Abstract
DNA mismatch recognition is performed in eukaryotes by two heterodimers known as MutSα (Msh2/Msh6) and MutSβ (Msh2/Msh3) that must reside in the nucleus to function. Two putative Msh2 nuclear localization sequences (NLS) were characterized by fusion to green fluorescent protein (GFP) and site-directed mutagenesis in the context of Msh2. One NLS functioned in GFP targeting assays and both acted redundantly within Msh2. We examined nuclear localization of each of the MutS monomers in the presence and absence of their partners. Msh2 translocated to the nucleus in cells lacking Msh3 and Msh6; however, cells lacking Msh6 showed significantly decreased levels of nuclear Msh2. Furthermore, the overall protein levels of Msh2 were significantly diminished in the absence of Msh6, particularly if Msh2 lacked a functional NLS. Msh3 localized in the absence of Msh2, but Msh6 localization depended on Msh2 expressing functional NLSs. Overall, the nuclear levels of Msh2 and Msh6 decline when the other partner is absent. The data suggest a stabilization mechanism to prevent free monomer accumulation in the cytoplasm.
1. Introduction
The most common hereditary colorectal cancer syndrome and certain sporadic cancers have been directly linked to defects in DNA mismatch repair [reviewed in 1]. DNA mismatch repair is a highly conserved biological process contributing to the accurate preservation of genetic material. The mechanism of repair includes recognition and binding of a mismatch in the DNA helix, followed by cleavage, unwinding, and degradation of the error-containing strand. After the error is removed, a new DNA strand with correct base pairing is synthesized [reviewed in 2].
In eukaryotes, nuclear DNA mismatch recognition involves three homologs (Msh2, Msh3, and Msh6) of the prokaryotic MutS DNA mismatch repair protein [3]. Msh6 and Msh3 interact with Msh2 to form the heterodimers MutSα and MutSβ, respectively [4,5]. Like bacterial MutS, these complexes are thought to bind nonspecifically to DNA until encountering a mismatch, at which point the dimer undergoes a conformational change allowing stable binding [6–8]. Fully functional yeast mismatch repair requires both heterodimers. Specifically, MutSα associates with single base mispairs and insertion-deletion loops of one nucleotide, whereas MutSβ binds small and intermediate-sized insertion-deletion loops [9], including single nucleotide loops [10,11].
As in yeast, the human MutS homologs, Msh2, Msh6, and Msh3, form analogous heterodimers to recognize and bind nuclear genomic mismatches [12,13]. The binding specificities of the heterodimers, however, differ slightly from yeast. Single base mis-pairings are similarly recognized only by human MutSα; however, both dimers detect insertion-deletion loops between one and eight nucleotides [14]. This specificity overlap, in conjunction with the finding that 90% of nuclear Msh2 is associated with Msh6 [14], suggests that MutSα is the primary mismatch recognition heterodimer in humans. Downstream events in eukaryotic DNA mismatch repair occur after formation of a higher order complex between MutSα or MutSβ and the heterodimer MutLα, comprised of Mlh1 and Pms2 (Pms1 in yeast) [15], or MutLβ, consisting of Mlh1 and Mlh3 [16].
Considerable research has delineated the components and mechanisms of DNA repair; however, less is known about the regulation. For example, the post-translational trafficking of mismatch recognition proteins to the nucleus is a relatively unexplored area of research. In eukaryotes, passage through the nuclear envelope presents an opportunity for repair regulation by controlling access to the DNA. Entry into the nucleus by passive diffusion is prohibited for molecules larger than ~60 kDa [17–19]. The sizes of the mismatch recognition subunits (Msh2, 105 kDa; Msh3, 127 kDa; Msh6, 160 kDa) dictate that they must be actively transported through the nuclear pores. In addition, facilitated nuclear transport can accommodate macromolecules with a diameter of 39 nm (nearly 50 MDa) without disassembly [20]. Thus, the size of the nuclear pore complexes would permit mismatch recognition heterodimers formed in the cytoplasm to localize to the nucleus. Given these parameters, the mismatch repair proteins could localize to the nucleus in either their monomeric or dimeric states.
Investigation of mouse mismatch repair heterodimer MutLα indicates that the subunits not only can, but must, undergo dimerization for nuclear import [21]. Interaction of the proteins seemed to promote transport by altering conformations to expose nuclear localization sequences (NLS). Import of human Msh2 and Msh6 was additionally postulated to occur after dimer formation because Msh2 contains no classical NLS and the nuclear levels of Msh2 drop in cell lines not expressing Msh6; it was thus assumed to rely upon the NLS of Msh6 for nuclear translocation [22].
The research described in this paper was conducted in the S. cerevisiae eukaryotic mismatch repair model organism and focused on characterizing the nuclear import of the mismatch recognition proteins of MutSα and MutSβ. We found that the Msh2 and Msh6 MutSα heterodimer partners are dependent upon one another for efficient nuclear localization and stabilization of Msh2 levels. The Msh2 and Msh3 MutSβ heterodimer partners do not appear to significantly regulate one another; however this likely reflects the need for fewer MutSβ molecules to scan the genome for insertion/deletion loops compared with the much more frequent single-base mispairs and single nucleotide insertion/deletion loop mismatches recognized by MutSα.
2. Materials and Methods
2.1. Microbial and Molecular Techniques
Strains (Table 1) and plasmids (Table 2) were manipulated using standard microbial and molecular techniques [23,24]. Primers (Table 3) were synthesized by Integrated DNA Technologies, Inc (Coralville, IA). Restriction endonuclease digestions and Polymerase Chain Reactions (PCR) were performed using manufacturer recommended reaction conditions (New England Biolabs; Beverly, MA).
Table 1.
Yeast strains used in this study*
| Name | Genotype | Plasmids | Source |
|---|---|---|---|
| AGY75 | MATα ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2Δ::LEU2 | pSH44 | [33] |
| AGY292 | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGBD-MSH2 | [33] |
| AGY293 | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGBD-C2 | [33] |
| AGY298 | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGBD-MSH2-R542P | [33] |
| AGY333 | MATα trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGAD-MSH6 | [33] |
| AGY341 | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal43 gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGBD-MSH2-C345R | [33] |
| AGY881 | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGBD-MSH2-L521P | [33] |
| AGY908 | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | pGBD-MSH2-S762Y | [33] |
| MY10029 | MATα msh2Δ::URA3 his3-11,15 ade2-1 trp1-1 ura3-1 | pMSH2-Δ552 | This study |
| MY10030 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 lys2Δ RAD5 msh2Δ::LEU2 msh3Δ::kanMX4 | pMSH2-Δ552 | This study |
| MY10031 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 msh2Δ::URA3 msh6Δ::kanMX4 | pMSH2-Δ552 | This study |
| MY10032 | MATα ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2Δ::LEU2 msh3Δ::kanMX4 msh6Δ::kanMX4 | pMSH2-Δ552 | This study |
| MY10081 | MATα MSH3-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pMSH2 | This study |
| MY10082 | MATα MSH3-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pMSH2-Δ552Δ525 | This study |
| MY10083 | MATα MSH3-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pRS413 | This study |
| MY10084 | MATα MSH3-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pMSH2-Δ525 | This study |
| MY10085 | MATα MSH3-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pMSH2-Δ552 | This study |
| MY10086 | MATα MSH6-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL+ pMSH2 | This study |
| MY10087 | MATα MSH6-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL+ pMSH2-Δ552Δ525 | This study |
| MY10088 | MATα MSH6-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pRS413 | This study |
| MY10089 | MATα MSH6-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pMSH2-Δ525 | This study |
| MY10091 | MATα MSH6-RFP::kanMX6 msh2Δ::URA3 his3-11,15 trp1-1 ura3-1 | p3xGFP-HDEL + pMSH2-Δ552 | This study |
| MY10102 | MATα trp1-1 his3-11,15 ura3-1 msh2Δ::URA3 MSH6-RFP::kanMX6 | p3xGFP-HDEL + pRS413 | This study |
| MY10103 | MATα trp1-1 his3-11,15 ura3-1 msh2Δ::URA3 MSH6-RFP::kanMX6 | p3xGFP-HDEL + pMSH2 | This study |
| MY10106 | MATα trp1-1 his3-11,15 ura3-1 msh2Δ::URA3 MSH6-RFP::kanMX6 | p3xGFP-HDEL + pMSH2-C345R | This study |
| MY10108 | MATα trp1-1 his3-11,15 ura3-1 msh2Δ::URA3 MSH6-RFP::kanMX6 | p3xGFP-HDEL + pMSH2-L521P | This study |
| MY10127 | MATα trp1-1 his3-11,15 ura3-1 msh2Δ::URA3 MSH6-RFP::kanMX6 | p3xGFP-HDEL + pMSH2-R542P | This study |
| MY10197 | MATα ade2-1 his3-11,15 trp1-1 ura3-1 msh2Δ::URA3 | pMSH2-Δ525 | This study |
| MY10198 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 lys2Δ msh2Δ::LEU2 msh3Δ::kanMX4 | pMSH2-Δ525 | This study |
| MY10199 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 msh2Δ::URA3 msh6Δ::kanMX4 | pMSH2-Δ525 | This study |
| MY10200 | MATα ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2Δ::LEU2 msh3Δ::kanMX4 msh6Δ::kanMX4 | pMSH2-Δ525 | This study |
| MY10246 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 msh2Δ::LEU2 msh3Δ::kanMX4 | pMSH2 | This study |
| MY10247 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 msh2Δ::URA3 msh6Δ::kanMX4 | pMSH2 | This study |
| MY10248 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 msh2Δ::LEU2 msh3Δ::kanMX4 msh6Δ::kanMX4 | pMSH2 | This study |
| MY10283 | MATα trp1-1 his3-11,15 ura3-1 msh2Δ::URA3 MSH6-RFP::kanMX6 | pMSH2-S762Y + p3xGFP-HDEL | This study |
| MY10299 | MATa his3-11,15 trp1-1 ura3-1 | p3xGFP + pCFP-HDEL | This study |
| MY10300 | MATa his3-11,15 trp1-1 ura3-1 | p3xGFP-NLSSV40 + pCFP-HDEL | This study |
| MY10301 | MATa his3-11,15 trp1-1 ura3-1 | p3xGFP-NLS525 + pCFP-HDEL | This study |
| MY10302 | MATa his3-11,15 trp1-1 ura3-1 | p3xGFP-NLS552 + pCFP-HDEL | This study |
| MY9741 | MATα ade2-1 his3-11,15 trp1-1 ura3-1 msh2Δ::URA3 | pMSH2 | This study |
| MY9742 | MATα ade2-1 his3-11,15 trp1-1 ura3-1 msh2Δ::URA3 | pRS413 | This study |
| MY9820 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 msh2Δ::LEU2 msh3Δ::kanMX4 msh6Δ::kanMX4 | pMSH2-Δ525Δ552 | This Study |
| MY9821 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 msh2Δ::URA3 msh6Δ::kanMX4 | pMSH2-Δ525Δ552 | This Study |
| MY9822 | MATα ade2-1 his3-11,15 trp1-1 ura3-1 msh2Δ::URA3 | pMSH2-Δ525Δ552 | This study |
| MY9825 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 lys2Δ msh2Δ::LEU2 msh3Δ::kanMX4 | pMSH2-Δ525Δ552 | This study |
All strains are derived from W303 except for the yeast 2-hybrid strains (AGY292, 293, 298, 333, 341, 881, and 908). The strains were confirmed to be wild-type at the RAD5 locus by PCR and at the CAN1 locus by canavanine resistance assays.
