Abstract
Analysis of suppressors that alleviate the acute envelope stress phenotype of a ΔbamBΔdegP strain of Escherichia coli identified a novel protein MzrA and pleiotropic envZ mutations. Genetic evidence shows that overexpression of MzrA – formerly known as YqjB and EcfM – modulates the activity of EnvZ/OmpR similarly to pleiotropic EnvZ mutants and alter porin expression. However, porin expression in strains devoid of MzrA or overexpressing it is still sensitive to medium osmolarity, pH and procaine, all of which modulate EnvZ/OmpR activities. Thus, MzrA appears to alter the output of the EnvZ/OmpR system but not its ability to receive and respond to various environmental signals. Localization and topology experiments indicate that MzrA is a type II membrane protein, with its N-terminus exposed in the cytoplasm and C-terminus in the periplasm. Bacterial two-hybrid experiments determined that MzrA specifically interacts with EnvZ but not with OmpR or the related membrane sensor kinase, CpxA. This and additional genetic and biochemical evidence suggest that the interaction of MzrA with EnvZ would either enhance EnvZ's kinase activity or reduce its phosphatase activity, thus elevating the steady state levels of OmpR∼P. Furthermore, our data show that MzrA links the two-component envelope stress response regulators, CpxA/CpxR and EnvZ/OmpR.
Introduction
The outer membrane of Gram-negative bacteria is the first defence barrier between the often hostile milieu in which the bacteria live and their intracellular contents. Consequently, integrity of the outer membrane is of utmost importance for bacterial survival and pathogenesis. A unique glycolipid, lipopolysaccharide, coats the outer leaflet of the outer membrane, while phospholipids are partitioned exclusively in the inner leaflet of the outer membrane (Nikaido and Vaara, 1985). The outer membrane is also home to β-barrel-forming outer membrane proteins (OMPs) as well as lipoproteins (Nikaido and Vaara, 1985).
The regulation of β-barrel OMP synthesis has been extensively studied. In particular, the EnvZ/OmpR-mediated regulation of OmpF and OmpC, the two major pore-forming OMPs, has been a paradigm for understanding gene regulation by two-component signal transduction systems (for reviews, see Hoch and Silhavy, 1995). EnvZ is a membrane-bound sensor kinase, although the signal it senses is not known. OmpR is a cytosolic response regulator, which binds to the promoter region of the porin genes. Changes in medium osmolarity profoundly affect OmpF and OmpC expression: OmpC is preferentially expressed in high osmolarity, whereas OmpF expression is favoured in low osmolarity (van Alphen and Lugtenberg, 1977). Although the level of OmpR∼P in vivo has not been determined, it is presumed that the [OmpR∼P] increases with increasing osmolarity, promoting activation and then repression of ompF and activation of ompC. Other factors, including local anaesthetics (Villarejo and Case, 1984), pH (Heyde and Portalier, 1987; Sato et al., 2000) and nutrition limitation (Liu and Ferenci, 1998), also influence ompF and ompC transcription in an EnvZ/OmpR-dependent manner.
In recent years, biogenesis of OMPs has received renewed attention owing to the discovery of the β-barrel OMP assembly machine (Bam), comprised of four lipoproteins (BamBCDE) and a β-barrel OMP, BamA (Wu et al., 2005; Misra, 2007; Sklar et al., 2007). In the event of OMP misassembly or other envelope aberrations, cells invoke regulatory responses to minimize the envelope stress. Two principal regulators that control these responses are a specialized sigma factor σE (Alba and Gross, 2004) and the CpxA/CpxR two-component system (Raivio and Silhavy, 2001). An inner membrane protein, RseA, normally sequesters σE but misassembly of OMPs triggers two-step proteolysis of RseA, releasing σE. Free σE activates transcription of factors involved in OMP biogenesis, including periplasmic foldases SurA, Skp; a periplasmic foldase/protease, DegP; members of the Bam complex; genes involved in lipopolysaccharide biogenesis; and small RNAs that inhibit OMP synthesis (Rhodius et al., 2006). Thus genes activated by the σE regulon help maintain a homoeostasis between OMP synthesis and OMP assembly. In contrast to the activation of σE, a diverse group of envelope stresses appear to activate the CpxA/CpxR system (DiGiuseppe and Silhavy, 2003). Once activated, the CpxA/CpxR system induces the synthesis of periplasmic folding/degradation factors, including DsbA, PpiA and DegP, indicating that regardless of the source of envelope aberration, the ultimate targets of the CpxA/CpxR system include misfolded envelope proteins.
The robustness of the envelope homeostasis loop can be compromised if cells lack one or more OMP folding and/or degradation factors (Rizzitello et al., 2001; Onufryk et al., 2005). An Escherichia coli mutant simultaneously lacking DegP and BamB displays a conditional lethal (temperature-sensitive) phenotype, i.e. unable to grow at 37°C, but forms small but homogeneous colonies at 30°C (Charlson et al., 2006). The absence of BamB also causes antibiotic hypersensitivity to both hydrophobic (e.g. rifampin) and hydrophilic (e.g. vancomycin) antibiotics (Eggert et al., 2001; Vuong et al., 2008). In the present study, the temperature and antibiotic sensitivity phenotypes of ΔbamBΔdegP were exploited to isolate multicopy and mutational suppressors.
Isolation of multicopy suppressors identified a novel gene, mzrA (yqjB), while mutational suppressors identified pleiotropic envZ mutations. These mutations affect the expression of genes that are archetypal members of the EnvZ/OmpR regulon, such as ompF and ompC, and also influence the expression of genes, including lamB and malE, normally not controlled by the EnvZ/OmpR regulon (Wandersman et al., 1980; Case et al., 1986). Characterization of these suppressors showed that overexpression of mzrA (yqjB) modulates the EnvZ/OmpR two-component signal transduction regulon in a manner similar to the envZ suppressor mutations. The discovery of this property of yqjB led to its new designation, mzrA–modulator of EnvZ and OmpRA. The phenotypic convergence of the two classes of suppressors suggests that they work through a similar mechanism to reduce envelope stress; in part, this entails reduction in OMP synthesis. Consistent with this view, Guillier and Gottesman (2006) previously showed that multicopy plasmid clones carrying exuT′exuRyqjABC′ activate omrA and omrB via EnvZ/OmpR.
Results
Overexpression of mzrA (yqjB) mitigates envelope stress
To identify potentially new genes involved in the response to envelope stress, we introduced a random plasmid gene library, prepared from the MC4100 chromosome, into the ΔbamBΔdegP strain and sought transformants that improved growth at 37°C. DNA sequence analysis of the insert DNA from four positive clones showed that three contained the intact yfgM-bamB genes while one clone carried a contiguous chromosomal region containing exuT′, exuR, yqjA, mzrA (yqjB), yqjC, yqjD and yqjE′ genes. We focused on the exuT-yqj clone because the yfgM-bamB clone most likely improved growth by complementing the ΔbamB mutation of the recipient strain. Even though the exuT-yqj plasmid was isolated among clones that improved growth of the ΔbamBΔdegP strain at 37°C, transformants purified at 37°C formed small colonies. At 30°C, they formed colonies that were much larger than those formed by the parental ΔbamBΔdegP strain. Subsequent subcloning experiments narrowed the gene responsible for improved growth of the ΔbamBΔdegP strain to mzrA (yqjB).
To gain insight into the mechanism by which overexpression of MzrA (YqjB) improves the growth of the ΔbamBΔdegP strain, we first examined the OMP profile. We have previously shown that reduced OMP levels partially suppress the temperature-sensitive phenotype of ΔbamBΔdegP (Charlson et al., 2006). The presence of the original exuT-yqj clone in the ΔbamBΔdegP strain showed a marked reduction of OmpF from cell envelopes (Fig. 1A). In the subsequent experiment, we used a plasmid clone expressing MzrA (YqjB) from an IPTG-inducible promoter. Overexpression of MzrA (YqjB) from an IPTG-inducible promoter dramatically reduced levels of OmpF and LamB, and to some extent MalE, but not OmpC and OmpA. The effects of MzrA (YqjB) were just as pronounced in a bamB+degP+ strain (Fig. 1B and C; lanes 1 and 2) as they were in a strain deleted for these genes (data not shown). Thus, mitigation of envelope stress was independent of other known pathways and was remarkably similar to a class of suppressor mutations identified through our antibiotic selection of ΔbamBΔdegP that localized to envZ (see below; Fig. 8). This result prompted us to examine the relationship between MzrA (YqjB) and the EnvZ/OmpR two-component regulatory system in influencing OMP and MalE levels.
Fig. 1.
