Abstract
Sphingolipidomics, a branch of lipidomics, focuses on the large-scale study of the cellular sphingolipidomes. In the current review, two main approaches for the analysis of cellular sphingolipidomes (i.e. LC-MS- or LC-MS/MS-based approach and shotgun lipidomics-based approach) are briefly discussed. Their advantages, some considerations of these methods, and recent applications of these approaches are summarized. It is the authors’ sincere hope that this review article will add to the readers understanding of the advantages and limitations of each developed method for the analysis of a cellular sphingolipidome.
Keywords: Intrasource separation, Lipidomics, Multi-dimensional mass spectrometry, Sphingolipidomics, Shotgun lipidomics
1 Introduction
It is the era of “omics” due to the development of technology and the demand for a systems biology approach to study life science [1–4]. Lipidomics, defined as the large-scale study of the pathways and networks of cellular lipids, is one of the emerging and rapidly expanding research fields in systems biology [5–7]. Although lipidomics has only emerged as a distinct field within the past few years [5–7], numerous new discoveries and/or advances have already been made [7–17]. It has been increasingly recognized that lipidomics plays essential roles in identifying the biochemical mechanisms of lipid metabolism/trafficking/homeostasis, investigating the functions of an individual gene of interest, identifying novel biomarkers, and evaluating drug efficacy, among others [7–18].
Sphingolipidomics, a branch of lipidomics that appeared in peer-reviewed articles around 2005 [19, 20], focuses on the large-scale study of the cellular sphingolipidomes. The emergence of this subfield in lipidomics is very natural for multiple reasons. First, sphingolipids play essential and diverse roles in cellular functions. Besides serving as important structural components of cell membranes and lipoproteins, sphingolipids are the regulators of cell proliferation/differentiation/apoptosis, cell migration, cellular signaling, cell membrane trafficking, cell interactions with their neighbors, cell morphology, etc. [19, 21, 22]. For example, complex glycosphingolipids play crucial roles in cell-cell communication and cell-matrix/protein interactions [23, 24]. Ceramide predominantly inhibits cell growth and induces apoptosis while its metabolites, sphingosine-1-phosphate (S1P) and ceramide-1-phosphate (C1P), promote cell growth and survival [21]. As the agonists of the lysophosphatidic acid (lysoPtdH) family of receptors, S1P, sphinganine-1-phosphate, and psychosine are important lipid second messengers in cellular signaling [19, 25]. Therefore, again, it is critical to determine the altered levels of both ceramide and its metabolites when one studies cellular signaling in cell growth or cell death.
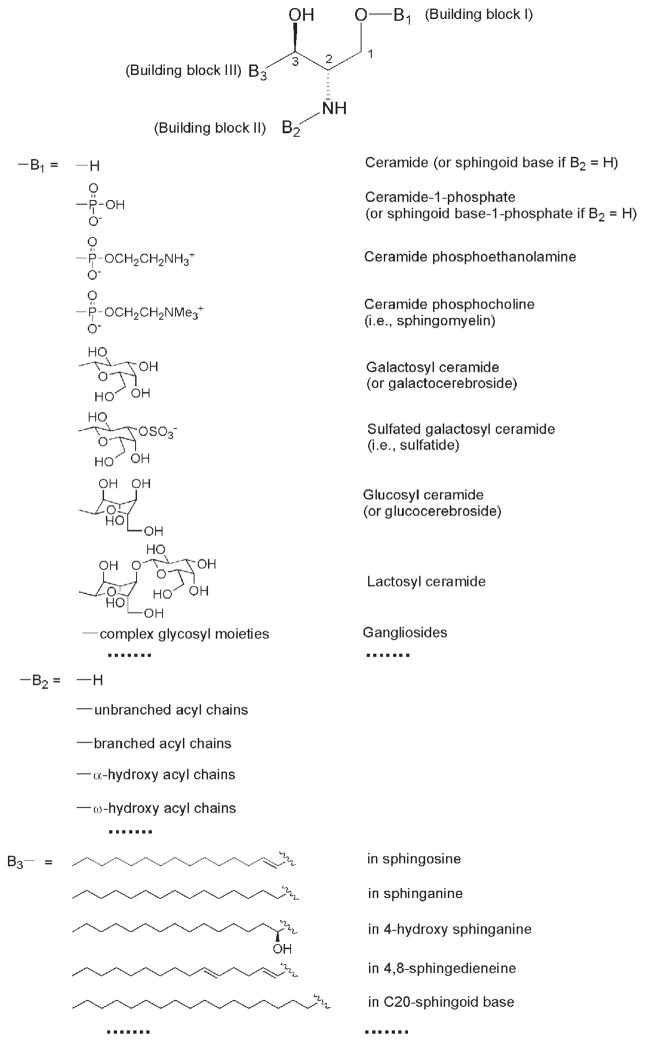
Second, the chemical structures of molecular species in the cellular sphingolipidome are very unique relative to other categories of lipids in a cellular lipidome. All sphingolipids contain a sphingoid-based backbone although the aliphatic moiety of this backbone can vary widely (Fig. 1). The derivatives from the sphingoid base are relatively inert to an acid or base environment in comparison to the glycerol-based lipids. Therefore, either the sphingoid base or the entire sphingolipidome can be isolated after treatment of a lipid extract from a biological sample under either strong or mild basic conditions, respectively [20, 26].
Figure 1.
General structure of sphingoid-based lipids. The building block B1 represents a different polar moiety (linked to the oxygen at the C1 position of the sphingoid base). The building block B2 represents fatty acyl chains (acylated to the primary amine at the C2 position of the sphingoid base) with or without the presence of a hydroxyl group which is usually located at the α- or ω-position. The building block B3 represents the aliphatic chains in all of the possible sphingoid bases, which are carbon-carbon linked to the C3 position of sphingoid bases and vary with the aliphatic chain length, degree of unsaturation, the presence of branches, and the presence of an additional hydroxyl group. This illustration has been modified from ref. [11] with permission.
Third, the variations of sphingolipid molecular species are very complex. Tens of thousands of sphingolipid molecular species are potentially present in a cellular sphingolipidome, depending on the cut edge of the content of each species, which can be detected using the available technologies. A comprehensive classification and nomenclature of this complex sphingolipidome can be found at the website of www.sphingomap.com. Alternatively, this complex sphingolipidome can also be represented through a simplified general structure with three building blocks (Fig. 1). The building block I (B1) represents a different polar moiety (linked to the oxygen at the C1 position of the sphingoid base) including hydrogen, phosphoethanolamine, phosphocholine, galactose, glucose, lactose, sulfated galactose, and other complex sugar groups [corresponding to ceramide, ceramide phosphoethanolamine, sphingomyelin (CerPCho), galactosylceramide (GalCer), glucosylceramide (GluCer), lactosylceramide, sulfatide, and other glycosphingolipids such as gangliosides, respectively] (Fig. 1). Each of these polar groups represents a class of sphingolipids in a cellular sphingolipidome. We can readily list over 20 sphingolipid classes that are commonly present in a biological sample. The building block II (B2) represents fatty acyl chains (acylated to the primary amine at the C2 position of the sphingoid base) with or without the presence of a hydroxyl group, which is usually located at the α-or ω-position (Fig. 1). Over 100 types of acyl chains that are commonly present in a cellular sphingolipidome can be readily counted. These acyl chains vary from 14 to 26 carbons with a certain degree of unsaturation (containing 0–6 double bonds depending on the chain length) in each, and with or without the presence of a hydroxyl group which is usually located at the α- or ω-position. The presence of the branched or modified (oxygenated, nitrated, etc.) fatty acyl chains further complicates the family of fatty acyl chains. The building block III (B3) represents the aliphatic chains in all possible sphingoid bases, which are carbon-carbon linked to the C3 position of sphingoid bases and vary with the aliphatic chain length, the degree of unsaturation, branching, and the presence of an additional hydroxyl group (Fig. 1). Over 100 types of this aliphatic chain can also be readily counted as similarly discussed above. Therefore, a combination of these three factors would yield at least 200,000 sphingolipid molecular species. At the current stage, tens to hundreds of sphingolipid molecular species are readily analyzed by using different approaches [20, 26, 27].
