Abstract
Mast cells play a central role in type I hypersensitivity reactions and allergic disorders such as anaphylaxis and asthma. Activation of mast cells, through a cascade of phosphorylation events, leads to the release of mediators of the early phase allergic response. Understanding the molecular architecture underlying mast cell signaling may provide possibilities for therapeutic intervention in asthma and other allergic diseases. Although many details of mast cell signaling have been described previously, a systematic, quantitative analysis of the global tyrosine phosphorylation events that are triggered by activation of the mast cell receptor is lacking. In many cases, the involvement of particular proteins in mast cell signaling has been established generally but the precise molecular mechanism of the interaction between known signaling proteins often mediated through phosphorylation is still obscure. Using recently advanced methodologies in mass spectrometry, including automation of phosphopeptide enrichments and detection, we have now substantially characterized, with temporal resolution as short as 10 s, the sites and levels of tyrosine phosphorylation across 10 min of FcεRI-induced mast cell activation. These results reveal a far more extensive array of tyrosine phosphorylation events than previously known, including novel phosphorylation sites on canonical mast cell signaling molecules, as well as unexpected pathway components downstream of FcεRI activation. Furthermore, our results, for the first time in mast cells, reveal the sequence of phosphorylation events for 171 modification sites across 121 proteins in the MCP5 mouse mast cell line and 179 modification sites on 117 proteins in mouse bone marrow-derived mast cells.
Keywords: Mast Cells/Basophils, Fc Receptors, Signal Transduction, Protein Kinases/Phosphatases
Introduction
Mast cells are regarded as crucial effector cells in allergic reactions and IgE associated immune responses (1). Activation of mast cells, a critical feature of type I hypersensitive reactions, leads to the release of a wide range of chemical mediators and cytokines that recruit inflammatory cells and regulate inflammatory responses, such as mucus secretion, vasodilation, and bronchoconstriction (2). During activation, the mast cell high-affinity IgE receptor FcεRI is crosslinked by allergens through bound IgE, leading to a cascade of signaling events and the release of preformed inflammatory mediators localized in specialized granules, the de novo synthesis and secretion of pro-inflammatory lipid mediators, and the synthesis and secretion of cytokines and chemokines (3).
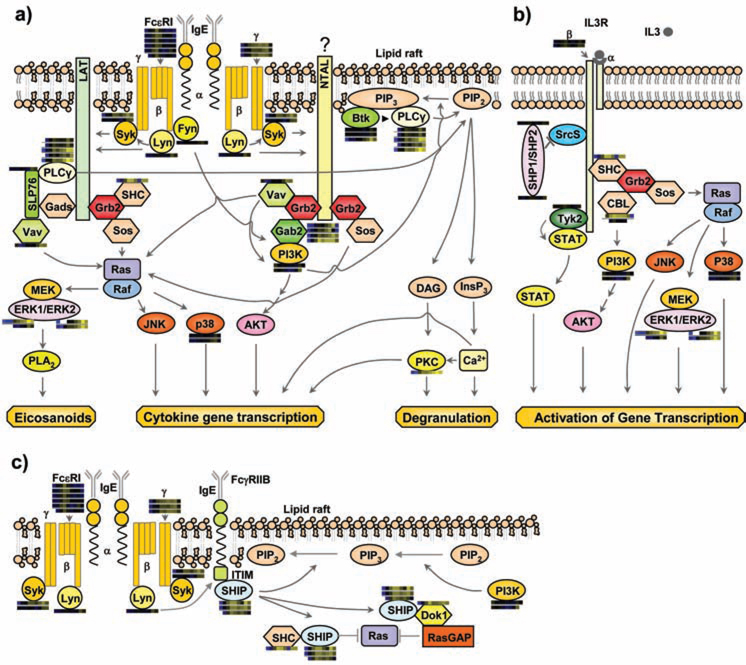
Some aspects of FcεRI-mediated mast cell activation and inhibition pathways have been described previously (4) (Figure 1). FcεRI receptor expressed on mast cells is comprised of three subunits: an IgE-binding α subunit, a signal-amplifying β subunit, and two disulfide-linked signal-initiating γ subunits (5). Following FcεRI aggregation, the activation of β subunit-bound protein tyrosine kinase Lyn and, subsequently, tyrosine phosphorylation of ITAM regions in the β and γ subunits of FcεRI, initiates a complex series of intracellular signaling events (6–10). Phosphorylated ITAMs provide docking sites for SH2 domain-containing cytoplasmic tyrosine kinases such as Syk (11), Lyn, and Fyn (12), leading to the activation of Syk (11). The subsequent phosphorylation of LAT results in the recruitment and activation of several proteins including Gads, PLCγ, SLP76, VAV, Grb2, SHC, and SOS leading ultimately to mast cell degranulation, cytokine gene transcription, and synthesis of lipid mediators (4). The phosphorylation of NTAL might also participate in mast-cell signaling by binding Grb2 (4). In mast cells, coaggregation of FcεRI with the low affinity receptor for IgG (FcγRIIb), leads to the inhibition of antigen-induced mast cell degranulation and cytokine production (Figure 1c) (13). Inhibitory pathways triggered by FcεRI/FcγRIIb coaggregation include the Lyn kinase mediated phosphorylation of ITIM tyrosine residues in FcγRIIb leading to recruitment of SHIP followed by its association with SHC and Dok1, eventually culminating in inactivation of Ras (3).
Figure 1.
Multiple previously established signaling cascades in activated mast cells with quantitative proteomic data from MCP5 cells represented as heatmap bars beside individual proteins. These heatmap bars represent the change of abundance of phosphorylation on known mast cell signaling proteins through a timecourse of Fc receptor stimulation (as additionally portrayed in Figure 4a). a) FcεRI-mediated activation pathways. b) IL-3 receptor mediated pathway. c) A representative inhibitory pathway. Syk: spleen tyrosine kinase; LAT: linker for activation of T cells; Grb2: growth-factor-receptor-bound protein 2; Sos: son of sevenless homologue; SHC: SH2-domain-containing transforming protein C; Gads: Grb2-related adaptor protein 2; SLP76: SH2-domain-containing leukocyte protein of 65 kDa; PLCγ: phospholipase C γ; NTAL: non T cell activation linker; Gab2: Grb2-associated binding protein 2; ERK: extracellular-signal-regulated kinase; PLA2: phospholipase A2; p38: Mitogen activated protein kinase 14; PIP2: phosphatidylinositol-4,5-bisphosphate; PIP3: phosphatidylinositol-3,4,5-triphosphate; BTK: bruton's tyrosine kinase; DAG: diacylglycerol; InsP3: inositol-1,4,5-triphosphate; PKC: protein kinase C; SHP1/SHP2: SH2 containing protein tyrosine phosphatase 1/2; Srcs: Src family kinases ; Dok1: docking protein, 62 KD; RasGAP: GTPase activating protein .
Recent advances in the area of phosphorylation analysis utilizing emerging mass spectrometric proteomic technologies, have created the possibility of simultaneous, quantitative monitoring in total cellular lysates of large numbers of phosphorylation sites following receptor stimulation (14–16). To facilitate the quantitative analysis of tyrosine phosphorylation sites in FcεRI activated mast cells, peptide phosphotyrosine immunopreciptiation (14, 17) was employed to enrich complex cellular lysates for tyrosine phosphorylated peptides prior to immobilized metal affinity chromatography (IMAC) and MS/MS analysis using a label-free quantitation method (18) (Figure 2).
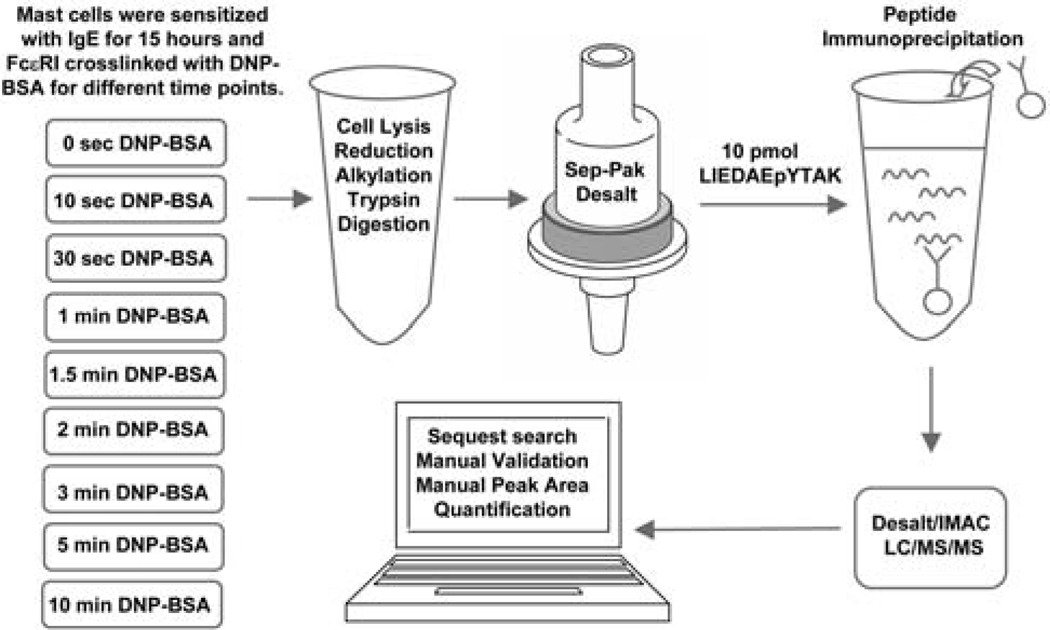
Figure 2.
Experimental design. Mast cells were sensitized with anti-DNP IgE, FcεRI receptors were crosslinked with DNP-BSA through bound IgE for the indicated time. Cells were lysed, proteins were reduced, alkylated, and trypsin digested to peptides. Peptides were desalted by Sep-Pak cartridge, enriched through immunoprecipitation and IMAC, and then subjected to LC-MS/MS analysis. The SIC peak area of a copurified synthetic peptide LIEDAEpYTAK was used to normalize cell-derived phosphopeptide SIC peak areas in a label free quantitation method.
A plethora of quantitative proteomic methods have recently been developed utilizing the incorporation of isotopic labels into proteins or through “label-free” methodologies (19). Highly reproducible chromatographic enrichment and detection of cell-derived phosphopeptides is essential for the successful implementation of a label-free quantitation method. To provide the necessary chromatographic performance and sensitive detection of phosphopeptides, an automated phosphoproteomic platform for the enrichment of phosphotyrosine immunoprecipitated peptides was utilized (20). This system employs automated desalting, IMAC enrichment and reversed-phase separation of peptides in a highly reproducible fashion as all column elutions and loading steps are precisely replicated with computer controls. This system provides highly reproducible retention times and peak areas as well as highly sensitive detection of cell-derived phosphopeptides as described previously (20).
FcεRI-mediated mast cell signaling pathways, including many components, their interactions and their phosphorylation sites, have been intensively studied. However, a systematic, quantitative analysis of global mast cell phosphorylation has never been performed. To generate a comprehensive map of mast cell phosphorylation events, a quantitative proteomic analysis of mast cell signaling through FceRI was performed. This study provides a starting foundation for the detailed examination of the biological role of these newly discovered phosphorylation sites through mutagenesis, leading ultimately to a better molecular understanding of the structure of the mast cell signaling pathway.