Table 2.
Plasmids used in this study
| Name | Relevant Markers | Strain Number | Source |
|---|---|---|---|
| pMR5484 | RFP-kanMX4 | MR5484 | S. Clark, Rose Lab |
| pSH44 | PLEU2-(GT)16.5-URA3 TRP1/ARS CEN ampr | [29] | |
| pMR3453 | PGAL-GFP CEN4 ARS1 LEU2 ampr | MR3453 | [61] |
| pCFP-HDEL | CFP-HDEL TRP1 CEN ARS ampr | MR5214 | P. Melloy, Rose Lab |
| p3xGFP-HDEL | 3xGFP-HDEL TRP1 CEN ARS ampr | MR5029 | [62] |
| pMSH2-Δ525 | msh2-ΔNLS525 HIS3 CEN ARS ampr | MR5210 | this study |
| pMSH2-Δ552 | msh2-ΔNLS552 HIS3 CEN ARS ampr | MR5658 | this study |
| pMSH2-Δ525Δ552 | msh2-ΔNLS525 NLS552 HIS3 CEN ARS ampr | MR5586 | this study |
| pMSH2 | MSH2 HIS3 CEN ARS ampr | AG17 | [33] |
| pRS413 | HIS3 CEN ARS ampr | [63] | |
| pMSH2-C345R | msh2-C345R HIS3 CEN ARS ampr | AG208 | [33] |
| pMSH2-R542P | msh2-R542P HIS3 CEN ARS ampr | AG29 | [33] |
| pMSH2-S762Y | msh2-S762Y HIS3 CEN ARS ampr | AG462 | [33] |
| pMSH2-L521P | msh2-L521P HIS3 CEN ARS ampr | AG420 | [33] |
| pGBD-C2 | GBD TRP1 2μ ampr | [38] | |
| pGBD-MSH2 | GBD-MSH2 TRP1 2μ ampr | AG124 | [33] |
| pGBD-MSH2-C345R | GBD-msh2-C345R TRP1 2μ ampr | AG235 | [33] |
| pGBD-MSH2-R542P | GBD-msh2-R542P TRP1 2μ ampr | AG132 | [33] |
| pGBD-MSH2-S762Y | GBD-msh2-S762Y TRP1 2μ ampr | AG455 | [33] |
| pGBD-MSH2-L521P | GBD-msh2-L521P TRP1 2μ ampr | AG447 | [33] |
| pGAD-MSH6 | GAD-MSH6 LEU2 2μ ampr | AG333 | [33] |
| pBS-3xGFP–TRP1 | 3xGFP TRP1 ampr | [64] | |
| pPMSH2-GFP | PMSH2-GFP HIS3 CEN ARS ampr | This study | |
| p3xGFP | PMSH2-3xGFP HIS3 CEN ARS ampr | MR5173 | This study |
| p3xGFP-NLSSV40 | PMSH2-3xGFP-NLS SV40 HIS3 CEN ARS ampr | MR5201 | This study |
| p3xGFP-NLS525 | PMSH2-3xGFP-NLS 525 HIS3 CEN ARS ampr | MR5202 | This study |
| p3xGFP-NLS552 | PMSH2-3xGFP-NLS 552 HIS3 CEN ARS ampr | MR5203 | This study |
Table 3.
Primers used in this study
| Name | Purpose | Sequence (5′ to 3′) |
|---|---|---|
| MSH6-5 | MSH6 disruption | GGCTTCTGGTTAAGTTTGGC |
| MSH6-3 | MSH6 disruption | GCTGAATCATAGGTCAAG |
| MSH3-5 | MSH3 disruption | GTGCAATGACGATTTGAGCGGCC |
| MSH3-3 | MSH3 disruption | GCAACTTGTACAAGGCCAAGGC |
| PR648 | Verification of 3′ kanMX junction | TATGGAACTGCCTCGGTGAG |
| PR649 | Verification of 5′ kanMX junction | TCGATAGATTGTCGCACCTG |
| MSH6F2+ | MSH6::RFP fusion | CGTTTTGAAAAGTCTATTTAGTATAATTTGATGATTTACAAATCCGGTGGTGGTGGTGGTGGTCGGATCCCCGGGTTAATTAA |
| MSH6R1− | MSH6::RFP fusion | AAATAAGTAAAAATCTTACATACATCGTAAATGAAAATACGGATTCGAGCTCGTTTAAAC |
| MSH3F2+ | MSH3::RFP fusion | GCAACGGATAAACTCGCGAAATTACTATCATTGGATATCCACGGTGGTGGTGGTGGTCGGATCCCCGGGTTAATTAA |
| MSH3R1− | MSH3::RFP fusion | GTCTGATAATGCTGCATTTAGAACATACGTACCATCCGCAGGATTCGAGCTCGTTTAAAC |
| MSH2GFP 5′ | Amplify GFP | CCTAACATCAAAATCCTCAGATTAAAAGTATGAGTAAAGGAGAAGAACTT |
| vecGFP 3′ | Amplify GFP | GTGGCGGCCGCTCTAGAACTAGTGGATCCTTATTTGTATAGTTCATCCATGCC |
| vecMSH2 5′ | Amplify PMSH2 | ATCGAATTCCTGCAGCCCGGGGGATCCACCTCAACAGCTACACATTC |
| MSH2GFP 3′ | Amplify PMSH2 | AAGTTCTTCTCCTTTACTCATACTTTTAATCTGAGGATTTTGATGTTAGG |
| SV40NLS+ | SV40 NLS 3xGFP fusion | GGCATGGATGAACTATACAAACCAAAGAAGAAGAGAAAGTAAACTAGTTCTAGAGCGGCCGCCAC |
| SV40NLS− | SV40 NLS 3xGFP fusion | GTGGCGGCCGCTCTAGAACTAGTTTACTTTCTCTTCTTCTTTGGTTTGTATAGTTCATCCATGCC |
| 2NLS525+ | NLS 525 3xGFP fusion | GGCATGGATGAACTATACAAACCGGACAAAAAACTGAAGTTGTAAACTAGTTCTAGAGCGGCCGCCAC |
| 2NLS525− | NLS 525 3xGFP fusion | GTGGCGGCCGCTCTAGAACTAGTTTACAACTTCAGTTTTTTGTCCGGTTTGTATAGTTCATCCATGCC |
| 2NLS552+ | NLS 552 3xGFP fusion | GGCATGGATGAACTATACAAACGTAAACATAAGAAGTAAACTAGTTCTAGAGCGGCCGCCAC |
| 2NLS552− | NLS 552 3xGFP fusion | GTGGCGGCCGCTCTAGAACTAGTTTACTTCTTATGTTTACGTTTGTATAGTTCATCCATGCC |
| PR724 | Sequencing NLS GFP fusions | GGAAACAGCTATGACCATG |
| ΔNLS 525 | Mutagenesis of MSH2 NLS525 | GCTGAAGATCTAGGATTCGATCCGGCTGCTGCTCTGGCTTTGGAGAACCATCATCTGCATGG |
| ΔNLS 552 | Mutagenesis of MSH2 NLS552 | GACACGTAATGACGCCAAGGAGTTACGTGCTGCTGCTGCTTACATTGAGTTGTCGACAGTAAAAGC |
| MSH2-7 | Sequencing of NLSs | AAGTTGAGTTTAATGAGG |
2.2. Strain Construction
Kanamycin marked deletions of MSH6 (msh6Δ::kanMX4) and MSH3 (msh3Δ::kanMX4) were engineered in W303 RAD5 CAN1 derived strains to create single, double and triple mismatch recognition gene deletions using a single-step PCR-mediated gene disruption [25,26]. Specifically, the deletion loci were amplified from strains obtained from the Yeast Deletion Consortium [27]. The msh6Δ::kanMX4 locus was amplified with primers MSH6-5 and MSH6-3, and msh3Δ::kanMX4 was amplified with MSH3-5 and MSH3-3. Molecular confirmation of proper gene replacement was achieved through PCR of the MSH6 and MSH3 loci as well as the 5′ and 3′ junctions of the integrated cassette. Primer pairs MSH6-5, MSH6-3 and MSH3-5, MSH3-3 were used to amplify the loci, while the pairing of primers MSH6-5 and MSH3-5 with PR649 and primers MSH6-3 and MSH3-3 with PR648 confirmed the 5′ and 3′ kanMX4 junctions, respectively. Gene disruptions in constructed strains were further verified by an observed loss of DNA mismatch repair in a canavanine drug sensitivity assay for msh6Δ [28] and dinculeotide instability assays for msh3Δ [29].
The C terminal-coding regions of MSH3 and MSH6 were fused to a kanamycin marked red fluorescent protein gene (RFP::kanMX6) using a PCR-based method described previously [30]. A 20-cycle reaction profile of 1 minute at 94°C, 1 minute at 50°C, and 2.5 minutes at 68°C, followed by a final 10 minute extension at 68°C was used to amplify plasmid DNA encoding RFP::kanMX6 (pMR5484). Primers to amplify the fusion locus for MSH3 were MSH3R1− and MSH3F2+. Primers to amplify the fusion locus for MSH6 were MSH6R1− and MSH6F2+. The fusions were designed to encode a flexible glycine linker between the protein and RFP coding sequences. The strains were confirmed by PCR of the fusion junctions and the functionality of the fusion proteins were tested in a canavanine drug sensitivity assay for MSH6-RFP::kanMX6 and a dinculeotide instability assay for MSH3-RFP::kanMX6 as described above. The Msh6 fusion protein was fully functional; however, the Msh3 fusion protein showed diminished mismatch repair efficiency.
