Abstract
Background
RNF139/TRC8 is a potential tumor suppressor gene with similarity to PTCH, a tumor suppressor implicated in basal cell carcinomas and glioblastomas. TRC8 has the potential to act in a novel regulatory relationship linking the cholesterol/lipid biosynthetic pathway with cellular growth control and has been identified in families with hereditary renal (RCC) and thyroid cancers. Haploinsufficiency of TRC8 may facilitate development of clear cell-RCC in association with VHL mutations, and may increase risk for other tumor types. We report a paternally inherited balanced translocation t(8;22) in a proposita with dysgerminoma.
Methods
The translocation was characterized by FISH and the breakpoints cloned, sequenced, and compared. DNA isolated from normal and tumor cells was checked for abnormalities by array-CGH. Expression of genes TRC8 and TSN was tested both on dysgerminoma and in the proposita and her father.
Results
The breakpoints of the translocation are located within the LCR-B low copy repeat on chromosome 22q11.21, containing the palindromic AT-rich repeat (PATRR) involved in recurrent and non-recurrent translocations, and in an AT-rich sequence inside intron 1 of the TRC8 tumor-suppressor gene at 8q24.13. TRC8 was strongly underexpressed in the dysgerminoma. Translin is underexpressed in the dysgerminoma compared to normal ovary.
TRC8 is a target of Translin (TSN), a posttranscriptional regulator of genes transcribed by the transcription factor CREM-tau in postmeiotic male germ cells.
Conclusion
A role for TRC8 in dysgerminoma may relate to its interaction with Translin. We propose a model in which one copy of TRC8 is disrupted by a palindrome-mediated translocation followed by complete loss of expression through suppression, possibly mediated by miRNA.
Background
Dysgerminomas are rare ovarian tumors most common in adolescent women, representing 5–10% of all malignant ovarian tumors in the first two decades of life. They arise from germ cells within the gonad and represent the ovarian counterpart of testicular seminoma[1]. Histologically and clinically dysgerminomas are classified as type II Germ Cell Tumors (GCT).
The female germ cells enter meiosis during intrauterine development (11–12 weeks of gestation), whereas for the male germ cells this only happens after the onset of puberty. This could explain the difference in incidence of the type II gonadal GCTs between females and males and the median age of clinical manifestation. In fact, the number of target cells (PGCs/gonocytes) for initiation is significantly lower in females compared with males [2].
RNF139/TRC8 (NM_007218; henceforth, TRC8) is a potential tumor suppressor gene (TSG) with similarity to PTCH [3]. Mutations in PTCH result in predisposition to basal cell carcinoma and medulloblastoma, while its inactivation leads to cell proliferation [4-6]. TRC8 was identified in a family with the constitutional translocation t(3;8)(p14.2;q24.1), and hereditary renal cell carcinoma (RCC) and thyroid cancer[3,7,8]. Recently, a second, independent, family with hereditary kidney cancer was discovered that carries a cytogenetically indistinguishable translocation and a VHL mutation. [9]. Secondary loss of the wild type TRC8 allele was observed in a subset of tumor cells, consistent with tumor suppressor function. TRC8 is a membrane-bound E3 ubiquitin ligase that inhibits the growth of cells in a ubiquitylation-dependent manner [10]. Interestingly, its level and stability are modulated by cholesterol and it interacts with components of the lipid homeostatic machinery, especially INSIG and the lipid-regulated transcription factors, SREBP1/2 [11]. A frequent genetic alteration in both hereditary and sporadic RCC is mutation of VHL, which encodes the targeting subunit of another ubiquitin ligase complex responsible for degrading hypoxia inducible factors, HIF1/2 alpha. TRC8 interacts with VHL and, in fruit flies, knockdown of either gene led to a similar embryonic phenotype [12]. Thus alterations in TRC8 have the potential to affect cellular growth control and possibly its linkage to lipid homeostasis and/or hypoxia responses.
TRC8 has been identified as a target of the Translin (TSN) gene in postmeiotic male germ cells, and testis is known to contain higher levels of TRC8 mRNA than other tissues[3]. TSN, formerly known as testis brain-RNA binding protein, is found in the cytoplasm and functions as a posttranscriptional regulator of a group of genes transcribed by the transcription factor CREM-tau. Cho et al.[13] identified four new TSN target mRNAs encoding diazepam-binding inhibitor-like 5, arylsulfatase A, a tetratricopeptide repeat structure-containing protein, and TRC8. These observations suggest an important function for TRC8 in germ cells. Here, we describe a paternally inherited balanced translocation t(8;22) in a proposita with dysgerminoma. The breakpoints of the translocation are located within the LCR-B low copy repeat on chromosome 22q11.21, containing the palindromic AT-rich repeat (PATRR) involved in recurrent and non-recurrent translocations and in an AT-rich sequence inside intron 1 of the TRC8 tumor-suppressor gene at 8q24.13. This translocation raises the possibility that disruption of TRC8 may contribute to development of the proposita's dysgerminoma.
Methods
Clinical report
The proband is the only one child born to healthy non-consanguineous parents with an unremarkable family history. Neither cancers nor miscarriages were reported among her relatives. She was born at term after an uneventful pregnancy. At birth, her growth parameters were normal. The neonatal period was normal. She was referred to our Institute at the age of 11 years because of persistent abdominal pain with fever and evidence of a pelvic solid mass discovered with ultrasonography in another hospital.
A total body CT scan upon admission showed a large (17 × 12 × 10 cm) inhomogeneous and necrotic mass arising from the pelvis with important displacement of the urinary tracts and bilateral hydronephrosis; a concomitant peritoneal fluid was also present in the pouch of Douglas.
No distant lesions were detected in other abdominal organs, CNS and lungs. The Tc-99 scintigraphy was negative for bone lesions. Among serum markers, only LDH (12980 U/L) (N.V. 84–362 U/L) and β-hCG (113 U/L) (N.V. <5 U/L) were abnormal for age, suggesting the hypothesis of a germinal tumor. A surgical approach was decided as first treatment. During surgery, a tumoral mass arising from the left ovary was completely resected with consensual monolateral annessectomy; a biopsy of the contra lateral ovary and a sample of peritoneal fluid were also sampled for histological evaluation.