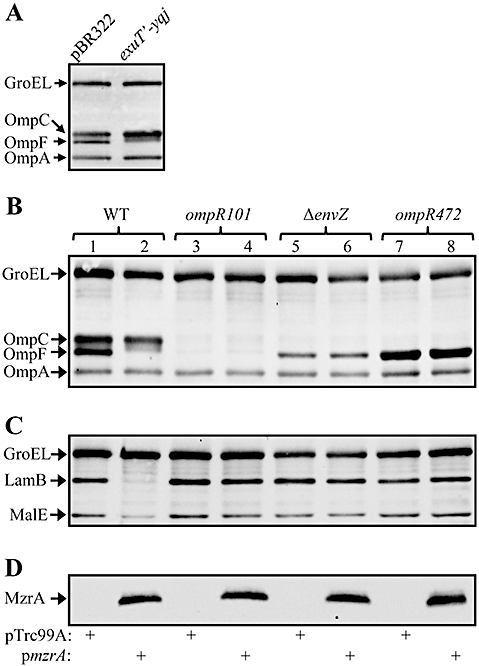
A. Effect of the exuT-yqj clone on OMPs in a ΔbamBΔdegP background. OMPs from cells grown overnight at 30°C were examined by Western blots using OmpF/OmpC/OmpA antibodies. B and C. Effect of MzrA (YqjB) overexpression on OmpA, OmpC and OmpF (B), and LamB and MalE (C) in different genetic backgrounds: envZ+/ompR+ (lanes 1 and 2), envZ+/ompR101 (lanes 3 and 4), ΔenvZ/ompR+ (lanes 5 and 6) and envZ+/ompR472 (lanes 7 and 8). Indicated proteins were identified by Western blot analysis from freshly grown (37°C) cells using (B) GroEL and OmpF/OmpC/OmpA antibodies or (C) GroEL, LamB and MalE antibodies. GroEL serves as a gel loading control. Leaky (without the addition of IPTG) expression of mzrA from a pTrc99A-mzrA plasmid was sufficient to observe the effects of MzrA. D. Detection of MzrAFLAG from cells analysed in (B) and (C) by FLAG antibodies. The presence of pTrc99A or pTrc99A-mzrA plasmid is indicated by the plus signs.
MzrA (YqjB) functions through EnvZ/OmpR
Overexpression of MzrA (YqjB) produces phenotypes similar to that conferred by pleiotropic envZ alleles (Wanner et al., 1979; Wandersman et al., 1980). Pleiotropic envZ alleles are dependent on OmpR (Slauch et al., 1988). Thus, we determined whether the effects of MzrA (YqjB) were also OmpR-dependent. For this experiment, we employed a non-polar null allele of ompR, ompR101 (Sarma and Reeves, 1977). As expected, expression of both OmpF and OmpC was abolished in an envZ+ompR101 background, but LamB and MalE were expressed normally (Fig. 1B and C). In contrast to the effect seen in an envZ+ompR+ background (Fig. 1C, lane 2), overexpression of MzrA (YqjB) in an envZ+ompR101 background failed to reduce LamB and MalE levels (Fig. 1C, lane 4), indicating that OmpR is required for the effect of MzrA (YqjB). To further evaluate a requirement of OmpR for the negative effect of MzrA (YqjB) overexpression on OMPs and MalE, we employed another well-studied ompR allele, ompR472 (Hall and Silhavy, 1981), which produces an OmpF+ OmpC- phenotype without influencing LamB and MalE expression (Fig. 1B and C, lane 7). Again, we failed to see any negative effect of MzrA (YqjB) overexpression on OmpF, LamB or MalE (Fig. 1B and C, lane 8), thus emphasizing the need for wild type OmpR to observe the effects of MzrA (YqjB).
Next we tested the requirement for EnvZ in MzrA (YqjB)-mediated effects. In the absence of EnvZ, OmpC is not expressed while OmpF is expressed at diminished levels (Fig. 1B, lane 5). The low OmpF expression seen in an envZ null background is thought to be due to the phosphorylation of OmpR by acetyl phosphate (Hsing and Silhavy, 1997). Although expression of OmpC and OmpF is affected, LamB and MalE expression is not significantly altered in an envZ null background (Fig. 1C, lane 5). If MzrA (YqjB) directly influences OmpR, regardless of how OmpR gets phosphorylated, we may still observe the negative effects of MzrA (YqjB) on OmpF, LamB and MalE in an envZ null background. On the other hand, if MzrA (YqjB) modulates EnvZ activity, then in an envZ null background overexpression of MzrA (YqjB) will fail to reduce OmpF, LamB or MalE levels. The data in Fig. 1 showed that overexpression of MzrA (YqjB) in an envZ null background was unable to exert a negative effect on OmpF, LamB or MalE (compare lanes 5 and 6 in B and C). It is important to note that MzrA expression from the pTrc99A clone was unaffected in various mutant OmpR and EnvZ backgrounds (Fig. 1D), demonstrating that a lack of negative effect on OMPs or MalE was not the result of reduced MzrA expression. These results support our hypothesis that MzrA (YqjB) influences EnvZ activity that then transforms OmpR into a form responsible for the pleiotropic phenotype. Thus, YqjB was renamed MzrA, for modulator of EnvZ and OmpRA and will be henceforth referred to by this designation.
MzrA influences steady-state expression of porins independently of the known modulators
As overexpression of MzrA affects porin expression in an EnvZ/OmpR-dependent manner, we investigated whether MzrA plays a role in the osmoregulation of porins. Envelopes were isolated from wild type and ΔmzrA strains carrying either an empty vector pBAD33 or pBAD33-mzrAFLAG. Cultures were grown in medium A (low osmolarity) and medium A supplemented with 15% sucrose (high osmolarity) (Kawaji et al., 1979). OMPs were analysed from envelopes by SDS (urea)-PAGE and protein bands were visualized by coomassie blue staining. In the wild type strain carrying just the empty vector, porin expression responded to medium osmolarity in an expected manner, i.e. high OmpF and low OmpC under low osmolarity and low OmpF and high OmpC under high osmolarity (Fig. 2A, lanes 1 and 3). In contrast, a significantly different porin expression profile was observed in the absence of MzrA, with OmpF being preferentially expressed regardless of the medium osmolarity (Fig. 2A, lanes 5 and 7). When MzrA was overexpressed, the porin expression profile switched in favour of OmpC, regardless of medium osmolarity (Fig. 2A, lanes 2, 4, 6 and 8). The fact that porin expression still responds to medium osmolarity in cells lacking or overexpressing MzrA suggets that MzrA is not the primarily receptor of medium osmolarity, rather, it appears to bias EnvZ/OmpR activity to favour OmpC expression over OmpF. In agreement with a recent report (Batchelor et al., 2005), we confirmed that deletion of cpxR, which only weakly reduces mzrA expression (see below), does not significantly influence porin expression or their osmoregulation (data not shown).
Fig. 2.
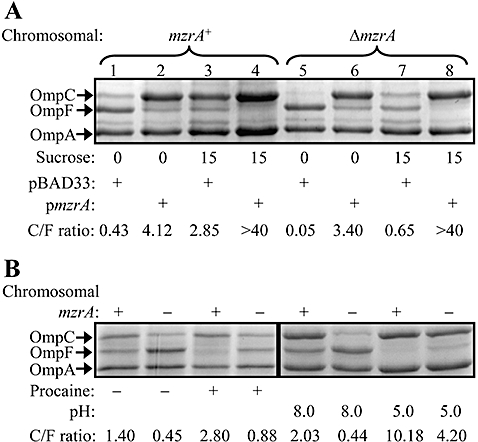
Effects of osmolarity, procaine and pH on porin expression in the presence or absence of MzrA. A. Envelopes from cells grown in medium A with (15%) or without (0) sucrose were analysed by SDS(urea)-PAGE and proteins were visualized by Coomassie blue stain. Strains used either had mzrA intact (lanes 1–4) or deleted (lanes 5–8) from the chromosome. Plus signs indicate presence of either a vector plasmid or an mzrA plasmid clone. All cultures were grown in the presence of 0.01% L-arabinose. B. For procaine's effect on porins, cells were grown in LB without salt and with (+) or without (−) 5 mM procaine. To assess the effect of pH on porins, cells were grown in 3-(N-morpholino)propanesulphonic acid (pH 8.0) or 2-(N-morpholino)ethanesulfonic acid (pH 5.0) buffered LB without salt. Envelopes from these cells were analysed by SDS(urea)-PAGE and protein bands were visualized after Coomassie staining. Plus and minus signs on top of the gel refer to the presence or absence, respectively, of the chromosomal mzrA gene. OmpC : OmpF ratios are shown.
The local anaesthetic, procaine, is known to alter porin expression in an EnvZ/OmpR-dependent manner (Villarejo and Case, 1984) similar to pleiotropic envZ mutations (Villarejo and Case, 1984; Case et al., 1986) and MzrA overexpression (Fig. 1). Porin expression in a strain deleted for mzrA still responded to the presence of procaine in the same fashion as did the MzrA+ parental strain (Fig. 2B). Thus, MzrA does not mediate the effect of procaine on porin expression. Medium pH is also known to influence porin expression in an EnvZ/OmpR-dependent manner (Heyde and Portalier, 1987; Sato et al., 2000). At pH 8.0, OmpF to OmpC ratio in an mzrA strain was significantly higher than in the parental mzrA+ strain, but at pH 5.0 OmpC was the predominant porin in both strains (Fig. 2B), showing that pH-dependent porin regulation is independent of MzrA. Together, these data suggest that MzrA modulates the output activity of the EnvZ/OmpR system without influencing its ability to respond to various environmental signals.
Evidence for MzrA's specificity
To test MzrA specificity for EnvZ and not other related membrane kinases, we used four different operon fusions whose activities are dependent on PhoP/PhoQ (mgtA::lacZ; mgrB::lacZ; Kato et al., 1999), CpxA/CpxR (ppiA::lacZ; Pogliano et al., 1997) or EnvZ/OmpR (ompF::lacZ; Hall and Silhavy, 1981). Overexpression of MzrA had no significant effect on mgtA::lacZ, mgrB::lacZ or ppiA::lacZ activities (Fig. 3A). We also used cpxP::lacZ as a second and perhaps a more sensitive target of the CpxA/CpxR regulon and found no appreciable change in its activity (a 1.13-fold increase) when MzrA was overexpressed. In contrast, overexpression of MzrA significantly reduced the activity of an EnvZ/OmpR-regulated ompF::lacZ fusion (Fig. 3A). Thus, the actions of MzrA appear to be specific to the EnvZ/OmpR regulon.