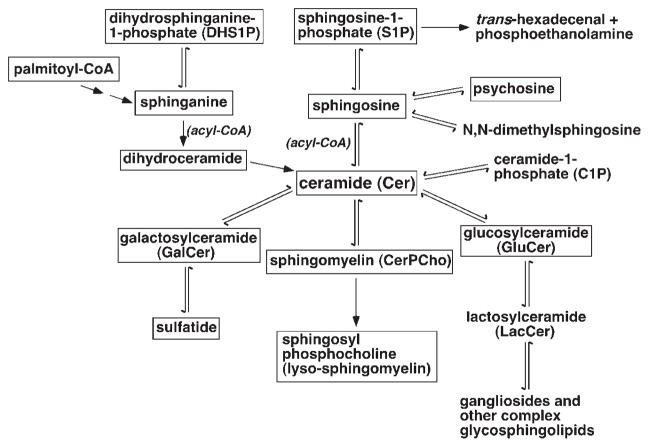
Fourth, the sphingolipid classes and molecular species in a cellular sphingolipidome are interwoven in their metabolism and homeostasis. Figure 2 shows a simplified network of the common sphingolipid classes and other related lipids in the mammalian sphingolipidome. In the network, ceramide serves as a core component for the homeostasis of all sphingolipid classes including CerPCho, GalCer, sulfatide, GluCer, gangliosides, and other complex glycosphingolipids through different pathways. Therefore, changes in expression levels and/or enzyme activities in any pathway in the network induced by a physiological or pathological factor will result in a new homeostasis of the cellular sphingolipidome to a certain degree. No doubt, we could better understand the underlying biochemical mechanisms if we were able to determine the entire sphingolipidome.
Figure 2.
A simplified network of the common sphingolipid classes and other related lipids in the mammalian sphingolipidome. The sphingolipid classes with frames are those that can be quantitatively analyzed by shotgun (sphingo)lipidomics at its current stage.
Finally, the content of sphingolipid molecular species among a class and/or between the classes varies broadly. Many membrane structural sphingolipid classes (e.g. GalCer, sulfatide, CerPCho, and gangliosides) are quite abundant in many organs, particularly in the brain. They are at the level of tens to hundreds of nmol/mg of tissue protein. In contrast, many of the signaling sphingolipid classes (e.g. sphingosine, S1P, C1P, psychosine, etc.) are in extremely low abundance under normal physiological conditions (at or below the level of a few pmol/mg of tissue protein, only representing ≪0.01 mol-% of the total cellular lipids). Therefore, quantitative analyses of these compounds have become very difficult by conventional chromatographic methods due to the sensitivity issue [28]. This issue has been substantially improved with the application of soft ionization techniques in mass spectrometry (MS) [e.g. electrospray ionization MS (ESI/MS)] which will be the focus of this review.
Collectively, these, among other factors like differential distribution and function of sphingolipids in different cellular membranes (e.g. lipid rafts) and compartments, have led to the emergence of sphingolipidomics. However, as an initial step in the emerging field, methods used for the quantitative analyses of the cellular sphingolipidome at an in-depth and large-scale level have been rapidly developing, but still remain challenging. There are two basic approaches that have been employed for such applications, i.e. the liquid chromatography (LC)-MS- or LC-MS/MS-based methods and the direct infusion-based shotgun lipidomics approach. Both approaches are briefly discussed in this review. However, it should be emphasized that many excellent reviews that focused on the particular methods have previously been published and should also be consulted for a better understanding of these methods [20, 29–32].
2 LC-MS-based analysis of a cellular sphingolipidome
High-performance LC (HPLC) with a variety of columns (e.g. normal phase, reversed phase, chiral separation, ultra performance, ion exchange, etc.) has been coupled to MS for a variety of purposes [33]. LC-MS has demonstrated extreme power in the analysis of the complex isomeric eicosanoids [34]. Although LC-MS provides structural information about the analytes, MS essentially serves as a mass detector (i.e. similar to a UV detector or other detectors which are connected to an HPLC system) in quantitation. The determined total ion current (TIC) can be used to reconstitute the TIC chromatograph for the purpose of lipid quantitation if a standard curve of a compound is established under identical experimental conditions. The combination of ESI/MS detection with HPLC separation and the sensitivity of TIC compared to other detection modalities make LC-MS an obvious choice for lipid profiling and for quantitation if great care is taken in establishing the relationship between instrument response and the known concentrations of an analyte. Indeed, LC-MS has been employed in many applications for the identification of lipid molecular species (see www.lipidlibrary.co.uk/lit_surv/general/h_pl_msp.htm for a list of publications). However, large-scale lipid quantitation using this modality on a level of global analysis is quite limited [35], although quantitative analysis of a small number of lipids for which standard curves can be generated is quite common [36].
For example, LC-MS has been successfully employed to analyze the isolated ceramide molecular species of human hair. By combination of reversed-phase HPLC separation with selected ion extraction after MS detection, Masukawa and colleagues [37] have detected and identified 73 hair ceramide molecular species. The investigators have determined the presence of both sphingosine-type and sphinganine-type sphingoid bases containing 16, 18, 19, and 20 carbon numbers. They have also found various acyl amides with and without a hydroxyl moiety at the α-position. There is no doubt that this study represents one of the most comprehensive analyses of ceramide molecular species in hair samples and other mammalian tissue samples.
By using normal-phase LC coupled to atmospheric pressure chemical ionization (APCI)-MS and nano-ESI-MS/MS, Sandhoff and colleagues [38] have investigated the complex ceramide molecular species of human skin. They have identified a total of 67 ceramide molecular species wherein the chain lengths of the sphingoid bases ranged from C12 to C22 and the chain lengths of the fatty amides varied between C28 and C36. They have also found ω-hydroxy fatty acid in the skin ceramide molecular species. Colsch and colleagues [39] have identified the presence of sphingadienine in the mammalian brain through product ion analysis of sphingolipid molecular species which were initially classified by using normal phase LC.
It should be recognized that when a reversed-phase HPLC column is used to resolve individual lipid molecular species, the relatively polar mobile phase that is commonly employed could cause difficulties with solubility in a molecular species-dependent manner. If an elution gradient is employed to resolve individual molecular species by a reversed-phase HPLC column, changes in the components of the mobile phase as the solvent gradient progresses may also cause an ionization stability problem and may alter the ionization efficiency. If a normal-phase HPLC column is employed for separation of different lipid classes in general, different lipid molecular species in a class are not uniformly distributed in the eluted peak (i.e. each individual molecular species of a class may possess its own distinct retention time and peak shape due to differential interactions with the stationary phase). This trailing effect may alter the ionization efficiency of an analyte in a concentration-dependent manner.