Histological analysis confirmed a classic dysgerminoma of the ovary without evidence of tumoral cells in other tissue samples. No chemotherapy was administered after surgery according to Italian protocol (TCG 2004) for stage 1 ovarian dysgerminoma and the patient was discharged with a follow-up program. Nine months after the diagnosis the patient is healthy and shows normal β-hCG values for age.
Hystology and Immunohistochemistry
Formalin-fixed paraffin-embedded tissue specimens were cut from paraffin blocks and stained with hematoxylin and eosin for microscopic examination. Immunohistochemical labelling was performed by a three-step indirect immunoperoxidase technique. Formalin fixed, paraffin embedded, 3 micron serial tissue sections were incubated at room temperature for 30 minutes with the antibodies reported in Table 1. Additional sections were incubated overnight at 4°C with a rabbit polyclonal antibody against the C-terminus of the p145 human c-kit protein (Oncogene, Boston, MA, USA) and with hypoxia-related antibodies: anti-HIF-2α mAb (clone ep190p, Abcam; Cambridge, UK), anti-VEGF polyclonal Ab (Santa Cruz Biotechnology; Santa Cruz, CA), anti-FGFR (clone VBS1, Chemicon; Temecula, CA), anti-VHL (clone 3F391, Abcam), anti-CA IX polyclonal Ab (Abcam), anti-COX2 polyclonal Ab (Abcam), anti-IGF1R (clone SPM138, Abcam). A monoclonal antibody anti-RNF139/TRC8 (Abcam) was used on normal fallopian tube and dysgerminoma specimens from our proposita.
Table 1.
Antibodies
| Antibody | Source |
| anti-MNF116 cytokeratin mAb) | (clone MNF116, Dako; Glostrup, Denmark |
| anti-AE1/AE3 cytokeratin mAb | (clone AE1/AE3, Dako) |
| anti-CD66e mAb | (clone 12-140-10, Novocastra; Newcastle, UK) |
| anti-EMA mAb | (clone GP1.4, Novocastra) |
| anti-vimentin mAb | (clone V9, Novocastra) |
| anti-PLAP mAb | (clone 8A9, Novocastra) |
| anti-beta HCG mAB | (Novocastra) |
| anti-NSE mAb | (clone 5E2, Novocastra) |
| anti-GFAP mAb | (clone GA5, Novocastra) |
| anti-desmin mAb | (clone DE-R-11, Novocastra) |
| anti-Ki 67 mAb | (clone MIB-1, Dako) |
| anti-CD31 mAb | (clone JC70A, Dako) |
| anti-CD45 | (clone T29/33, Dako) |
| anti-CD68 mAb | (clone KP1, Dako) |
| anti-CD117 | polyclonal Ab (Dako) |
| p145 human c-kit protein | (Oncogene, Boston, MA, USA) |
| CD30 | (Clone BER-H2)Dako |
| S100 | Polyclonal Ab, Dako |
| CD3 | Polyclonal Ab, Dako |
| CD20 | Clone L26, Dako |
| CD45 | Clone T29/33, Dako |
| CD56 | Clone CD564, Novocastra |
| Synaptophysin | Clone SY38, Dako |
| Chromogranin A | Clone DAK-A3, Dako |
Sections were subsequently incubated at room temperature with anti-mouse or anti-rabbit Ig antibodies conjugated to peroxidase-labelled dextran polymer (Dako). The chromogen 3, 3-diaminobenzidine was used in the presence of hydrogen peroxide. Slides were counterstained with Mayer's haematoxylin.
Negative controls were performed using blocking serum in place of primary antibody. Positive and negative controls were run in parallel with each batch, and appropriate results were obtained. Finally, histochemical reaction of cryosectioned tumor was used to evaluate lipid deposition. Cryosections were fixed in 10% neutral buffered formalin. Samples were stained with Oil Red "O" kit (Diapath, Martinengo, Italy).
Cytogenetics investigations
Chromosome preparations were made from cultured lymphocytes from the proposita and her parents using standard high-resolution techniques. To define the breakpoint of the translocation we used fluorescent in situ hybridization (FISH) with BAC clones spanning the chromosomal 8q24.1 and 22q11.2 regions selected according to the University of California Santa Cruz (UCSC) Human Genome Assembly (March 2006 assembly).
FISH analysis on paraffin embedded tumoral tissue
To prepare paraffin-embedded tissue sections fixed on positively charged slides, we cut 4 – 5 μm thick paraffin sections using a microtome. Floating sections were mounted on positively charged slides. To deparaffinize specimens, slides were treated by Paraffin Pretreatment Kit (Abbott Molecular Inc., IL, USA) and treated with protease, then hybridization was performed with the appropriate Vysis protocol.
Array – CGH analysis
Molecular karyotyping was performed using the Human Genome CGH Microarray Kits 44B and 244A (Agilent Technologies, Palo Alto, CA, USA) covering the whole genome with a resolution of ~100 kb and ~35 Kb, respectively. Briefly, 1 μg of patient and sex-matched pooled reference DNAs were processed according to the manufacturer's protocol. Fluorescence was scanned in a dual-laser scanner and the images were extracted and analyzed with Agilent Feature Extraction software (v9.5.3.1) and CGH Analytics software (v3.5.14) respectively. Changes in test DNA copy number at a specific locus are observed as the deviation of the log ratio value from a modal value of 0.
Generation of somatic cell hybrids
Somatic cell hybrid clones were generated by fusing the HPRT-negative RJK88 Chinese hamster cell line with lymphoblastoid cell lines (LCLs) from the patient[14].
DNA extraction and genotyping
Genomic DNA from fresh and frozen samples was extracted using a standard proteinase K digestion, followed by phenol/chloroform extraction, and resuspended in water. FFPE material from rolled sections was extracted using the QiaAmp DNA Mini Kit (Qiagen) according to manufacturer's instructions, with minor modifications. DNA quantitation was determined by spectrophotometry (NanoDrop, Thermo Scientific, Wilmington, DE). Other genomic DNAs from tissues and cell lines were extracted with DNAzol (MRC, Inc, Cincinnati, OH). Genotyping of polymorphic loci was performed by amplification with primers labeled with fluorescent probes (ABI 5-Fam, Hex and Tet) followed by analysis on an ABI 310 Genetic Analyzer (Applied Biosystems). Non-polymorphic loci were assayed by electrophoresis on agarose gels. All primers were purchased from MWG Biotech AG (Ebersberg, Germany). The sequences of all primers used are available from the authors.