Fig. 3.
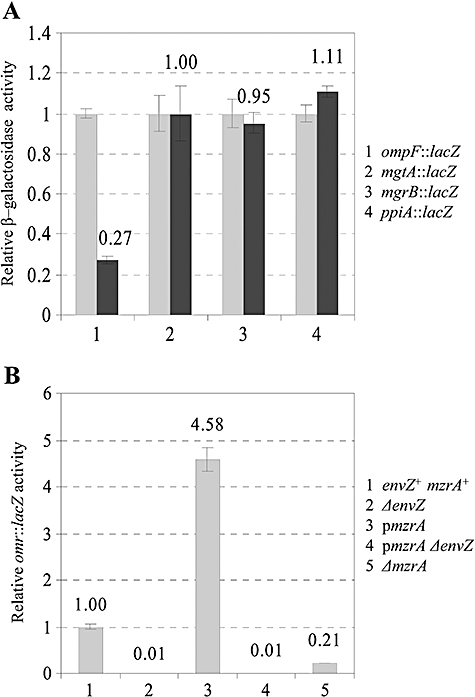
Specificity of MzrA. A. Effects of MzrA on ompF, mgtA, mgrB and ppiA expression were measured by assaying β-galactosidase activities of the chromosomal lacZ fusions to these genes. MzrA's expression from a plasmid was either not induced (light grey bars) or induced with 0.2% arabinose (dark grey bars). B. MzrA modulates the expression of omr genes through the EnvZ/OmpR system. Expression of omr was measured from a chromosomal omr::lacZ fusion. In some instances (B; 3 and 4), expression of MzrA from a pBAD24 clone was induced by the addition of 0.2% arabinose.
The omrAB genes were recently described as additional downstream targets of the EnvZ/OmpR regulon (Guillier and Gottesman, 2006). If MzrA activates the EnvZ/OmpR regulatory system then omrAB expression is expected to increase when MzrA is overproduced and decrease in the absence of MzrA. Consistent with this, omr::lacZ activity increased 4.5-fold in response to MzrA overexpression and this increase was absolutely dependent on the EnvZ/OmpR system (Fig. 3B). Moreover, in a strain deleted for mzrA, omr::lacZ activity decreased fivefold (Fig. 3B), showing that chromosomal expression of MzrA is sufficient to modulate the EnvZ/OmpR regulon.
Together, these results suggest that overexpression of MzrA activates the EnvZ/OmpR regulatory system, presumably by directly modulating EnvZ enzymatic activity. The activated EnvZ/OmpR system then influences the expression of a number of downstream target genes, such as ompF, ompC and omrAB, archetypal members of the EnvZ/OmpR regulon. Other affected genes include lamB and malE, which are normally not influenced by EnvZ/OmpR unless MzrA is overexpressed or in the presence of pleiotropic envZ mutations (Wanner et al., 1979; Wandersman et al., 1980).
MzrA links CpxA/CpxR and EnvZ/OmpR regulons
The yqjA and mzrA open reading frames are separated by three nucleotides and are thought to be cotranscribed from σ70(p1)- and σE(p2)-dependent promoters located upstream of yqjA (Dartigalongue et al., 2001). Moreover, the yqjAmzrA operon has been shown to be regulated by the CpxA/CpxR two-component regulatory system (Yamamoto and Ishihama, 2006), which responds to envelope stress (Raivio and Silhavy, 2001). To corroborate the previous results, we measured the β-galactosidase activity of a chromosomal yqj::lacZ fusion. In a constitutively activated CpxA* mutant background (Cosma et al., 1995) the activity increased 5.6-fold (Fig. 4A). Interestingly, in the absence of CpxR, yqj::lacZ activity decreased only modestly (27%), indicating that additional regulator(s) may play a role in activating yqjAmzrA. As mentioned above, yqjAmzrA was reported to be regulated by a specialized sigma factor, σE. We found that yqj::lacZ activity was slightly increased (1.5-fold) in a ΔrseA background in which σE is fully active (Alba and Gross, 2004).
Fig. 4.
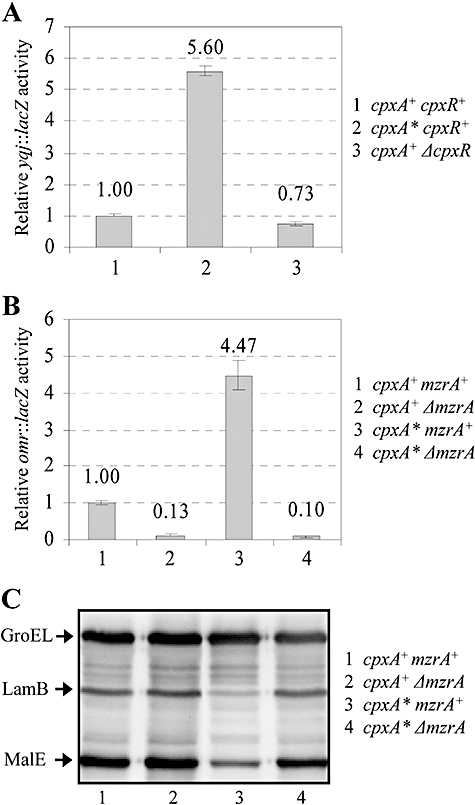
Expression of omr, yqjAmzrA, lamB and malE in various genetic backgrounds. Expression of omr (A) and yqjAmzrA (B) was measured by assaying the activities of the respective chromosomal lacZ fusions. LamB and MalE levels were measured by Western blot analysis (C) as described in the Fig. 1 legend.
As MzrA functions through EnvZ/OmpR (Fig. 1), it stands to reason that the activated CpxA/CpxR system may influence the EnvZ/OmpR regulon via MzrA. To test this possibility, we used an omr::lacZ chromosomal fusion whose expression is dependent on EnvZ/OmpR (Fig. 4B). In a constitutively activated CpxA*/CpxR background, the omr::lacZ activity rose more than fourfold over that present in a CpxA+ CpxR+ background, and this increase was dependent on MzrA (Fig. 4B). Furthermore, it can be seen in Fig. 4C that activated CpxA*/CpxR downregulates LamB and MalE expression in an MzrA-dependent manner, thus providing further evidence for an MzrA-mediated link between these two-component systems. The absence of MzrA did not alter the effect of CpxR on ompF and ompC (data not shown). This is not surprising because CpxR has been shown to directly influence ompF and ompC (Batchelor et al., 2005).
MzrA is an inner membrane protein
MzrA is a 127-residue protein with a theoretical pI of 9.51. It contains a single predicted trans-membrane (TM) domain extending from residues 11 to 31 of the polypeptide chain (Fig. S1). To determine the cellular localization of MzrA, we constructed a fully functional variant of MzrA containing a C-terminal FLAG tag. Fractionation of cells into the cytoplasm, the periplasm and the envelopes showed that MzrA is an envelope protein (Fig. 5A). SurA and DnaK were present only in the periplasmic and cytoplasmic fractions respectively (Fig. 5A and B), thus confirming the purity of various fractions.
Fig. 5.
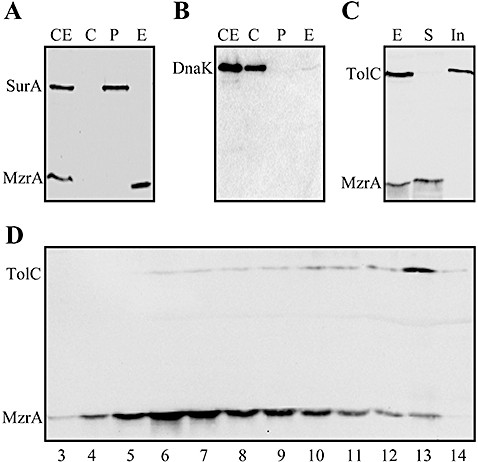
Localization of MzrA. Freshly grown cultures expressing MzrAFLAG from pBAD24 plasmid were fractionated into various cellular compartments and proteins from various fractions were visualized through Western blot analysis. Proteins samples from whole-cell extract (CE), cytoplasm (C), periplasm (P) and envelopes (E) fractions were probed with FLAG- and SurA- (A) or DnaK (B) specific antibodies. C. Whole-cell envelopes (E) were treated with 0.3% sarcosyl and proteins from sarcosyl soluble (S) and sarcosyl insoluble pellet (In) were analysed by a Western blot using TolC- and FLAG-specific antibodies. D. Sucrose density gradient analysis of cell envelopes (E) obtained from a strain expressing MzrAFLAG. Protein samples from fractions 3–14 were analysed by Western blot analysis using TolC- and FLAG-specific antibodies. MzrAFLAG peaks with the inner membrane fractions 6 and 7, while TolC peaks with an outer membrane fraction 13.