Materials and Methods
Cell Culture, FcεRI Stimulation and Cell Lysis
The mouse bone marrow-derived mast cell line MCP5 was cultured in RPMI 1640 (Sigma, St. Louis, MO) supplemented with 10% heat-inactivated FBS (Sigma), 2 mM L-glutamine, 100 U/ml penicillinG, 100 µg/ml streptomycin (Cellgro, Herndon, VA) and 5% conditioned media from mouse IL-3 expressing D11 fibroblasts grown in 5% CO2 at 37 °C. Bone marrow cells from C57BL/6 mice were cultured in IL3-containing medium for 4–6 wk to generate immature mast cells (bone marrow-derived mast cells (BMMCs)) with > 95% purity (c-Kit+ and FcεRI+ by flow cytometry). Animal experiments were approved by the Brown University Institutional Animal Care and Use Committee and performed in accordance with the guidelines of the National Institutes of Health. MCP5 cells and BMMCs were sensitized with 0.5 µg/ml mouse monoclonal anti-DNP IgE (clone SPE-7, Sigma) overnight at a density of 2×106 cells/ml in a CO2 incubator at 37 °C. Cells were then washed with Tyrode's buffer and treated at 2.5×107 cells/ml in Tyrode's buffer with 100 ng/ml DNP[27]-BSA (Biosearch Technologies, Novato, CA) at 37 °C, for varying durations. The reaction was stopped by the addition of a final concentration of 8 M urea, 1 mM Na3VO4, 100 mM NH4HCO3, pH 8.0 lysis buffer. After incubation on ice for 20 min, lysates from all time points were centrifuged at 14,000xg for 15 min at 4 °C. Lysate protein concentration was measured by the DC Protein Assay (Biorad, Hercules, CA).
Protein Reduction, Alkylation and Digestion
Proteins were reduced with 10 mM DTT for 1 h in a 56 °C water bath followed by alkylation with 55 mM indoacetamide for 1 h at room temperature in the dark. Cell lysates were diluted five times with 100 mM NH4HCO3, pH 8.9. Proteins were digested with affinity purified, TPCK treated trypsin (Promega, Promega, WI) at a trypsin:protein ratio 1:100 (w/w) overnight at 37 °C. Tryptic peptides were desalted using Sep-Pak C18 Cartridges (Waters, Milford, MA) as described (21) and dried in a Speed Vac plus (Thermo Savant, Holbrook, NY).
Peptide Immunoprecipitaton
Dry peptides from each time point (1×108 cells/time point) were reconstituted in 1 ml cold IP buffer (30 mM Tris, 30 mM NaCl, and 0.3% NP40, PH 7.4) and 10 pmol synthetic peptide LIEDAEpYTAK was added to each time point as a control for label-free quantitation, accompanying the cellular phosphopeptides through the peptide immunoprecipitation, subsequent peptide purification steps, and reversed-phase elution of peptides into the mass spectrometer. Bead-conjugated anti-phosphotyrosine antibody was prepared by coupling monoclonal anti-phosphotyrosine antibody clone pTyr100 (Cell Signaling Technology, Danvers, MA) to protein G agarose beads (Roche, Basel, Switzerland) noncovalently at 2 mg/ml overnight at 4 °C. Anti-phosphotyrosine beads were then added at 15 µl resin/ 1×108 cells overnight at 4 °C with gentle shaking. Beads were washed and eluted as described (14).
Automated Desalt-IMAC/nano-LC/ESI-MS
Tryptic peptides were analyzed by a fully automated phosphoproteomic technology platform incorporating peptide desalting via reversed-phase chromatography, gradient elution to an Fe3+-loaded IMAC column, reversed-phase separation of peptides followed by tandem mass spectrometry with static peak parking as described previously (20). Briefly, tryptic peptides were loaded onto a desalting reversed-phase column (360 µm OD × 200 µm ID fused silica (Polymicro Technologies, Phoenix, AZ), fritted with an inline microfilter (M520, Upchurch Scientific, Oak Harbor, WA), packed on a pressure bomb (GNF Commercial Systems, San Diego, CA) with 12 cm SelfPack POROS 10 R2 resin (Applied Biosystems, Foster City, CA)). Peptides were rinsed with 0.1 M acetic acid/MilliQ water (solvent A) at a flow rate of 10 µl/min and then eluted to an Fe3+-activated IMAC column (360 µm OD × 200 µm ID fused silica, bomb packed with 15 cm SelfPack POROS 20 MC resin (Applied Biosystems)) with an HPLC gradient of 0–70% solvent B (0.1M acetic acid/acetonitrile) in 17 min at a flow rate of 1.8 µl/min. The column was washed with ∼40 µl of a 25:74:1 acetonitrile/water/acetic acid mixture containing 100 mM NaCl followed by 4 min solvent A at a flow rate of 0.035 ml/min. Enriched phosphopeptides were eluted to a precolumn (360 µm OD × 75 µm ID) containing 2 cm of 5 µm Monitor C18 resin (Column Engineering, Ontario, CA) with 40 µl of 25 mM potassium phosphate (pH 9.0) and rinsed with solvent A for 30 min. Peptides were eluted into the mass spectrometer (LTQ-FT, Thermo Electron, San Jose, CA) through an analytical column (360 µm OD × 75 µm ID fused silica with 12 cm of 5 µm Monitor C18 particles with an integrated ∼4 µm ESI emitter tip fritted with 3 µm silica; Bangs Labs, Fishers, IN) with an HPLC gradient (0–70% solvent B in 30 min). Static peak parking was performed via flow rate reduction from the initial 200 nl/min to ∼20 nl/min when peptides began to elute as judged from a BSA peptide scouting run as described previously (20). The electrospray voltage of 2.0 kV was applied in a split flow configuration as described (20). Spectra were collected in positive ion mode and in cycles of one full MS scan in the FT (m/z: 400–1800) (∼1 s each) followed by MS/MS scans in the LTQ (∼0.3 s each) sequentially of the five most abundant ions in each MS scan with charge state screening for +1, +2, and +3 ions and dynamic exclusion time of 30 s. The AGC was 1,000,000 for the FTMS MS scan and 10,000 for the LTMS MS/MS scans. The max ion time was 100 ms for the LTMS MS/MS scan and 500 ms for the FTMS full scan. FTMS resolution was set at 100,000.
Database analysis
MS/MS spectra were automatically searched against the mouse NCBI non-redundant protein database using the SEQUEST algorithm provided with Bioworks 3.2SR1 (22). Search parameters specified a differential modification of phosphorylation (+79.9663 Da) on serine, threonine, and tyrosine residues and a static modification of carbamidomethylation (+57.0215 Da) on cysteine. Prior to manual spectral validation, SEQUEST results were filtered by xcorr (+1> 1.5; +2>2.0; +3>2.5), precursor mass error (<20 ppm), minimum repetition of 5 of 9 total time points containing an MS/MS spectra for each individual peptide, nonredundant within each time point, and phosphotyrosine containing. For all the peptides exceeding these thresholds (labeled “high stringency” throughout this manuscript), SEQUEST peptide sequence assignments and the position of the phosphorylation site were manually validated as described previously (22). Manually validated spectra for all reported phosphopeptides are available (http://mastcellpathway.com).
Quantitation of Relative Phosphopeptide Abundance
Relative quantitation was performed on all manually validated peptides via calculation of selected ion chromatogram (SIC) peak areas, normalization with the SIC peak area of the copurified synthetic peptide LIEDAEpYTAK, and generation of a heatmap representation. SIC peak areas were calculated using newly developed software programmed in Microsoft Visual Basic 6.0 based on Xcalibur Development Kit 2.0 SR1 (Thermo Electron Co., San Jose, CA). Manually validated SIC peak areas for all reported phosphopeptides are available (Supplemental Material 1). Peak areas determined with this tool were consistent with areas determined manually through the manufacturer's software, Xcalibur. Peak areas were represented as heatmaps. Phosphopeptide abundance was represented as black when a peptide's peak area was approximately the geometric mean for that peptide across all time points. A blue color represented abundance less than this average and a yellow color represented abundance more than this average. The magnitude of change of the heatmap color was based on the log of the fold change of each individual peptide peak area compared to the geometric mean for that peptide across all time points. All heatmap representations were based upon SIC peak areas normalized in each time point to LIEDAEpYTAK control peptide that was spiked in an equal amount to each time point sample and copurified and analyzed with the cell-derived peptides. Blanks in the heatmap indicated that both a manually validated MS/MS spectra and SIC peak were not observed for that phosphopeptide in that time point.
Clustering of Phosphorylation Time Series
Peptide peak areas were normalized and clustered according to their temporal profiles. Peptide peak areas were normalized to have a mean of 0 and a standard deviation of 1 in order to capture the temporal profile of each peptide. Peptides with missing data across 9 time points or whose max fold change was less than 4 fold were discarded prior to clustering. Normalized peak areas were clustered using a fuzzy k-means clustering algorithm called MFuzz (23). MFuzz assigns a membership value to each peptide for every cluster, allowing partial membership of a peptide to more than one group. Peptides with less than 70% membership to the best cluster were removed. Since the number of clusters, k, was specified as a parameter to the algorithm, the optimal number of clusters was determined by running the clustering algorithm 100 times with k = 3, ‥, 9. The best run was chosen for each k as the smallest sum of all proteins to their assigned cluster center. Phosphoprotein sequences were searched against the Minmotif Miner (MnM) dataset for identification of sequence motifs containing phosphorylated tyrosine residues (24).
Results
To clearly define the temporal dimension of mast cell signaling as revealed through protein phosphorylation, a phosphoproteomic time course experiment was conducted (Figure 2). Anti-DNP IgE sensitized mouse bone marrow-derived mast cells (BMMCs) and a mast cell line MCP5 were stimulated through DNP-BSA crosslinking. Within the mast cell time course experiment described here, the automated chromatographic system performed as expected with highly reproducible retention times and peak areas for each phosphopeptide amongst the various time points. The LIEDAEpYTAK phosphopeptide normalization standard accompanied the true cellular phosphopeptides through peptide immunoprecipitation, desalt, IMAC, and reversed-phase elution and detection in the mass spectrometer. The average SIC peak area of this peptide standard was 2.4 ×107 with a standard of deviation of 0.7 ×107. Additionally, the standard deviation of retention times of each cellular phosphopeptide was compared to the peptide's average half maximal peak width amongst the nine time points. The magnitude of the variation of retention time amongst the time points for each detected phosphopeptide was well within the width of the peaks on average, indicating coelution of phosphopeptides in the separate time point analyses. The average standard deviation of 6.3 s for peptide retention time amongst the nine time points, the average maximal variation in retention time of 19 s amongst the nine time points, and the average half-maximal SIC peak width of 22 s indicated the highly reproducible acquisition of data.