Southern blot analysis was conducted on 5 μg aliquots of genomic DNA cut with SacI and PstI restriction enzymes. The DNAs were separated on 0.8% agarose/0.5× TBE gels, transferred to Hybond-N+ (GE Healthcare, Italy), hybridized to non-radioactively labelled probes (Gene Images Random-Prime DNA Labeling kit, GE Healthcare, Italy) and detected with CDP-Star detection reagent (GE Healthcare, Italy).
Expression analysis
Total RNA was extracted from all tissues and cell lines with Trizol (Invitrogen, Milano, Italy) following manufacturer's protocols. Additional Human Tissue Total RNA was purchased from Stratagene. cDNA synthesis was performed with Ready-To-Go You-Prime First strand beads (Amersham) and random hexamers; Non-quantitative RT-PCR was performed in 25 μl reactions, using JumpStart Red ACCUTaq LA DNA polymerase (Sigma) and the following protocol: 1 min at 96°C; 30 cycles of 30 sec at 94°C/30 sec at 58°C/2 min at 68°C; 5 min at 68°C final elongation time; PCR products were analysed on 1.5% agarose TAE gels. G3PDH amplification primers and protocol were from Clontech.
Quantitative gene expression was assessed by Real-Time Quantitative PCR (RT-Q-PCR) on a 7900 HT Sequence Detection System (Applied Biosystems) using SYBR®Green PCR Master Mix (Applied Biosystems). Validation experiments demonstrated that amplification efficiencies of the control and all target amplicons were approximately equal (not shown); accordingly, relative quantification of DNA amount was obtained using the Comparative CT method (described in Applied Biosystems User Bulletin #2, December 11, 1997: ABI PRISM 7700 Sequence Detection System).
TRC8 mutation and CpG island methylation analysis
Primer pairs were selected covering the promoter region (1 pair), exon 1 (1 pair), and the protein coding region of exon 2 (4 pairs) of the TRC8 gene. PCRs were performed in 25 μl reactions, using JumpStart Red ACCUTaq LA DNA polymerase (Sigma) and the following protocol: 30 sec at 96°C; 35 cycles of 30 sec at 94°C/30 sec at 60°C/2 min at 68°C; 10 min at 68°C final elongation time; 5% DMSO was added to promoter and exon 1 PCRs. All PCR products were analysed on 1.5% agarose TAE gels.
We also analyzed CpG methylation of the TRC8 promoter-associated CpG island. Bisulphite treatment was carried out as described by Kubota et al.[15]. Methylation levels were tested by sequencing and restriction mapping of the fragments amplified from bisulphite-treated DNA with TaqI restriction enzyme. The VHL gene was sequenced from tumor and normal DNA according to the methods described by Poland et al. [9].
Results
Histology and Immunohystochemistry
The left ovary measured 17 × 12 × 10 cm and its weight was 1,115 g. The cut surface was solid, partly nodular, and fleshy, with gray/rose coloration and some necrotic areas. Histologically, the tumor was composed of uniform cells consistent with a diagnosis of dysgerminoma. No metastases were demonstrated microscopically in any of the retroperitoneal lymph nodes examined. The right ovary was apparently normal with a light elongated shape.
The tumor showed strong positive staining (with membrane enhancement) for PLAP, CD117 and CD68, focal positivity for MNF116, while the balance of antibodies were negative. TRC8 was positive in normal Fallopian tube but negative in the dysgerminoma (Fig. 1A, B). Expression profiles of hypoxia-related tissue factors were as follows: HIF-2α and FGFR expression was detectable only in a minority of cells, while VHL and CA IX were consistently expressed in the context of CD117 positive vital tumor tissue (Fig. 1C–G); in the same areas, immunolabeling with the C-terminus of the p145 human c-kit protein was negative. Histochemical reaction with Oil Red "O" on cryosectioned tissue tumor showed a moderate deposition of lipids in selected areas (Fig 1H).
Figure 1.
Immunohistochemical analysis of antigens in dysgerminoma. (A) normal Fallopian tube; (B-H) dysgerminoma. (A) intense TRC8 immunoperoxidase expression in control tissue. (B) negative immunoperoxidase TRC8 reaction in dysgerminoma; C, CD117 strong membrane staining in dysgerminoma; D, diffuse and intense VHL expression; E, FGFR cytoplasmic expression; F, diffuse and intense CAIX expression; G, HIF2alpha nuclear and cytoplasmic expression in sporadic cells; H, moderate lipid deposition detected by Oil Red O. Original magnification A-H ×100.
Cytogenetic analysis and mapping of the translocation breakpoints
Giemsa staining identified a chromosomal translocation (8;22)(q24.13;q11.21) in the proband (Fig. 2A, B). The translocation was also present in her apparently normal father, while her mother had a normal karyotype. No additional relative in the paternal lineage was available for testing. FISH analysis using genomic clones localized the translocation breakpoints to within RP11-562F10 (GenBank Accession No. AZ537723) at 22q11.21 (Fig. 2C) and RP11-158K1 (GenBank Accession No. AF313041) at 8q24.13 (Fig. 2D). Five known genes have been mapped to the RP11-158K1 clone (TRMT12, TRC8, TATDN1, NDUFB9, MTSS1) and three to RP11-562F10 (ZNF74, SCARF2, KLHL22). This latter BAC clone also contains two segmental duplications, DC3315 and DC3316.
Figure 2.

Cytogenetics and FISH results from the proposita's lymphocytes, A, cut-out and B, idiogram of the normal (left) and derivative (right) chromosomes 8 and 22. C, FISH with BAC RP11-562F10 (red signals) spanning the breakpoint on the long arm of chromosome 22. D, FISH with BAC RP11-158K1 demonstrates that the chromosome 8 breakpoint is within this clone. Arrowheads indicate the signal on the normal chromosome while arrows mark derivative chromosomes.
To further characterize the breakpoints, we generated somatic cell hybrids from the lymphoblastoid cell line of the proposita. From 100 hybrid clones, one contained only a normal chromosome 8, six retained a normal chromosome 22, seven retained the der(8) and four retained the der(22) chromosome. Based on the UCSC (March 2006 freeze) map, we typed the hybrids for informative chromosome 8 markers (see Table 2) and localized the breakpoint to a 700 bp region in intron 1 of the TCR8 gene. This region contains an AluSp and a 180 bp (AT)n simple repeat (Fig. 3A, B).