2.3. Plasmid Construction
To construct GFP NLS fusions, the GFP coding sequence (GFP CS) was amplified from pMR3453 (Table 2) using PCR primers MSH2GFP 5′ and vecGFP 3′ (Table 3), and the MSH2 promoter, (PMSH2) was amplified with primers vecMSH2 5′ and MSH2GFP 3′. Wild-type yeast cells were transformed with GFP CS, PMSH2, and BamHI-linearized pRS413 to create pPMSH2-GFP. The 3xGFP coding sequence (3xGFP CS) on pBS-3xGFP–TRP1 (John Cooper, University of North Carolina, Chapel Hill) was liberated with BamHI and NotI and purified from the gel slice (GENECLEAN Kit, Bio101, Vista, CA). Plasmid p3xGFP was created by homologous recombination [31] between 3xGFP CS and NcoI-linearized pPMSH2-GFP.
Putative NLS in MSH2 starting at codon 525 (NLS 525) and at codon 552 (NLS552) as well as the SV40 large T antigen NLS (NLS SV40) were fused in frame to the C-terminus of the third GFP in p3xGFP to create p3xGFP-NLS525, p3xGFP-NLS552, and p3xGFP-NLSSV40 respectively. The fusions were accomplished as follows: complementary oligonucleotides encoding the NLSs (Table 3) with flanking sequences homologous to p3xGFP were annealed, mixed with SpeI-linearized p3xGFP and used to transform wild-type yeast. Nucleotide sequencing (GENEWIZ, Inc, South Plainfield, NJ) of extracted plasmid DNA with primer PR724 confirmed successful recombinants, and digestion with PvuII confirmed that the repeated GFP coding sequence was intact.
2.4. Site-Directed Mutagenesis of Msh2 NLSs
Putative NLSs were mutagenized in the context of the MSH2 coding sequence using a plasmid-encoded MSH2 gene with multiple hemagglutinin epitopes inserted (MSH2::HA) as the template DNA (pMSH2, Table 2) according to the dut− ung− method [32]. The potential NLS 525 (PDKKLKL) was mutagenized to PAAALAL (pMSH2-Δ525) using the ΔNLS 525 mutagenic primer (Table 3) and the putative NLS 552 (RKHKK) was mutagenized to AAHAA (pMSH2-Δ552) using the ΔNLS 552 mutagenic primer (Table 3). A double NLS knock-out (pMSH2-Δ525Δ552) was created using the two mutagenic primers in a single reaction. The mutations were verified by nucleotide sequencing with primer MSH2-7 (GENEWIZ, Inc.).
2.5 Mismatch Repair Functionality
DNA mismatch repair assays were performed to test for MutSα/β separation of function caused by the mutagenesis of NLS 525. Constructs pMSH2-Δ525, pMSH2-Δ552, pMSH2-Δ525Δ552, pMSH2, and the pRS413 were used to transform AGY75 a msh2Δ strain [33] harboring the pSH44 dinucleotide instability reporter construct [29]. Colonies from each of the transformations were tested for DNA mismatch repair using qualitative assays using previously detailed experiments [3,29,34].
2.6. Indirect Immunofluorescence of Msh2 and Msh2 NLS Mutant Proteins
The strains used in the analysis were as follows: msh2Δ + pMSH2 (MY9741), msh2Δmsh3Δ + pMSH2 (MY10246), msh2Δmsh6Δ + pMSH2 (MY10247), msh2Δmsh3Δmsh6Δ + pMSH2 (MY10248), msh2Δ + pMSH2-Δ525 (MY10197), msh2Δmsh3Δ + pMSH2-Δ525 (MY10198), msh2Δmsh6Δ + pMSH2-Δ525 (MY10199), msh2Δmsh3Δmsh6Δ + pMSH2-Δ525, (MY10200), msh2Δ + pMSH2-Δ552 (MY10029), msh2Δmsh3Δ + pMSH2-Δ552 (MY10030), msh2Δmsh6Δ + pMSH2-Δ552 (MY10031), msh2Δmsh3Δmsh6Δ + pMSH2-Δ552 (MY10032), msh2Δ + pMSH2-Δ525Δ552 (MY9822), msh2Δmsh3Δ + pMSH2-Δ525Δ552 (MY9825), msh2Δmsh6Δ + pMSH2-Δ525Δ552 (MY9821), and msh2Δmsh3Δmsh6Δ + pMSH2-Δ525Δ552 (MY9820). In addition, MY9742 cells (msh2Δ + a vector control, pRS413) served as a negative control for background fluorescence.
Cultures of exponentially growing yeast strains were processed for immunofluorescence as recommended [35] with the following specifications: the cells were fixed for 30 min, incubated for 30 min at room temperature with mouse α-HA 12CA5 antibody (Princeton Monoclonal Facility) diluted 1:400, and incubated at room temperature for 45 min with goat α-mouse IgG Alexa Fluor 488 (Molecular Probes, Invitrogen Corporation) diluted 1:200. After the appropriate washes, the cells were stained with the DNA-specific dye, DAPI (4′,6′-diamidino-2-phenylindole, Molecular Probes) as recommended. Cellular morphology was observed with differential interference contrast (DIC) optics and Msh2 localization and cellular DNA were visualized by fluorescence microscopy using a Nikon Eclipse E600 microscope (Nikon, Inc., Melville, NY) equipped with a DIC H Plan Fluor 100x oil immersion objective lens (NA=1.3). Fluorescence was visualized using Nikon filter sets: UV-2A (for DAPI stained nuclei) and B-2A (for Msh2 localization). Images are recorded using a Nikon DXM1200 digital camera and ACT-1 software, version 2 and stored as electronic files.
The nuclear and cytoplasmic fluorescence intensities were determined for equal areas using the ImageJ [36]. The ratio of nuclear/cytoplasmic fluorescence, the standard error of the mean, and p values based on a two-tailed TTEST, two samples with equal variance were calculated were calculated using Microsoft Excel or Synergy Software KaleidaGraph version 4.03.
2.7. Fluorescence Microscopy
For localization of 3xGFP and the 3xGFP-NLS fusion proteins yeast strains MY10299 (p3xGFP + pCFP-HDEL), MY10300 (p3xGFP-NLSSV40 + pCFP-HDEL), MY10301 (p3xGFP-NLS525 + pCFP-HDEL), and MY10302 (p3xGFP-NLS552 + pCFP-HDEL) cells were grown to exponential phase, concentrated, and analyzed with a Delta Vision® RT Restoration Imaging System (Applied Precision, LLC) deconvolution microscope. Images were taken using a Cool-SNAP CCD camera (Photometrics, Roper Scientifics, Inc., Tucson, AZ) and softWoRx® version 3.3.6 software. Quantification of nuclear and cytoplasmic fluorescence was as described for the indirect immunofluorescence except that the nuclei were determined by the a nuclear envelope (NE) endoplasmic reticulum (ER) fluorescent marker (Cyan Fluorescent Protein fused to the HDEL ER retention signal, CFP-HDEL) co-expressed in each strain.
Localization of Msh3-RFP and Msh6-RFP co-expressed with 3xGFP-HDEL and either no Msh2 (pRS413), wild-type Msh2 (pMSH2) or Msh2 NLS variants (pMSH2-Δ525, pMSH2-Δ552, or pMSH2-Δ525Δ552) was conducted as above with the following strains: MY10083 (MSH3-RFP::kanMX6 + p3xGFP-HDEL + pRS413), MY10081 (MSH3-RFP::kanMX6 + p3xGFP-HDEL + pMSH2), MY10084 (MSH3-RFP::kanMX6 + p3xGFP-HDEL + pMSH2-Δ525), MY10085 (MSH3-RFP::kanMX6 + p3xGFP-HDEL + pMSH2-Δ552), MY10082 (MSH3-RFP::kanMX6 + p3xGFP-HDEL + pMSH2-Δ525Δ552), MY10088 (MSH6-RFP::kanMX6 + p3xGFP-HDEL + pRS413), MY10086 (MSH6-RFP::kanMX6 + p3xGFP-HDEL + pMSH2), MY10089 (MSH6-RFP::kanMX6 + p3xGFP-HDEL + pMSH2-Δ525), MY10091 (MSH6-RFP::kanMX6 + p3xGFP-HDEL + pMSH2-Δ552), and MY10087 (MSH6-RFP::kanMX6 + p3xGFP-HDEL + pMSH2-Δ525Δ552).
2.8. Immunoblot Analysis
Approximately 3 × 107 cells of the strains described for the indirect immunofluorescence were used to prepare proteins extracts [24,37]. Samples were fractionated on a 7% resolving gel using standard discontinuous SDS-PAGE and immunoblotting techniques [23]. Detection of HA-tagged Msh2 and Msh2 NLS mutant protein was conducted according to the Amersham ECL™ Western Blotting System (GE Healthcare Life Sciences, Piscataway, NJ). The primary antibody used was mouse 12CA5 monoclonal antibody specific for the HA epitope (Princeton Monoclonal Facility). The secondary antibody was α-mouse IgG horse radish peroxidase (HRP) conjugated secondary antibody (GE Healthcare Life Sciences). Both antibodies were used at a 1:2500 dilution. After visualization of Msh2, the membrane was re-probed with rabbit α-Kar2 polyclonal (1:50,000 dilution) and α-rabbit IgG HRP (GE Healthcare Life Sciences, 1:2,500 dilution) antibodies to assay for equal protein concentrations and loadings of the samples.
2.9. Yeast 2-hybrid Assays
The MATa yeast 2-hybrid reporter strain PJ69-4A [38] harboring pGBD-C2 (AGY293), pGBD-MSH2 (AGY292), pGBD-MSH2-L521P (AGY881), pGBD-MSH2-S762Y (AGY908), pGBD-MSH2-R542P (AGY298), or pGBD-MSH2-C345R (AGY341) were crossed to a yeast 2-hybrid MATα strain PJ69-4α [38] harboring pGAD-MSH6 (AGY333) as described previously [33]. Diploid yeast strains were mated for 24 hours at 30°C and replica-printed to selective plates. For semi-quantitative assays, diploid cultures were grown in liquid medium lacking leucine and tryptophan (–LEU –TRP) to saturation. Strains were spotted onto –LEU –TRP plates and plates lacking leucine, tryptophan and histidine ( –LEU –TRP –HIS) to select for the 2-hybrid interaction.