We then examined whether MzrA localizes to the inner or outer membrane by employing two methods. In the first method, which involves differential solubilization of membrane proteins, we found that a 0.3% solution of sarcosyl fully solubilized MzrAFLAG from the envelopes, whereas TolC, an OMP, remained insoluble (Fig. 5C). This indicated that MzrA is an inner membrane protein. The location of MzrA was confirmed by sucrose density gradient analysis (Fig. 5D), which showed that MzrA localizes with the lighter density inner membrane fractions (buoyant density of around 1.15 g cm−3). In contrast, TolC fractionated with the heavier density outer membrane fractions (buoyant density of around 1.22 g cm−3). As a control marker for the inner membrane proteins, we excised and sequenced a protein band whose presence peaked in the same fractions as MzrA and determined it to be AtpD, the inner membrane-localized β-subunit of ATPase. Together, these analyses established that MzrA is an inner membrane protein.
The C-terminus of MzrA is exposed to the periplasm
The presence of a single putative N-terminal TM domain renders the possibility that the C-terminal domain of MzrA is either exposed in the cytoplasm or in the periplasm. To determine its topology, we first constructed a C-terminal MzrA-reporter gene fusion with PhoA in which codons corresponding to the mature PhoA protein were fused downstream of the MzrA open reading frame. The alkaline phosphatase activity of PhoA is functional in the oxidizing environment of the periplasm (Boyd et al., 1987). The resulting MzrA::PhoA fusion was bi-functional, indicating that its expression not only stimulated alkaline phosphatase activity (an 80-fold increase in alkaline phosphatase activity over background chromosomal activity; Fig. 6A), but also conferred MzrA-specific phenotypes, such as inhibition of OmpF (Fig. 6B). MzrA::PhoA activity in a dsbA background, deficient in periplasmic oxidoreductase activity, was reduced over eightfold compared with a dsbA+ background (Fig. 6A), confirming the periplasmic orientation of MzrA::PhoA. An in-frame deletion removing residues 13–28 of the putative TM domain of MzrA from the MzrA::PhoA fusion completely abolished its alkaline phosphatase activity and failed to reduce OmpF level (Fig. 6B), indicating a requirement of the TM domain for membrane localization and activity. We also constructed MzrA::αLacZ fusions and used them in conjunction with the LacZ ω fragment, to test the orientation of the C-terminus of MzrA. The Lac complementation data obtained from these experiments corroborated the conclusion drawn from the MzrA::PhoA data (data not shown). These results provided strong genetic evidence that the MzrA C-terminus is exposed in the periplasm. Evidence that the N-terminus of MzrA is exposed in the cytoplasm came from Cya fusion studies described below.
Fig. 6.
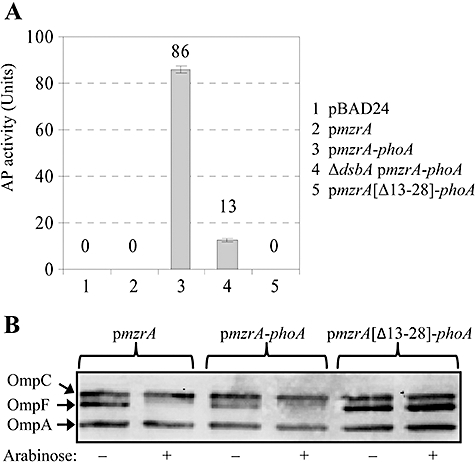
Bi-functional nature of MzrA::PhoA. A. Alkaline phosphatase (AP) activities of cells expressing various plasmid clones in the presence (lanes 1–3 and 5) or the absence (lane 4) of the periplasmic disulphide bond isomerase DsbA. B. Functionality of MzrA of MzrA::PhoA, with or without the putative TM domain of MzrA, was assessed by analysing OmpF levels. Cells carrying various plasmids constructs, as shown on top of the gel, were grown without (−) or with (+) arabinose (0.2%) to induce the expression of plasmid-borne MzrA or fusion proteins. Protein extracts from these cells were analysed by Western blots using antibodies to detect OmpF, OmpC and OmpA. Expression of plasmid-borne proteins was induced with 0.2% arabinose.
MzrA interacts with EnvZ
The genetic data presented thus far suggested that MzrA modulates the EnvZ/OmpR regulon by influencing EnvZ (Fig. 1). To determine if MzrA interacts with EnvZ in vivo, we employed a bacterial two-hybrid reporter system based on functional complementation between Bordetella pertussis adenylate cyclase fragments, T18 and T25, expressed separately from two compatible plasmid replicons (Karimova et al., 1998). Adenylate cyclase activity is restored only when proteins fused to T18 and T25 interact. Functional reconstitution of B. pertussis adenylate cyclase in an E. coliΔcya lac+ strain is monitored by assaying the activity of a cAMP-CRP-dependent lac promoter of a chromosomally encoded lac operon. Thus, measurement of the β-galactosidase activity provides a convenient and sensitive means of detecting in vivo protein–protein interactions. Fusions were constructed with T18 and T25 present at the N- or C-terminal of EnvZ (both termini of EnvZ are exposed to the cytoplasm). Only N-terminal MzrA T18/T25 fusions were constructed because the topology data above indicated that the C-terminus of MzrA is exposed in the periplasm and therefore, C-terminal T18 and T25 fusions are not expected to produce functional complementation. We also constructed OmpR fusions to detect their interactions with MzrA. Lastly, to test the specificity of this in vivo assay, we made N- and C-terminal T18 fusions with the membrane kinase protein, CpxA. All plasmids expressing fusion constructs were transformed into a Δcya lac+ strain and functional complementation was assessed by measuring β-galactosidase activity of the chromosomally encoded lacZ gene (Fig. 7). Formation of MzrA and EnvZ homo-dimers or homo-oligomers was evident by a 33- and 12-fold (lanes 3 and 10) increase in β-galactosidase activity over the control strain (lane 2).
Fig. 7.
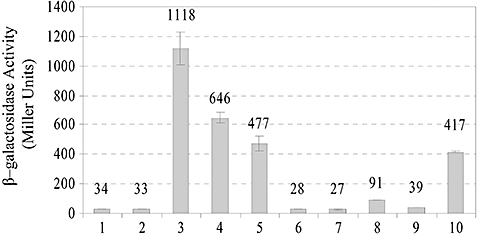
Interactions between MzrA and EnvZ. A bacterial two-hybrid system was used to detect in vivo protein–protein interactions. Values of β-galactosidase activities, calculated in Miller Units, with standard errors are shown: (1) pT25/pT18, (2) pT25::MzrA/pT18, (3) pT25::MzrA/pT18::MzrA, (4) pT25::MzrA/pEnvZ::T18, (5) pT25::MzrA/pT18::EnvZ, (6) pT25::MzrA/pT18::OmpR, (7) pT25::MzrA/pOmpR::T18, (8) pT25::MzrA/pCpxA::T18, (9) pT25::MzrA/pT18::CpxA and (10) pT25::EnvZ/pEnvZ::T18.
Importantly, formation of MzrA-EnvZ heteromers was readily apparent from two different EnvZ constructs resulting in a 19- and 14-fold increase in β-galactosidase activity (lanes 4 and 5). In contrast, no significant MzrA–OmpR interactions (lanes 6 and 7), or MzrA–CpxA interactions (lanes 8 and 9) were detected. Thus, MzrA interacts with EnvZ, but not with OmpR or CpxA.
Pleiotropic envZ mutations suppress ΔbamB ΔdegP
Concurrent with the multicopy suppressor analysis, we carried out a genetic analysis to identify mutational suppressors of ΔbamBΔdegP. These suppressors were isolated at 30°C and 37°C on Luria broth (LB) agar (LBA) medium supplemented with vancomycin. Subsequent characterization showed that, as expected, they displayed reduced sensitivity towards vancomycin, but also to other antibiotics, such as rifampin, to which the parental strain is also sensitive. Curiously, even though some suppressors of this class were isolated at 37°C, they failed to form regular-sized colonies at 37°C; however, at 30°C all suppressors formed large homogeneous colonies similar to the bamB+degP+ strain.
Examination of OMPs and MalE by Western blot analysis from revertants showed that they had significantly reduced levels of OmpF, LamB and MalE, but the levels of OmpC and OmpA were not reduced. These characteristics persisted even when the ΔbamBΔdegP alleles were replaced by the corresponding wild type alleles (Fig. 8A and B). To assess whether the reduction in OmpF levels was due to an effect on ompF transcription, we introduced a chromosomal ompF::lacZ operon fusion via a linked Tn10 (Tcr) marker into a strain carrying the suppressor mutation. In the suppressor strain, ompF::lacZ activity was found to be extremely low (light blue colonies on Xgal plates) compared with that of the parental strain (dark blue colonies on Xgal plates). We exploited this ompF::lacZ-down phenotype of the suppressor mutation to isolate plasmid clones carrying a wild type copy of the suppressor gene. DNA sequence analysis of plasmid clones partially reversing the Lac-down phenotype revealed that they contained a contiguous chromosomal region with intact greB, ompR and envZ genes. Direct DNA sequencing of this region of the chromosome from suppressor isolates revealed that they carried envZ mutations, resulting in a W134C, F284C or R397L substitution or a five-amino-acid insertion after residue 26 of EnvZ. Introduction of one of the mutant envZ alleles, causing a R397L substitution, in a strain expressing a chromosomal omr::lacZ fusion resulted in an over ninefold increase in the β-galactosidase activity (Fig. 8C), showing the constitutively activated nature of this envZ allele (and presumably the other three envZ alleles as well). Based on OMP and MalE profiles, this class of envZ suppressor mutations resembles the previously described pleiotropic envZ alleles (Wanner et al., 1979; Wandersman et al., 1980). Therefore, we also tested the previously characterized pleiotropic envZ11 and envZ473 alleles and found that they too suppressed the temperature and drug sensitivity phenotypes of the ΔbamBΔdegP strain (data not shown).