Table 2.
Genomic typing of somatic cell hybrid clones with chromosome 8 and 22 markers.
| Locus | Position (Kb) | Genomic | Chr. 8 | Chr. 22 | der(8) | der(22) | |
| Chromosome 8 | D8S1784 | 106172 | 287 | 287 | 287 | ||
| D8S514 | 123811 | 221/223 | 223 | 221 | |||
| D8S1832 | 125517 | 180/186 | 180 | 186 | |||
| RH1927 | 125534 | + | + | + | |||
| RNF139 E1 | 125556 | + | + | + | |||
| Chr8-2-1 | 125559 | + | + | + | |||
| Chr8-2-2 | 125562 | + | + | + | |||
| Chr8-3-1 | 125563 | + | + | + | |||
| Chr8-3-2 | 125564 | + | + | + | |||
| Chr8-2-3 | 125565 | + | + | + | |||
| RNF139 E2 | 125567 | + | + | + | |||
| TATDN1 E10 | 125570 | + | + | + | |||
| TATDN1 E6 | 125590 | + | + | + | |||
| NDUFB9 E2 | 125625 | + | + | + | |||
| NDUFB9 E4 | 125631 | + | + | + | |||
| MTSS1 E14 | 125635 | + | + | + | |||
| MTSS1 E13 | 125637 | + | + | + | |||
| MTSS1 E11 | 125639 | + | + | + | |||
| D8S1799 | 125646 | 234 | 234 | 234 | |||
| D8S272 | 137804 | 241/255 | 255 | 241 | |||
| Chromosome 22 | D22S427 | 16971 | 100 | 100 | 100 | ||
| RH78249 | 18480 | + | + | + | |||
| RH57884 | 18609 | + | + | + | |||
| RH102235 | 19092 | + | + | + | |||
| RH93744 | 19126 | + | + | + | |||
| D22S280 | 31539 | 210/212 | 210 | 212 | |||
Figure 3.
Map of the translocated regions. (A) schematic of the genomic region rearranged on chromosome 8 showing the translocation breakpoint (black arrowhead), the BAC clones used to refine the cytogenetic characterization (colored bars), and the molecular localization of the breakpoint to an AT-rich simple repeat (light grey box) in intron 1 of the TRC8 gene. The position of the SacI restriction sites (S) and of the probe used for Southern analysis are indicated. (B), Southern blot showing SacI-cleaved fragments of the first intron containing the AT-rich repeat in the proband (P), her father (F), several normal controls (C1–3) and the BAC RP11-158K1 (Bac). Positions of the proband's alleles (arrows) and two molecular weight markers are shown. (C) Localization of the chromosome 22 breakpoint, contained within BAC RP11-562F10.
The 8q24 breakpoint region was refractory to amplification (not shown). We examined the repeat-rich segment by digestion with SacI followed by Southern blot analysis and observed two bands both in the proband and her father; the larger shared fragment presumably contains the rearrangement's breakpoint. One of the normal control samples also contained two alleles; in all cases, the fragments were several hundred bases larger than in the RP11-158K1 clone (Fig. 3B), consistent with polymorphism of the AT-rich VNTR.
On chromosome 22, the breakpoint was localized to the LCR-B [16]/LCR22-3a [17] which is the same region involved in the recurrent t(11;22)(q23;q11.2) [16,18,19] as well as t(17;22)(q11.2;q11.2) and several other translocations [17] (Fig. 3C). Since most of the 22q11 breakpoints involved in these translocations occur at the center of PATRR22, we postulated that this was also the most likely breakpoint in our case, but the breakpoint junctions could not be amplified using the method described by Gotter et al[20].
To investigate the presence of the translocated chromosomes in the tumoral tissue, we performed a dual colour hybridization experiment on deparaffinized slices from the dysgerminoma (summarized in Fig. 4). We used a probe specific for the centromere of chromosome 8 (CEP8; Vysis) to visualize its copy number in single cells. To detect the normal and derivative chromosomes 8 and 22 we used dual FISH with BACs RP11-383J24 (chromosome 8) and CTD-2319K10 (chromosome 22) both adjacent to the breakpoints, to unambiguously identify the derivative 8 (Fig. 4C). Dual FISH was also performed with BACs CTD-3107D22 (chromosome 22) and RP11-532M24 (chromosome 8) to detect the derivative 22 (Fig. 4D). This experiment revealed that ~40% of tumor cells contained two copies of the derivative 8 while while the remaining 60% contained one copy. Similar results were obtained for the derivative 22; all cells also contained one normal chromosome 8 and one normal chromosome 22.
Figure 4.
Interphase FISH on deparaffinized sections from dysgerminoma. a) schematic representation of the results of the interphase FISH. b) Nuclei show two or three red signals (CEP8) indicating the presence of nuclei with two or three chromosomes 8, respectively.c) Dual color hybridization with BAC RP11-383J24 (red signal) and BAC CTD-2319K10 (green signal) indicate the presence of a normal chromosome 8 and a normal chromosome 22 (single signals); nuclei show one or two derivative chromosome 8 (arrows). d) Dual color hybridization with BAC CTD-3107D22 (red signal) and BAC RP11-532M24 (green signal) indicate the presence of a normal chromosome 8 and a normal chromosome 22 (single signals) and one or two derivative chromosome 22 (arrows).
Array-CGH results
We carried out array-CGH analysis on the dysgerminoma's DNA to map all chromosomal imbalances at ~100 kb average resolution. Non-mosaic gain of an entire chromosome was detected for chromosomes 7 and 15, while mosaicism for a whole-chromosome gain was detected for chromosomes 1, 6, 16, 19, 20, and 21 (not shown). A structural chromosomal imbalance was detected for chromosomes 2, 4, 8, 11, 12, 17, and 22, both as gain or loss (see Table 3). Array comparative hybridization profiles of the abnormal chromosomes are shown in Fig. 5. Array-CGH analysis with 8.9 KB overall median probe spacing, performed on the genomic DNA of our proband and her parents, excluded a concomitant deletion or duplication.
Table 3.