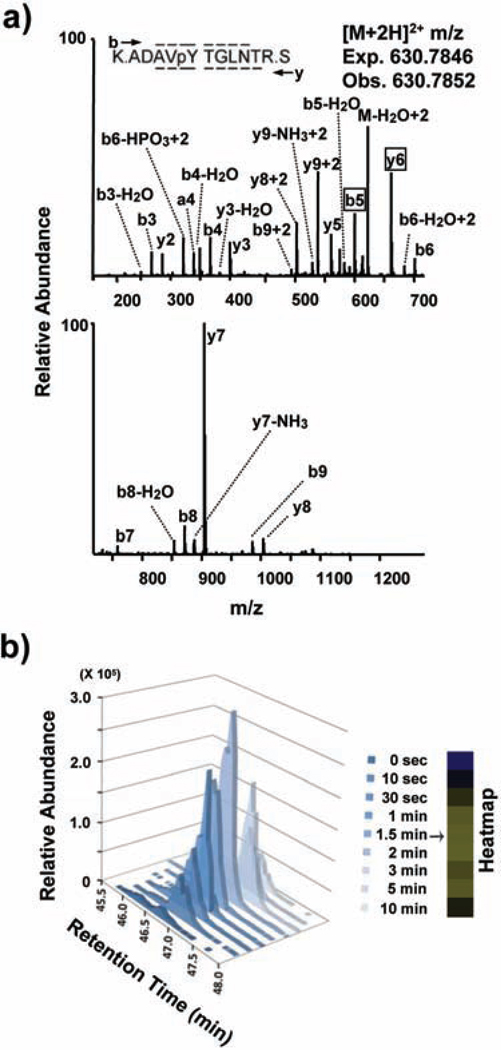
For every phosphopeptide, spectra were manually validated for both the correct sequence assignment as well as the position of the phosphorylation site as illustrated for the FcεRIγ receptor ITAM-containing phosphopeptide ADAVpYTGLNTR (Figure 3a). Although 549 unique tyrosine phosphorylation sites on 426 proteins in MCP5 cells and 450 unique tyrosine sites on 342 proteins in BMMCs were uncovered with high quality sequence assignments (Xcorr +1>1.5; +2>2.0; +3>2.5; precursor mass error less than 20 ppm), only peptides with repeated observations of MS/MS spectra in at least five of the nine time points in the MCP5 dataset and two of the three time points in BMMC dataset were manually validated and quantified (high stringency). We also defined a medium stringency threshold which was identical to the high stringency threshold with only the removal of the requirement for repeated observations of MS/MS spectra in separate time points. Medium stringency threshold was used only in the comparison of BMMC to MCP5 data sets (Figure 4c only). The quantitation for the representative FcεRIγ receptor ITAM-containing phosphopeptde ADAVpYTGLNTR is shown (Figure 3b). After manual validation and quantitation, the temporal variation of 171 phosphorylation sites on 121 proteins was revealed in FcεRI-stimulated MCP5 cells while 179 phosphorylation sites on 117 proteins was revealed in stimulated BMMCs (Supplemental Material 1).
Figure 3.
Manual validation and quantitation of the FcεRIγ ITAM-containing phosphopeptide. A) MS/MS spectrum of the FcεRIγ-derived phosphopeptide ADAVpYTGLNTR. A single MS/MS spectrum is displayed on two axes to enhance detail. Peaks are assigned to theoretical a, b, or y type ions or corresponding neutral loss of ammonia or water from b, or y ions. Sequence coverage across the peptide is represented by overlines or underlines on the peptide sequence. Boxed phosphorylation site-determining peaks (y6 & b5) and the absence of an assigned M-H3PO4 peak definitively indicate that the phosphorylation site is on Tyr65 and not on Ser66. B) Selected-ion chromatogram (SIC) or plot of the ion current over time for m/z 930.7846 corresponding to the (M+2H)2+ of the FcεRIγ-derived phosphopeptide ADAVpYTGLNTR for all the 9 time points. The corresponding heatmap representing the same peptide's relative abundance across the 9 time points is shown.
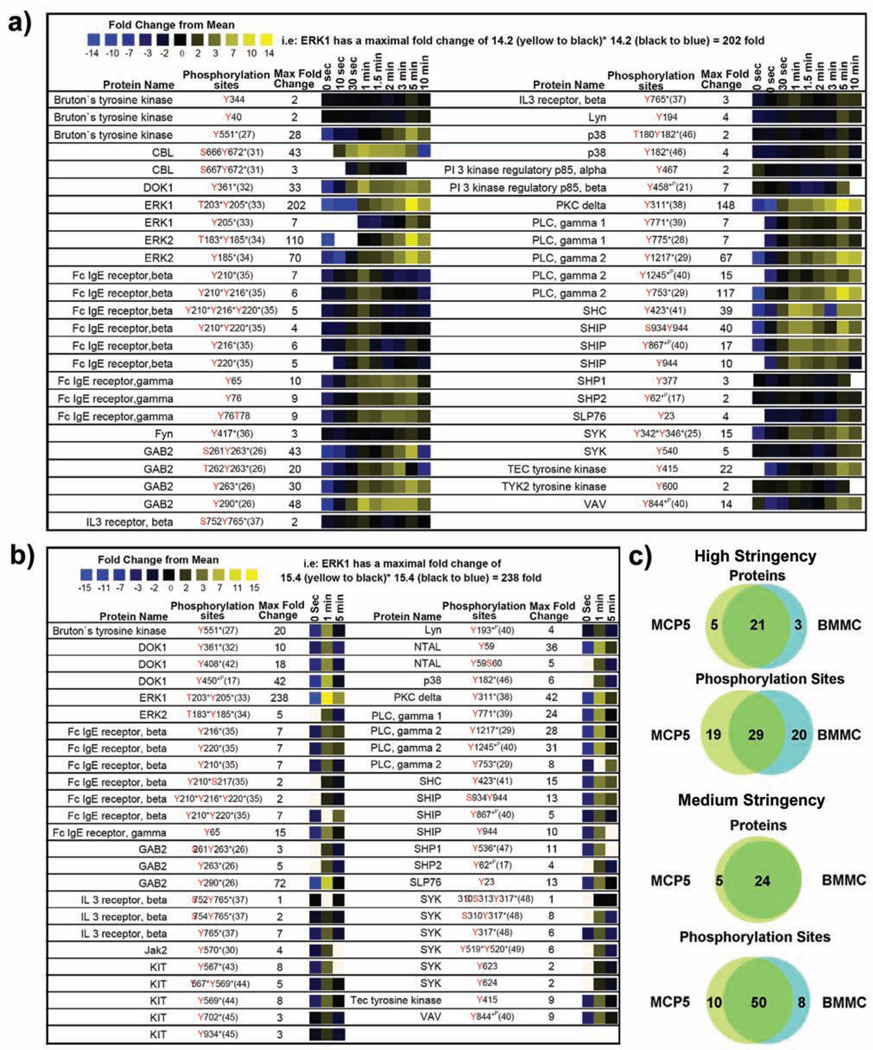
Figure 4.
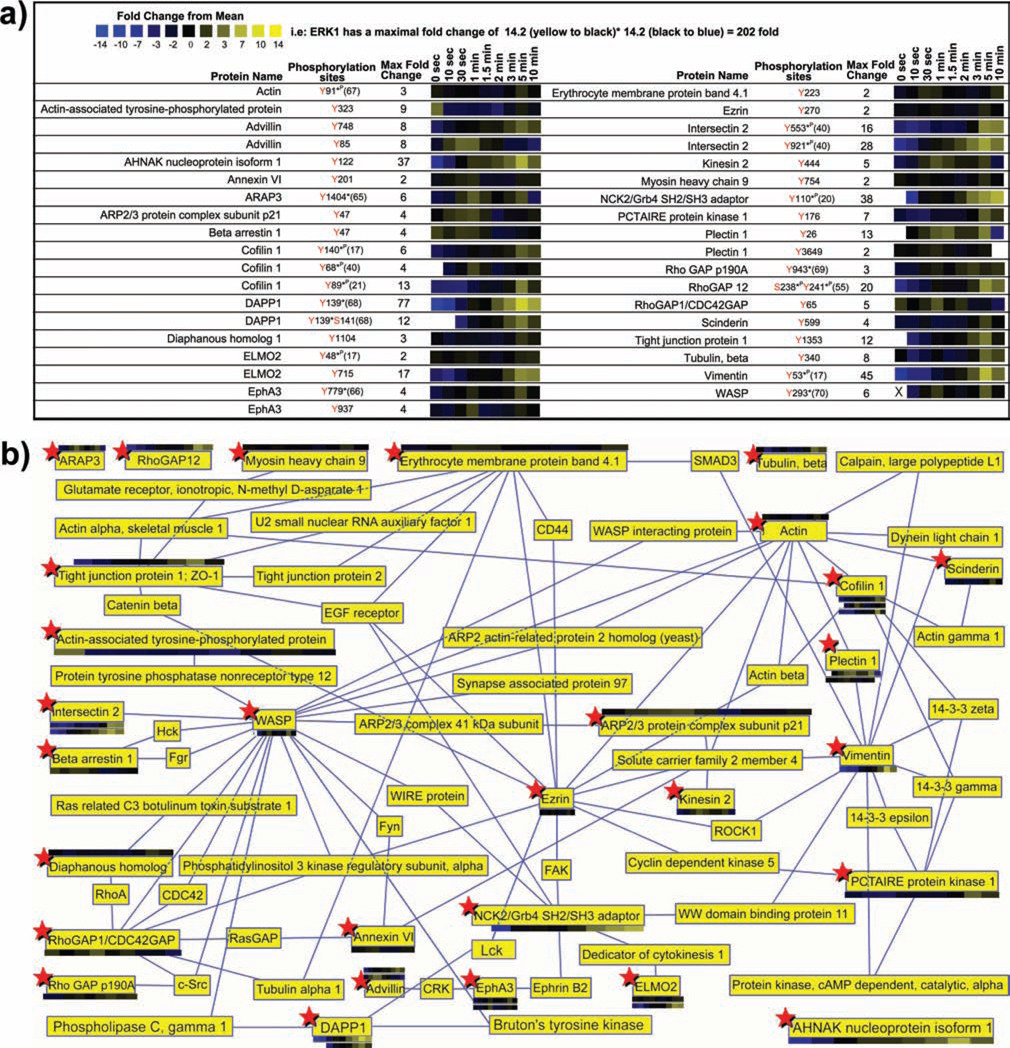
Quantitative proteomic analysis of canonical mast cell signaling proteins. A heatmap representation of temporal changes in tyrosine phosphorylation of proteins previously established to be involved in mast cell signaling observed following FcεRI aggregation in a) MCP5 cells or b) bone marrow derived mast cells (BMMC) from C57BL6 mice. In the heatmap, black color represents a peptide abundance equal to the geometric mean for that peptide across all time points. Blue color represents a peptide abundance less than the mean while yellow color corresponds to an abundance more than the mean. c)Comparison between the overall pattern of tyrosine phosphorylation of canonical mast cell signaling proteins in MCP5 cells and that of BMMCs is shown. Two different data thresholds were applied to the proteomic data to understand the true differences between the proteins and phosphorylation sites observed in BMMC and MCP5. A high stringency comparison required high quality SEQUEST matches with Xcorr (+1> 1.5; +2>2.0; +3>2.5), precursor mass error (<20 ppm), minimum of 5 of 9 (for MCP5) or 2 of 3 (for BMMC) total time points containing an MS/MS spectra for each individual peptide, phosphotyrosine containing, and manually validated. A medium stringency comparison was identical to the high stringency thresholds without minimum repetition of peptide observations in separate time points. Phosphorylation sites discussed in the literature previously are marked with (*P) if identified using phosphoproteomic method alone or (*) if identified using traditional approaches such as site-directed mutagenesis.
Temporal quantitative analysis of canonical mast cell signaling protein phosphorylation
Dynamic tyrosine phosphorylation of proteins associated with the canonical mast cell signaling pathway was observed (Figure 1 and 4) (17, 21, 25–49). Quantitative analysis of tyrosine phosphorylation of proteins involved in the known mast cell signaling pathway for both MCP5 cells (Figure 4a) and BMMCs (Figure 4b) is shown for comparison. The high similarity of tyrosine phosphorylated proteins observed between the MCP5 dataset and the BMMC dataset (89% identical; Figure 4c) argues for the validity of the MCP5 cell line as a model system for mast cell activation through FcεRI. All of the remaining three proteins observed in BMMC but absent from the MCP5 dataset were actually observed in the MCP5 dataset at medium stringency. Furthermore, 59% of identical phosphorylation sites were observed both in MCP5 and BMMC (Figure 4c). Of the 41% of sites observed only in BMMC, over half of those missing sites were also observed in MCP5 at medium stringency.
The small differences in specific phosphorylation sites observed in MCP5 compared to BMMC in the known mast cell signaling pathway could be explained by subtle variations in the absolute abundance of each phosphopeptide, or variations in the level of induction of phosphorylation after FceRI activation. For example, it is quite possible that sites observed in BMMC but seemingly absent in MCP5 are actually present at levels slightly below the sensitivity limit of our proteomic methodology. Also the MCP5 cell line was derived from AD12-SV40 infected BMMC from Balb/C mice while the BMMC analyzed in this experiment were obtained from C57BL6 mice (50). The subtle differences observed in specific phosphorylation sites could also arise from the different strains of mice compared in this experiment.