3. Results
3.1. Msh2 translocates to the nucleus in the absence of Msh6 and Msh3
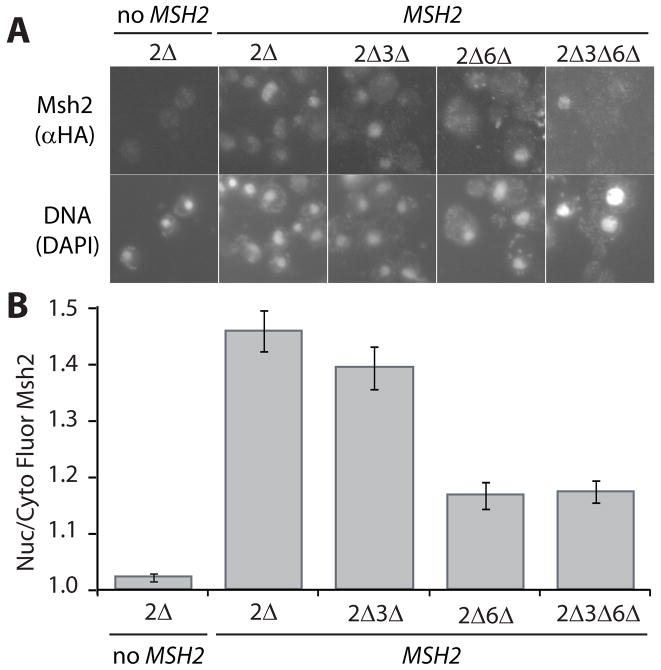
Current models for the nuclear import of both human MutSα and mouse MutLα suggest that dimerization is a requirement for transport [21,22]. We hypothesized that the nuclear transport of yeast Msh2 might similarly rely upon heterodimer associations. Localization of Msh2 was evaluated in the absence of Msh3, Msh6, and both Msh3 and Msh6 by immunofluorescence (Fig. 1A). In these experiments, yeast strains lacking MSH2 (2Δ) alone or also MSH6 (2Δ6Δ), MSH3 (2Δ3Δ), or all three (2Δ3Δ6Δ) were transformed with a centromere-based plasmid (pMSH2) expressing functional hemaglutinin (HA) epitope tagged Msh2 (MSH2) from the endogenous MSH2 promoter. These conditions mimic chromosomal expression of MSH2. As a negative control for background fluorescence the msh2Δ strain was transformed with a vector control pRS413 (no MSH2). The cells were processed for immunofluorescence to visualize Msh2 and stained with DAPI to determine the position of the nuclei (Fig. 1A). For each cell, the fluorescence intensities of Msh2 in the nucleus and the cytoplasm were measured within an equal area and the ratio of nuclear/cytoplasmic fluorescence was calculated (Fig. 1B).
Fig. 1.
Indirect Immunofluorescence of Msh2 in the presence and absence of heterodimer partners Msh3 and Msh6. Yeast deletion strains (see Materials and Methods) lacking MSH2 (2Δ), MSH2 and MSH3 (2Δ3Δ), MSH2 and MSH6 (2Δ6Δ), and MSH2, MSH3 and MSH6 (2Δ3Δ6Δ) were transformed with a plasmid, pMSH2, expressing a hemagluttinin (HA) epitope-tagged Msh2 (MSH2). Cells lacking MSH2 transformed with a plasmid vector served as a negative control for background fluorescence (2Δ, no MSH2). Exponentially growing cells were prepared for immunofluorescence, incubated with mouse anti-hemaglutinin (α-HA) monoclonal primary antibody and goat α-mouse IgG Alexa Fluor 488 secondary antibody. (A) Representative Images of Msh2 in the presence and absence of heterodimer partners. Top panels are of Msh2 localization. Bottom panels show the nuclear staining of the cells in the top panels with the DNA specific fluorescent dye DAPI. (B) Quantitative Measurements of Nuclear Msh2. The nuclear and cytoplasmic fluorescence of Msh2 were determined using the ImageJ public domain Java image processing program [36]. The ratio of nuclear/cytoplasmic fluorescence (Nuc/Cyto Fluor Msh2) is plotted for each strain. The error bars signify the standard error of the mean. Approximately 50 cells per sample were analyzed.
In the presence of Msh3 and Msh6, the Msh2 protein showed significant (p = 2 × 10−19) localization to the nucleus with a nuclear/cytoplasmic fluorescence intensity (N/C) of 1.46 ± 0.04 compared to the “no Msh2” background fluorescence control ratio of 1.02 ± 0.01. The absence of Msh3 reduced, but did not significantly diminish nuclear localization of Msh2 (N/C of 1.39 ± 0.04, p = 0.2), suggesting that the formation of MutSβ is not necessary for trafficking the bulk of Msh2 to the nucleus. Some Msh2 localized to the nucleus in cells lacking Msh6 cells (N/C of 1.16 ± 0.02, p = 3 × 10−7 compared to the no Msh2 control); however the localization was statistically lower than that observed in cells expressing both heterodimer partners (p = 4 × 10−10). Thus, Msh2 nuclear import is significantly diminished, but not blocked in the absence of its MutSα heterodimer partner, Msh6.
If the nuclear import of Msh2 exclusively depends upon heterodimer interactions, Msh2 should fail to localize to the nucleus when Msh3 and Msh6 are both absent. However, Msh2 was concentrated in the nucleus in cells lacking both Msh3 and Msh6 to the same extent as cells lacking only Msh6 (N/C of 1.17 ± 0.02). In summary, Msh6 influences Msh2 nuclear accumulation; however, Msh2 does not have a strict dependency upon interaction with its heterodimer partners for transport into the nucleus. The fact that Msh2 is able to translocate in the absence of its heterodimer partners led to the premise that Msh2 possesses a nuclear localization sequence(s) sufficient for import into the nucleus.
3.2. Msh2’s nuclear localization sequence starting at codon 525 directs GFP to the nucleus
Typically, macromolecule cargoes destined for the nucleus contain an NLS enabling them to be bound by import receptors [reviewed in 39]. Although not all nuclear proteins contain the same NLS motif, the classic NLS is a short lysine- and arginine-rich amino acid sequence originally identified in SV40 large T antigen [40]. The motif-finding PSORT II prediction program [41] identified two putative monopartite NLSs in Msh2, PDKKLKL beginning at amino acid 525 (NLS 525) and RKHKK beginning at amino acid 552 (NLS 552).
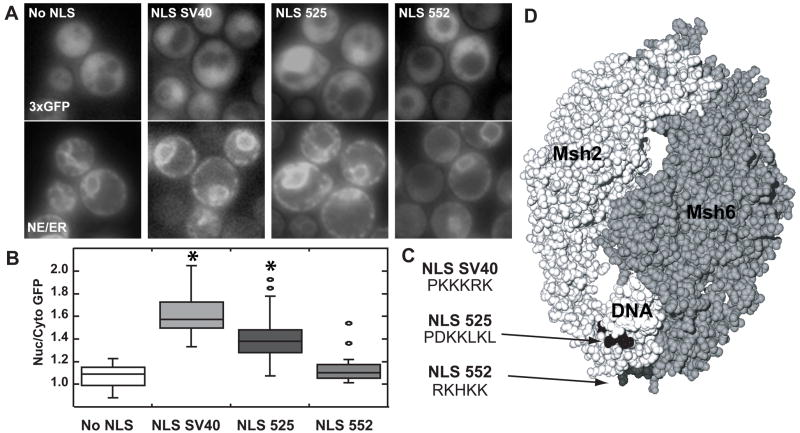
To determine whether the identified NLS sequences were sufficient to mediate nuclear import, each Msh2 putative NLS was fused in frame to the C-terminus of plasmid-encoded triple green fluorescent protein (3xGFP). The triple GFP cargo was used to increase fluorescence intensity and ensure dependence on NLS-mediated transport. Localization of 3xGFP was quantified employing the same methods described above. In these experiments the nuclear position was determined using an endoplasmic reticulum/nuclear envelope (ER/NE) fluorescent marker consisting of a four amino acid endoplasmic reticulum retention sequence (HDEL) fused to cyan fluorescent protein (CFP-HDEL). Representative images are shown in Fig. 2A.
Fig. 2.
Assessing the functionality of Msh2’s putative NLSs. Msh2 putative NLSs starting at codon 525 (NLS 525) and starting at codon 552 (NLS 552) as well as the SV40 NLS (NLS SV40) were each fused in frame to the C-terminus of plasmid encoded triple green fluorescent protein (3xGFP). Triple GFP with no NLS (no NLS) served as a negative control. (A) Representative Images of GFP Nuclear Localization. Using fluorescence microscopy, nuclear localization of 3xGFP was assessed (top panels). Nuclear position was determined using an endoplasmic reticulum/nuclear envelope (ER/NE) fluoresent marker consisting of an endoplasmic reticulum retention sequence fused to cyan fluorescent protein (bottom panels). (B) Quantitative Measurements of Nuclear 3xGFP. The nuclear and cytoplasmic fluorescence of 3xGFP were determined using ImageJ [36]. Box plots of the ratio of nuclear/cytoplasmic fluorescence (Nuc/Cyto GFP) were plotted using Synergy Software KaleidaGraph Version 4.03. The horizontal lines in the boxes indicate the median ratio of nuclear to cytoplasmic GFP in cells of a given strain. The boxes signify the range of values encompassing half of the data. The bars show the range of the entire data set. Individual circles represent outliers. The asterisks indicate the data sets statistically different from the No NLS control. Approximately 50–100 cells were counted for each strain. (C) The amino acid sequences of the NLSs used. The sequences using the single amino acid code for the Msh2 putative NLSs starting at codon 525 (NLS 525) and starting at codon 552 (NLS 552) as well as the SV40 NLS (NLS SV40) are shown. (D) Msh2 putative yeast NLSs map to the DNA binding region of human MutSα. The image is of the MutSα structure [42] with the Msh2 subunit in light grey (left) and the Msh6 subunit in a darker grey (right). The mismatched DNA molecule is in white (DNA). The arrows highlight the putative NLS positions enhanced in black (NLS525) and dark grey (NLS 552). Images were generated by manipulating 208C.pdb [42] using the Swiss PDB Viewer version 3.7 [65] and POV Ray Tracer program, version 3.6.
Localization of 3xGFP fused at its C-terminus to the SV40 large T antigen NLS (NLS SV40) established a standard for nuclear 3xGFP accumulation (Fig. 2A and 2B). This positive control showed significant nuclear localization (N/C of 1.63 ± 0.04) compared to the no NLS negative control (N/C of 1.07 ± 0.03, p <0.0001). Nuclear localized 3xGFP-NLS SV40 confirmed that import machinery can access a NLS at the C-terminus of 3xGFP.
When Msh2’s putative NLS starting at codon 525 (PDKKLKL) was fused to 3xGFP, the fluorescent protein also accumulated in the nucleus (N/C = 1.40 ± 0.03, Fig. 2B). The nuclear localization of 3xGFP-NLS 525 was significantly different from 3xGFP alone (p <0.0001), indicating that NLS 525 is sufficient to target macromolecular cargoes to the nucleus. Conversely, fusion of NLS 552 (RKHKK) to 3xGFP did not significantly facilitate nuclear transport (N/C = 1.14 ± 0.03). The localization of 3xGFP fused to NLS 552 was not different from the localization of 3xGFP without an NLS (p = 0.08, Fig. 2B). Thus, of the two putative NLSs, only NLS 525 is sufficient to direct nuclear import when separated from the native Msh2 protein.