List of the imbalances found in the analysis of the Dysgerminoma.
| Chr | Chromosome Band | Gain/Loss* | Position | Size (Mb) | Notes |
| Chr 2 | 2p24.1 → 2q37.1 | enh | Chr2:24,134,000..233,241,000 | 209.10 | mosaic 30% |
| Chr 2 | 2q37.1 → qtel | enh | Chr2:233,241,000..242,951,149 | 9.71 | More copies |
| Chr 4 | 4q21.21 → qtel | dim | Chr4:80,599,000..191,273,063 | 110.70 | mosaic 30% |
| Chr 8 | 8ptel → q24.13 | enh | Chr8:1..125,719,000 | 125.70 | |
| Chr 8 | 8q24.13 → qtel | enh | Chr8:125,719,000..146,274,826 | 20.56 | mosaic 30% |
| Chr 11 | 11p15.3 → 11q23.3 | dim | Chr11:11,570,000..119,487,000 | 107.90 | mosaic 30% |
| Chr 11 | 11q23.3 → qtel | dim | Chr11:119,487,000..134,452,384 | 14.97 | |
| Chr 12 | 12ptel → q23.1 | enh | Chr12:1..94,196,000 | 94.20 | More copies |
| Chr 12 | 12q23.1 → qtel | enh | Chr12:94,196,000..132,349,534 | 38.15 | |
| Chr 17 | 17q21.3 → qtel | enh | Chr17:38,922,000..78,774,742 | 39.85 | mosaic 30% |
Regions interested by the deletions/duplications/triplications are reported together with their mapping position and cytogenetic bands (hg 17/Build 35, May 2004), The size of each imbalance is always related to the platform resolution having 100 kb average distribution of the clones along the genome. The size of the intervening regions is shown, although it should be considered that breakpoints may fall within a segment of 100 kb (average) proximal and distal to the starting and ending clones. * Enh = enhanced log2 ratio (gain); dim = diminished log2 ratio (loss).
Figure 5.

Array comparative genome hybridization profiles of the chromosomes involved in rearrangements in the dysgerminoma.
VHL gene mutations
The proband was heterozygous for multiple VHL SNPs in her blood DNA, while her father was homozygous for all of them. We amplified by long-range PCR and sequenced all exons of the VHL gene on 3p25.3 in both directions from tumor DNA, finding no evidence of mutations or loss of heterozygosity.
Expression analysis
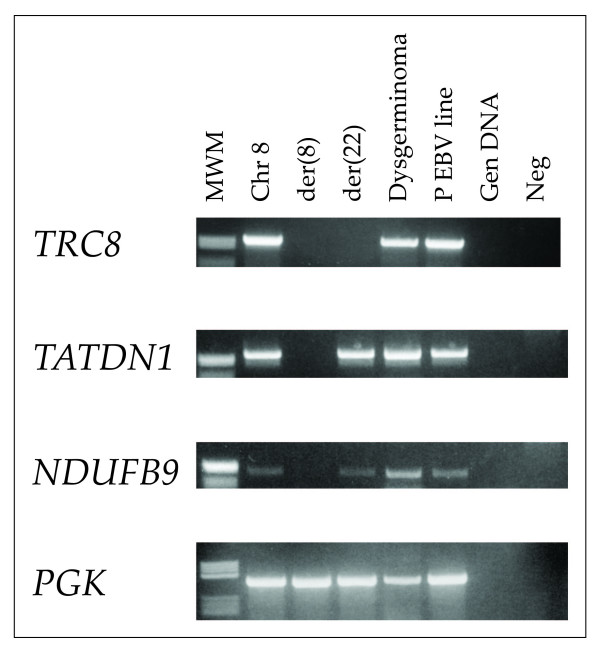
Non-quantitative RT-PCR analysis of TRC8 showed that the gene was expressed in the Epstein-Barr Virus (EBV) line and in the hybrid clone containing a normal chromosome 8, but not in hybrids carrying the der(8) or der(22) chromosomes (Fig. 6). In contrast, the neighbouring genes TATDN1 and NDUFB1 were expressed in the der(22) hybrid. Expression of all three genes was detectable in the dysgerminoma.
Figure 6.
Non-quantitative RT-PCR analysis for expression of TRC8, TATDN1, NDUFB9 in cell lines and dysgerminoma. RNA isolated from indicated hybrids and proband's nomal or tumor tissues was reverse transcribed and used for PCR amplification with the indicated gene-specific primers. The housekeeping gene PGK was used as a control; MWM, molecular weight marker V (Roche); Neg, no DNA control.
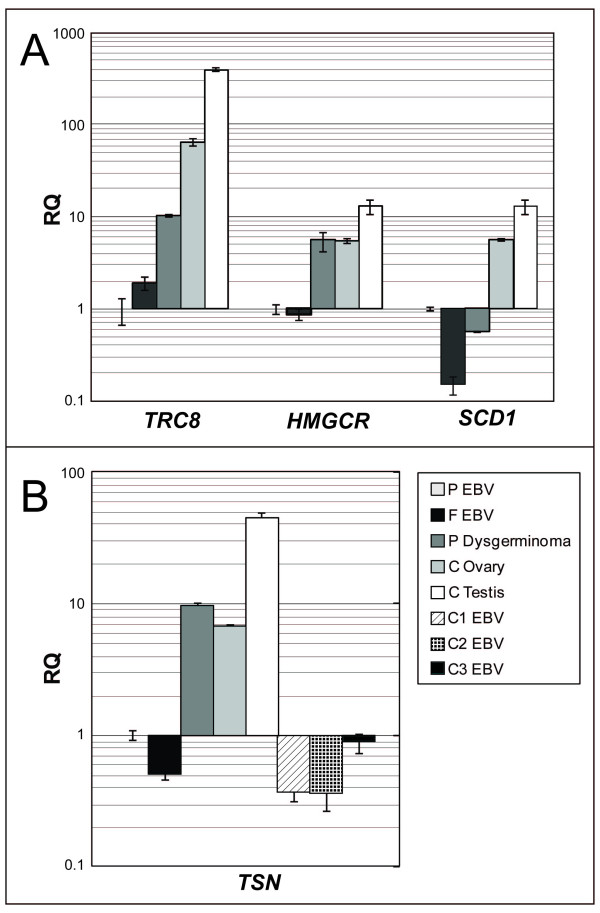
Using Real-Time RT-PCR, we quantitated expression of TRC8 in normal human ovary and testis, in the dysgerminoma, in EBV lines from the proband and her father, and in three additional EBV lines from normal subjects. Ovary and testis RNA come from pooled samples (ovary: 15 caucasian females, aged 20–60; testis: 39 caucasian males, aged 14–64) and consequently can be considered "averaged" tissue samples. We also analyzed the expression of HMGCR and SCD1, two SREBP target genes potentially down-regulated by TRC8 in RCC cells, and of the housekeeping gene PGK (Fig. 7A). Compared to normal ovary, TRC8 was expressed approximately 10 times less in the dysgerminoma. In contrast, it was expressed 4-fold more in testis. All lymphoblastoid lines expressed TRC8 at very low levels, and there was no difference in expression between the subjects carrying the translocation and the control lines. Because of low TRC8 expression in the tumor, we anticipated that SREBP target genes, including HMGCR and SCD1 might be overexpressed. In contrast, we observed that SCD1 was strongly underexpressed while HMGCR was not affected in the dysgerminoma compared to normal ovary. Perhaps the complex regulation of each gene makes it difficult to discern the effect of reduced TRC8, or the whole ovary is not a good control tissue for germ cell expression. In addition, regulation of lipid homeostatic genes by TRC8 may be specific for some tissues or tumors. The level of PGK expression was comparable in all tissues.