Recently, LC-MS/MS analysis with a triple quadrupole mass spectrometer in multiple reaction monitoring (MRM) mode has become a very popular technique for the quantitative analysis of sphingolipid molecular species, due to its increased efficiency and potential accuracy for quantitative analysis. In MRM mode, the first quadrupole serves as a mass selector to pass a specific molecular ion, and the third quadrupole serves as a mass detector to detect at least one specific product ion resulting from the molecular ion while the second quadrupole is used as a collision cell. The transition pair or pairs of molecular ion/product ion(s) is established after characterization of a class of lipids of interest through product ion analysis. The key point for the successful performance of the MRM technique is the specificity of the paired product ion to the molecular ion in a lipid class of interest.
Bielawski and colleagues [31] have reported a detailed protocol on how the MRM technique coupled with a C8 reversed-phase column is successfully used to analyze 40 sphingolipid molecular species (including internal standards) from a biological lipid extract without separation of individual sphingolipid classes. The investigators have established multiple standard curves for quantitative analysis. However, consideration of the effects of differential 13C isotopic distribution on the quantitation of different molecular species related to the selected internal standard may simplify the protocol and result in more accurate quantitative results for those without standard curves than the current protocol. It should be pointed out that Bielawski and colleagues have used the MRM pair transition for analysis of sphingosine and dihydrosphingosine, based on a fragment ion resulting from the loss of water, but this seems not very specific to sphingoid bases.
The protocol based on LC-MS/MS in MRM mode that has been reported by Merrill and colleagues is the most comprehensive one for the analyses of sphingolipid molecular species among all the published methods using the MRM technique. Therefore, this approach has been termed as “sphingolipidomics” [20]. In the protocol, the authors have listed the ion transition pairs of many different sphingolipid classes for which the protocol is able to cover. Moreover, the authors have given multiple instrumental settings that are related to the MRM experiment on different mass spectrometers. These parameters should be very helpful to investigators in setting up their own experiments to determine sphingolipid molecular species by using the MRM technique. However, it would have been better if the following aspects had been discussed in the protocol, including (1) how the internal standards are used for quantitation of the monitored molecular ions; (2) how to overcome the effects of differential 13C isotopic distribution on the quantitation of different molecular species related to the selected internal standard; and (3) how to overcome the effects of solvent change in a gradient on the ionization efficiency of molecular species eluted at the different time frame as mentioned above if there is no standard curve for each molecular species.
LC-tandem MS in MRM mode has also been broadly employed for quantitation of the levels of individual molecular species in a sphingolipid class of interest. For example, Lieser and colleagues have developed a method for quantitation of sphingosine and sphinganine contents in lipid extracts by MRM analyses of three pairs of molecular ion/product ions of each analyte to increase the specificity [40]. Quantitation has been performed in comparison to an internal standard (i.e. C17 sphingosine analog). Fuller and colleagues [41, 42] have employed the MRM technique to determine the altered glycosphingolipid levels in the pathological state of Gaucher’s disease. Masukawa and colleagues [43] have employed the selected ion monitoring technique to quantitatively analyze 73 ceramide molecular species in human hair.
Collectively, LC-MS and LC-MS/MS have been playing very important roles in the analysis of sphingolipid molecular species, classes, and/or the entire cellular sphingolipidome. Enrichment of low- or very low-abundance molecular species and elimination of the ion suppression effects between different lipid molecular species or different classes are among the lists of the roles. However, some considerations present in this approach for quantitative analysis on a large scale are also apparent and should be recognized. For example, first, it is very difficult to determine a standard curve for each individual lipid molecular species from a particular biological preparation. Second, even if it were practical to have a standard curve for each lipid compound, the determined standard curve is externally measured and the involvement of multiple steps of sample preparation, separation, and analysis can introduce experimental errors in each step, which can be propagated for the entire process. Third, differential loss of lipids on the column is also not unusual [44]. Fourth, while separation of molecular species by using a reversed-phase column prevents lipid-lipid interactions of one lipid with another molecular species (for the most part) there is a large, up to a 1000-fold increase in the amount of lipid-lipid interactions with the same lipid (homodimer formation) since reversed-phase HPLC is typically used to concentrate samples. Lipids in sufficient concentrations tend to aggregate. Finally, carryover is always a concern in LC-MS [45, 46].
3 Shotgun lipidomics-based analysis of a cellular sphingolipidome
3.1 Introduction
Different from LC-based lipid analysis, the analytical platforms without direct coupling with any chromatography for separation of lipid classes and/or lipid molecular species have been referred to as “shotgun lipidomics”. In shotgun lipidomics, lipid extracts from biological samples are carefully prepared to eliminate the salt or other aqueous phase-soluble components and properly diluted to a concentration at which lipid aggregation is minimal under the selected experimental conditions (e.g. solvents, temperature, etc.). The former factor is to reduce the chemical noise while the latter one is mainly to eliminate the lipid aggregation since lipid aggregation can alter the ionization efficiency. Analysis of lipids under the aggregation state results in an apparent ionization efficiency that is dependent on the physical properties (i.e. the number of carbons and the degree of unsaturation) of individual molecular species [30, 47]. It should be pointed out that the effect of lipid aggregation on quantitation is also severely present in the LC-MS-based platforms although establishment of a standard curve for an individual lipid species of interest may reduce this effect. Two somewhat different platforms of shotgun lipidomics have been developed. The principles and applications of both platforms will be briefly discussed in the following sections.
3.2 Sphingolipid analysis by using the data-dependent shotgun lipidomics approach
Shevchenko and colleagues have recently developed a shotgun lipidomics platform in a data-dependent manner [16, 32, 48–50]. The efficient acquisition of a mass spectrum and the high mass accuracy/resolution of the instrument inherent in a hybrid quadrupole time-of-flight (QqTOF) or linear ion trap-orbitrap (LTQ Orbitrap) mass spectrometer are the advantages of these instruments. Therefore, product ion analysis of a selected ion using these instruments can be more rapidly performed than using a conventional triple-quadrupole mass spectrometer. Shevchenko and colleagues have exploited the advantages of these instruments and performed the product ion analyses of all selected ions of lipid molecular species in a data-dependent manner after direct infusion of individual lipid samples. After collecting the fragments from the arrayed product ion analyses, computer-aided analysis enables one to extract a particular fragment of interest that is specific to a class or a group of lipids in the precursor ion (PI) format or a neutral loss (NL) manner.
The high mass accuracy present in the instruments enables one to accurately establish the relationship between the fragments and the molecular ions, thereby eliminating any contribution and/or confusion of the isobaric ions during qualitative and quantitative analysis of lipid molecular species. With the aid of their developed software program (i.e. Lipid Profiler™), the group has readily applied the approach to identify and profile many biological samples. It should be recognized that both PI scanning and NL scanning in shotgun lipidomics are equivalent to MRM, which is commonly used in LC-MS as discussed above. Furthermore, by employing multiple PI and/or NL, the specificity of analyzing a lipid class of interest can be substantially increased. However, applications to the analysis of the very low-abundance lipid classes and/or molecular species are still limited by using this shotgun lipidomics platform. Moreover, the presence of the differential ion transmission in a mass-dependent manner in the instruments that have been employed as previously described [51] should be considered in the quantitative analyses by employing this shotgun lipidomics platform.