3.3. The putative NLSs map to the DNA binding domain of Msh2
To better understand Msh2’s putative NLSs in the context of the protein, NLS 525 and NLS 552 were modeled in three dimensions using the human MutSα heterodimer crystal structure [42]. When human MutSα is bound to mismatched DNA, the regions that correspond to yeast NLS 525 and NLS 552 map adjacent to one another on the periphery of the protein in the DNA binding domain (Fig. 2D). Although both putative NLSs are exposed, the side chains of nearby amino acids may render NLS 525 less accessible. If the structure of yeast Msh2 is similar, we predict that the import machinery would have difficulty accessing and binding the NLS525 motif. The putative NLS starting at codon 552, however, is predicted to be accessible. Crystallographic analyses showed that the region of the MutS dimer containing the putative NLSs is disordered in the absence of DNA [7,42], therefore the accessibilities of the NLSs in the cytoplasmic form are uncertain.
3.4. Localization of Msh2 lacking a functional NLS is dependent on Msh6
Although a putative NLS may have the capacity to target exogenous cargo to the nucleus, it does not prove that the NLS functions in the context of the original protein. In addition, an inefficient NLS may be more functional in the structural context of the native protein. To test the efficiencies of nuclear targeting, the NLSs 525 and 552 were mutagenized both separately and together in the Msh2 coding sequence. NLS 525, PDKKLKL, was mutagenized to PAAALAL and NLS 552, RKHKK, was mutagenized to AAHAA. Alanines have been shown to inactivate NLSs in previous experiments [43–45]. Though acidic and thus atypical for a canonical NLS, the aspartic acid residue in NLS 525 was also mutagenized to alanine in case its polarity influenced the functionality of the signal. The proline residue in NLS 525 was unaltered, despite its theorized ability to enhance NLS function, because of its potential importance in folding or maintaining tertiary structure. The leucines were also retained in the mutagenized NLS 525 because they are shown by crystallographic studies to be crucial for structural integrity of MutS [7]. The regions were changed by site-directed mutagenesis of plasmid-based, epitope-tagged MSH2 (expressing Msh2) to create msh2-ΔNLS525 (expressing Msh2Δ525), msh2-ΔNLS552 (expressing Msh2Δ552) and msh2-ΔNLS525ΔNLS552 (expressing Msh2Δ525Δ552).
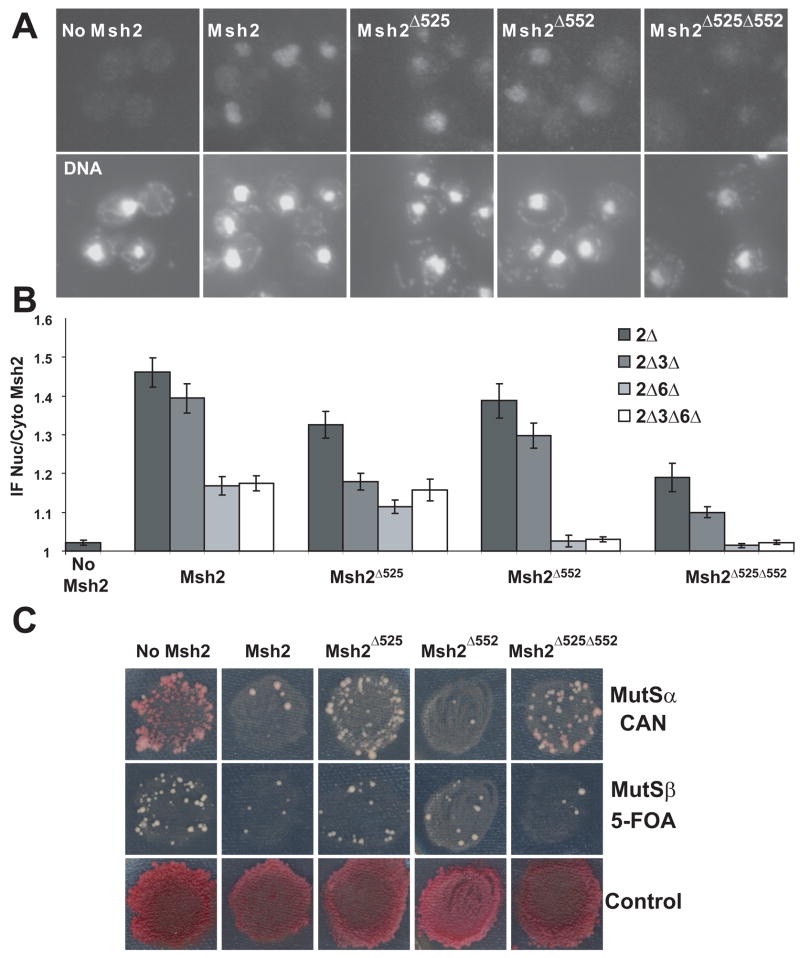
The localization of the wild-type and NLS variant Msh2 proteins were observed by immunofluorescence in the various mismatch recognition gene deletion strains described above for Fig. 1. The nuclear to cytoplasmic ratios of Msh2 NLS variants were calculated as before (Fig. 3B). All three NLS mutant proteins were found in the nucleus in the presence of heterodimer partners Msh3 and Msh6 (p values < 0.001 compared to the No Msh2 control) (Fig. 3A, 3B). Msh2Δ552 localized with an efficiency similar to wild-type Msh2 (p = 0.2); however, the Msh2Δ525 and the Msh2Δ525Δ552 NLS mutant protein nuclear accumulations were significantly lower than the wild-type Msh2 protein (p= 0.008 and p= 8 × 10−7, respectively). The fact that the double NLS mutant protein was less efficiently localized than either of the single NLS mutant proteins suggested that both NLSs are operative in the context of the protein and that there is a redundancy of function.
Fig. 3.
Indirect Immunofluorescence of Msh2 and Msh2 NLS Mutant Proteins in the Presence and Absence of Heterodimer Partners Msh3 and Msh6. Yeast deletion strains (see Materials and Methods and Figure legend 1) were transformed with plasmids, pMSH2, pMSH2-Δ525, pMSH2-Δ552, and pMSH2-Δ525Δ552, all expressing a hemagluttinin (HA) epitope-tagged Msh2 variants. The wild-type protein (Msh2), the NLS 525 mutant protein (Msh2Δ525), the NLS 552 mutant protein (Msh2 Δ552), and the double NLS mutant protein (Msh2 Δ525 Δ552) were visualized using mouse anti-hemaglutinin (α-HA) as described in the Materials and Methods and in the legend for Fig. 1. Cells lacking MSH2 transformed with a plasmid vector served as a negative control for background fluorescence (No Msh2). (A) Representative Images of Msh2 and Msh2 NLS mutant proteins in the presence of heterodimer partners, Msh6 and Msh3. Top panels are of plasmid expressed Msh2 or Msh2 NLS mutant protein localization in yeast strain lacking chromosomal MSH2 (2Δ), but expressing MSH6 and MSH3. Bottom panels show the nuclear staining of the cells in the top panels with the DNA specific fluorescent dye DAPI. (B) Quantitative Measurements of Nuclear Msh2. The nuclear and cytoplasmic fluorescence of Msh2 (shown also in Fig. 1) and Msh2 NLS mutant proteins were determined using the ImageJ [36]. The ratio of nuclear/cytoplasmic fluorescence (IF Nuc/Cyto Msh2) is plotted for each strain. The error bars signify the standard error of the mean. Approximately 100 cells were measured for each strain. The data shown for No Msh2 and Msh2 is also depicted in Fig. 1 and is shown here for comparative purposes. (C) Mismatch repair assay of the NLS variant strains. DNA mismatch repair assays were was carried out by replica-printing patches of msh2Δ stains with the pSH44 dinucleotide instability reporter [29] expressing either no Msh2 (no Msh2), the wild-type protein (Msh2) or one of the Msh2 NLS variants (MshΔ525, Msh2Δ552, Msh2Δ525Δ552) on medium containing canavanine (CAN) to assay for single-base pair mismatch repair defects primarily a consequence of impaired Msh2/Msh6 (MutSα) function, as well as on medium containing 5-FOA to assay for dinucleotide instability caused by defects in Msh2/Msh3 (MutSβ), and finally on medium allowing for cell growth to assess uniformity of plating and growth (Control).
As with the wild-type Msh2 protein (Fig. 1, 3B), the absence of Msh3 diminished, but did not significantly alter the nuclear localization of the Msh2Δ552 (p = 0.09 when comparing Msh2Δ552 expressed in 2Δ and 2Δ3Δ). However, Msh2Δ525 and Msh2Δ525Δ552 showed strongly reduced nuclear localization in the absence of Msh3 (p = 0.0002, comparing Msh2Δ525 expressed in 2Δ and 2Δ3Δ; p = 0.03, comparing Msh2Δ525Δ552 expressed in 2Δ and 2Δ3Δ). One explanation for this result is the possibility that mutation of the charged residues in the putative NLS 525 might lessen Msh2’s ability to interact specifically with Msh6, causing a drop in localization efficiency similar to that observed when Msh6 is deleted. We reasoned that the failure to interact would be evidenced in a mismatch repair assay specific for Msh2/Msh6 (MutSα) function. We evaluated Msh2/Msh3 (MutSβ) function using a microsatellite instability assay [29], and Msh2/Msh6 (MutSα) function using canavanine (CAN) resistance [11]. We found that replacing the charged residues with alanines at NLS 525 caused a MutSα (Msh2/Msh6) specific loss of activity (Fig. 3C), while mutagenesis of NLS 552 did not cause a mismatch repair defect. This finding suggests that the decrease in nuclear localization efficiency of Msh2Δ525 and Msh2Δ525Δ552 in the absence of Msh3 is attributable to a decreased interaction with Msh6.
As before with wild-type Msh2 (Fig. 1 and Fig. 3), the absence of Msh6 had a significant effect on the nuclear localization of Msh2 NLS mutant proteins. Msh6 is particularly important for proteins with NLS 552 mutagenized (Fig. 3B). In the absence of Msh6, the nuclear localization of Msh2Δ552 and Msh2Δ525Δ552 resembled the No Msh2 controls (all p values greater than 0.4). Furthermore, wild-type Msh2 and the NLS mutant proteins failed to concentrate in the nucleus in cells lacking Msh3 and Msh6 to the same extent as with cells only lacking Msh6. Thus, the heterodimer partner Msh6 is important for Msh2 and Msh2Δ525 nuclear localization (Fig. 1 and 3) and is required for the Msh2 proteins lacking NLS 552 (Fig. 3).
