Winslow RM. Solution structure of poly(ethylene) glycol-conjugated hemoglobin revealed by small-angle x-ray scattering: Implications for a new oxygen therapeutic. Biophys J. 2008;94(1):173–181. doi: 10.1529/biophysj.107.114314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topchieva IN, Efremova NV, Khvorov NV, Magretova NN. Synthesis and Physicochemical Properties of Protein Conjugates with Water-Soluble Poly(Alkylene Oxides) Bioconjugate Chem. 1995;6(4):380–388. doi: 10.1021/bc00034a007. [DOI] [PubMed] [Google Scholar]
- Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discovery Today. 2005;10(21–24):1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- Wang W. Instability, stabilization, and formulation of liquid protein pharmaceuticals. Int J Pharm. 1999;185(2):129–188. doi: 10.1016/s0378-5173(99)00152-0. [DOI] [PubMed] [Google Scholar]
- Wang W. Lyophilization and development of solid protein pharmaceuticals. Int J Pharm. 2000;203(1–2):1–60. doi: 10.1016/s0378-5173(00)00423-3. [DOI] [PubMed] [Google Scholar]
- Yang Z, Domach M, Auger R, Yang FX, Russell AJ. Polyethylene glycol-induced stabilization of subtilisin. Enzyme Microb Technol. 1996;18(2):82–89. [Google Scholar]
- Zavodszky P, Kardos J, Svingor A, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA. 1998;95(13):7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]