This approach has been successfully employed for the analysis of ceramide phosphatidylinositol species in yeast [52]. Specifically, Ejsing and colleagues [52] have employed this approach to profile yeast sphingolipid molecular species in total lipid extracts, including inositolphosphoceramide (IPC), mannosyl-inositolphosphoceramide (MIPC), and mannosyl-diinositolphosphoceramide (M(IP)2C). Through characterization of these sphingolipid classes in yeast, Ejsing et al. [52] have demonstrated the specific product ions for each individual class, including m/z 241.0 ([IP–H2O]−) and 259.0 ([IP]−) from deprotonated IPC; m/z 403.1 ([MIP–H2O]−) and 421.1 ([MIP]−) from deprotonated MIPC, and m/z 241.0 ([IP–H2O]−), 331.0 ([M(IP)2]2−) and 583.1 ([M(IP)2–P]−) from doubly charged negative-ion M(IP)2C. Through determination of the PI of these specific fragments, Ejsing et al. [52] have profiled 12 IPC molecular species, 10 MIPC molecular species, and 6 M(IP)2C molecular species in a Pichia pastoris strain.
3.3 Shotgun sphingolipidomics
3.3.1 Introduction of intrasource separation and multi-dimensional MS-based shotgun lipidomics
One of the major analytical platforms in current lipidomics practice is multi-dimensional MS (MDMS)-based shotgun lipidomics [30, 53, 54]. This platform has now evolved into a mature technology that includes a series of simple steps such as multiplexed extractions, intrasource separation/selective ionization of a specific category of lipids, identification of those individual lipid molecular species that have been selectively ionized using MDMS and array analyses, and quantitation of the identified lipid molecular species using a two-step procedure in conjunction with data processing. The underlying principle of shotgun lipidomics is to maximally exploit differences or uniqueness in the physical and chemical properties of a class of lipids of interest.
Therefore, the differential solubility of the different lipid classes in various solvents under varying pH conditions is utilized as an initial but critical step to maximally separate and enrich the lipid class(es) of interest. For example, many lipid classes (e.g. S1P, lysoPtdH, acylcarnitine, etc.) can be efficiently extracted under acidic conditions [8]. Gangliosides and acyl-CoAs are highly soluble in polar solvents and are partitioned into the aqueous phase during chloroform extraction [55–57]. Thus, these lipid classes can be reverse-extracted by using butanol under acidic conditions. Moreover, very hydrophobic lipid classes (e.g. cholesterol and its esters, triacylglycerol, free fatty acids, etc.) can be extracted and enriched with hexane. Fmoc chloride can be added to quickly tag the amine-containing lipids and increase the sensitivity for analysis of these lipids through NL scanning of the tagged Fmoc moiety [58].
Next, in MDMS-based shotgun lipidomics, the differential acidic or basic properties of lipid classes in an extracted lipid solution are exploited to selectively ionize different lipid classes in the positive- or negative-ion modes (i.e. intrasource separation) and to achieve maximal ionization sensitivity [59]. Therefore, the lipid classes containing phosphate, sulfate, and carboxylate (e.g. anionic phospholipids, ethanolamine-containing phospholipid (e.g. PtdEtn), acyl-CoA, sulfatide, gangliosides, free fatty acids, and S1P) can be selectively ionized in the negative-ion mode, particularly under basic conditions (i.e. in the presence of NH4OH or LiOH) whereas lipid classes containing amine (e.g. acylcarnitine) can be readily ionized in the positive-ion mode under acidic conditions [8]. Molecular species of other lipid classes can be ionized as either alkaline or anion (e.g. chloride, acetate, or formate) adducts in the positive- or negative-ion mode, respectively. It should be emphasized that this intrasource separation is equivalent to the separation of analytes employing electrophoresis or ion exchange chromatography, which can be used to replace the general LC separation to a certain degree.
Finding a sensitive and unique fragment after collision-induced dissociation (CID), which is specific to a class or a group of lipids of interest, is the third key step for successfully profiling and quantifying individual molecular species in the class. Specifically, either NL scanning or PI scanning at the mass or m/z ratio of the fragment of interest, respectively, can be performed to “isolate” a given class or a group of lipids from which individual lipid molecular species can be identified in an MDMS analysis fashion. Each of these fragments represents a building block of the class or the group of lipids (Fig. 1) [30].
Finally, quantitation by shotgun lipidomics is performed in a two-step procedure [30, 60, 61] after considering the uniqueness of lipids that undergo aggregation above a certain concentration and recognizing the extension of a linear dynamic range through analysis of the building blocks. Specifically, the abundant and non-overlapping molecular species of a class are quantified by comparison with a pre-selected internal standard of the class after 13C de-isotoping [8, 53] from a survey scan. Next, some or all of these determined molecular species of the class (plus the pre-selected internal standard) are used as standards to determine the content of other low-abundance or overlapping molecular species using one or multiple NL and/or PI scans as described above. Through this second step in the quantitation process, the linear dynamic range of quantitation can be dramatically extended by eliminating background noise and by filtering the overlapping molecular species through an MDMS approach [8].
Although the advantages of shotgun lipidomics are obvious, we have recognized a few limitations. For example, due to signal overlaps of low-abundance molecular species of a class of interest (e.g. CerPCho) with potential isomeric/isobaric lipid molecular species in other lipid class(es) [e.g. glycerophosphocholine (PtdCho)] in both the first and second steps of quantitation in shotgun lipidomics, identification and quantitation of these molecular species become inaccessible. Moreover, in the worst case, all molecular species of a class of interest are buried in the survey scan mass spectrum and no single ion peak can be quantitated by the first step of quantitation due to the presence of other major lipid classes, which is generally called “ion suppression”. Class-specific MS/MS analysis by employing two or more internal standards can be used to assess individual molecular species of the class, as previously described, and is widely used [62, 63]. However, the linear dynamic range of quantitation by comparison of ion peak intensities with a pre-selected internal standard can be reduced by the presence of other major lipids. It should be noted that the MRM technique coupled with LC-MS has been well recognized for its power for quantitation of the targeted and not overlapped molecular species from well-characterized lipid extract samples (see above). However, shotgun lipidomics with PI scanning and NL scanning in MDMS format is very useful and efficient to quantitatively analyze the molecular species of the entire lipid class of interest in a non-targeted manner from any unknown sample.
3.3.2 Sample preparation in shotgun sphingolipidomics
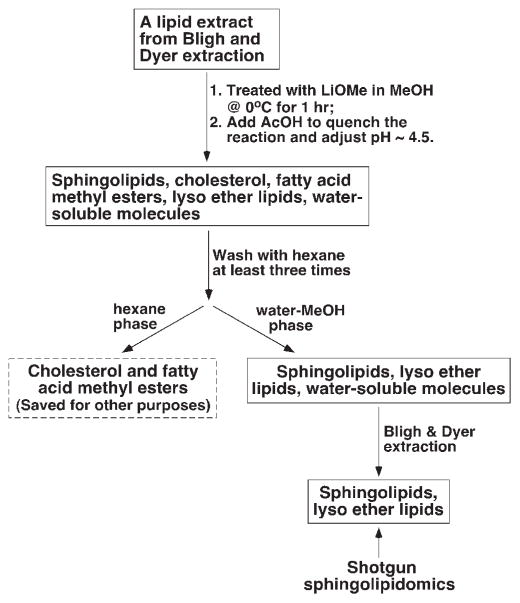
Very recently, by exploiting two distinct chemical characteristics of sphingolipids, we have extended the shotgun lipidomics platform and developed an approach for the analysis of those sphingolipid classes and/or molecular species that have previously been difficult for shotgun lipidomics to analyze. First, we exploited the base-resistant character of all sphingolipids under mildly basic conditions in comparison to the esterified glycerolipids. Although this base-resistant feature has been widely used previously for isolation of sphingolipids prior to chromatographic separation (see refs. [29, 31, 64–66] for examples), we have modified the protocol and treated the lipid extracts with alkaline methanolysis (e.g. lithium methoxide in methanol). Thus, the esterified glycerolipids, cholesterol and its esters are removed and sphingolipids are enriched by lithium methoxide-catalyzed ester exchange reaction followed by liquid-liquid extraction (Fig. 3).