Fig. 8.
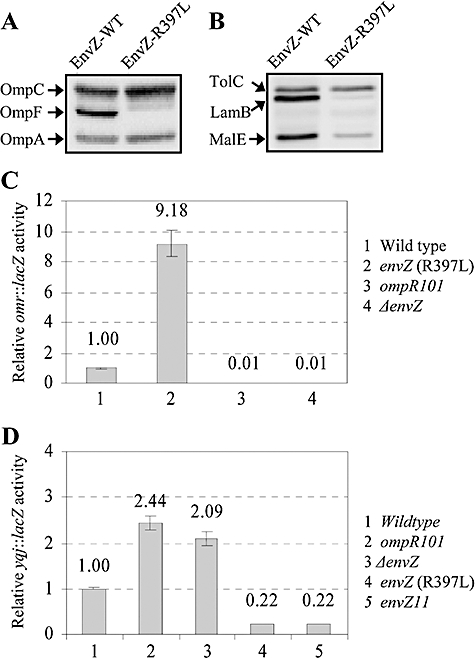
Effect of an envZ suppressor allele on OMPs, MalE and omr. Protein extracts expressing wild type (WT) or mutant (EnvZ-R397L) EnvZ were analysed by Western blots using (A) OmpF/OmpC/OmpA antibodies or (B) TolC, LamB and MalE antibodies. OmpA and TolC serve as gel loading controls. omr (C) or yqjAmzrA (D) expression were assessed by measuring the activity of the respective chromosomal lacZ fusions, in genetic backgrounds shown in the figures.
mzrA expression is negatively controlled by EnvZ/OmpR
Kato et al. (2003) discovered a negative regulatory feedback loop in which the expression of pmrD was shown to be negatively regulated by PmrA, the response regulator of the PmrB/PmrA two-component system; PmrD has been shown to promote phosphorylation of PmrA (Kato and Groisman, 2004). We asked whether such feedback-negative regulatory loop also exists in case of mzrA-EnvZ/OmpR. In the absence of EnvZ or OmpR, the expression of yqj::lacZ went up twofold (Fig. 8D). Moreover, in constitutively activated EnvZ mutant backgrounds expressing either EnvZ11 or EnvZ (R397L), the expression of yqj::lacZ was reduced almost fivefold (Fig. 8D). These results are consistent with a notion that the expression of mzrA, whose product modulates EnvZ/OmpR, is feedback-inhibited by these regulators.
The mechanistic basis of the pleiotropic envZ phenotype
In order to understand the underlying effect of the EnvZ substitution on its activity, we overexpressed and purified the cytoplasmic domain of wild type EnvZ (EnvZc) and one of the EnvZ mutants (R397L) for which we have already presented in vivo data in Fig. 8. We compared their autophosphorylation, phosphotransfer to OmpR and dephosphorylation properties in vitro.
We first examined EnvZ autophosphorylation and phosphotransfer to OmpR by incubating EnvZc and EnvZ R397Lc with [γ-32P]-ATP for various times and analysing the products via autoradiography after separation on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 9A). The wild type and mutant activities are directly compared over 10 min time-course. It is apparent that the wild type and the mutant have quite similar autophosphorylation levels. We selected 10 min time point for EnvZ autophosphorylation to compare the phosphotransfer to OmpR (Fig. 9B). After addition of OmpR, the phosphoryl group from EnvZ∼P is transferred to OmpR and the mass of the labelled band shifts (lanes 2–4 and 6–8). By visual inspection, the phosphotransfer activities of the wild type and the mutant appear somewhat similar. However, densitometric analysis of OmpR∼P (produced by EnvZ∼P or EnvZcR397L∼P) indicated that at every time point, there was more OmpR∼P present when phosphorylated by EnvZcR397L compared with the wild type (Fig. 9C). This is most dramatic at 15 min, where there was nearly 50% more OmpR∼P present. We then measured EnvZ ATPase activity. EnvZ, phosphorylated directly from ATP, transfers its histidyl phosphate to OmpR and subsequently stimulates OmpR∼P dephosphorylation. This results in the net breakdown of ATP and the subsequent release of inorganic phosphate from OmpR∼P. The sum of these reactions can therefore be analysed by measuring ATP hydrolysis (Kenney, 1997; Mattison and Kenney, 2002). We measured inorganic phosphate (Pi) production over time at a ratio of 2:1 EnvZ to OmpR (Fig. 9D). It is apparent from the figure that the EnvZ R397L mutant has an approximately 10-fold lower turnover rate (0.12 μmol Pi mg−1 h−1) compared with the wild type (1.48 μmol Pi mg−1 h−1). As we have already shown that the EnvZ mutant is not defective in its autophosphorylation (Fig. 9A) and early phosphotransfer appears to be similar (data not show) (Fig. 9B and C), a dramatic reduction in the turnover observed in the presence of the EnvZ R397L mutant in the ATPase assay (Fig. 9D) may be due to the diminished ability of the mutant EnvZ to dephosphorylate OmpR∼P, thus increasing its half-life. It is worth noting that previously characterized pleiotropic EnvZ mutants were also shown to have diminished phosphatase activity, thus resulting in an elevated half-life of OmpR∼P (Aiba et al., 1989; Tokishita et al., 1991; Hsing et al., 1998). Presumably in vivo, this increased stability promotes OmpR binding at promoters that it does not normally regulate, resulting in a pleiotropic phenotype. Alternatively, constitutive activation of normally OmpR-regulated promoters ultimately leads to secondary regulatory cascades that are responsible for the phenotypes observed.
Fig. 9.
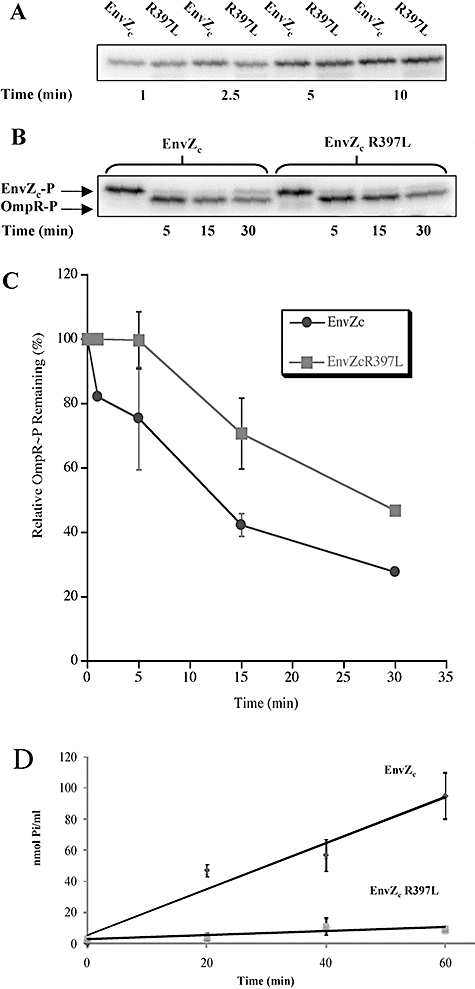
A. EnvZc and EnvZc R397L display similar rates of autophosphorylation. EnvZc (and EnvZc R397L) were added at a final concentration of 4 μM. Time of incubation is as follows: lanes 1 and 2 (1 min), lanes 3 and 4 (2.5 min), lanes 5 and 6 (5 min) and lanes 7 and 8 (10 min). Each reaction was stopped with 3 μl of denaturing sample buffer and 10 μl of the 20 μl reaction was loaded on the gel. B. OmpR∼P persists after phosphorylation by EnvZc R397L. EnvZc or EnvZc R397L was added at a final concentration of 4 μM. OmpR was added at a final concentration of 2 μM. Lanes 1 and 5 contain the results of a 10 min autophosphorylation reaction with EnvZc or EnvZc R397L respectively. Phosphotransfer to OmpR is shown in lanes 2–4 and 6–8 with the time of each reaction indicated below each lane. Reactions were stopped by the addition of 3 μl of denaturing sample buffer, and 10 μl of the 20 μl reaction was loaded on the gel. C. Densitometric analysis. Densitometry on the experiment shown in (B) (and a repeat experiment) was performed using Molecular Dynamics ImageQuant Software, version 5.0. The results are plotted as a per cent of the EnvZ∼P determined as a function of time, the symbol represents the mean and the error bars indicate the range of two separate experiments. The absence of error bars at 1 and 30 min was due to the elimination of that time point in the second experiment. D. The EnvZc R397L ATPase is reduced by 10-fold compared with the wild type EnvZc. ATPase assays were performed with EnvZc (2 μM) and the EnvZc R397L mutant (2 μM) in the presence of OmpR (1 μM). Reactions were performed in triplicate and the standard deviation at each time point is indicated in parentheses. For EnvZc, the values at 20, 40 and 60 min are 47.01 (3.86), 56.90 (9.95) and 95.08 (14.85) respectively. For EnvZc R397L, the values at the same time points are 4.42 (2.26), 11.09 (5.51) and 9.10 (0.59) respectively. The ATPase activity was expressed as nanomoles of Pi liberated per ml (see Experimental procedures for details).