Lyn binds to FcεRIβ in resting mast cells and once activated following receptor aggregation, it phosphorylates the ITAM tyrosine residues on FcεRIβ and FcεRIγ (6–10). Phosphorylation on both FcεRIβ and FcεRIγ was extremely rapid with increased phosphorylation as early as 10 s and substantial phosphorylation at 1 min. FcεRIγ phosphorylation was sustained at a high level from 1 min to 5 min whereas FcεRIβ phosphorylation declined rapidly after reaching its maximum at 1 min. All ITAM phosphorylation sites on FcεRI were observed in BMMC with the exception of FcεRIγ Tyr76.
The phosphorylation of ITAMs on FcεRI leading to the recruitment and activation of SH2 domain-containing proteins, such as Syk, results in a cascade of phosphorylation of downstream signaling molecules including LAT, Gab2, PI3K, Btk/Tec, PLCγ, and SLP76 (4). We observed novel phosphorylation sites (MCP5 Tyr194, BMMC Tyr193) within the SH2 domain of Lyn and the known kinase activation site (MCP5 Tyr416) within the tyrosine kinase domain of Fyn (51). Mutations of both tyrosine residues (Tyr342Tyr346) of Syk are known to induce a significant reduction in FcεRI-mediated mast cell degranulation and calcium flux (25). We observed an increase in phosphorylation of Syk at 1 min post-receptor stimulation followed by maximal phosphorylation increase of 15 fold overall at 5 min on the two autophosphorylation sites (Tyr342Tyr346) in MCP5. A 6 fold induction of phosphorylation at 1 min on the autophosphorylation sites Tyr317, Tyr519, and Tyr520 of Syk was observed in BMMC. Novel phosphorylation sites (Tyr540, Tyr623, and Tyr624) on Syk were also observed.
Dynamic phosphorylation of known signaling proteins was observed beyond the initial phosphorylation of the receptor and receptor-associated kinases. Adaptor protein Gab2 can be phosphorylated by Fyn tyrosine kinase, creating binding sites for Grb2, PI3K, SHP2, and Crkl (26). We observed strong induction of phosphorylation on the known Crkl binding sites in MCP5 and BMMC (Tyr263, Tyr290) on Gab2. Phosphorylation of biologically uncharacterized sites with strong Scansite (52) motifs for SH2 interaction with the Src kinase Fyn and phosphorylation by Src kinase was observed on the PI3K p85 regulatory subunit beta (Tyr458) and the p85 regulatory subunit alpha (Tyr467) in MCP5 cells. Formation of PIP3 from PIP2, through the action of PI3K, leads to activation of Btk and Tec tyrosine kinases, which results in the phosphorylation and activation of PLCγ (4). Syk or Lyn mediated phosphorylation of Btk (Tyr551 observed in both BMMC and MCP5) leads to activation of Btk tyrosine kinase activity and phosphorylation of PLCγ (27). The location of novel phosphorylation sites observed within the PH domain (Tyr40) and SH2 domain (Tyr344) of Btk in MCP5 suggests that these sites might play a regulatory role in binding of Btk to PIP3 or interactions with other tyrosine phosphorylated signaling molecules. Similarly, phosphorylation of the Btk homologue Tec was observed in both BMMC and MCP5 (Tyr415, homologous to Tyr551 of Btk) within its tyrosine kinase domain. Both Tyr771and Tyr775 on PLCγ1 are known to be activation-induced tyrosine phosphorylation sites and Tyr775 is established as a critical phosphorylation site for PLCγ1 activation (28, 53). Phosphorylation of these two tyrosine sites on PLCγ1 in MCP5 (Tyr771 and Tyr775) and in BMMC (Tyr771) was observed. Phosphorylation of Tyr753 on PLCγ2, which has been previously characterized to regulate the lipase activity of PLCγ2 (54), was highly elevated upon receptor stimulation in both MCP5 and BMMC. PLCγ2 phosphorylation on Tyr1217, and on the biologically uncharacterized site Tyr1245 was also observed in both MCP5 and BMMC (55). Ras can be activated through Syk mediated phosphorylation of SHC and binding of SHC to Grb2 and SOS (56). We observed strong and rapid induction of phosphorylation of SHC on the Syk phosphorylated residue Tyr423 in both MCP5 and BMMC (57). The largest change in phosphorylation after receptor aggregation was observed on the autophosphorylation sites Thr203/Tyr205 of ERK1 (202 fold induced in MCP5 and 238 fold induced in BMMC). Strong elevation of ERK2 autophosphorylation on Thr183/Tyr185 was also observed in both BMMC and MCP5 (58).
Dynamic tyrosine phosphorylation was observed on inhibitory signaling molecules that participate in the regulation of mast cell signaling after coaggregation of FcεRI and FcγRIIB. Although IgG was not added in our experiment, IgE also binds and activates FcγRIIB signaling in mouse mast cells (59). There was rapid and strong induction of tyrosine phosphorylation at previously uncharacterized sites on SHIP (Ser934/Tyr944, Tyr867, Tyr944) in both BMMC and MCP5, an enzyme whose activation leads to cleavage of PIP3 and inhibition of Btk mediated Ca2+ release (3). Following receptor coaggregation, studies have shown that phosphorylated SHIP associates with SHC and Dok1 through their phosphotyrosine binding (PTB) domains (13). Phosphorylation of Dok1 at Tyr361 (induced in both BMMC and MCP5), the major binding site of NCK and RasGAP, leads to its association with RasGAP and inhibition of FcεRI-induced ERK1/2 activation and calcium mobilization (13). Induction of phosphorylation on biologically uncharacterized sites was also observed on Dok1 in BMMC (Tyr408, Tyr450). We observed a profound increase in tyrosine phosphorylation of the Src kinase regulated site Tyr311 of PKCδ (in both MCP5 and BMMC), a negative regulator of antigen-induced mast cell degranulation (60, 61). Clathrin-mediated endocytic recycling of activated FcεRI occurs through interaction between the receptor and the tyrosine phosphorylated scaffold/E3 ubiquitin ligase Cbl and CIN85 complex (62). Strong induction of tyrosine phosphorylation was observed on Cbl in MCP5 after receptor crosslinking (Ser666Tyr672).
Temporal analysis of protein phosphorylation associated with the actin cytoskeleton
In mast cells, actin cytoskeleton is crucially involved in the processes of cytosolic granule exocytosis and receptor endocytosis. Prior to cell stimulation, granules are kept apart from their fusion sites by the cytoskeletal barrier composed predominantly of actin and myosin. In response to stimulation, the actin cytoskeleton serves as a scaffold for signaling molecule recruitment and provides pathways for vesicle transportation (63). A dramatic reorganization of the cytoskeleton is essential for mast cell granule exocytosis (64). Many phosphorylated components of the actin cytoskeleton were observed in FcεRI stimulated MCP5 mast cells (Figure 5) (17, 20, 21, 40, 55, 65–70). Although phosphorylation on cytoskeleton-associated proteins was also observed in BMMC (and available in Supplemental Material 1), the remaining discussion is concentrated on the MCP5 dataset. Phosphorylation of the cytoskeletal components actin (Tyr91), beta tubulin (Tyr340), advillin (Tyr85, Tyr748), cofilin 1 (Tyr68, Tyr140, Tyr89), plectin (Tyr26, Tyr3649), and vimentin (Tyr53) was induced after mast cell receptor activation. Conversely, a rapid reduction of phosphorylation on a previously undescribed phosphorylation site Tyr323 of an “actin-associated tyrosine-phosphorylated protein” was observed immediately following receptor aggregation.
Figure 5.
Quantitative proteomic analysis of proteins associated with the actin cytoskeleton. a) A heatmap representation of temporal changes in tyrosine phosphorylation of proteins associated with the actin cytoskeleton including proteins with roles in receptor endocytosis, vesicle movement or exocytosis, or cellular adhesion following FcεRI aggregation on MCP5 cells. b) Quantitative phosphoproteomic data represented in the context of a protein interaction map built upon the proteins observed in Figure 5A (marked with red stars) and known protein-protein interactions contained in the Human Protein Reference Database (HPRD; http://www.hprd.org/). The interaction map is constrained to a maximum of two degrees of separation between experimentally observed phosphoproteins.
Dynamic cytoskeletal rearrangements are also regulated through the action of receptor tyrosine kinases such as the Ephrin receptors. There is now much evidence that Ephrin receptor activation leads to signaling pathways regulating cell adhesion and plasticity of the actin cytoskeleton (66). We detected phosphorylation of the previously established autophosphorylation sites within the tyrosine kinase domain of EphA3 at Tyr779 as well as a novel site at Tyr937, adjacent to a known SH2 interaction site critical in regulating downstream signaling events via interactions with cytosolic proteins (71, 72). One of the cytosolic adaptor molecules that interact with phosphorylated Ephrin receptors is Nck. Nck proteins modulate actin cytoskeletal dynamics by linking proline-rich effector molecules to tyrosine kinases or phosphorylated signaling intermediates (73). A strong elevation of phosphorylation was observed on NCK2/Grb4 SH2/SH3 adaptor at Tyr110.
The Rho family of GTPases, including Rac, are critically important in the regulation of dynamic processes within the actin cytoskeleton downstream of receptor protein tyrosine kinases in mast cells (74). We observed phosphorylation of sites on a collection of proteins that regulate the activity of Rho GTPases. A collection of GTPase activating proteins (GAPs) showed elevated tyrosine phosphorylation as a result of receptor activation in mast cells. ARAP3 is a dual GAP for RhoA and Arf6 that controls dynamic actin rearrangements and vesicular trafficking events (75). Elevation of tyrosine phosphorylation was observed on ARAP3 at a site (Tyr1404) that was previously shown to negatively regulate cell spreading through inhibition of ARAP3 activation of Rho GTPases (65). Another RhoGAP, p190RhoGAP stably associates with RasGAP through a Src mediated phosphorylation site at Tyr943 (76). An increase in phosphorylation was observed on this critical tyrosine residue in stimulated mast cells. The RhoGAP family member, RhoGAP12, also became tyrosine phosphorylated after mast cell receptor activation (Ser238/Tyr241). ELMO2 forms a ternary complex with RhoG and Crk2 to activate Rac1 (77, 78). A strong elevation of ELMO2 tyrosine phosphorylation was observed on a novel site Tyr715 after mast cell receptor aggregation.
The activation of Rho family GTPases after mast cell receptor activation triggers exocytosis of histamine-containing cytoplasmic granules within seconds of stimulation (79). We observed rapid tyrosine phosphorylation of many proteins associated with exocytosis after receptor aggregation. The interaction between WASP and Cdc42 induces a conformational change, stimulating Arp2/3 protein complex to nucleate actin polymerization (74). Arp2/3 protein is associated with secretory granules at the point of contact between the vesicle and the cellular membrane, providing an actin structure that enhances the efficiency of the exocytotic process (80). In B cells, WASP phosphorylation by Src-type kinases at Tyr293 stimulates actin polymerization indepedently of Cdc42 (81). We observed a rapid increase in tyrosine phosphorylation at this site on WASP. A rapid increase in tyrosine phosphorylation was also observed on Arp2/3 subunit p21 after mast cell receptor stimulation (Tyr47). The motor protein kinesin has been proposed to transport intracellular organelles and vesicles to the cell periphery preceding exocytosis of secretory vesicles (82). We detected rapid phosphorylation at a novel tyrosine site on kinesin 2 in stimulated mast cells (Tyr448). Rapid tyrosine phosphorylation of annexin VI, a negative regulator of membrane exocytosis (83), was also observed at a novel site Tyr201.