Figure 3.
A schematic illustration of sample preparation for shotgun sphingolipidomics (adapted from ref. [26] with permission).
Briefly, when a lipid extract was treated with lithium methoxide/methanol solution, all ester-linked fatty acyl moieties were converted to fatty acid methyl esters which, along with cholesterol and free fatty acids if they were present in the lipid extract, were readily removed from the reaction mixture by extraction with hexane. Next, the sphingolipidome present in the reaction mixture could be recovered by Bligh and Dyer extraction through which the majority of polar compounds generated by methanolysis could be removed. During the development of this method, we examined a variety of reaction conditions including the base used for cleavage of esters, temperature, and reaction time. We found that the current protocol gives the best recovery of the entire sphingolipidome. It should be pointed out that, although the recovery of each individual sphingolipid class may be different during the process of sample preparation, this does not present a problem for quantitation since an internal standard for each class is added during the original lipid extraction. The difference in recovery of the internal standard from the endogenous molecular species of the corresponding sphingolipid class is relatively small and only causes a negligible effect on the accurate quantitation of these molecular species.
In addition to removing potential overlapping non-sphingolipid molecular species, treatment of lipid extracts with lithium methoxide provides two other advantages. First, the procedure can enrich sphingolipids without the use of column chromatography(s). Second, this approach dramatically increases the effective dynamic range of quantitation for sphingolipid molecular species since many of the co-existing lipids (glycerolipids, cholesterol, etc.) have been eliminated. Therefore, the advantages of this shotgun sphingolipidomics approach allow us to establish a foundation to analyze many sphingolipid classes (e.g. sphingosine, lysoCerPCho, psychosine, etc.) in the extremely low-abundance region without prior enrichment by column chromatography. Under the experimental conditions where the concentration of infused solution is kept below 100 pmol of total lipids/μL in 1: 1 CHCl3/MeOH to avoid lipid aggregation, a 10–50-fold enrichment after base treatment could be easily achieved since the composition of sphingolipids in crude lipid extracts is commonly only a few percent of the total lipids.
3.3.3 Advantages and applications of shotgun sphingolipidomics
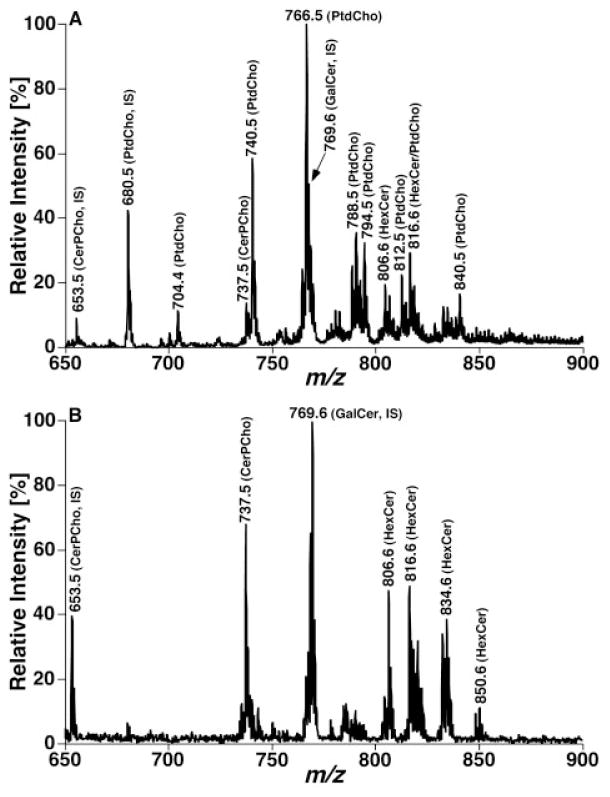
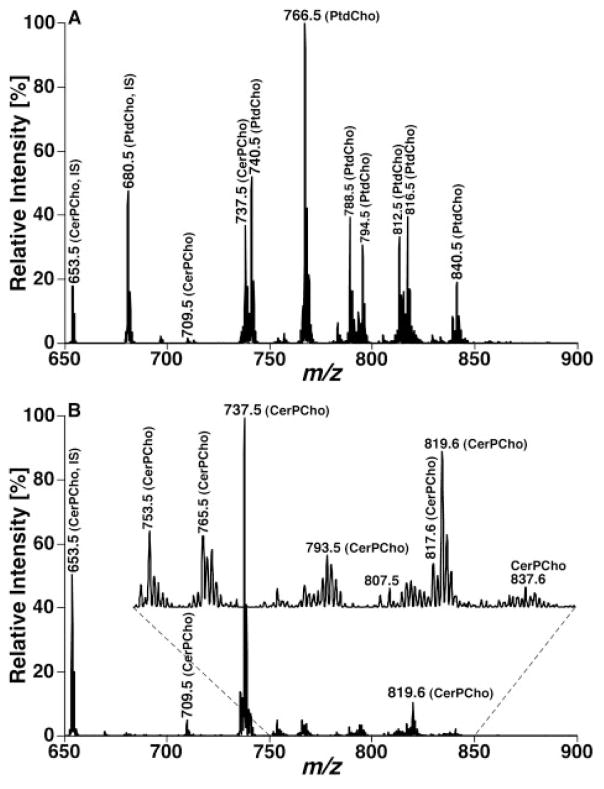
One prominent advantage of the developed shotgun sphingolipidomics is that it enables us to determine the levels of many low-abundance or overlapped molecular species in a relative abundant sphingolipid class [e.g. CerPCho and hexosylceramide (HexCer)] in comparison to original shotgun lipidomics. For example, ions of CerPCho and HexCer molecular species in original shotgun lipidomics have been either suppressed to a certain degree by the presence of much more abundant PtdCho molecular species or overlapped with M+1 13C isotopologues of the abundant PtdCho molecular species. Therefore, quantitative analysis of these classes of sphingolipids can be more accurate and more comprehensive in comparison to the original shotgun lipidomics. Figure 4 exemplifies a comparison between MS analyses of a lipid extract from mouse cortex before and after treatment of the lipid extract with lithium methoxide. This comparison clearly demonstrates the CerPCho and HexCer molecular ions after eliminating ion suppression from PtdCho molecular species. Figure 5 shows the comparison between the MS/MS analyses of phosphocholine-containing molecular species in a lipid extract of mouse cortex before and after the treatment of the lipid extract with lithium methoxide through NL scanning of 183.1 u (i.e. phosphocholine) which is specific for PtdCho and CerPCho molecular species in this mass region. The mass spectra clearly show that many additional CerPCho molecular species in low abundance could be readily identified and quantified by shotgun sphingolipidomics (Fig. 5B).
Figure 4.
Shotgun lipidomics analyses of sphingolipid molecular species before and after treatment of a mouse cortex lipid extract with lithium methoxide in the positive-ion mode in the presence of a small amount of LiOH. The mass spectra (A) and (B) were acquired directly from a lipid extract of mouse cortex before and after treatment with lithium methoxide, respectively, as illustrated in Fig. 3. IS denotes internal standard. Both spectra are displayed after being normalized to the base peak in each spectrum.