Figure 7.
A, Quantitative RT-PCR analysis of TRC8, HMGCR, and SCD1 expression in the proband's dysgerminoma, in normal tissues and in EBV lines from the proband (P) and her father (F). The housekeeping gene PGK was used as endogenous control, while proband's EBV line was the reference tissue. B, Expression analysis of translin (TSN) in tissues and EBV-transformed cell lines. PGK was again used as endogenous control. Relative quantity (RQ) is shown on a logarithmic scale.
We quantified expression of the TSN gene encoding translin, a protein involved in post-transcriptional regulation of a number of genes, including TRC8, during spermatogenesis (Fig. 7B). TSN was expressed approximately 3 times more in the testis than in normal ovary, while it was expressed approximately two times less in the dysgerminoma. Expression was homogeneously lower in all EBV lines. Due to the rarity of dysgerminomas, our findings are limited to the one sample we were able to collect and analyze; whether or not these expression levels are characteristic of dysgerminomas or unique to this translocation case is unknown.
Analysis of tumor-derived TRC8 for genetic and epigenetic changes
Genomic DNA sequences, including the TRC8 promoter and exons 1 and 2, were examined from both the proband's EBV line and the dysgerminoma. There were no differences between the two sequences, and both were identical to the human reference sequence (NC_000008, from 125556189 to 125570040). Sequences of the TRC8 transcript reverse-transcribed from the EBV line and dysgerminoma were also identical to the database. Lastly, we examined promoter methylation patterns of TRC8, and found it was completely unmethylated in both tissues (not shown)
Discussion
We report a girl who developed a dysgerminoma at the age of 10 years and carries a balanced translocation t(8;22)(q24.13;q11.21). The breakpoints of our proposita may be identical to those recently described by Gotter et al., [20]. In that case, the proband showed an unbalanced translocation with a supernumerary der(22) derived as a result of 3:1 meiotic malsegregation of the paternal balanced translocation 46, XY, t(8;22)(q24.13;q11.21). The proband showed an abnormal phenotype, while his father was apparently normal. Interestingly, a breakpoint within the first intron of the TRC8 gene has been described as an inherited rearrangement associated with renal cell carcinoma in two unrelated families[3,9,21,22]. In our case, we have localized the breakpoint to a 700 bp region in intron 1 of the TCR8 gene, containing an AluSp and a 180 bp (AT)n simple repeat. The breakpoint is probably positioned, as in the family described by Gotter et al.[20], in a palindrome within the simple repeat. In our case and in Gotter et al.[20], the breakpoint on chromosome 22 lies within the PATRR on 22q11. The analysis of many unrelated t(11;22)(q23;q11) cases revealed that the breakpoints occur within palindromic PATRRs on 11q23 and 22q11 (PATRR11 and PATRR22). The majority of the breakpoints, including the case described by Gotter et al.[20], are localized to the centre of the PATRRs, suggesting that the palindrome mid-point is susceptible to double-strand breaks (DSBs); this would occur as the result of a cruciform extrusion promoting the DSBs that initiate stabilizing rearrangements or recombination events[23]. The PATRR22 appears to be highly unstable in the human genome[24]. Palindrome-mediated genomic instability contributes to a variety of genome rearrangements including not only constitutional translocations, but also large germ line deletions[25]. Cancer-related gross chromosomal rearrangements and genomic amplification are also reported to be associated with palindromic DNA[26,27]. Recent studies have produced a detailed map showing that palindromes tend to cluster at specific regions, some of which undergo gene amplification[28]. Individual tumors seem to have a non-random distribution of palindromes in their genomes, and a subset of palindromic loci is associated with gene amplification. Inverted repeats or palindromes are known to generate hairpins or cruciform structures, facilitated by intrastrand pairing of complementary single-strand DNA sequences assembled in the lagging strand during DNA replication. A double-strand break (DSB) introduced in these hairpins can result in deletions or in either inter- or intrachromosomal recombination events mediated by the homologous recombination machinery[26,28].
TRC8 encodes an endoplasmic reticulum-resident E3-ubiquitin ligase with multiple transmembrane segments. TRC8 contains a C-terminal RING-H2 domain shown to catalyze in vitro ubiquitylation reactions[10,11]. In Drosophila and mammalian cells, over-expressed TRC8 suppressed growth[11,12,29]. In HEK293 cells, TRC8 induced G2/M arrest and accumulation of sub-G1 cells indicative of apoptosis[11]. These effects were dependent upon an ubiquitylation-competent RING domain, suggesting that specific substrates were critical to growth inhibition. Tumor formation in a nude mouse model was also inhibited by TRC8 in a RING-dependent manner. TRC8 contains a putative sterol-sensing domain (SSD) which in other proteins confers sensitivity to sterols, affecting either stability or trafficking between membrane compartments[3]. Moreover, knock-down of endogenous TRC8 increased levels of the lipid homeostasis transcription factors, the SREBPs and their target genes, including enzymes of cholesterol and lipid metabolism [11]. However, these effects were only evident in cells simultaneously depleted of sterols, since cholesterol-replete cells contain very low amounts of TRC8. Importantly, expression of activated SREBP-1 partially restored the growth of TRC8-inhibited cells [11], indicating that changes in lipid regulation underlie some of the growth inhibitory actions of this tumor suppressor. Since these data suggested that TRC8 activity linked growth control to the cholesterol/lipid homeostasis pathway, we analyzed two SREBP target genes, HMGCR and SCD1, in control and tumor tissue (Fig. 6). However, expression levels were depressed compared to normal ovarian tissues. This could be due to lack of proper control tissues (germ cells as opposed to whole ovary) or perhaps more likely to the presence of plentiful lipids, since the effects of TRC8 loss are most readily observed following sterol-depletion [11].