Phosphorylation of proteins previously associated with exocytosis was also observed on a collection of proteins late after receptor stimulation. Scinderin, a Ca2+-dependent actin filament-severing protein, regulates exocytosis by affecting the organization of the microfilament network proximal to the plasma membrane (84). We identified a novel tyrosine phosphorylation site Tyr599 on scinderin. AHNAK/desmoyokin activates PLCγ1 through PKCα mediated arachidonic acid release and is known to be associated with neural exocytotic vesicles (85, 86). We observed strong induction of tyrosine phosphorylation on AHNAK (Tyr122, Lyn kinase ITAM-like motif D/E-X-X-Y-X-X-I/L).
We observed phosphorylation of proteins that are known to play roles in regulating receptor internalization after stimulation of mast cells. Mast cell FcεRI receptor endocytosis occurs through clathrin-coated pits (87). Intersectin 2 functions cooperatively with WASP and cdc42 to link the clathrin endocytic machinery to WASP-mediated actin polymerization and receptor endocytosis (88). Phosphorylation of two biologically undefined sites was induced after mast cell receptor activation on intersectin 2 (Tyr553 and Tyr921). Beta arrestin regulates receptor internalization of G-protein coupled receptors as well as receptor protein tyrosine kinases such as EGFR (89). We observed a decrease in phosphorylation of beta arrestin on a previously undescribed site Tyr47. Our experiment revealed strong induction of phosphorylation on DAPP1/Bam32 at the Src family kinase-mediated phosphorylation site Tyr139, as well as a previously uncharacterized site Ser141 (90). Although DAPP1/Bam32 has never been previously described in mast cells, it is known to regulate B cell receptor internalization through activation of Rac1 and actin reorganization subsequent to PH domain mediated interaction with PIP2 produced through SHIP cleavage of PIP3 (90).
Phosphorylation of known signaling molecules not previously associated with canonical mast cell signaling
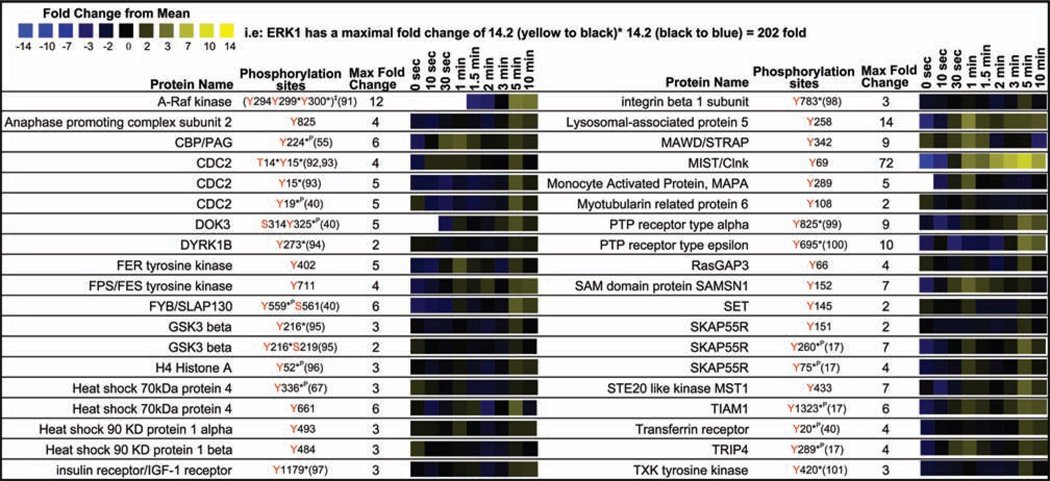
We detected phosphorylation of many known signaling proteins not previously associated with the canonical pathway in stimulated mast cells (Figure 6) (17, 40, 55, 67, 91–101). In receptor-activated mast cells, we observed rapid induction of tyrosine phosphorylation on MAWD at a novel site (Tyr342). MAWD protein is a scaffolding, signaling molecule that activates ERK signaling (102). Rapid Lyn-mediated phosphorylation of Fps and Fer tyrosine kinases causes phosphorylation of platelet-endothelial cell adhesion molecule 1 (Pecam-1) leading to activation of SHP1 and SHP2, limiting mast cell activation (103). Tyrosine phosphorylation at a previously undescribed site Tyr402 was observed on Fer kinase. FPS phosphorylation was also observed at a novel phosphorylation site Tyr711.
Figure 6.
Quantitative proteomic analysis from MCP5 cells of known signaling proteins not previously associated definitively with the canonical FcεRI mast cell signaling cascade in mast cells.
We saw strong induction of tyrosine phosphorylation on the established Lyn kinase phosphorylation site Tyr69 of MIST/Clnk (104). The SLP76 homologue MIST/Clnk is tyrosine phosphorylated by Lyn and Fyn kinase through a complex of SKAP55 and FYB/SLAP130 leading to binding of PLCγ, VAV, Grb2, and LAT (105, 106). Tyrosine phosphorylation on other members of this protein complex was observed (SKAP55R Tyr75, Tyr151, and Tyr260; FYB/SLAP130 Tyr559, Ser561) (107).
Phosphorylation of proteins not previously associated definitively with any signaling cascade
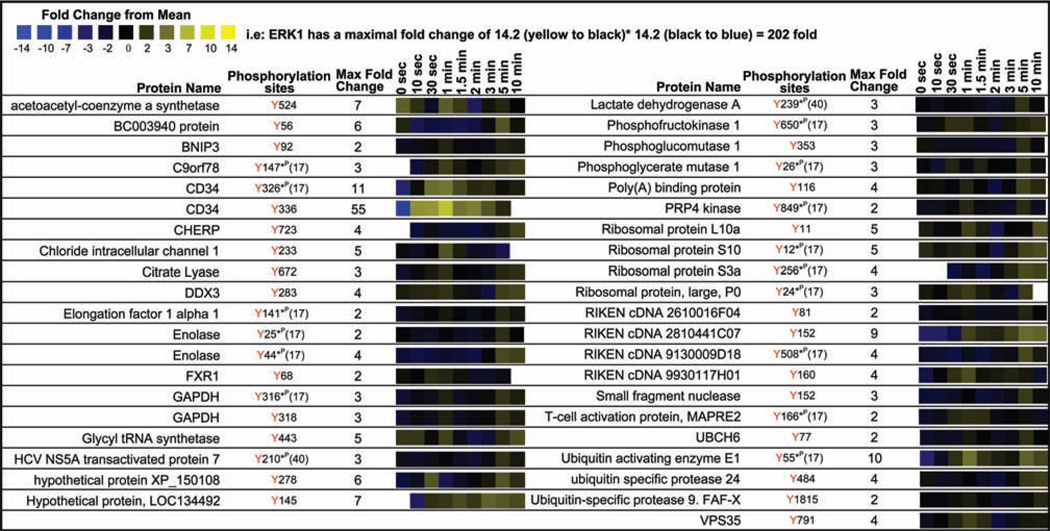
A collection of protein tyrosine phosphorylation was revealed in our quantitative proteomic analysis on proteins not previously associated with any signaling pathway (Figure 7) (17, 40) including a group of metabolic enzymes, RNA interacting proteins, unnamed proteins, and proteins associated with ubiquitination. None of these phosphorylation sites have yet been characterized biologically. With a few exceptions, the magnitude of change in phosphopeptide abundance within this group of proteins after receptor stimulation was relatively small compared to the actin cytoskeleton associated proteins, canonical mast cell signaling proteins, and signaling proteins not associated with mast cell groups.
Figure 7.
Quantitative proteomic analysis of proteins not previously associated definitively with any signaling cascade but observed in MCP5 cells. This heatmap represents the temporal changes in tyrosine phosphorylation of proteins with undefined function in any signaling pathway following FcεRI aggregation in MCP5 cells.
The largest change in phosphopeptide abundance amongst this group of proteins was observed on the mast cell adhesion glycoprotein CD34 (108). CD34 is expressed both on hematopoietic stem cells and mature BMMCs (108). We observed strong phosphorylation of two tyrosine residues (Tyr326, Tyr336) within the short cytosolic portion of CD34 after FcεRI receptor aggregation. Induction of phosphorylation was also observed on the chloride intracellular channel 1 Tyr233, hypothetical proteins XP_150108 Tyr278, LOC134492 Tyr145, RIKEN cDNA 2810441C07 Tyr152, RIKEN cDNA 9930117H01 Tyr160, and the ubiquitin activating enzyme E1 Tyr55.
Discussion
A comprehensive molecular understanding of mast cell signaling will allow the development of improved therapies for treatment of acute allergic disorders such as anaphylaxis and asthma as well as a fundamental understanding of the associated cellular pathways. Our study for the first time provides a view, in unprecedented detail, of the dynamic tyrosine phosphoproteome triggered after FcεRI-mediated mast cell activation.
The temporal arrangement of phosphorylation events observed after mast cell receptor stimulation is suggestive of the placement of these events within signaling pathways, as illustrated by the sequential phosphorylation at known sites observed amongst proteins in the canonical pathway (Figure 1). A rapid increase in phosphorylation was observed at 10 s after receptor crosslinking on all expected ITAM phosphorylation sites of FcεRIβ and FcεRIγ. Phosphorylation of FcεRIβ was transient while FcεRIγ phosphorylation was sustained for 5 minutes. The timing of phosphorylation of SYK at previously established autophosphorylation sites Tyr342/Tyr346 paralleled phosphorylation on FcεRIγ, suggesting rapid activation of Syk following FcεRIγ phosphorylation. The phosphorylation of Btk on Tyr551 was increased later at 2 min, further replicating the expected structure of the canonical mast cell signaling pathway. Phosphorylation of PLCγ2 at the established Btk phosphorylation site Tyr1217 and the Btk and Lyn phosphorylation site Tyr753 increased concurrently with Btk activation, suggesting rapid phosphorylation of these site by Btk or phosphorylation of these sites by another kinase such as Lyn or Fyn (54, 109). PLCγ2 is known to be phopshorylated by Btk, Lyn, and Fyn (54, 109). ERK1 and ERK2 phosphorylation was maximal very late at 5 min consistent with the placement of these downstream signaling proteins within the known canonical mast cell pathway.
It is clear from the observation of overlapping pattern of phosphorylation of some proteins, that the timing of phosphorylation alone is insufficient for defining the precise placement of proteins within the signaling cascade. Firstly, phosphorylation is a rapid modification requiring a high density of time points to define the precise sequence of phosphorylation. Secondly, examination of phosphorylation in individual cells such as with recently developed FACS techniques will be necessary to eliminate errors introduced by heterogeneities within bulk populations of cells necessary for proteomic analysis (110). Thirdly, the mast cell signaling pathway has many branch points and similar patterns of phosphorylation are not necessarily indicative of the coincident placement of proteins within a pathway. Nonetheless, a general indication of the relative pathway placement of newly discovered phosphorylation sites is afforded by this temporal phosphoproteomic analysis. Also, an indication of whether a phosphorylation event is an active participant in the signaling pathway is provided quantitatively by the magnitude of change in its phosphorylation. Phosphorylation sites that have been previously characterized to be biologically meaningful tended to have a higher fold change of phosphorylation (compare Figure 4 to 7). There was also clear evidence of signal amplification in this dataset as downstream signaling proteins such as ERK and PLCγ displayed greater induction of phosphorylation (>100 fold maximally induced) compared to FcεRI (<10 fold maximally induced).
Phosphorylation of proteins previously associated with exocytosis, such as ARP2/3 p21 subunit, annexin VI, and kinesin 2, was observed to be induced rapidly after receptor crosslinking. Conversely, phosphorylation was also observed on the exocytosis-associated protein AHNAK late after receptor activation. Although the precise role that these early and late phosphorylation events play in exocytosis remains to be determined, the timing is suggestive of a role in initiation or completion of exocytosis.