Figure 5.
Shotgun lipidomics analyses of sphingolipid molecular species before and after treatment of a mouse cortex lipid extract with lithium methoxide in the NL mode in the presence of a small amount of LiOH. The mass spectra in (A) and (B) were acquired by using the NL scanning of 183.1 u (i.e. phosphocholine) from the lipid extract of mouse cortex before and after treatment with lithium methoxide, respectively, as illustrated in Fig. 3. IS denotes internal standard. The ion peaks in (B) represent lithiated CerPCho molecular species. Both spectra are displayed after being normalized to the base peak in each spectrum.
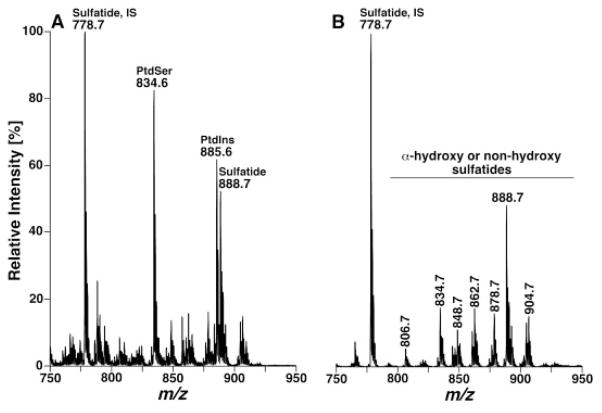
Similarly, analyses of sulfatide molecular species in the original shotgun lipidomics have heavily relied on the PI scan of 97 Th (i.e. sulfatide) due to the presence of ion suppression from other anionic lipid classes and potential overlapping of sulfatide molecular species with glycerophosphoinositol (PtdIns) isotopologues (Fig. 6A). The quantitation of low-abundance sulfatide molecular species is questionable in some cases [67, 68]. By using shotgun sphingolipidomics, the overlap of sulfatide molecular species with PtdIns isotopologues has been cleaned up and the negative-ion ESI mass spectrum in the m/z range of 600–1000 only shows deprotonated sulfatides (Fig. 6B). Therefore, the contents of many sulfatide molecular species were determined in the MS survey scan in the first step of quantitation (Fig. 6A). The contents of other low-abundance sulfatide molecular species can be assessed through the second step of quantitation by using PI scanning of 97 Th (i.e. sulfate) and using the determined abundant sulfatide molecular species as internal standards.
Figure 6.
Shotgun lipidomics analyses of sulfatide molecular species before and after treatment of a mouse cortex lipid extract with lithium methoxide in the negative-ion mode. The mass spectra in (A) and (B) were acquired directly from a lipid extract of mouse cortex before and after treatment with lithium methoxide, respectively, as illustrated in Fig. 3, by using a nanomate device. IS denotes internal standard. Both mass spectra are displayed after being normalized to the base peak in each spectrum.
Another prominent advantage of the developed shotgun sphingolipidomics is that it enables us to identify and quantify very minor sphingolipid classes without enrichment by chromatography. For example, it was demonstrated that sphingosine (and sphinganine) can be readily ionized in positive-ion mode in the presence of a small amount of formic acid (e.g. 0.1%) in 1: 1 CHCl3/MeOH [26]. Quantitation of sphingosine and sphinganine can be readily achieved through NL scanning of 48.0 u (corresponding to a formaldehyde molecule and a water molecule) in the presence of two internal standards with a limit of detection at the level of 0.1 fmol/μL [26]. The contents of sphingosine (and sphinganine) in mouse brain and plasma samples were determined with this developed method, and these results are within the range of the values published in the literature [69, 70]. Very recently, this method has also been applied to study the sphingosine levels in the lysosome to understand the sulfatide metabolism pathways after neuronal cell treatment with sulfatides [71]. This methodology has been employed to determine the levels of other low-abundance sphingolipid metabolites (e.g. lyso-CerPCho, psychosine, S1P, and sphinganine) in a variety of biological samples [26] (Jiang and Han, unpublished data). Furthermore, by using this method, we have readily identified and quantitated over 40 ceramide molecular species. These species contained d18:1 (sphingosine backbone), d18:0 (sphinganine backbone), and d20:1 with a variety of fatty acyl amides with or without a hydroxyl moiety in addition to other sphingolipid molecular species and glycerolipid molecular species from a lipid extract of post-mortem human temporal cerebellar white matter (Han, unpublished data). Figure 7 shows a representative two-dimensional MS analysis of these ceramide molecular species from a lipid extract of human brain temporal cerebellar white matter. This represents the most comprehensive determination of the ceramide molecular species of human brain samples and indicates the power of shotgun sphingolipidomics in the analysis of sphingolipid molecular species in a non-targeted manner. Collectively, by using this newly developed shotgun sphingolipidomics, identification of alterations in metabolic pathways and networks of the sphingolipidome induced by any biological perturbation from a limited material resource shall be greatly facilitated.
Figure 7.
Two-dimensional mass spectrometric analyses of ceramide molecular species from a lipid extract of human brain temporal cerebellar white matter in a shotgun sphingolipidomics approach. The lipid sample from human temporal white matter for shotgun sphingolipidomics was prepared as illustrated in Fig. 3 in the presence of 1 nmol C17:1 ceramide/mg protein. A conventional ESI mass spectrum in the negative-ion mode was acquired prior to analysis of the building blocks of ceramide molecular species by NL scanning. These building blocks of ceramide molecular species include sphingoid bases of sphingosine (NL 256.2 and NL 327.3), sphinganine (NL 258.2 and NL 329.3), and C20-sphingoid base (NL 284.3 and NL 355.3) with or without the presence of a hydroxyl group in the fatty amide chains as previously described [74]. IS denotes internal standard. All mass spectra are displayed after normalization to the base peak in each individual spectrum.
One of the major difficulties of shotgun sphingolipidomics at its current stage is the absence of proper sphingolipid analogs that can serve as internal standard(s) for a sphingolipid class of interest. Although this difficulty can be overcome by the efforts of our chemical design and synthesis, method development in the shotgun sphingolipidomics approach for some sphingolipid classes is hindered. Another limitation of shotgun sphingolipidomics at its current stage is the definitive analysis of isomeric GalCer and GluCer molecular species.
4 Conclusion/perspective
In this review, we have discussed the significance of sphingolipidomics as well as two main approaches for the analyses of cellular sphingolipidomes (i.e. LC-MS- or LC-MS/MS-based approach and shotgun lipidomics-based approach). The advantages and some considerations of these approaches have also been briefly discussed. Due to the space limitation, some other recently developed methods or applications for the analyses of sphingolipids (e.g. comparative lipidomics approach [66, 72] and direct profiling of gangliosides [55]) have been omitted in the discussion. It is apparent that great efforts are still warranted for the technological development of the sphingolipidomics, especially in the area of automation (including software and devices) and penetration (i.e. the very low-abundance sphingolipid classes and molecular species) [73]. In the long term in sphingolipidomics, the development of technologies that enable us to study cellular sphingolipid molecular species temporally, spatially, and dynamically are crucial. With the development of technologies for sphingolipidomics, the role of the sphingolipidome in cellular functions and the underlying causes of the altered sphingolipidomes in a disease state can be addressed. Collectively, sphingolipidomics should bring us to a new level of understanding of a cellular sphingolipidome.
Acknowledgments
This work was supported by NIA Grant R01 AG23168, NIA Grant R01 AG31675, and the Neurosciences Education and Research Foundation.