Interestingly, TRC8 expression appeared down-regulated in tumor tissue (Fig. 6) while TRC8 protein was not detectable in tumor tissue by IHC (Fig. 1). This was despite the absence of any evidence for loss, genetic alteration or promoter methylation of the remaining TRC8 allele. We surmise that additional mechanisms reducing TRC8 expression may be involved in this dysgerminoma. For example, the 3' UTR of TRC8 contains target sites for multiple microRNAs (miR), several of which are highly conserved among human, mouse, rat, dog and chicken (miR-218, -101, -27, -128, -375). In addition, the coding sites for some of these miRs are found on the aneuploid chromosomes observed in this tumor (i.e., miR-218-1 is on chromosome 4). It is possible that miRNAs are mediating the additional loss of expression for this gene. Recently, the stabilization of specific miRNAs by Translin in male germ cells has been reported[30].
In our patient Translin (TSN), a protein involved in post-transcriptional regulation of TRC8 and of a number of genes during spermatogenesis, is expressed two times less in the dysgerminoma than in normal ovary.
Translin, formerly known as testis brain-RNA binding protein (TB-RBP), is both a DNA-binding and RNA-binding protein. As a DNA binding protein, it interacts with the breakpoint junctions of chromosomal translocations[31,32]. Translin also mediates intracellular and intercellular mRNA transport, possibly controlling the temporal and spatial translation of specific mRNAs in postmeiotic germ cells [33-35]. Kasai et al. [36] have proposed that human Translin may play a role in DNA recombination, repair, and metabolism, but deficiencies in somatic recombination or DNA repair are not readily detectable in mice lacking Tsn [37]. However, the marked increase in apoptosis of spermatocytes in Tsn-null mice suggests a role for TSN during meiosis, perhaps functioning as a post-transcriptional regulator in spermatocytes [37]. Recently, Cho et al. [13] identified four meiotic mRNA targets of Translin, one of which is TRC8. They hypothesized that disruption of the precise regulation of TRC8 may cause the growth retardation and the increased apoptosis in pachytene spermatocytes observed in Tsn-null mice [37]. To our knowledge, there are no studies of the role of the TSN gene in female meiosis.
TRC8 is a potent TSG involved in different tissue-specific pathways. Its haploinsufficiency may facilitate the development of CC-RCC in association with VHL mutations, or otherwise lead to increased risk for other types of tumor. Any role in dysgeminoma may relate to its interaction with translin. We envision a model whereby one copy of TRC8 is disrupted by palindrome-mediated translocation (step 1), then, in a second step, translin, miRNA, or another yet undiscovered mechanism leads to further loss of TCR8 expression, setting the stage for deregulated proliferation.
Abbreviations
(RNF139): Ring Finger Protein 139; (TRC8): Translocation in Renal Carcinoma on chromosome 8; (PGC): Primordial Germ Cell; (BAC): Bacterial Artificial Chromosome; (array-CGH): Comparative Genomic Hybridization; (IHC): Immunohystochemistry.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RMG, RG, GG, OZ, and CG contributed to the writing of this paper. Experimental work was done by SG, SB, HAD, PF and AG. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by Telethon Grant GGP06208A to R.G.
Contributor Information
Stefania Gimelli, Email: stefania.gimelli@gmail.com.
Silvana Beri, Email: silvana.beri@bp.lnf.it.
Harry A Drabkin, Email: harry.drabkin@uchsc.edu.
Claudio Gambini, Email: claudiogambini@ospedale-gaslini.ge.it.
Andrea Gregorio, Email: andreagregorio@ospedale-gaslini.ge.it.
Patrizia Fiorio, Email: patriziafiorio@ospedale-gaslini.ge.it.
Orsetta Zuffardi, Email: zuffardi@unipv.it.
Robert M Gemmill, Email: gemmill@musc.edu.
Roberto Giorda, Email: roberto.giorda@bp.lnf.it.
Giorgio Gimelli, Email: giorgiogimelli@ospedale-gaslini.ge.it.