Phosphorylation of proteins previously associated with receptor endocytosis occurred late after receptor activation. Intersectin 2 is a known participant in mast cell receptor endocytosis, linking WASP-mediated actin polymerization and the clathrin endocytic machinery. Highly induced phosphorylation of biologically uncharacterized sites (Tyr553 and Tyr921) was observed to be maximally induced at 5 min after receptor activation on intersectin 2. Tyrosine phosphorylation at Tyr139 was maximally elevated on the receptor endocytosis-related protein DAPP1/Bam32 at 5 min.
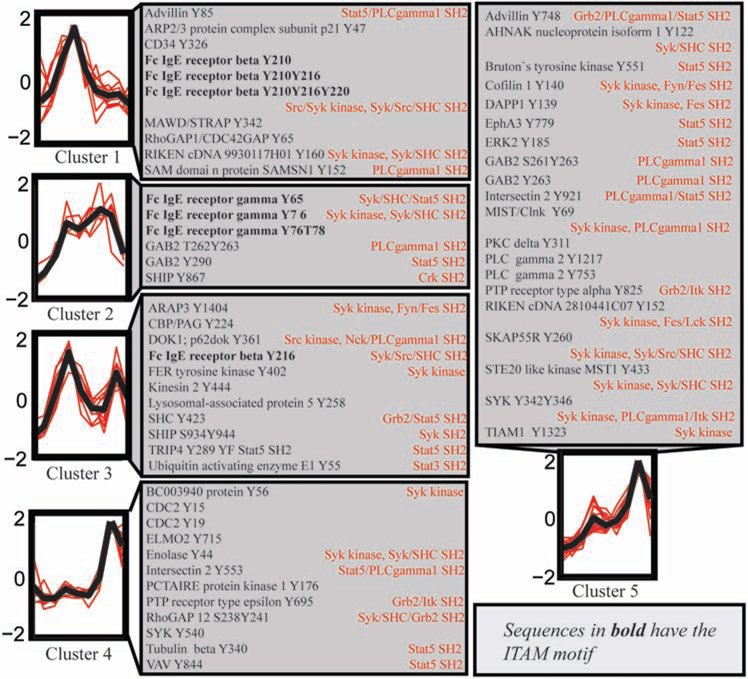
Clustering of quantitative proteomic data allows for the identification of groups of peptides with similar temporal responses (Figure 8). While it is not possible to infer the structure of the signaling pathway directly from these clusters, they provide a general indication of the placement of signaling proteins within the pathway as described in the results section. Cluster 1 contains the earliest and most transient phosphorylation events including phosphorylation of the ITAM motif of FcεRIβ. Cluster 2, including all the FcεRIγ ITAM phosphorylated peptides, displays a similar pattern of early phosphorylation but this phosphorylation is sustained for a longer period of time. The binding of Syk to the FcεRIγ ITAM phosphorylated residues may protect these phosphorylation sites from dephosphorylation by a phosphatase. Cluster 4 and 5 display the late induction of phosphorylation and include a large number of previously established downstream signaling molecules within the mast cell pathway.
Figure 8.
Clustering of phosphoproteomic time series from MCP5 cells. Peptide peak areas were normalized and clustered according to their temporal profiles using k-means clustering. Five clusters are shown with the mean temporal profile (black line) and individual peptide temporal profiles (red lines) for each cluster. Protein sequences were searched against the Minimotif Miner (http://sms.engr.uconn.edu/) database to find sequence motifs surrounding phosphorylated tyrosine residues (indicated in red text).
Cluster 3 displays a bimodal pattern of phosphorylation. Included within cluster 3 is a tyrosine phosphorylation site at Tyr216 contained within the linker region of the FcεRIβ ITAM motif (YXXL-X6-YXXL). Although the canonical ITAM phosphorylation sites at Tyr210 and Tyr220 of FcεRIβ show rapid and transient phosphorylation, singly phosphorylated Tyr216 within the ITAM linker region displays a remarkable bimodal pattern. This noncanonical ITAM tyrosine phosphorylation site at Tyr216 has been suggested to play an inhibitory role in mast cell signaling, probably through its association with the inositol phosphatase SHIP and the protein tyrosine phosphatase SHP1/SHP2 (35). The bimodal pattern of phosphorylation on FcεRIβ at Tyr216 is shared by a collection of inhibitory mast cell signaling proteins including SHIP, SHC, and Dok1, possibly indicating phosphorylation by a common kinase. A bimodal pattern of phosphorylation could indicate the presence of two different kinases that could alter the phosphorylation of a single site and/or the existence of two separate pools of signaling proteins within the cell. Tyr423 of SHC is phosphorylated by an assortment of different tyrosine kinases including Lck, Fyn, and Syk in T cells (111, 112). In mast cells it is possible that this site is phosphorylated initially by Lyn and then by newly activated Syk.
The results described here offer many encouraging leads for further detailed analysis of the mast cell signaling pathway. The elucidation of the role of each phosphorylation site will require the use of traditional biochemical techniques, such as creation of site-directed mutants and signaling protein disruptions. The traditional paradigm for unraveling the structure of signaling pathways by the sequence motif-inspired creation of site-directed mutants is greatly enhanced by knowledge of true cellular phosphorylation sites prior to creation of the site directed mutants, minimizing the futile creation of mutants of sites that do not exist in mast cells. Quantitative phosphoproteomic profiling of mast cells harboring site-directed mutants of newly discovered phosphorylation sites could greatly complement traditional phenotypic assays of mast cell activation such as release of histamine, western blotting for specific proteins or specific sites, or cytokine release assays.
Supplementary Material
Acknowledgments
We wish to thank Dr. Laurent Brossay and Dr. Karsten Sauer for reviewing this manuscript.
Support: This work was supported by NIH grant #2P20RR015578 and by a Beckman Young Investigator Award.
References
- 1.Wedemeyer J, Tsai M, Galli SJ. Roles of mast cells and basophils in innate and acquired immunity. Curr Opin Immunol. 2000;12:624–631. doi: 10.1016/s0952-7915(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 2.Sim AT, Ludowyke RI, Verrills NM. Mast cell function: regulation of degranulation by serine/threonine phosphatases. Pharmacol Ther. 2006;112:425–439. doi: 10.1016/j.pharmthera.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Kawakami T, Galli SJ. Regulation of mast-cell and basophil function and survival by IgE. Nat Rev Immunol. 2002;2:773–786. doi: 10.1038/nri914. [DOI] [PubMed] [Google Scholar]
- 4.Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nat Rev Immunol. 2006;6:218–230. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- 5.Kinet JP. The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu Rev Immunol. 1999;17:931–972. doi: 10.1146/annurev.immunol.17.1.931. [DOI] [PubMed] [Google Scholar]
- 6.Jouvin MH, Adamczewski M, Numerof R, Letourneur O, Valle A, Kinet JP. Differential control of the tyrosine kinases Lyn and Syk by the two signaling chains of the high affinity immunoglobulin E receptor. J Biol Chem. 1994;269:5918–5925. [PubMed] [Google Scholar]
- 7.Eiseman E, Bolen JB. Engagement of the high-affinity IgE receptor activates src protein-related tyrosine kinases. Nature. 1992;355:78–80. doi: 10.1038/355078a0. [DOI] [PubMed] [Google Scholar]
- 8.Hutchcroft JE, Geahlen RL, Deanin GG, Oliver JM. Fc epsilon RI-mediated tyrosine phosphorylation and activation of the 72-kDa protein-tyrosine kinase, PTK72, in RBL-2H3 rat tumor mast cells. Proc Natl Acad Sci USA. 1992;89:9107–9111. doi: 10.1073/pnas.89.19.9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver JM, Burg DL, Wilson BS, McLaughlin JL, Geahlen RL. Inhibition of mast cell Fc epsilon R1-mediated signaling and effector function by the Syk-selective inhibitor, piceatannol. J Biol Chem. 1994;269:29697–29703. [PubMed] [Google Scholar]
- 10.Yamashita T, Mao SY, Metzger H. Aggregation of the high-affinity IgE receptor and enhanced activity of p53/561yn protein-tyrosine kinase. Proc Natl Acad Sci USA. 1994;91:11251–11255. doi: 10.1073/pnas.91.23.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benhamou M, Ryba NJ, Kihara H, Nishikata H, Siraganian RP. Protein-tyrosine kinase p72syk in high affinity IgE receptor signaling. Identification as a component of pp72 and association with the receptor gamma chain after receptor aggregation. J Biol Chem. 1993;268:23318–23324. [PubMed] [Google Scholar]
- 12.Parravicini V, Gadina M, Kovarova M, Odom S, Gonzalez-Espinosa C, Furumoto Y, Saitoh S, Samelson LE, O'Shea JJ, Rivera J. Fyn kinase initiates complementary signals required for IgE-dependent mast cell degranulation. Nat Immunol. 2002;3:741–748. doi: 10.1038/ni817. [DOI] [PubMed] [Google Scholar]
- 13.Ott VL, Tamir I, Niki M, Pandolfi PP, Cambier JC. Downstream of kinase, p62(dok), is a mediator of Fc gamma IIB inhibition of Fc epsilon RI signaling. J Immunol. 2002;168:4430–4439. doi: 10.4049/jimmunol.168.9.4430. [DOI] [PubMed] [Google Scholar]
- 14.Ji-Eun K, White FM. Quantitative Analysis of Phosphotyrosine Signaling Networks Triggered by CD3 and CD28 Costimulation in Jurkat Cells. J Immunol. 2006;176:2833–2843. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 16.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 18.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, Sauer K, Brock A, Horn DM, Schultz PG, Peters EC. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc Natl Acad Sci USA. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 20.Ficarro SB, Salomon AR, Brill LM, Mason DE, Stettler-Gill M, Brock A, Peters EC. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Eng J, McCormack A, Yates JR. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spec. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 23.Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 24.Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, Maciejewski MW, Gryk MR, Piccirillo B, Schiller SR, Schiller MR. Minimotif Miner: a tool for investigating protein function. Nat Methods. 2006;3:175–177. doi: 10.1038/nmeth856. [DOI] [PubMed] [Google Scholar]
- 25.Simon M, Vanes L, Geahlen RL, Tybulewicz VL. Distinct roles for the linker region tyrosines of Syk in FcepsilonRI signaling in primary mast cells. J Biol Chem. 2005;280:4510–4517. doi: 10.1074/jbc.M410326200. [DOI] [PubMed] [Google Scholar]
- 26.Crouin C, Arnaud M, Gesbert F, Camonis J, Bertoglio J. A yeast two-hybrid study of human p97/Gab2 interactions with its SH2 domain-containing binding partners. FEBS Lett. 2001;495:148–153. doi: 10.1016/s0014-5793(01)02373-0. [DOI] [PubMed] [Google Scholar]
- 27.Baba Y, Hashimoto S, Matsushita M, Watanabe D, Kishimoto T, Kurosaki T, Tsukada S. BLNK mediates Syk-dependent Btk activation. Proc Natl Acad Sci U S A. 2001;98:2582–2586. doi: 10.1073/pnas.051626198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano CJ, Graham L, DeBell K, Rawat R, Veri MC, Bonvini E, Rellahan BL, Reischl IG. A new tyrosine phosphorylation site in PLC gamma 1: the role of tyrosine 775 in immune receptor signaling. J Immunol. 2005;174:6233–6237. doi: 10.4049/jimmunol.174.10.6233. [DOI] [PubMed] [Google Scholar]