Abbreviations
- Cer
ceramide
- CerPCho
sphingomyelin
- C1P
ceramide-1-phosphate
- ESI
electrospray ionization
- GalCer
galactosylceramide or galactocerebroside
- GluCer
glucosylceramide
- HexCer
mono-hexosylceramide
- HPLC
high-performance liquid chromatography
- IP
inositolphosphate
- IPC
inositolphosphoceramide
- LC
liquid chromatography
- PtdCho
glycerophosphocholine
- PtdGro
glycerophosphoglycerol
- PtdH
phosphatidic acid
- PtdIns
glycerophosphoinositol
- PtdSer
glycerophosphoserine
- MDMS
multi-dimensional mass spectrometry
- MIPC
mannosyl-inositolphosphoceramide
- M(IP)2C
mannosyl-diinositolphosphoceramide
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- NL
neutral loss
- PI
precursor ion
- S1P
sphingosine-1-phosphate
- TIC
total ion current
Footnotes
Conflict of interest statement
The authors have declared no conflict of interest.
References
- 1.Kiechle FL, Zhang X, Holland-Staley CA. The -omics era and its impact. Arch Pathol Lab Med. 2004;128:1337–1345. doi: 10.5858/2004-128-1337-TOEAII. [DOI] [PubMed] [Google Scholar]
- 2.Altman RB, Rubin DL, Klein TE. An “omics” view of drug development. Drug Dev Res. 2004;62:81–85. [Google Scholar]
- 3.Vemuri GN, Aristidou AA. Metabolic engineering in the -omics era: Elucidating and modulating regulatory networks. Microbiol Mol Biol Rev. 2005;69:197–216. doi: 10.1128/MMBR.69.2.197-216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan MW. Omics and its 15 minutes. Exp Biol Med. 2007;232:471–472. [PubMed] [Google Scholar]
- 5.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Lagarde M, Geloen A, Record M, Vance D, Spener F. Lipidomics is emerging. Biochim Biophys Acta. 2003;1634:61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 8.Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 9.Walker JM, Krey JF, Chen JS, Vefring E, Jahnsen JA, Bradshaw H, Huang SM. Targeted lipidomics: Fatty acid amides and pain modulation. Prostaglandins Other Lipid Mediat. 2005;77:35–45. doi: 10.1016/j.prostaglandins.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Serhan CN. Mediator lipidomics. Prostaglandins Other Lipid Mediat. 2005;77:4–14. doi: 10.1016/j.prostaglandins.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Han X. Neurolipidomics: Challenges and developments. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanova PT, Milne SB, Forrester JS, Brown HA. LIPID arrays: New tools in the understanding of membrane dynamics and lipid signaling. Mol Interv. 2004;4:86–96. doi: 10.1124/mi.4.2.6. [DOI] [PubMed] [Google Scholar]
- 13.Welti R, Shah J, Li W, Li M, Chen J, Burke JJ, Fauconnier ML, Chapman K, Chye ML, Wang X. Plant lipidomics: Discerning biological function by profiling plant complex lipids using mass spectrometry. Front Biosci. 2007;12:2494–2506. doi: 10.2741/2250. [DOI] [PubMed] [Google Scholar]
- 14.Schiller J, Suss R, Fuchs B, Muller M, Zschornig O, Arnold K. MALDI-TOF MS in lipidomics. Front Biosci. 2007;12:2568–2579. doi: 10.2741/2255. [DOI] [PubMed] [Google Scholar]
- 15.Albert CJ, Anbukumar DS, Monda JK, Eckelkamp JT, Ford DA. Myocardial lipidomics. Developments in myocardial nuclear lipidomics. Front Biosci. 2007;12:2750–2760. doi: 10.2741/2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 17.Kuksis A. Lipidomics in triacylglycerol and cholesteryl ester oxidation. Front Biosci. 2007;12:3203–3246. doi: 10.2741/2307. [DOI] [PubMed] [Google Scholar]
- 18.Watson AD. Thematic review series: Systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: A global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Maceyka M, Milstien S, Spiegel S. Sphingosine kinases, sphingosine-1-phosphate and sphingolipidomics. Prostaglandins Other Lipid Mediat. 2005;77:15–22. doi: 10.1016/j.prostaglandins.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH., Jr Ceramides and other bioactive sphingolipid backbones in health and disease: Lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Hoetzl S, Sprong H, van Meer G. The way we view cellular (glyco)sphingolipids. J Neurochem. 2007;103(Suppl 1):3–13. doi: 10.1111/j.1471-4159.2007.04721.x. [DOI] [PubMed] [Google Scholar]
- 23.Spiegel S, Milstien S. Sphingosine-1-phosphate: An enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 24.Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer zu Heringdorf D, Jakobs KH. Lysophospholipid receptors: Signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim Biophys Acta. 2007;1768:923–940. doi: 10.1016/j.bbamem.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Cheng H, Yang K, Gross RW, Han X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low abundance regime of cellular sphingolipids. Anal Biochem. 2007;371:135–145. doi: 10.1016/j.ab.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng H, Jiang X, Han X. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: A shotgun lipidomics study. J Neurochem. 2007;101:57–76. doi: 10.1111/j.1471-4159.2006.04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caligan TB, Peters K, Ou J, Wang E, Saba J, Merrill AH., Jr A high-performance liquid chromatographic method to measure sphingosine 1-phosphate and related compounds from sphingosine kinase assays and other biological samples. Anal Biochem. 2000;281:36–44. doi: 10.1006/abio.2000.4555. [DOI] [PubMed] [Google Scholar]
- 29.Sullards MC, Merrill AH., Jr Analysis of sphingosine 1-phosphate, ceramides, and other bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Sci STKE. 2001:PL1. doi: 10.1126/stke.2001.67.pl1. 2001. [DOI] [PubMed] [Google Scholar]
- 30.Han X, Gross RW. Shotgun lipidomics: Multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 31.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Schwudke D, Liebisch G, Herzog R, Schmitz G, Shevchenko A. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol. 2007;433:175–191. doi: 10.1016/S0076-6879(07)33010-3. [DOI] [PubMed] [Google Scholar]
- 33.Ackermann AL, Berna MJ, Eckstein JA, Ott LW, Chaudhary AK. Current applications of liquid chromatography/mass spectrometry in pharmaceutical discovery after a decade of innovation. Annu Rev Anal Chem. 2008;1:12.11–12.40. doi: 10.1146/annurev.anchem.1.031207.112855. [DOI] [PubMed] [Google Scholar]
- 34.Lee SH, Williams MV, DuBois RN, Blair IA. Targeted lipidomics using electron capture atmospheric pressure chemical ionization mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2168–2176. doi: 10.1002/rcm.1170. [DOI] [PubMed] [Google Scholar]
- 35.Hermansson M, Uphoff A, Kakela R, Somerharju P. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- 36.Liebisch G, Drobnik W, Reil M, Trumbach B, Arnecke R, Olgemoller B, Roscher A, Schmitz G. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS) J Lipid Res. 1999;40:1539–1546. [PubMed] [Google Scholar]