In summary, the data lead us to conclude that both NLS 525 and NLS 552 function redundantly in the context of the Msh2 protein and that Msh6, but not Msh3, plays a significant role in the nuclear localization of Msh2, particularly in the absence of fully functional NLSs. A possible mechanism to explain these findings is that MutSα dimerization stabilizes Msh2. If this model is correct, deleting the Msh6 heterodimer partner should destabilize Msh2 and lead to lower levels in the cell.
3.5. The steady-state levels of Msh2 and Msh2 NLS mutant proteins are lower in strains lacking Msh6
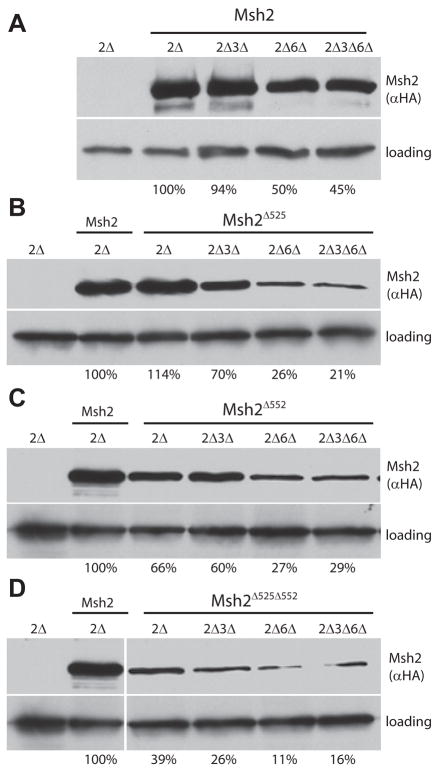
To test the dimerization stabilization hypothesis, we performed immunoblotting analyses to measure the steady-state levels of the wild-type Msh2 (Fig. 4A), Msh2Δ525 (Fig. 4B), Msh2Δ552 (Fig. 4C), and Msh2Δ525Δ552 (Fig. 4D) mutant proteins in the various deletion strains. NLS mutant protein levels in the presence and absence of heterodimer partners were calculated as a percentage of the wild-type Msh2 protein level, included as a control in each experiment.
Fig. 4.
Steady-state protein levels of Msh2 and Msh2 NLS variants in the presence and absence of Msh3 and Msh6. Proteins extracts from the strains described for Fig. 3 were analyzed using immunoblotting methods (see Materials and Methods). The nomenclature for the strains and expressed wild-type and NLS mutant proteins is the same as was described in the Fig. 3 legend panels A and B except that the msh2Δ strain with the pRS413 vector is simply designated 2Δ. After visualization of Msh2 with 12CA5 mouse α-hemaglutinin (αHA) and α-mouse IgG HRP antibodies, the membrane was re-probed with rabbit α-Kar2p polyclonal and α-rabbit IgG HRP antibodies (loading). Protein levels were quantified by densitometry using ImageJ [36]. The intensities were normalized to the loading control. The values are presented below the lanes and represent the percentage of wild-type Msh2 levels. The Msh2 Δ552 and the Msh2Δ525 Δ552 immunoblotting experiments were conducted on the same gel and membrane. Thus, the positive and negative controls are the same images reproduced for reference on the bottom panel.
For Msh2 and the Msh2 NLS mutant proteins, a significant reduction in steady-state levels was detected when the MSH6 gene was deleted (Fig. 4A–4C, compare 2Δ to 2Δ6Δ lanes). Furthermore, the additional deletion of MSH3 in strains lacking MSH6 did not change the protein levels when compared to the MSH6 single deletion (Fig. 4A–4C, compare 2Δ6Δ to 2Δ3Δ6Δ lanes). Thus, consistent with the immunofluorescence results, the wild-type and the Msh2 NLS mutant protein levels are higher in the presence of Msh6, supporting the MutSα dimerization stabilization model.
As with the immunoflurescence results, the absence of Msh3 did not alter the protein levels of wild-type Msh2 (Fig. 4A, compare 2Δ and 2Δ3Δ) or Msh2Δ552 (Fig. 4C, compare 2Δ and 2Δ3Δ). Additionally, Msh2Δ525 and Msh2Δ525Δ552 showed diminished levels in the absence of Msh3 (Fig. 4B and 4D, compare 2Δ and 2Δ3Δ), reinforcing the conclusion that the NLS 525 mutant proteins have decreased ability to interact with Msh6 (Fig. 3C).
Apart from supporting the hypothesis that Msh6 stabilizes Msh2, these observations strengthen the conclusions derived from the nuclear localization experiments. For example, in the presence of both heterodimer partners, the protein level of Msh2Δ525 is approximately the same as wild-type Msh2 (~114%, Fig. 4B) yet the nuclear localization is significantly diminished (Fig. 3B, p = 0.008, compared to wild-type Msh2), confirming the validity of the conclusion that NLS 525 influences the efficiency of nuclear localization. Conversely, in the presence of Msh3 and Msh6, Msh2Δ552 is present at 66% wild-type levels (Fig. 4C) yet the mutant protein localized to the nucleus with wild-type efficiency (Fig. 3B). Thus, differences in the cellular Msh2 levels do not account for the nuclear localization results. In fact, nuclear localization significantly above background is detectable even when the NLS mutant protein is found at 21% of wild-type (Fig. 4B, Msh2Δ525 expressed in 2Δ3Δ6Δ).
Taken together, these findings show that Msh6, but not Msh3, is important for the localization and protein levels of Msh2, particularly when Msh2 does not possess NLS 552. Because Msh2 functions as part of a heterodimer, we wanted to test the hypothesis whether Msh2 similarly alters the nuclear localization of Msh3 and/or Msh6.
3.6. Msh6, but not Msh3, is dependent on Msh2 for nuclear localization
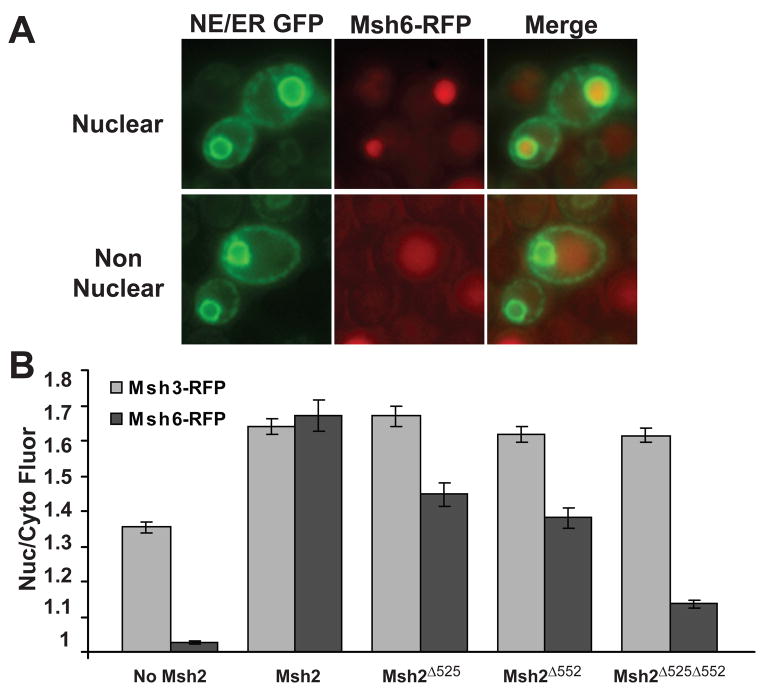
To test the possibility that Msh2 regulates the localization of its heterodimer partners, the chromosomally encoded MSH6 and MSH3 genes were fused with the coding sequence for red fluorescent protein (RFP). To assay the effects of Msh2 and the NLS mutant proteins on nuclear localization of Msh6-RFP and Msh3-RFP, the RFP fusion strains also expressed no Msh2, wild-type Msh2, or the Msh2 NLS variants, Msh2Δ525, Msh2Δ552, or Msh2Δ525Δ552. The fluorescence intensities of the RFP tagged proteins were recorded and the nuclear to cytoplasmic ratios were calculated as before, utilizing 3xGFP-HDEL fluorescence to define the nuclei. Representative images shown in Fig. 5A provide examples of nuclear localized (top panels) and non-nuclear localized Msh6-RFP.
Fig. 5.
Assessment of Msh3 and Msh6 Nuclear localization in the presence and absence of Msh2 and Msh2 NLS variants. Strains expressing Red Fluorescent Protein tagged Msh3 (Msh3-RFP) or Msh6 (Msh6-RFP) were transformed with the nuclear envelope, endoplasmic reticulum triple GFP marker to visualize the nuclei as well as with plasmids expressing no Msh2, Msh2, or Msh2 NLS variants. Specifically, strains msh2Δ MSH6-RFP or msh2Δ MSH3-RFP harboring p3xGFP-HDEL and either pRS413 (no Msh2), pMSH2 (Msh2) or pMSH2-Δ525 (Msh2Δ525), pMSH2-Δ552 (Msh2Δ552), or pMSH2-Δ525Δ552 (Msh2Δ525Δ552) were viewed by deconvolution fluorescence microscopy. (A) Representative images of nuclear localized Msh6-RFP (top panels) and non nuclear Msh6-RFP (bottom panels). Nuclear envelope/endoplasmic reticulum 3xGFP (NE/ER GFP) fluorescence is shown in the left panels. Msh6-RFP fluorescence is shown in the middle panels and a combined image with 3xGFP and Msh6-RFP (Merge) is shown in the right panels. (B) Quantitative Measurements of Nuclear Msh3-RFP and Msh6-RFP. The nuclear and cytoplasmic fluorescence of Msh3-RFP (light grey) and Msh6-RFP (darker grey) were determined using the ImageJ [36]. The ratio of nuclear/cytoplasmic fluorescence (Nuc/Cyto Fluor) is plotted for each strain. The error bars signify the standard error of the mean. Approximately 100 cells were measured for each strain.
The quantified results revealed a striking difference between the heterodimer partners as to their dependency upon Msh2 for nuclear localization (Fig. 5B). Msh3-RFP showed diminished, but significant nuclear accumulation in the absence of Msh2 (N/C = 1.40 ± 0.01); whereas Msh6-RFP failed to show nuclear accumulation in the absence of Msh2 (N/C = 1.026 ± 0.005). Given Msh3’s lack of dependency on wild-type Msh2 it is not surprising that Msh3-RFP localized to the nucleus in the strains expressing the Msh2 NLS mutant proteins (p values all > 0.4, compared to wild-type Msh2). Conversely, Msh6-RFP showed decreasing levels of nuclear accumulation in the Msh2Δ525 (N/C = 1.45 ± 0.03), Msh2Δ552 (N/C = 1.38 ± 0.03), and Msh2Δ525Δ552 (N/C = 1.14 ± 0.01) NLS mutant protein expressing strains. Msh6 is estimated to reside in the cell at a concentration seven times greater than Msh3 [46], therefore, the data cannot be explained simply by low levels of Msh6 compared to Msh3.