- 29.Kim YJ, Sekiya F, Poulin B, Bae YS, Rhee SG. Mechanism of B-cell receptor-induced phosphorylation and activation of phospholipase C-gamma2. Mol Cell Biol. 2004;24:9986–9999. doi: 10.1128/MCB.24.22.9986-9999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argetsinger LS, Kouadio JL, Steen H, Stensballe A, Jensen ON, Carter-Su C. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol Cell Biol. 2004;24:4955–4967. doi: 10.1128/MCB.24.11.4955-4967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teckchandani AM, Panetti TS, Tsygankov AY. c-Cbl regulates migration of v-Abl-transformed NIH 3T3 fibroblasts via Racl. Exp Cell Res. 2005;307:247–258. doi: 10.1016/j.yexcr.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Woodring PJ, Meisenhelder J, Johnson SA, Zhou GL, Field J, Shah K, Bladt F, Pawson T, Niki M, Pandolfi PP, Wang JY, Hunter T. c-Abl phosphorylates Dok1 to promote filopodia during cell spreading. J Cell Biol. 2004;165:493–503. doi: 10.1083/jcb.200312171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butch ER, Guan KL. Characterization of ERK1 activation site mutants and the effect on recognition by MEK1 and MEK2. J Biol Chem. 1996;271:4230–4235. doi: 10.1074/jbc.271.8.4230. [DOI] [PubMed] [Google Scholar]
- 34.Her JH, Lakhani S, Zu K, Vila J, Dent P, Sturgill TW, Weber MJ. Dual phosphorylation and autophosphorylation in mitogen-activated protein (MAP) kinase activation. Biochem J. 1993;296(Pt 1):25–31. doi: 10.1042/bj2960025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.On M, Billingsley JM, Jouvin MH, Kinet JP. Molecular dissection of the FcRbeta signaling amplifier. J Biol Chem. 2004;279:45782–45790. doi: 10.1074/jbc.M404890200. [DOI] [PubMed] [Google Scholar]
- 36.Maksumova L, Le HT, Muratkhodjaev F, Davidson D, Veillette A, Pallen CJ. Protein tyrosine phosphatase alpha regulates Fyn activity and Cbp/PAG phosphorylation in thymocyte lipid rafts. J Immunol. 2005;175:7947–7956. doi: 10.4049/jimmunol.175.12.7947. [DOI] [PubMed] [Google Scholar]
- 37.van Dijk TB, Caldenhoven E, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. Multiple tyrosine residues in the intracellular domain of the common beta subunit of the interleukin 5 receptor are involved in activation of STAT5. FEBS Lett. 1997;412:161–164. doi: 10.1016/s0014-5793(97)00768-0. [DOI] [PubMed] [Google Scholar]
- 38.Murugappan S, Shankar H, Bhamidipati S, Dorsam RT, Jin J, Kunapuli SP. Molecular mechanism and functional implications of thrombin-mediated tyrosine phosphorylation of PKCdelta in platelets. Blood. 2005;106:550–557. doi: 10.1182/blood-2004-12-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tvorogov D, Wang XJ, Zent R, Carpenter G. Integrin-dependent PLC-gammal phosphorylation mediates fibronectin-dependent adhesion. J Cell Sci. 2005;118:601–610. doi: 10.1242/jcs.01643. [DOI] [PubMed] [Google Scholar]
- 40.Goss VL, Lee KA, Moritz A, Nardone J, Spek EJ, MacNeill J, Rush J, Comb MJ, Polakiewicz RD. A common phosphotyrosine signature for the Bcr-Abl kinase. Blood. 2006;107:4888–4897. doi: 10.1182/blood-2005-08-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Camerini V, Bender TP, Ravichandran KS. A nonredundant role for the adapter protein Shc in thymic T cell development. Nat Immunol. 2002;3:749–755. doi: 10.1038/ni820. [DOI] [PubMed] [Google Scholar]
- 42.Kashige N, Carpino N, Kobayashi R. Tyrosine phosphorylation of p62dok by p210bcr-abl inhibits RasGAP activity. Proc Natl Acad Sci U S A. 2000;97:2093–2098. doi: 10.1073/pnas.040547997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayle J, Letard S, Frank R, Dubreuil P, De Sepulveda P. Suppressor of cytokine signaling 6 associates with KIT and regulates KIT receptor signaling. J Biol Chem. 2004;279:12249–12259. doi: 10.1074/jbc.M313381200. [DOI] [PubMed] [Google Scholar]
- 44.Price DJ, Rivnay B, Fu Y, Jiang S, Avraham S, Avraham H. Direct association of Csk homologous kinase (CHK) with the diphosphorylated site Tyr568/570 of the activated c-KIT in megakaryocytes. J Biol Chem. 1997;272:5915–5920. doi: 10.1074/jbc.272.9.5915. [DOI] [PubMed] [Google Scholar]
- 45.Thommes K, Lennartsson J, Carlberg M, Ronnstrand L. Identification of Tyr-703 and Tyr-936 as the primary association sites for Grb2 and Grb7 in the c-Kit/stem cell factor receptor. Biochem J. 1999;341(Pt 1):211–216. [PMC free article] [PubMed] [Google Scholar]
- 46.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorenz U, Ravichandran KS, Pei D, Walsh CT, Burakoff SJ, Neel BG. Lck-dependent tyrosyl phosphorylation of the phosphotyrosine phosphatase SH-PTP1 in murine T cells. Mol Cell Biol. 1994;14:1824–1834. doi: 10.1128/mcb.14.3.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong JJ, Yankee TM, Harrison ML, Geahlen RL. Regulation of signaling in B cells through the phosphorylation of Syk on linker region tyrosines. A mechanism for negative signaling by the Lyn tyrosine kinase. J Biol Chem. 2002;277:31703–31714. doi: 10.1074/jbc.M201362200. [DOI] [PubMed] [Google Scholar]
- 49.Keshvara LM, Isaacson CC, Yankee TM, Sarac R, Harrison ML, Geahlen RL. Syk- and Lyn-dependent phosphorylation of Syk on multiple tyrosines following B cell activation includes a site that negatively regulates signaling. J Immunol. 1998;161:5276–5283. [PubMed] [Google Scholar]
- 50.Arora N, Min KU, Costa JJ, Rhim JS, Metcalfe DD. Immortalization of mouse bone marrow-derived mast cells with Adl2-SV40 virus. Int Arch Allergy Immunol. 1993;100:319–327. doi: 10.1159/000236432. [DOI] [PubMed] [Google Scholar]
- 51.Cheng SH, Espino PC, Marshall J, Harvey R, Merrill J, Smith AE. Structural elements that regulate pp59c-fyn catalytic activity, transforming potential, and ability to associate with polyomavirus middle-T antigen. J Virol. 1991;65:170–179. doi: 10.1128/jvi.65.1.170-179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law CL, Chandran KA, Sidorenko SP, Clark EA. Phospholipase C-gammal interacts with conserved phosphotyrosyl residues in the linker region of Syk and is a substrate for Syk. Mol Cell Biol. 1996;16:1305–1315. doi: 10.1128/mcb.16.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozdener F, Dangelmaier C, Ashby B, Kunapuli SP, Daniel JL. Activation of phospholipase Cgamma2 by tyrosine phosphorylation. Mol Pharmacol. 2002;62:672–679. doi: 10.1124/mol.62.3.672. [DOI] [PubMed] [Google Scholar]
- 55.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 56.Jabril-Cuenod B, Zhang C, Scharenberg AM, Paolini R, Numerof R, Beaven MA, Kinet JP. Syk-dependent phosphorylation of Shc. A potential link between FcepsilonRI and the Ras/mitogen-activated protein kinase signaling pathway through SOS and Grb2. J Biol Chem. 1996;271:16268–16272. doi: 10.1074/jbc.271.27.16268. [DOI] [PubMed] [Google Scholar]
- 57.Marchetto S, Fournier E, Beslu N, Aurran-Schleinitz T, Dubreuil P, Borg JP, Birnbaum D, Rosnet O. SHC and SHIP phosphorylation and interaction in response to activation of the FLT3 receptor. Leukemia. 1999;13:1374–1382. doi: 10.1038/sj.leu.2401527. [DOI] [PubMed] [Google Scholar]
- 58.Seger R, Ahn NG, Boulton TG, Yancopoulos GD, Panayotatos N, Radziejewska E, Ericsson L, Bratlien RL, Cobb MH, Krebs EG. Microtubule-associated protein 2 kinases, ERK1 and ERK2, undergo autophosphorylation on both tyrosine and threonine residues: implications for their mechanism of activation. Proc Natl Acad Sci U S A. 1991;88:6142–6146. doi: 10.1073/pnas.88.14.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takizawa F, Adamczewski M, Kinet JP. Identification of the low affinity receptor for immunoglobulin E on mouse mast cells and macrophages as Fc gamma RII and Fc gamma RIII. J Exp Med. 1992;176:469–475. doi: 10.1084/jem.176.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leitges M, Gimborn K, Elis W, Kalesnikoff J, Hughes MR, Krystal G, Huber M. Protein kinase C-delta is a negative regulator of antigen-induced mast cell degranulation. Mol Cell Biol. 2002;22:3970–3980. doi: 10.1128/MCB.22.12.3970-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blake RA, Garcia-Paramio P, Parker PJ, Courtneidge SA. Src promotes PKCdelta degradation. Cell Growth Differ. 1999;10:231–241. [PubMed] [Google Scholar]
- 62.Molfetta R, Belleudi F, Peruzzi G, Morrone S, Leone L, Dikic I, Piccoli M, Frati L, Torrisi MR, Santoni A, Paolini R. CIN85 regulates the ligand-dependent endocytosis of the IgE receptor: a new molecular mechanism to dampen mast cell function. J Immunol. 2005;175:4208–4216. doi: 10.4049/jimmunol.175.7.4208. [DOI] [PubMed] [Google Scholar]
- 63.Pendleton A, Koffer A. Effects of latrunculin reveal requirements for the actin cytoskeleton during secretion from mast cells. Cell Motil Cytoskeleton. 2001;48:37–51. doi: 10.1002/1097-0169(200101)48:1<37::AID-CM4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 64.Lang T, Wacker I, Wunderlich I, Rohrbach A, Giese G, Soldati T, Aimers W. Role of actin cortex in the subplasmalemmal transport of secretory granules in PC-12 cells. Biophys J. 2000;78:2863–2877. doi: 10.1016/S0006-3495(00)76828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.I ST, Nie Z, Stewart A, Najdovska M, Hall NE, He H, Randazzo PA, Lock P. ARAP3 is transiently tyrosine phosphorylated in cells attaching to fibronectin and inhibits cell spreading in a RhoGAP-dependent manner. J Cell Sci. 2004;117:6071–6084. doi: 10.1242/jcs.01526. [DOI] [PubMed] [Google Scholar]
- 66.Lawrenson ID, Wimmer-Kleikamp SH, Lock P, Schoenwaelder SM, Down M, Boyd AW, Alewood PF, Lackmann M. Ephrin-A5 induces rounding, blebbing and deadhesion of EphA3-expressing 293T and melanoma cells by CrkII and Rho-mediated signalling. J Cell Sci. 2002;115:1059–1072. doi: 10.1242/jcs.115.5.1059. [DOI] [PubMed] [Google Scholar]
- 67.Walters DK, Goss VL, Stoffregen EP, Gu TL, Lee K, Nardone J, McGreevey L, Heinrich MC, Deininger MW, Polakiewicz R, Druker BJ. Phosphoproteomic analysis of AML cell lines identifies leukemic oncogenes. Leuk Res. 2006;30:1097–1104. doi: 10.1016/j.leukres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 68.Stephens LR, Anderson KE, Hawkins PT. Src family kinases mediate receptor-stimulated, phosphoinositide 3-kinase-dependent, tyrosine phosphorylation of dual adaptor for phosphotyrosine and 3-phosphoinositides-1 in endothelial and B cell lines. J Biol Chem. 2001;276:42767–42773. doi: 10.1074/jbc.M107194200. [DOI] [PubMed] [Google Scholar]