References
- Ulbright TM. Germ cell tumors of the gonads: a selective review emphasizing problems in differential diagnosis, newly appreciated, and controversial issues. Mod Pathol. 2005;18:S61–79. doi: 10.1038/modpathol.3800310. [DOI] [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LH. Testicular germ-cell tumors in a broader perspective. Nat Rev Cancer. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- Gemmill RM, West JD, Boldog F, Tanaka N, Robinson LJ, Smith DI, Li F, Drabkin HA. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc Natl Acad Sci USA. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/S0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr, Scott MP. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Cohen AJ, Li FP, Berg S, Marchetto DJ, Tsai S, Jacobs SC, Brown RS. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med. 1979;301:592–595. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- Li FP, Decker HJ, Zbar B, Stanton VP, Jr, Kovacs G, Seizinger BR, Aburatani H, Sandberg AA, Berg S, Hosoe S, Brown RS. Clinical and genetic studies of renal cell carcinomas in a family with a constitutional chromosome 3;8 translocation. Genetics of familial renal carcinoma. Ann Intern Med. 1993;118:106–111. doi: 10.7326/0003-4819-118-2-199301150-00005. [DOI] [PubMed] [Google Scholar]
- Poland KS, Azim M, Folsom M, Goldfarb R, Naeem R, Korch C, Drabkin HA, Gemmill RM, Plon SE. A constitutional balanced t(3;8)(p14;q24.1) translocation results in disruption of the TRC8 gene and predisposition to clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2007;46:805–812. doi: 10.1002/gcc.20466. [DOI] [PubMed] [Google Scholar]
- Brauweiler A, Lorick KL, Lee JP, Tsai YC, Chan D, Weissman AM, Drabkin HA, Gemmill RM. RING-dependent tumor suppression and G2/M arrest induced by the TRC8 hereditary kidney cancer gene. Oncogene. 2007;26:2263–2271. doi: 10.1038/sj.onc.1210017. [DOI] [PubMed] [Google Scholar]
- Lee JP, Brauweiler A, Rudolph M, Hooper JE, Drabkin HA, Gemmill RM. Proceedings of the 100th Annual Meeting of the American Association for Cancer Research. Denver, CO. Philadelphia (PA): AACR; 2009. TRC8/RNF139, a kidney cancer-associated tumor suppressor gene and E3-ubiquitin ligase, is sterol-regulated and interacts with lipid and protein biosynthetic pathways [abstract] p. 1287. [Google Scholar]
- Gemmill RM, Bemis LT, Lee JP, Sozen MA, Baron A, Zeng C, Erickson PF, Hooper JE, Drabkin HA. The TRC8 hereditary kidney cancer gene suppresses growth and functions with VHL in a common pathway. Oncogene. 2002;21:3507–3516. doi: 10.1038/sj.onc.1205437. [DOI] [PubMed] [Google Scholar]
- Cho YS, Iguchi N, Yang J, Handel MA, Hecht NB. Meiotic messenger RNA and noncoding RNA targets of the RNA-binding protein Translin (TSN) in mouse testis. Biol Reprod. 2005;73:840–847. doi: 10.1095/biolreprod.105.042788. [DOI] [PubMed] [Google Scholar]
- Giorda R, Cerritello A, Bonaglia MC, Bova S, Lanzi G, Repetti E, Giglio S, Baschirotto C, Pramparo T, Avolio L, Bragheri R, Maraschio P, Zuffardi O. Selective disruption of muscle and brain-specific BPAG1 isoforms in a girl with a 6;15 translocation, cognitive and motor delay, and tracheo-oesophageal atresia. J Med Genet. 2004;41:e71. doi: 10.1136/jmg.2003.012260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Das S, Christian SL, Baylin SB, Herman JG, Ledbetter DH. Methylation-specific PCR simplifies imprinting analysis. Nat Genet. 1997;16:16–17. doi: 10.1038/ng0597-15. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, McCain N, Goldberg R, Pandita RK, Duong S, Fox J, Blumenthal D, Lalani SR, Shaffer LG, Morrow BE. A common breakpoint on 11q23 in carriers of the constitutional t(11;22) translocation. Am J Hum Genet. 1999;65:1608–1616. doi: 10.1086/302689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiteri E, Babcock M, Kashork CD, Wakui K, Gogineni S, Lewis DA, Williams KM, Minoshima S, Sasaki T, Shimizu N, Potocki L, Pulijaal V, Shanske A, Shaffer LG, Morrow BE. Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum Mol Genet. 2003;12:1823–1837. doi: 10.1093/hmg/ddg203. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Emanuel BS. Alu-mediated PCR artifacts and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2000;9:2727–2732. doi: 10.1093/hmg/9.18.2727. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Gotter AL, Nimmakayalu MA, Jalali GR, Hacker AM, Vorstman J, Conforto Duffy D, Medne L, Emanuel BS. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog FL, Gemmill RM, Wilke CM, Glover TW, Nilsson AS, Chandrasekharappa SC, Brown RS, Li FP, Drabkin HA. Positional cloning of the hereditary renal carcinoma 3;8 chromosome translocation breakpoint. Proc Natl Acad Sci USA. 1993;90:8509–8513. doi: 10.1073/pnas.90.18.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/S0092-8674(00)81034-X. [DOI] [PubMed] [Google Scholar]
- Ashley T, Gaeth AP, Inagaki H, Seftel A, Cohen MM, Anderson LK, Kurahashi H, Emanuel BS. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am J Hum Genet. 2006;79:524–538. doi: 10.1086/507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Inagaki H, Ohye T, Kogo H, Kato T, Emanuel BS. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Hoglund M, Johansson B, Hagemeijer A, Park SS, Mitelman F, Lupski JR, Fioretos T. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- Kato T, Inagaki I, Kogo H, Ohye T, Yamada K, Emanuel BS, Kurahashi H. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum Mol Genet. 2008;17:1184–1191. doi: 10.1093/hmg/ddn008. [DOI] [PubMed] [Google Scholar]
- Gemmill RM, Lee JP, Chamovitz DA, Segal D, Hooper JE, Drabkin HA. Growth suppression induced by the TRC8 hereditary kidney cancer gene is dependent upon JAB1/CSN5. Oncogene. 2005;24:3503–3511. doi: 10.1038/sj.onc.1208509. [DOI] [PubMed] [Google Scholar]
- Yu Z, Hecht NB. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J Androl. 2008;29:572–579. doi: 10.2164/jandrol.108.005090. [DOI] [PubMed] [Google Scholar]
- Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- Wu XQ, Gu W, Meng X, Hecht NB. The RNA-binding protein, TBRBP, is the mouse homologue of translin, a recombination protein associated with chromosomal translocations. Proc Natl Acad Sci USA. 1997;94:5640–5645. doi: 10.1073/pnas.94.11.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales CR, Wu XQ, Hecht NB. The DNA/RNA-binding protein, TB-RBP, moves from the nucleus to the cytoplasm and through intercellular bridges in male germ cells. Dev Biol. 1998;201:113–123. doi: 10.1006/dbio.1998.8967. [DOI] [PubMed] [Google Scholar]
- Morales CR, Lefrancois S, Chennathukuzhi V, El-Alfy M, Wu X, Yang J, Gerton GL, Hecht NB. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev Biol. 2002;246:480–494. doi: 10.1006/dbio.2002.0679. [DOI] [PubMed] [Google Scholar]
- Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB. The kinesin KIF17b and RNA binding protein TB-RBP transport specific cAMP responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci USA. 2003;100:15566–15571. doi: 10.1073/pnas.2536695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M, Matsuzaki T, Katayanagi K, Omori A, Maziarz RT, Strominger JK, Aoki K, Suzuki K. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J Biol Chem. 1997;272:11402–11407. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- Chennathukuzhi V, Stein JM, Abel T, Donlon S, Yang C, Miller J, Allman D, Simmons RA, Hecht NB. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol. 2003;23:6419–6434. doi: 10.1128/MCB.23.18.6419-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]