Discussion
Characterization of suppressors that overcome envelope stress caused by ΔbamBΔdegP led to the discovery of a novel protein, MzrA, and pleiotropic mutations in envZ. Their suppressive effects are possibly due in part to reduced synthesis of two OMPs, OmpF and LamB, thereby reducing the OMP load on compromised assembly machinery. As both suppressors also lower the ΔbamB drug sensitivity phenotype, which cannot be fixed by deleting the ompF and lamB genes, we suspect additional alterations in the membrane structure are responsible for mending the outer membrane permeability breach of the ΔbamBΔdegP strain.
The EnvZ/OmpR two-component regulatory system has long been studied for its involvement in the osmoregulation of porins. Although an exhaustive amount of research has been carried out in this area, it remains unclear as to how EnvZ senses environmental stimuli and how this sensing alters its autokinase, phosphotransferase and phosphatase activities. Results presented in this study showed that MzrA, a small inner membrane protein, is an upstream regulator of the EnvZ/OmpR system. Although the porin expression profile in cells either lacking or overexpressing MzrA was drastically different compared with that seen in wild type MzrA+ cells, porin expression still responded to changes in medium osmolarity, pH or the presence of procaine in the medium, suggesting that MzrA is not the primary receptor of these three environmental cues that influence porin expression via EnvZ/OmpR. Based on the porin expression profile when MzrA is overexpressed (high OmpC, low OmpF regardless of the medium osmolarity), it appears that MzrA modulates the EnvZ/OmpR system to favour the accumulation of OmpR∼P (Fig. 9). This could be achieved by an elevated autokinase activity, diminished phosphatase activity or a combination of the two. Because OmpR has a high non-specific binding component and lacks highly conserved binding sites, it is a good global regulator (Rhee et al., 2008). Increasing the half-life of OmpR∼P (through envZ mutations or MzrA overexpression) would facilitate higher-affinity interaction with DNA (Head et al., 1998) and would enable OmpR to bind to promoters it does not normally regulate.
Using a bacterial two-hybrid system, we showed that MzrA interacts with EnvZ. Exactly how the binding of MzrA to EnvZ modulates EnvZ enzymatic activity remains to be determined, but we demonstrated that it does so by increasing OmpR∼P stability. MzrA binding might promote a conformation of EnvZ that exists in the presence of procaine or when EnvZ is constitutively altered by pleiotropic envZ mutations found here and isolated previously (Wanner et al., 1979; Wandersman et al., 1980). Although at present there is no biochemical evidence showing MzrA increases OmpR∼P stability, the observation that overexpression of MzrA produces a phenotype that is almost identical to that seen in a strain expressing a pleiotropic envZ allele, indicates that the biochemical state of EnvZ/OmpR in these two genetic backgrounds is also very similar; i.e. both backgrounds likely increase OmpR∼P stability.
The MzrA TM region is not very conserved among its homologues, which are found in at least nine members of the Enterobacteriaceae family (Fig. S1). The role of the TM domain may be to target the protein to the inner membrane and this was evident from the MzrA::PhoA data. In contrast, the central region of MzrA, which is exposed to the periplasm, contains conserved residues that may make direct contact with the periplasmic domain of EnvZ. We suggest that residues of the MzrA periplasmic domain play an important role in modulating EnvZ enzymatic activity. An earlier search for a periplasmic protein ligand that interacts with the periplasmic domain of EnvZ was unsuccessful (Egger and Inouye, 1996). Our finding that MzrA is an inner membrane protein would explain why MzrA was not identified in the previous biochemical screen where only soluble periplasmic proteins were probed.
We have provided evidence that MzrA links two major signal transduction regulatory systems: CpxA/CpxR and EnvZ/OmpR, which respond to different environmental and cellular signals to regulate outer membrane composition. The network of genes regulated by the CpxA/CpxR and EnvZ/OmpR systems is not entirely unique; for example, both systems independently regulate transcription of csgD, ompC and ompF (Batchelor et al., 2005; Jubelin et al., 2005). However, the environmental cues to which they respond are distinct; whereas alkaline pH or misfolded pilus subunits activate CpxA/CpxR, medium osmolarity is the best-characterized effector of EnvZ/OmpR. Thus, despite responding to different stimuli, MzrA-mediated linking of the CpxA/CpxR and EnvZ/OmpR regulatory systems would allow cells to amplify their response to appropriately adjust the envelope composition (see Fig. 10).
Fig. 10.
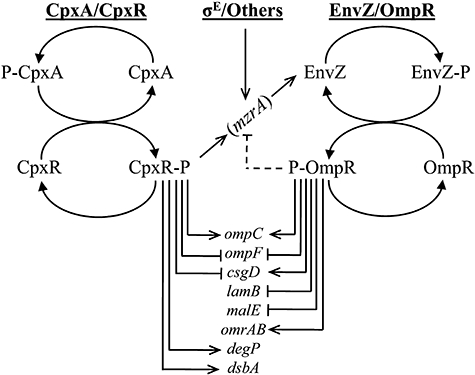
MzrA-mediated bridging of the CpxA/CpxR and EnvZ/OmpR regulons. The activated CpxA/CpxR and σE regulons turn on the synthesis of MzrA, which, in turn through its interaction with EnvZ, modulates EnvZ/OmpR's activity to favour the accumulation of OmpR∼P. High levels of OmpR∼P upregulates the expression of ompC, csgD and omrAB, but downregulates the expression of ompF, lamB and malE. High OmpR∼P also downregulates mzrA, thus establishing a negative-regulatory feedback loop. Also shown are genes that are independently or uniquely affected by the CpxA/CpxR regulon. Not all genes that may be affected by the activated CpxA/CpxR and EnvZ/OmpR regulons are shown. Others refer to unknown factors that may regulate MzrA's synthesis.
Bridging of separate two-component regulatory systems by a membrane protein is not unique to MzrA. Recently, a small protein, B1500, of E. coli has been shown to connect the EvgS/EvgA and PhoQ/PhoP two-component signal transduction pathways (Eguchi et al., 2007). B1500 is a relatively weakly conserved protein, as it is only found in E. coli and highly related species of Shigella. In contrast, MzrA, which is unrelated to YqjB from Bacillus, is a relatively well-conserved protein of the Enterobacteriaceae family (Fig. S1), thus signifying its widespread physiological role. Although there is no sequence homology between MzrA and B1500, and MzrA is almost twice as large as B1500, their membrane topology and possible mode of action on cognate sensor kinases appear to be very similar.
Modulators of bacterial two-component systems are not limited to inner membrane-localized proteins, such as MzrA and B1500. For example, two soluble proteins, CpxP and PmrD, have also been shown to modulate the CpxA/CpxR (Isaac et al., 2005) and PmrB/PmrA regulons (Kato and Groisman, 2004) respectively. CpxP is a periplasmic protein that is proposed to interact with the periplasmic domain of the sensor kinase CpxA to downregulate CpxA autokinase activity (Fleischer et al., 2007). On the other hand, cytosolic PmrD was shown to bind to the phosphorylated form of the response regulator, PmrA and stabilize PmrA∼P (Kato and Groisman, 2004). Interestingly, we discovered here that like the case with pmrD-PmrB/PmrA, mzrA expression is negatively regulated by EnvZ/OmpR that are post-translationally modulated by MzrA. Such negative feedback-regulatory loop appears to be well suited to re-establish homeostasis when the ‘signals’ that activate the mzrA-EnvZ/OmpR cascade have dissipated (Fig. 10).
The discovery of MzrA as a communicator between CpxA/CpxR and EnvZ/OmpR gives physiological credence to earlier claims of ‘cross-talk’ among different sensor kinases/response regulators (Laub and Goulian, 2007). Despite the existence of various cross-regulatory strategies, it is reasonable to consider that the genes affected by the primary regulon are the principal responders of a given stimulus and the genes influenced through cross-communicators are response amplifiers. For example, under envelope stress conditions, products of the genes including degP, ppiA and dsbA that are regulated by CpxA/CpxR would directly respond to help reduce envelope stress (Raivio and Silhavy, 2001). However, the MzrA-mediated connection of CpxA/CpxR to EnvZ/OmpR would allow participation of omrAB to decrease envelope stress by reducing the synthesis of certain OMPs (Lundrigan and Earhart, 1981; Guillier and Gottesman, 2006), thus helping to maintain homeostasis between OMP assembly and synthesis.
Both the pleiotropic envZ mutations and overexpression of MzrA reduce the drug hypersensitivity phenotype of ΔbamB, against which they were isolated, but also of unrelated ΔsurA and ΔtolA mutations. The root cause of the hypersensitivity phenotype conferred by the ΔbamB, ΔsurA and ΔtolA mutations is presently unknown, but it is likely due to a compromised outer membrane permeability barrier. Formation of phospholipid bilayer patches in the outer membrane is often thought to be associated with sensitivity to hydrophobic antibiotics, such as rifampin (Nikaido and Vaara, 1985), but sensitivity to a hydrophilic antibiotic, vancomycin, may be due to the formation of ‘cracks’ through which this large antibiotic can slip through (Ruiz et al., 2006). The ability to reduce sensitivity to both classes of antibiotics suggests that pleiotropic envZ mutations and the overexpression of MzrA employ a common mechanism to repair the outer membrane permeability breach. Interestingly, yqjA, the first gene of the yqjA-mzrA operon, has been shown to be involved in some aspect of membrane permeability (Shi et al., 2004) and phospholipid synthesis (Thompkins et al., 2008). We found that MzrA influences porin expression via EnvZ/OmpR but independent of YqjA (data not shown). Similarly, the ability of MzrA to reduce drug hypersensitivity of a ΔbamB strain was found to be independent of YqjA. Thus, a functional relationship, if any, between YqjA and MzrA remains unknown.