- 69.Tamura H, Fukada M, Fujikawa A, Noda M. Protein tyrosine phosphatase receptor type Z is involved in hippocampus-dependent memory formation through dephosphorylation at Y 1105 on p 190 RhoGAP. Neurosci Lett. 2006;399:33–38. doi: 10.1016/j.neulet.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 70.Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA. Fyn and PTP-PEST-mediated regulation of Wiskott-Aldrich syndrome protein (WASp) tyrosine phosphorylation is required for coupling T cell antigen receptor engagement to WASp effector function and T cell activation. J Exp Med. 2004;199:99–112. doi: 10.1084/jem.20030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Binns KL, Taylor PP, Sicheri F, Pawson T, Holland SJ. Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of Eph receptors. Mol Cell Biol. 2000;20:4791–4805. doi: 10.1128/mcb.20.13.4791-4805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smalla M, Schmieder P, Kelly M, Ter Laak A, Krause G, Ball L, Wahl M, Bork P, Oschkinat H. Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci. 1999;8:1954–1961. doi: 10.1110/ps.8.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frese S, Schubert WD, Findeis AC, Marquardt T, Roske YS, Stradal TE, Heinz DW. The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J Biol Chem. 2006;281:18236–18245. doi: 10.1074/jbc.M512917200. [DOI] [PubMed] [Google Scholar]
- 74.Burridge K, K Wennerberg. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 75.Krugmann S, Andrews S, Stephens L, Hawkins PT. ARAP3 is essential for formation of lamellipodia after growth factor stimulation. J Cell Sci. 2006;119:425–432. doi: 10.1242/jcs.02755. [DOI] [PubMed] [Google Scholar]
- 76.Roof RW, Haskell MD, Dukes BD, Sherman N, Kinter M, Parsons SJ. Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of pl90 RhoGAP-pl20 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol Cell Biol. 1998;18:7052–7063. doi: 10.1128/mcb.18.12.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hong-Geller E, Cerione RA. Cdc42 and Rac stimulate exocytosis of secretory granules by activating the IP(3)/calcium pathway in RBL-2H3 mast cells. J Cell Biol. 2000;148:481–494. doi: 10.1083/jcb.148.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock 180-binding protein Elmo. Nature. 2003;424:461–464. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- 79.Nishida K, Yamasaki S, Ito Y, Kabu K, Hattori K, Tezuka T, Nishizumi H, Kitamura D, Goitsuka R, Geha RS, Yamamoto T, Yagi T, Hirano T. Fc{epsilon}RI-mediated mast cell degranulation requires calcium-independent microtubule-dependent translocation of granules to the plasma membrane. J Cell Biol. 2005;170:115–126. doi: 10.1083/jcb.200501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gasman S, Chasserot-Golaz S, Malacombe M, Way M, Bader MF. Regulated exocytosis in neuroendocrine cells: a role for subplasmalemmal Cdc42/N-WASP-induced actin filaments. Mol Biol Cell. 2004;15:520–531. doi: 10.1091/mbc.E03-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cory GO, Garg R, Cramer R, Ridley AJ. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. Wiskott-Aldrich Syndrome protein. J Biol Chem. 2002;277:45115–45121. doi: 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- 82.Bi GQ, Morris RL, Liao G, Alderton JM, Scholey JM, Steinhardt RA. Kinesin- and myosin-driven steps of vesicle recruitment for Ca2+-regulated exocytosis. J Cell Biol. 1997;138:999–1008. doi: 10.1083/jcb.138.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Donnelly SR, Moss SE. Annexins in the secretory pathway. Cell Mol Life Sci. 1997;53:533–538. doi: 10.1007/s000180050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trifaro JM. Scinderin and cortical F-actin are components of the secretory machinery. Can J Physiol Pharmacol. 1999;77:660–671. [PubMed] [Google Scholar]
- 85.Lee IH, You JO, Ha KS, Bae DS, Suh PG, Rhee SG, Bae YS. AHNAK-mediated activation of phospholipase C-gammal through protein kinase C. J Biol Chem. 2004;279:26645–26653. doi: 10.1074/jbc.M311525200. [DOI] [PubMed] [Google Scholar]
- 86.Borgonovo B, Cocucci E, Racchetti G, Podini P, Bachi A, Meldolesi J. Regulated exocytosis: a novel, widely expressed system. Nat Cell Biol. 2002;4:955–962. doi: 10.1038/ncb888. [DOI] [PubMed] [Google Scholar]
- 87.Frankel DJ, Pfeiffer JR, Surviladze Z, Johnson AE, Oliver JM, Wilson BS, Burns AR. Revealing the topography of cellular membrane domains by combined atomic force microscopy/fluorescence imaging. Biophys J. 2006;90:2404–2413. doi: 10.1529/biophysj.105.073692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGavin MK, Badour K, Hardy LA, Kubiseski TJ, Zhang J, Siminovitch KA. The intersectin 2 adaptor links Wiskott Aldrich Syndrome protein (WASp)-mediated actin polymerization to T cell antigen receptor endocytosis. J Exp Med. 2001;194:1777–1787. doi: 10.1084/jem.194.12.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim J, Ahn S, Guo R, Daaka Y. Regulation of epidermal growth factor receptor internalization by G protein-coupled receptors. Biochemistry. 2003;42:2887–2894. doi: 10.1021/bi026942t. [DOI] [PubMed] [Google Scholar]
- 90.Allam A, Marshall AJ. Role of the adaptor proteins Bam32, TAPP1 and TAPP2 in lymphocyte activation. Immunol Lett. 2005;97:7–17. doi: 10.1016/j.imlet.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 91.Marais R, Light Y, Paterson HF, Mason CS, Marshall CJ. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 92.Atherton-Fessler S, Parker LL, Geahlen RL, Piwnica-Worms H. Mechanisms of p34cdc2 regulation. Mol Cell Biol. 1993;13:1675–1685. doi: 10.1128/mcb.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawasaki H, Komai K, Ouyang Z, Murata M, Hikasa M, Ohgiri M, Shiozawa S. c-Fos/activator protein-1 transactivates wee1 kinase at G(1)/S to inhibit premature mitosis in antigen-specific Th1 cells. Embo J. 2001;20:4618–4627. doi: 10.1093/emboj/20.16.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee K, Deng X, Friedman E. Mirk protein kinase is a mitogen-activated protein kinase substrate that mediates survival of colon cancer cells. Cancer Res. 2000;60:3631–3637. [PubMed] [Google Scholar]
- 95.Cole A, Frame S, Cohen P. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem J. 2004;377:249–255. doi: 10.1042/BJ20031259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tasaka K, Mio M, Akagi M, Saito T. Histamine release induced by histone and related morphological changes in mast cells. Agents Actions. 1990;30:114–117. doi: 10.1007/BF01969013. [DOI] [PubMed] [Google Scholar]
- 97.Peterson JE, Kulik G, Jelinek T, Reuter CW, Shannon JA, Weber MJ. Src phosphorylates the insulin-like growth factor type I receptor on the autophosphorylation sites. Requirement for transformation by src. J Biol Chem. 1996;271:31562–31571. doi: 10.1074/jbc.271.49.31562. [DOI] [PubMed] [Google Scholar]
- 98.Wennerberg K, Armulik A, Sakai T, Karlsson M, Fassler R, Schaefer EM, Mosher DF, Johansson S. The cytoplasmic tyrosines of integrin subunit beta1 are involved in focal adhesion kinase activation. Mol Cell Biol. 2000;20:5758–5765. doi: 10.1128/mcb.20.15.5758-5765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lammers R, Moller NP, Ullrich A. Mutant forms of the protein tyrosine phosphatase alpha show differential activities towards intracellular substrates. Biochem Biophys Res Commun. 1998;242:32–38. doi: 10.1006/bbrc.1997.7906. [DOI] [PubMed] [Google Scholar]
- 100.Toledano-Katchalski H, Elson A. The transmembranal and cytoplasmic forms of protein tyrosine phosphatase epsilon physically associate with the adaptor molecule Grb2. Oncogene. 1999;18:5024–5031. doi: 10.1038/sj.onc.1202883. [DOI] [PubMed] [Google Scholar]
- 101.Chamorro M, Czar MJ, Debnath J, Cheng G, Lenardo MJ, Varmus HE, Schwartzberg PL. Requirements for activation and RAFT localization of the T-lymphocyte kinase Rlk/Txk. BMC Immunol. 2001;2:3. doi: 10.1186/1471-2172-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halder SK, Anumanthan G, Maddula R, Mann J, Chytil A, Gonzalez AL, Washington MK, Moses HL, Beauchamp RD, Datta PK. Oncogenic function of a novel WD-domain protein, STRAP, in human carcinogenesis. Cancer Res. 2006;66:6156–6166. doi: 10.1158/0008-5472.CAN-05-3261. [DOI] [PubMed] [Google Scholar]
- 103.Udell CM, Samayawardhena LA, Kawakami Y, Kawakami T, Craig AW. Fer and Fps/Fes participate in a Lyn-dependent pathway from FcepsilonRI to platelet-endothelial cell adhesion molecule 1 to limit mast cell activation. J Biol Chem. 2006;281:20949–20957. doi: 10.1074/jbc.M604252200. [DOI] [PubMed] [Google Scholar]
- 104.Goitsuka R, Tatsuno A, Ishiai M, Kurosaki T, Kitamura D. MIST functions through distinct domains in immunoreceptor signaling in the presence and absence of LAT. J Biol Chem. 2001;276:36043–36050. doi: 10.1074/jbc.M106390200. [DOI] [PubMed] [Google Scholar]
- 105.Fujii Y, Wakahara S, Nakao T, Hara T, Ohtake H, Komurasaki T, Kitamura K, Tatsuno A, Fujiwara N, Hozumi N, Ra C, Kitamura D, Goitsuka R. Targeting of MIST to Src-family kinases via SKAP55-SLAP-130 adaptor complex in mast cells. FEBS Lett. 2003;540:111–116. doi: 10.1016/s0014-5793(03)00234-5. [DOI] [PubMed] [Google Scholar]
- 106.Goitsuka R, Kanazashi H, Sasanuma H, Fujimura Y, Hidaka Y, Tatsuno A, Ra C, Hayashi K, Kitamura D. A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation. Int Immunol. 2000;12:573–580. doi: 10.1093/intimm/12.4.573. [DOI] [PubMed] [Google Scholar]
- 107.Curtis DJ, Jane SM, Hilton DJ, Dougherty L, Bodine DM, Begley CG. Adaptor protein SKAP55R is associated with myeloid differentiation and growth arrest. Exp Hematol. 2000;28:1250–1259. doi: 10.1016/s0301-472x(00)00537-3. [DOI] [PubMed] [Google Scholar]
- 108.Drew E, Merzaban JS, Seo W, Ziltener HJ, McNagny KM. CD34 and CD43 inhibit mast cell adhesion and are required for optimal mast cell reconstitution. Immunity. 2005;22:43–57. doi: 10.1016/j.immuni.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 109.Watanabe D, Hashimoto S, Ishiai M, Matsushita M, Baba Y, Kishimoto T, Kurosaki T, Tsukada S. Four tyrosine residues in phospholipase C-gamma 2, identified as Btk-dependent phosphorylation sites, are required for B cell antigen receptor-coupled calcium signaling. J Biol Chem. 2001;276:38595–38601. doi: 10.1074/jbc.M103675200. [DOI] [PubMed] [Google Scholar]
- 110.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6:146–155. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 111.Walk SF, March ME, Ravichandran KS. Roles of Lck, Syk and ZAP-70 tyrosine kinases in TCR-mediated phosphorylation of the adapter protein Shc. Eur J Immunol. 1998;28:2265–2275. doi: 10.1002/(SICI)1521-4141(199808)28:08<2265::AID-IMMU2265>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 112.Wary KK, Mariotti A, Zurzolo C, Giancotti FG. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell. 1998;94:625–634. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.