In summary, Msh2 and the Msh2 NLS mutant proteins influence the nuclear localization of Msh6, but do not alter the nuclear accumulation of Msh3. Taken together the data suggest a reciprocal regulation of the localization of MutSα, but not of MutSβ monomers. In principle, the observed localization of Msh6 might be influenced by Msh2’s steady-state levels, the ability to efficiently dimerize to form MutSα, and/or the capacity of the Msh2 protein to sequester MutSα in the nucleus via functional NLSs.
3.7. Dimerization of MutSα is important for efficient localization of Msh6 to the Nucleus
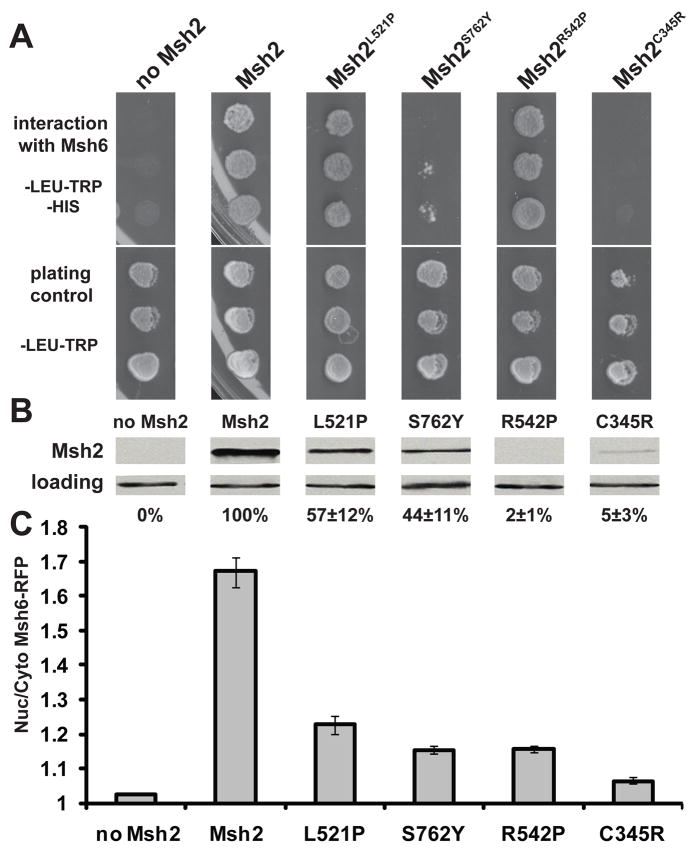
To distinguish between the effects of dimerization and protein levels of Msh2 on the nuclear accumulation of Msh6-RFP, we used previously characterized missense variants of Msh2 [33]. The variants included a leucine to proline substitution at yeast codon 521 (Msh2L521P) and an arginine to proline substitution at codon 542 (Msh2R542P), both residing near the DNA binding domain, effecting protein levels but not dimerization, a serine to tyrosine mutation at codon 762 (Msh2S762Y) located at the dimer interface effecting protein levels and subunit formation, and finally a cysteine to arginine alteration at codon 345 (Msh2C345R) in the central core region effecting protein levels and dimerization [33]. Pairs of missense variants were chosen to compare proteins with differing abilities to interact with Msh6 based on the yeast 2-hybrid assay (Fig. 6A), but with similar steady-state levels of Msh2 (Fig. 6B). Msh2L521P and Msh2S762Y represented variants with opposing abilities to interact with Msh6 (Fig. 6A), but with similarly decreased steady-state levels of Msh2 (~50% of wild-type levels in the Msh6-RFP strain, Fig. 6B). Msh2R542P and Msh2C345R likewise have differing capacities to interact with Msh6 (Fig. 6A), but, are found at similar low level in the cell (≤ 5 % of wild-type, Fig. 6B).
Fig. 6.
Nuclear Localization of Msh6-RFP is regulated by the dimerization capabilities of Msh2 as well as the Msh2 protein levels. (A) Two-hybrid assay to illustrate MutSα mismatch repair subunit formation with Msh2 missense variants. The MATa yeast 2-hybrid reporter strain was transformed with pGBD-C2 (negative control, No Msh2), pGBD-MSH2 (positive control, Msh2), pGBD-MSH2-L521P (Msh2L521P), pGBD-MSH2-S762Y (Msh2S762Y), pGBD-MSH2-R542P (Msh2S762Y), or pGBD-MSH2-C345R (Msh2C345Y). Transformants were mated with MATα yeast 2-hybrid reporter strains harboring pGAD-MSH6 (Msh6). Diploid cultures were spotted in triplicate onto agar plates and allowed to grow for 2 days at 30°C. Growth on selective medium also lacking histidine (–LEU –TRP –HIS) indicates a 2-hybrid interaction (interaction with Msh6). Medium lacking leucine and tryptophan (–LEU –TRP) selects for diploids harboring both pGAD and pGBD constructs (plating control). (B) Sample immunoblot showing the steady state levels of Msh2 missense variant proteins. Strains expressing Red Fluorescent Protein tagged Msh6 (Msh6-RFP) were transformed with the nuclear envelope, endoplasmic reticulum triple GFP marker to visualize the nuclei as well as with plasmids expressing no Msh2, Msh2, or Msh2 missense variants. Specifically, exponentially growing strains msh2Δ MSH6-RFP harboring p3xGFP-HDEL and either pRS413 (no Msh2), pMSH2 (Msh2) or pMSH2-L521P (L521P), pMSH2-S762Y (S762Y), pMSH2-R542P (R542P), or pMSH2-C345R (C345R) were processed for immunoblotting. After visualization of Msh2 (Msh2), the membrane was reprobed with rabbit α-Kar2p polyclonal and α-rabbit IgG HRP antibodies (loading). The bands were quantified with ImageJ [36] and normalized to the wild-type protein level. The percentages shown represent the results from the average of three immunoblots and the error (±) is the standard error of the mean. (C) Nuclear localization of Msh6-RFP in the presence of Msh2 missense variants. The strains described above were viewed by deconvolution fluorescence microscopy. The nuclei were visualized with 3xGFP-HDEL and cytoplasmic and nuclear fluorescence of Msh6-RFP were determined using ImageJ. The ratio of nuclear/cytoplasmic fluorescence (Nuc/Cyto Msh6-RFP) is plotted for each strain. The error bars signify the standard error of the mean. Approximately 100 cells were measured for each strain.
All of the Msh2 missense variants allowed for some Msh6-RFP accumulation in the nucleus (p <0.001 for all when compared to the No Msh2 negative control). However, nuclear accumulation was significantly lower than found in the strain expressing wild-type Msh2 (p <0.001 for all when compared to the Msh2 positive control). The ability to dimerize with Msh6 caused a significant difference in the missense pairs analyzed. Msh2L521P allowed nuclear accumulation of Msh6-RFP to a greater extent than Msh2S762Y (p = 0.008) in spite of the similar protein levels. Likewise, Msh2R542P showed higher nuclear localization of Msh6-RFP than Msh2C345R (p = 5 × 10−8). Interestingly, in spite of the different protein levels for Msh2S762Y (~44%) and Msh2R542P (~2%), the extent of nuclear accumulation of Msh6-RFP was the same (p = 0.9), suggesting that the ability of Msh2R542P to dimerize with Msh6 contributes significantly to the efficiency of nuclear accumulation. In summary, the levels of Msh2 and the ability to dimerize both appear to play a role in the efficient Msh6 nuclear localization.
4. Discussion
4.1. Summary
In this work, we delineated the nuclear transport parameters of the yeast MutSα (Msh2/Msh6) and MutSβ (Msh2/Msh3) DNA mismatch recognition complexes and found a pronounced reciprocal regulation of nuclear localization of MutSα, but not of MutSβ, subunits. The net effect would be the differential regulation of the stoichiometries of the MutS heterodimers, in balance with the types of mismatches generated during a typical round of DNA synthesis in yeast.
4.2. Nuclear localization capabilities of Msh2, Msh3, and Msh6
MutSα and MutSβ bind DNA mismatches as heterodimers before forming higher order repair complexes [47]. However, in principle, the relevant proteins may undergo nuclear transport are either as monomers or within MutSα or MutSβ. We found that yeast Msh2 is not strictly dependent upon interactions with its heterodimer partners for transport into the nucleus; although, Msh6 is required for efficient Msh2 nuclear localization and protein levels.
Of two potential NLSs identified in Msh2, only one conferred nuclear import capabilities when separated from the context of the Msh2 protein, but both functioned redundantly in the native protein. Involvement of NLS-mediated transport is supported by the finding that Msh2 co-purifies with Kap95 by tandem affinity purification [complex 17323, Biomolecular Object Network Databank, 48]. Kap95 is one of fourteen import receptors in the S. cerevisiae karyopherin-β superfamily, a group of proteins responsible for the transport and docking of macromolecules to the nuclear pore complexes [reviewed in 49,50]. The presence of Msh2 and Kap95 in the same complex strengthens our observations that the import of Msh2 is via a classical NLS. Interestingly, hMSH2 specifically forms a complex with importin α [51]. In yeast, Srp1, the yeast karyopherin α homolog, forms a dimer with karyopherin β Kap95 to mediate import of nuclear proteins [52].
Msh2 was not required for the nuclear localization of Msh3, which was surprising given that the yeast Msh3 protein does not contain a classical NLS motif [41]. However, not all proteins imported into the nucleus possess canonical NLSs [reviewed in 50]. The fact that Msh3 appears to have independent nuclear transport capability explains the observation that deleting both of Msh2’s NLSs in the presence of Msh3 does not result in a complete loss of nuclear Msh2. In contrast, Msh2 was required for the nuclear localization of Msh6. Consistent with this view, missense variants of Msh2 showed that the capacity to form stable MutSα heterodimers is critical for the efficient nuclear localization of Msh6. Tandem affinity purification analysis also identified Msh6 co-purifying with Msh2 and Kap95 [48], supporting the hypothesis that nuclear translocation of Msh6 occurs after heterodimerization.