- 37.Masukawa Y, Tsujimura H, Narita H. Liquid chromatography-mass spectrometry for comprehensive profiling of ceramide molecules in human hair. J Lipid Res. 2006;47:1559–1571. doi: 10.1194/jlr.D600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Farwanah H, Pierstorff B, Schmelzer CE, Raith K, Neubert RH, Kolter T, Sandhoff K. Separation and mass spectrometric characterization of covalently bound skin ceramides using LC/APCI-MS and nano-ESI-MS/MS. J Chromatogr B Anal Technol Biomed Life Sci. 2007;852:562–570. doi: 10.1016/j.jchromb.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 39.Colsch B, Afonso C, Popa I, Portoukalian J, Fournier F, Tabet JC, Baumann N. Characterization of the ceramide moieties of sphingoglycolipids from mouse brain by ESI-MS/MS: Identification of ceramides containing sphingadienine. J Lipid Res. 2004;45:281–286. doi: 10.1194/jlr.M300331-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Lieser B, Liebisch G, Drobnik W, Schmitz G. Quantification of sphingosine and sphinganine from crude lipid extracts by HPLC electrospray ionization tandem mass spectrometry. J Lipid Res. 2003;44:2209–2216. doi: 10.1194/jlr.D300025-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Hein LK, Meikle PJ, Hopwood JJ, Fuller M. Secondary sphingolipid accumulation in a macrophage model of Gaucher disease. Mol Genet Metab. 2007;92:336–345. doi: 10.1016/j.ymgme.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Fuller M, Rozaklis T, Lovejoy M, Zarrinkalam K, Hopwood JJ, Meikle PJ. Glucosylceramide accumulation is not confined to the lysosome in fibroblasts from patients with Gaucher disease. Mol Genet Metab. 2008;93:437–443. doi: 10.1016/j.ymgme.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Masukawa Y, Tsujimura H. Highly sensitive determination of diverse ceramides in human hair using reversed-phase high-performance liquid chromatography-electrospray ionization mass spectrometry. Lipids. 2007;42:275–290. doi: 10.1007/s11745-006-3012-6. [DOI] [PubMed] [Google Scholar]
- 44.DeLong CJ, Baker PRS, Samuel M, Cui Z, Thomas MJ. Molecular species composition of rat liver phospholipids by ESI-MS/MS: The effect of chromatography. J Lipid Res. 2001;42:1959–1968. [PubMed] [Google Scholar]
- 45.Esch SW, Williams TD, Biswas S, Chakrabarty A, Levine SM. Sphingolipid profile in the CNS of the twitcher (globoid cell leukodystrophy) mouse: A lipidomics approach. Cell Mol Biol. 2003;49:779–787. [PubMed] [Google Scholar]
- 46.Berdyshev EV, Gorshkova IA, Garcia JG, Natarajan V, Hubbard WC. Quantitative analysis of sphingoid base-1-phosphates as bisacetylated derivatives by liquid chromatography-tandem mass spectrometry. Anal Biochem. 2005;339:129–136. doi: 10.1016/j.ab.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Natl Acad Sci USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 49.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 50.Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem. 2007;79:4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- 51.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 52.Ejsing CS, Moehring T, Bahr U, Duchoslav E, Karas M, Simons K, Shevchenko A. Collision-induced dissociation pathways of yeast sphingolipids and their molecular profiling in total lipid extracts: A study by quadrupole TOF and linear ion trap-orbitrap mass spectrometry. J Mass Spectrom. 2006;41:372–389. doi: 10.1002/jms.997. [DOI] [PubMed] [Google Scholar]
- 53.Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electro-spray ionization tandem mass spectrometry. Anal Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 54.Han X, Yang J, Cheng H, Ye H, Gross RW. Towards fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Tsui ZC, Chen QR, Thomas MJ, Samuel M, Cui Z. A method for profiling gangliosides in animal tissues using electrospray ionization-tandem mass spectrometry. Anal Biochem. 2005;341:251–258. doi: 10.1016/j.ab.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 56.Kalderon B, Sheena V, Shachrur S, Hertz R, Bar-Tana J. Modulation by nutrients and drugs of liver acyl-CoAs analyzed by mass spectrometry. J Lipid Res. 2002;43:1125–1132. doi: 10.1194/jlr.m200060-jlr200. [DOI] [PubMed] [Google Scholar]
- 57.Golovko MY, Murphy EJ. An improved method for tissue long-chain acyl-CoA extraction and analysis. J Lipid Res. 2004;45:1777–1782. doi: 10.1194/jlr.D400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J Lipid Res. 2005;46:1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han X, Yang K, Yang J, Fikes KN, Cheng H, Gross RW. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J Am Soc Mass Spectrom. 2006;17:264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Han X, Cheng H, Mancuso DJ, Gross RW. Caloric restriction results in phospholipid depletion, membrane remodeling and triacylglycerol accumulation in murine myocardium. Biochemistry. 2004;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- 61.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 62.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes inArabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 64.Gatt S. Enzymatic hydrolysis of sphingolipids. I. Hydrolysis and synthesis of ceramides by an enzyme from rat brain. J Biol Chem. 1966;241:3724–3730. [PubMed] [Google Scholar]
- 65.Portman OW, Alexander M. Metabolism of sphingolipids by normal and atherosclerotic aorta of squirrel monkeys. J Lipid Res. 1970;11:23–30. [PubMed] [Google Scholar]
- 66.Guan XL, He X, Ong WY, Yeo WK, Shui G, Wenk MR. Non-targeted profiling of lipids during kainate-induced neuronal injury. FASEB J. 2006;20:1152–1161. doi: 10.1096/fj.05-5362com. [DOI] [PubMed] [Google Scholar]
- 67.Han X, Cheng H, Fryer JD, Fagan AM, Holtzman DM. Novel role for apolipoprotein E in the central nervous system: Modulation of sulfatide content. J Biol Chem. 2003;278:8043–8051. doi: 10.1074/jbc.M212340200. [DOI] [PubMed] [Google Scholar]
- 68.Cheng H, Xu J, McKeel DW, Jr, Han X. Specificity and potential mechanism of sulfatide deficiency in Alzheimer’s disease: An electrospray ionization mass spectrometric study. Cell Mol Biol. 2003;49:809–818. [PubMed] [Google Scholar]
- 69.te Vruchte D, Lloyd-Evans E, Veldman RJ, Neville DC, Dwek RA, Platt FM, van Blitterswijk WJ, Sillence DJ. Accumulation of glycosphingolipids in Niemann-Pick C disease disrupts endosomal transport. J Biol Chem. 2004;279:26167–26175. doi: 10.1074/jbc.M311591200. [DOI] [PubMed] [Google Scholar]
- 70.He X, Dagan A, Gatt S, Schuchman EH. Simultaneous quantitative analysis of ceramide and sphingosine in mouse blood by naphthalene-2,3-dicarboxyaldehyde derivatization after hydrolysis with ceramidase. Anal Biochem. 2005;340:113–122. doi: 10.1016/j.ab.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 71.Zeng Y, Cheng H, Jiang X, Han X. Endosomes and lysosomes play distinct roles in sulfatide-induced neuroblastoma apoptosis: Potential mechanisms contributing to abnormal sulfatide metabolism in related neuronal diseases. Biochem J. 2008;410:81–92. doi: 10.1042/BJ20070976. [DOI] [PubMed] [Google Scholar]
- 72.Guan XL, Wenk MR. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from. Sacchar omyces cerevisiae Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- 73.Han X. An update on lipidomics: Progress and application in biomarker and drug development. Curr Opin Mol Ther. 2007;9:586–591. [PubMed] [Google Scholar]
- 74.Han X. Characterization and direct quantitation of ceramide molecular species from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2002;302:199–212. doi: 10.1006/abio.2001.5536. [DOI] [PubMed] [Google Scholar]