Experimental procedures
Bacterial strains and chemicals
Escherichia coli K-12 strains used here were mostly derived from MC4100 (Casadaban, 1976) and are listed in the Table S1. The bacterial adenylate cyclase-based two-hybrid system strains and plasmids were purchased from Euromedex. JM109 (Promega) was used for LacZα complementation analysis. Immun-star HRP substrate was purchased from Bio-Rad. Enzyme-catalysed fluorescence substrate was purchased from GE Healthcare. Rabbit anti-FLAG polyclonal antibodies and goat anti-mouse IgG secondary antibodies were purchased from Sigma-Aldrich. Mouse anti-DnaK and goat anti-GroEL polyclonal antibodies were purchased from Stressgen. ONPG (2-ortho-Nitrophenyl-β-D-galactopyranoside) was from ACROS and ρ-NPP (ρ-Nitrophenyl Phosphate) was from Thermo Fisher Scientific. Procaine was purchased from MP Biomedicals. All other chemicals were of analytical grade.
The LB and LBA were prepared as described previously (Silhavy et al., 1984) and when required, supplemented with ampicillin (50 μg ml−1), chloramphenicol (12.5 μg ml−1), kanamycin (25 μg ml−1) or arabinose (0.01% or 0.2%). Low-osmolarity medium-A contains nutrient broth (7.0 g l−1), yeast extract (1.0 g l−1), K2HPO4 (3.7 g l−1), KH2PO4 (1.3 g l−1) and glycerol (2.0 g l−1) (Kawaji et al., 1979) and was supplemented with 15% sucrose for high osmolarity. For analysing procaine's response on porin expression, LB was prepared without NaCl and was supplemented with procaine (5 mM) when necessary. For analysing pH response on porin expression, LB without salt was buffered with 100 mM 2-(N-morpholino)ethanesulfonic acid or 100 mM 3-(N-morpholino)propanesulphonic acid, and pH was adjusted to 5.0 or 8.0, respectively, with HCl.
DNA methods
Standard bacterial genetic methods were carried out as described by Silhavy et al. (1984). Deletion of genes from their chromosomal locations and subsequent curing of the antibiotic-resistant marker at the deletion sites were done using the λ-red mediated gene deletion method as described previously (Datsenko and Wanner, 2000). Deletions were confirmed using PCR. In some instances lacZ was recombined at the deletion site by the method of Ellermeier et al. (2002). Primers used for deletions and cloning are listed in the Table S2. Chromosomal fragments containing envZ, ompR, cpxA, phoA or mzrA were amplified using gene-specific cloning primers. Restricted PCR-amplified fragments were cloned into appropriately digested pBAD24, pBAD33 (Guzman et al., 1995), pTrc99A (GE Healthcare), pKT25, pUT18 or pUT18C (Karimova et al., 1998). The mzrA::phoA plasmid clone was created by PCR-amplifying mzrA fragments, restricting them with EcoRI and XbaI and ligating into predigested pBAD24. phoA fragments, digested with XbaI and SalI, were ligated into the appropriately digested pBAD24-mzrA (EcoRI-XbaI) clone. The Quickchange Site-Directed Kit from Stratagene was used to delete the putative TM domain from MzrA from the mzrA-phoA plasmid clone as well as to introduce the R397L mutation into the EnvZc clone as per the manufacturer's instructions using the primers listed in the Table S2.
Protein methods
For Western blot analysis, overnight cultures were diluted 100-fold into appropriately supplemented media and grown 3 h. Cell pellets were resuspended in sample buffer and heated at 95°C for 5 min and analysed by SDS-PAGE. Urea (4 M) was added to the SDS-polyacrylamide running gel in order to better resolve OmpC and OmpF. Following electrophoresis proteins were transferred onto Immobilin-P (Millipore) using a mini-transblot (Bio-Rad) and incubated in primary antibody for 1.5 h. Primary rabbit antibodies and dilutions used were: OmpF/C/A (1:16 000), GroEL (1:50 000), LamB (1:10 000), MalE (1:10 000), FLAG (1:1000), TolC (1:5000). Goat anti-rabbit HRP-conjugated immunoglobulin G secondary antibodies were incubated for 1 h. Membranes were incubated with Immun-Star HRP substrate for 5 min and protein bands were visualized with a Molecular-Imager-ChemiDoc-XRS System from Bio-Rad. Protein bands were quantified using the Quantity One software from Bio-Rad. Primary mouse antibodies for DnaK (1:10 000) were incubated for 1.5 h followed by incubation with goat anti-mouse IgG alkaline phosphatase conjugate for 1 h. After 5 min exposure to enzyme-catalysed fluorescence substrate the membranes were scanned with a phosphorimager and analysed with the ImageQuant (Molecular Dynamics) program. For the identification of protein bands from SDS-PAGE gels, bands of interests were excised, and subjected to LC-MS/MS analysis at the University of Arizona Proteomics Consortium.
EnvZc purification
EnvZc and EnvZcR397L were produced from plasmid pPH001, grown in LB and purified as previously described (Park and Inouye, 1997). The protein concentration was determined using a Micro BCA Assay kit from Pierce, using BSA standards.
EnvZ phosphorylation and OmpR phosphotransfer assays
Phosphorylation and phosphotransfer experiments were carried out as described by Tran et al. (2000). Phosphorylation of EnvZc was performed in 20 μl reactions containing a (final concentration) of 50 mM Tris (pH 7.5), 50 mM KCl and 20 mM MgCl2. EnvZc and the EnvZc R397L were added at a final concentration of 4 μM, and the reactions were initiated by the addition of 2 μCi of [γ-32P]-ATP followed by incubation for 1, 2.5, 5 or 10 min at room temperature. For phosphotransfer reactions, after an initial 10 min autophosphorylation of EnvZc or EnvZc R397L, OmpR (2 μM final concentration) was added and incubation proceeded for 5, 15 or 30 min at room temperature. All reactions were stopped by the addition of 3 μl of denaturing sample buffer [124 mM Tris-HCl (pH 6.8), 20% (v/v) glycerol, 4% (w/v) SDS, 8% (v/v) β-mercaptoethanol and 0.025% (w/v) bromophenol blue]. Ten microlitres of each reaction was loaded on a 15% SDS-PAGE gel, and the gel was subsequently dried, exposed to a phosphorimager screen and visualized on a Molecular Dynamics Storm 860 Imager.
Measurement of OmpR-dependent EnvZc ATPase activity
The EnvZc-ATPase activity was assayed using a modification of the method of Brotherus et al. (1981) as described by Kenney (1997). Proteins were stored in a buffer containing 20 mM Tris, 0.1 mM EDTA, 750 mM NaCl and 0.9% glycerol at 4°C. The assay solution (pH of 7.2) was aliquoted to minimize freeze/thawing, and stored at −20°C. The reaction was performed in a total volume of 0.6 ml containing the following (mM): 0.5 EGTA, 130 NaCl, 20 KCl, 3 MgCl2, 3 Na2ATP, 50 imidazole and 0.03% albumin. EnvZc and OmpR were added at the following final concentrations: 2 and 1 μM respectively. The assay was initiated by the addition of the ATPase assay solution and stopped by the addition of 1 ml of a solution containing: 0.5% ammonium molybdate, 473 mM HCl, 1.5% SDS and 163 mM ascorbic acid. The samples were incubated for 10 min at 0°C, followed by the addition of 1.5 ml of a solution of 68 mM sodium citrate, 154 mM sodium arsenite and 2% glacial acetic acid and incubated at 37°C for 5 min. After 20 min at room temperature, the optical density of the samples at 850 nm was determined in a Beckman Coulter DU800 spectrophotometer and compared with Na2HPO4 standards. The ATPase activity was expressed as nanomoles of Pi liberated per ml and the nonenzymatic hydrolysis was corrected for by the inclusion of blanks that did not contain protein. EnvZc ATPase activity, which is independent of OmpR, was subtracted from the total ATPase activity when both proteins were present and the corrected values are indicated.
Enzymatic assays
β-Galactosidase and alkaline phosphatase activities were determined by published methods (Miller, 1992; Michaelis et al., 1983). These activities were measured from culture grown to mid-log phase (OD600 of 0.7–0.9). Kinetic analysis of enzyme activity was carried out using a VersaMax (Molecular Dynamics) microtiter plate reader in quadruplicate.
Cell fractionation
Overnight cultures were diluted 1:100 and grown 3 h with appropriate supplements. Equivalent amounts of cells, based on OD600, were pelleted and resuspended in a periplasmic extraction buffer (10 mM Tris-HCl pH 7.5, 0.5 M sucrose, 10 mM EDTA, 0.2 mg ml−1 lysozyme) and periplasm was extracted by the gentle osmotic shock method (Ariéet al., 2001). Following extraction of the periplasm cells were pelleted and washed before resuspension in a lysis buffer (100 mM Tris pH 7.5, 10 mM MgCl2, 0.25 μg ml−1 DNase I and 2 mM PMSF) and lysed using a French Press. Cell lysates were centrifuged for 1 h at 105 000 g, 4°C to separate soluble (cytoplasm and residual periplasm) and insoluble (inner and outer membranes) fractions. Cell fractions were then subjected to SDS (urea)-PAGE (as described above) followed by Coomassie staining.