The findings of this study suggest, together with data for human Msh2 nuclear import, a subtle lack of conservation between the nuclear import pathways in the human and yeast mismatch repair systems. Reports have shown that human Msh2 does not have a classical NLS motif and requires Msh6 for efficient nuclear localization [22]. Regardless of the precise mechanism of nuclear import, the dimerization requirement for MutSα is conserved from yeast to humans, but the Msh2 and Msh6 proteins have exchanged the presence of a canonical NLS.
4.3. Stabilization and translocation model
We propose a model to explain the observed MutSα reciprocal regulation. Msh2 and Msh6 would dimerize in the cytoplasm, stabilizing Msh2. Furthermore, translocation into the nucleus would also be somewhat protective. The stabilized proteins within the MutSα heterodimer would be efficiently transported into the nucleus using Msh2’s NLSs bound by the karyopherin β transport system. If Msh6 is not present, some Msh2 would be translocated into the nucleus using the canonical NLSs, thereby stabilizing a fraction of the Msh2 protein. This model is consistent with the finding that the absence of Msh6 and mutagenesis of Msh2’s NLSs caused a significant drop in Msh2 overall cellular levels.
We speculate that the stabilization mechanism may simply be sequestration away from a site of significant degradation. For example, the cytoplasm may contain factors such as cytoplasm-specific kinases/phosphatases or ubiquitin ligases that modify exposed residues in the unbound monomer. Both proposed cytoplasm-specific modifications could target the protein for degradation and ultimately lead to turnover of the Msh2 monomer at a higher rate than is seen in the nucleus. In support of this model, human MutSα has been shown to be a substrate for phosphorylation-regulated ubiquitination [53]. Additionally, human Msh2 stabilization of Msh6 and Msh3 has previously been observed and heterodimer association is the proposed mechanism of protection [14].
Part of our model posits that dimerization with Msh6 is stabilizing for Msh2 by blocking residues that signal for degradation. Given this, one might predict the same protective effect upon dimerization with Msh3. We observed a slight drop in nuclear localization efficiency (~15% decrease) and protein levels (~5% decrease) in the absence of Msh3. However this small effect could be explained by the fact that Msh3 is present at significantly lower levels than Msh6. Large-scale proteomic analyses predict that Msh3’s abundance is 736 molecules/cell, whereas Msh6’s abundance is 5,330 molecules/cell, a 7-fold difference [46]. Thus, although dimerization with Msh3 might be similarly protective, its low level in the cell renders the effect insignificant compared to Msh6.
4.4. Functional significance of modulating the levels of the MutS heterodimers during replication and oxidative damage
The regulation of Msh2 at the level of heterodimer formation and nuclear translocation has important consequences for the fidelity of the genome. In yeast, replication slippage at microsatellites is estimated to occur at a rate ~50 times higher than mis-incorporation of nucleotides [54]. However, these microsatellite loci are found at a frequency of only ~0.1% [55]. Thus, in spite of an elevated rate of insertion/deletion loop mismatch formation at such sites, the predicted requirement for MutSβ (Msh2/Msh3) is significantly less than for MutSα (Msh2/Msh6). We estimate that the overall rate of the different types of mutations leads to a requirement for MutSβ at a level of only 5–6 % relative to MutSα.
It is also likely that excess MutSβ would interfere with the efficiency of MutSα heterodimers scanning of the genome for single base pair mismatches. Interference has been observed recently in vitro where MutSα proteins scanning on DNA are impeded or caused to reverse direction upon encountering another MutSα heterodimer [56].
In summary, we suggest that a regulatory mechanism would be required to maintain the ratio of MutSα to MutSβ for appropriate and sufficient coverage of the genome to match the spectrum of mutations that arise during replication. The regulatory mechanism could simply be a consequence of stoichiometry of the monomers in the cytoplasm. Assuming the affinities between Msh2 and its heterodimer partners are the same, the stoichiometry of the heterodimer partners in yeast [7 Msh6 to 1 Msh3, 46] would dictate that ~85% of the MutS complexes are MutSα. The predicted relative abundance value is close to the 90% reported for human MutSα [14] and correlates well with the rate of error combined with the predicted quantity of single base pair and single nucleotide/insertion-deletion mismatch recognition needed during a typical yeast DNA replication cycle [54].
The human genome is estimated to be 1% microsatellite DNA, ten times greater than the amount in yeast [57]; however, as mentioned above, the human MutSα complex is able to recognize small insertion/deletion loops up to 8 nucleotides [17,18]. It is of interest that the relative abundance of MutSα vs. MutSβ is likely to be conserved from yeast to humans, but that the specificity of binding is not. However, the net effect is that the appropriate relative abundance exists for adequate surveillance of the genome during replication.
During replication, the frequencies of the different types of mismatches are likely to occur at a relatively constant level, dictated by the intrinsic error rate of DNA polymerase and the size and frequency of DNA repeat sequences [58]. However, DNA mispairings recognized by MutSα are also formed during oxidative damage [59 and references therein]. Because mispairings recognized by MutSα are generated from both replication and from oxidative damage, we postulate that the MutSα heterodimer levels would need to be responsive to environmental conditions. In contrast, because insertion/deletion loops arise only during replication slippage, MutSβ requirements would be more constant. Along these lines, it is of interest that the MSH6 transcript is induced 11-fold upon re-oxygenation after anaerobic growth [60]. Thus, the observed regulation of the nuclear localization of MutSα heterodimer partners would make sense for adaptation to DNA damaging oxidative stress.
Acknowledgments
We are grateful to Patricia Melloy, Sean Clark, Ruth Tennen, John Cooper and Thomas Petes for generously supplying plasmids. This research was supported by Princeton University, The Department of Molecular Biology at Princeton University, the New Jersey Commission on Cancer Research and a National Institute of Health grant GM037739 awarded to Mark D. Rose.
Abbreviations used in this paper
- HNPCC
hereditary non-polyposis colorectal cancer
- PCR
polymerase chain reaction
- 5-FOA
5-fluoro-ortic acid
- CAN
canavanine
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- NLS
nuclear localization sequence
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch Repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Reenan RA, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 5.Alani E, Sokolsky T, Studamire B, Miret JJ, Lahue RS. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol Cell Biol. 1997;17:2436–2447. doi: 10.1128/mcb.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J Biol Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 7.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 8.Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Mol Cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 9.Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 11.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 12.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan JJ, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 13.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 15.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci U S A. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci U S A. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 18.Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 19.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 20.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Platt JL, Cascalho M. Dimerization of MLH1 and PMS2 limits nuclear localization of MutLalpha. Mol Cell Biol. 2003;23:3320–3328. doi: 10.1128/MCB.23.9.3320-3328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christmann M, Kaina B. Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J Biol Chem. 2000;275:36256–36262. doi: 10.1074/jbc.M005377200. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel FM, Brent R, Kingston RE, Moore DD, Scidman JG, Smith RA, Struhl K. Current protocols in molecular biology. John Wiley & Sons; New York: 1994. [Google Scholar]
- 24.Burke D, Dawson T. Methods in yeast genetics : a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Plainview, N.Y.: 2000. Stearns and Cold Spring Harbor Laboratory. [Google Scholar]
- 25.Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 26.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 29.Henderson ST, Petes TD. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gammie AE, Erdeniz N, Beaver J, Devlin B, Nanji A, Rose MD. Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics. 2007;177:707–721. doi: 10.1534/genetics.107.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 35.Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 36.Rasband WS. Image J, National Institutes of Health. 1997–2008 http://rsb.info.nih.gov/ij/
- 37.Ohashi A, Gibson J, Gregor I, Schatz G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- 38.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 40.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 41.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 42.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 43.Kenna MA, Brachmann CB, Devine SE, Boeke JD. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo KQ, Elsasser S, Chang DC, Campbell JL. Regulation of the localization and stability of Cdc6 in living yeast cells. Biochem Biophys Res Commun. 2003;306:851–859. doi: 10.1016/s0006-291x(03)01082-9. [DOI] [PubMed] [Google Scholar]
- 45.Damelin M, Silver PA, Corbett AH. Nuclear protein transport. Methods Enzymol. 2002;351:587–607. doi: 10.1016/s0076-6879(02)51870-x. [DOI] [PubMed] [Google Scholar]
- 46.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 47.Kijas AW, Studamire B, Alani E. Msh2 separation of function mutations confer defects in the initiation steps of mismatch repair. J Mol Biol. 2003;331:123–138. doi: 10.1016/s0022-2836(03)00694-6. [DOI] [PubMed] [Google Scholar]
- 48.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 49.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 50.Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- 51.Knudsen NO, Nielsen FC, Vinther L, Bertelsen R, Holten-Andersen S, Liberti SE, Hofstra R, Kooi K, Rasmussen LJ. Nuclear localization of human DNA mismatch repair protein exonuclease 1 (hEXO1) Nucleic Acids Res. 2007;35:2609–2619. doi: 10.1093/nar/gkl1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 53.Hernandez-Pigeon H, Quillet-Mary A, Louat T, Schambourg A, Humbert O, Selves J, Salles B, Laurent G, Lautier D. hMutS alpha is protected from ubiquitin-proteasome-dependent degradation by atypical protein kinase C zeta phosphorylation. J Mol Biol. 2005;348:63–74. doi: 10.1016/j.jmb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Kruglyak S, Durrett R, Schug MD, Aquadro CF. Distribution and abundance of microsatellites in the yeast genome can Be explained by a balance between slippage events and point mutations. Mol Biol Evol. 2000;17:1210–1219. doi: 10.1093/oxfordjournals.molbev.a026404. [DOI] [PubMed] [Google Scholar]
- 55.Lim S, Notley-McRobb L, Lim M, Carter DA. A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet Biol. 2004;41:1025–1036. doi: 10.1016/j.fgb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, Greene EC. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma PC, Grover A, Kahl G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007;25:490–498. doi: 10.1016/j.tibtech.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 59.Holmes SF, Scarpinato KD, McCulloch SD, Schaaper RM, Kunkel TA. Specialized mismatch repair function of Glu339 in the Phe-X-Glu motif of yeast Msh6. DNA Repair (Amst) 2007;6:293–303. doi: 10.1016/j.dnarep.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai LC, Kosorukoff AL, Burke PV, Kwast KE. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1468–1489. doi: 10.1128/EC.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fitch PG, Gammie AE, Lee DJ, de Candal VB, Rose MD. Lrg1p Is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics. 2004;168:733–746. doi: 10.1534/genetics.104.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melloy P, Shen S, White E, McIntosh JR, Rose MD. Nuclear fusion during yeast mating occurs by a three-step pathway. J Cell Biol. 2007;179:659–670. doi: 10.1083/jcb.200706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guex N, Diemand A, Peitsch MC. Protein modelling for all. Trends Biochem Sci. 1999;24:364–367. doi: 10.1016/s0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]