Localization of MzrA-FLAG
Inner membrane proteins from envelopes of cells expressing MzrA-FLAG were solubilized at room temperature for 30 min in 0.3% solution of sarcosyl, and insoluble material was removed by centrifugation at 4°C for 1 h at 105 000 g. Cell envelopes were separated into inner and outer membranes by sucrose density gradients 30–55% (wt/vol) as described previously (Misra et al., 2000). Gradient fractions were subjected to SDS-PAGE, followed by Western blot analysis (see above) probing for FLAG and TolC. Buoyant densities were calculated by measuring the refractive index of each fraction at room temperature.
Acknowledgments
We are indebted to Tom Silhavy, Natacha Ruiz, Tracy Raivio and Yixin Shi for some bacterial strains used here and to Yixin Shi for insightful discussion. Plasmid pPH001 was a gift from the Inouye laboratory. We thank Leanne Misra for reading the manuscript. This work was supported by Grants R01-GM048167 (R.M.) and GM58746 (L.J.K.) from the National Institutes of Health and MCB-0613014 (L.J.K.) from the National Science Foundation. H.G. and E.S.C. were partly supported by the ARCS and School of Life Sciences Undergraduate Research fellowships respectively. We would like to express our gratitude to George Tsaprailis, Director of the Arizona Proteomics Consortium, which is supported by NIEHS, National Institutes of Health Grant ES06694 (to the South-west Environmental Health Sciences Center), NCI, National Institutes of Health Grant CA23074 (to the Arizona Cancer Center) and BIO5 Institute of the University of Arizona.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aiba H, Nakasai F, Mizushima S, Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- Alba BM, Gross CA. Regulation of the Escherichia coliσE-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- van Alphen W, Lugtenberg B. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J Bacteriol. 1977;131:623–630. doi: 10.1128/jb.131.2.623-630.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arié J-P, Sassoon N, Betton J-M. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol Microbiol. 2001;39:199–210. doi: 10.1046/j.1365-2958.2001.02250.x. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Walthers D, Kenney LJ, Goulian M. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins OmpF and OmpC. J Bacteriol. 2005;187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D, Guan C-D, Willard S, Wright A, Strauch K, Beckwith J. Enzymatic activity of alkaline phosphatase precursor depends on its cellular location. In: Torriani-Gorini A, Rothman FG, Silver S, Wright A, Yagil E, editors. Phosphate Metabolism and Cellular Regulation in Microorganisms. Washington, DC: American Society for. Microbiology Press; 1987. pp. 89–93. [Google Scholar]
- Brotherus JR, Moller JV, Jorgensen PL. Soluble and active renal Na,K-ATPase with maximum protein molecular mass 170,000 ± 9,000 daltons; formation of larger units by secondary aggregation. Biochem Biophys Res Comm. 1981;100:146–154. doi: 10.1016/s0006-291x(81)80075-7. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to select promoters in Escherichia coli using bacteriophage Lambda and Mu. J Mol Biol. 1976;141:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Case CC, Bukau B, Granett S, Villarejo MR, Boos W. Contrasting mechanisms of envZ control of mal and pho regulon genes in Escherichia coli. J Bacteriol. 1986;166:706–712. doi: 10.1128/jb.166.3.706-712.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on the biogenesis of Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma CL, Danese PN, Carlson JH, Silhavy TJ, Snyder WB. Activation of the Cpx two-component signal transduction pathway in Escherichia coli suppresses envelope associated stresses. Mol Microbiol. 1995;18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coliσE regulon. J Biol Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger LA, Inouye M. Purification and characterization of the periplasmic domain of EnvZ osmosensor in Escherichia coli. Biochem Biophys Res Commun. 1996;231:68–72. doi: 10.1006/bbrc.1996.6007. [DOI] [PubMed] [Google Scholar]
- Eggert US, Ruiz N, Falcone BV, Branstrom AA, Goldman RC, Silhavy TJ, Kahne D. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, et al. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:18712–18717. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Janakiraman A, Slauch JM. Construction of targeted single copy lac fusions using ë Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- Fleischer R, Heermann R, Jung K, Hunke S. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J Biol Chem. 2007;282:8583–8593. doi: 10.1074/jbc.M605785200. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodeling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MN, Silhavy TJ. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K-12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- Head CG, Tardy A, Kenney LJ. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- Heyde M, Portalier R. Regulation of major outer membrane porin proteins of Escherichia coli K12 by pH. Mol Gen Genet. 1987;208:511–517. doi: 10.1007/BF00328148. [DOI] [PubMed] [Google Scholar]
- Hoch JA, Silhavy TJ. Two-Component Signal Transduction. Washington, DC: American Society for Microbiology Press.; 1995. [Google Scholar]
- Hsing W, Silhavy TJ. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing W, Russo FD, Bernd KK, Silhavy TJ. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac DD, Pinkner JS, Hultgren SJ, Silhavy TJ. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc Natl Acad Sci USA. 2005;102:17775–17779. doi: 10.1073/pnas.0508936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, Vianney A, Beloin C, Ghigo J-M, Lazzaroni J-C, Lejeune P, Dorel C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Tanabe H, Utsumi R. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Latifi T, Groisman EA. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranslational activator PmrD. Proc Natl Acad Sci USA. 2003;100:4706–4711. doi: 10.1073/pnas.0836837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Mizuno T, Mizushima S. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins 0–8 and 0–9 of Escherichia coli K-12. J Bacteriol. 1979;140:843–847. doi: 10.1128/jb.140.3.843-847.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney LJ. Kinase activity of EnvZ, an osmoregulatory signal transducing protein of Escherichia coli. Arch Biochem Biophys. 1997;346:303–311. doi: 10.1006/abbi.1997.0315. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Liu X, Ferenci T. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J Bacteriol. 1998;180:3917–3922. doi: 10.1128/jb.180.15.3917-3922.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan M, Earhart CF. Reduction in three iron-regulated outer membrane proteins and protein a by the Escherichia coli K-12 perA mutation. J Bacteriol. 1981;146:804–807. doi: 10.1128/jb.146.2.804-807.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Kenney LJ. Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J Biol Chem. 2002;277:11143–11148. doi: 10.1074/jbc.M111128200. [DOI] [PubMed] [Google Scholar]
- Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A Laboratory Manual and Handbook for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. A short course in bacterial genetics; pp. 71–74. [Google Scholar]
- Misra R. First glimpse of the crystal structure of YaeT's POTRA domains. ACS Chem Biol. 2007;2:649–651. doi: 10.1021/cb700212p. [DOI] [PubMed] [Google Scholar]
- Misra R, Castillo-Keller M, Deng M. Overexpression of protease-deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol. 2000;182:4882–4888. doi: 10.1128/jb.182.17.4882-4888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufryk C, Marie-Laure C, Fang FC, Gross CA. Characterization of the six lipoproteins in the σE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Inouye M. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Lynch AS, Belin D, Lin ECC, Beckwith J. Regulation of Escherichia coli cell envelope and proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- Rhee JE, Sheng W, Morgan LK, Nolet R, Liao X, Kenney LJ. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem. 2008;283:8664–8677. doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006;4:43–59. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Advances in understanding of bacterial outer-membrane biogenesis. Nature Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Sarma V, Reeves P. Genetic locus (ompB) affecting a major outer-membrane protein in Escherichia coli K-12. J Bacteriol. 1977;132:23–27. doi: 10.1128/jb.132.1.23-27.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Machida K, Arikado E, Saito H, Kekegawa T, Kobayashi H. Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl Environ Microbiol. 2000;66:943–947. doi: 10.1128/aem.66.3.943-947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Cromie MJ, Hsu F-F, Turk J, Groisman EA. PhoP-regulated Salmonella resistance to the antimicrobial peptides megainin 2 and polymyxin B. Mol Microbiol. 2004;53:229–241. doi: 10.1111/j.1365-2958.2004.04107.x. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; Experiments with gene fusions. [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch JM, Garrett S, Jackson DE, Silhavy TJ. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol. 1988;170:439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompkins K, Chattopadhyay B, Xiao Y, Henk MC, Doerrler WT. Temperature sensitivity and cell division defects in an Escherichia coli strain with mutations in yghB and yqjA, encoding related and conserved inner membrane proteins. J Bacteriol. 2008;190:4489–4500. doi: 10.1128/JB.00414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokishita SI, Kojima A, Aiba H, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the periplasmic domain of the membrane-located protein kinase EnvZ. J Biol Chem. 1991;266:6780–6785. [PubMed] [Google Scholar]
- Tran VK, Oropeza R, Kenney LJ. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J Mol Biol. 2000;299:1257–1270. doi: 10.1006/jmbi.2000.3809. [DOI] [PubMed] [Google Scholar]
- Villarejo M, Case CC. envZ mediates transcriptional control by local anesthetics but is not required for osmoregulation in Escherichia coli. J Bacteriol. 1984;159:883–887. doi: 10.1128/jb.159.3.883-887.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C, Moreno F, Schwartz M. Pleiotropic mutations rendering Escherichia coli K-12 resistant to bacteriophage TP1. J Bacteriol. 1980;143:1374–1383. doi: 10.1128/jb.143.3.1374-1383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL, Sarthy A, Beckwith J. Escherichia coli pleiotropic mutant that reduces amounts of several periplasmic and outer membrane proteins. J Bacteriol. 1979;140:229–239. doi: 10.1128/jb.140.1.229-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ishihama A. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci Biotechnol Biochem. 2006;70:1688–1695. doi: 10.1271/bbb.60024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


