Abstract
In Escherichia coli, Rob activates transcription of the SoxRS/MarA/Rob regulon. Previous work revealed that Rob resides in 3–4 immunostainable foci, that dipyridyl and bile salts are inducers of its activity, and that inducers bind to Rob’s C-terminal domain (CTD). We propose that sequestration inactivates Rob by blocking its access to the transcriptional machinery and that inducers activate Rob by mediating its dispersal, allowing interaction with RNA polymerase. To test “sequestration-dispersal” as a new mechanism for regulating the activity of transcriptional activators, we fused Rob’s CTD to SoxS and used indirect immunofluorescence microscopy to determine the effect of inducers on SoxS-Rob’s cellular localization. Unlike native SoxS, which is uniformly distributed throughout the cell, SoxS-Rob is sequestered without inducer, but is rapidly dispersed when cells are treated with inducer. In this manner, Rob’s CTD serves as an anti-sigma factor in regulating the co-sigma factor-like activity of SoxS when fused to it. Rob’s CTD also protects its N-terminus from Lon protease, since Lon’s normally rapid degradation of SoxS is blocked in the chimera. Accordingly, Rob’s CTD has novel regulatory properties that can be bestowed on another E. coli protein.
Keywords: gene regulation, intracellular localization, immunofluorescence microscopy, anti-sigma factor, proteolysis
Introduction
The transcriptional activators SoxS, MarA, and Rob form a subset of the AraC/XylS family of proteins1 in that their amino acid sequences average 49% identity along the length of the shortest member of the subset2 and they are the direct transcription activators3–5 of a highly overlapping set of about 50 target genes6–8, the SoxS/MarA/Rob regulon. The three paralogous proteins recognize the same highly degenerate, 20 bp DNA binding site located within two classes of target promoters3–5,9–11, they bind DNA as monomers5,12,13 and they differentially regulate the transcription of target genes14 in response to various environmental stresses as described below.
Resistance to the oxidative stress imposed by superoxide-generating, redox-cycling agents like paraquat is carried out in two stages by two gene products, SoxR and SoxS. 15–19. Thus, upon oxidation of its 2Fe-2S clusters, constitutively expressed SoxR becomes activated and then it activates transcription of soxS20–24. In the second stage, molecules of de novo synthesized SoxS form binary complexes with RNA polymerase (RNAP); the complexes then scan the chromosome for SoxS binding sites located within SoxS-dependent promoters and activate transcription from them25,26. This mechanism of transcription activation by SoxS is called “pre-recruitment”25,26 and in it SoxS functions as a co-sigma factor27. Once the defense against the oxidative stress has been achieved, SoxR becomes reduced and SoxR-activated transcription of soxS ceases20–24. Then, the remaining SoxS protein is rapidly degraded by Lon protease and expression of the genes of the regulon returns to the basal level28.
A similar two-gene, two-stage system is induced by and confers resistance to multiple antibiotics and aromatic weak acids like salicylate. Upon exposure to an inducer, MarR repressor becomes inactivated and the marRAB operon is subsequently derepressed29–33. Then, newly synthesized MarA appears to activate transcription of the regulon’s genes by the same pre-recruitment mechanism (also called “DNA scanning”34) as that of SoxS. This system is reset by a process similar to that of SoxS: when the stress is relieved, MarR becomes active and represses transcription of the marRAB operon; then, synthesis of MarA ceases, the residual MarA is rapidly degraded by Lon protease, and transcription of the regulon’s genes returns to the pre-induced level28.
Recently, TetD was identified as a fourth member of the subset of the AraC/XylS family of proteins because of its extensive amino acid sequence identity with the sequences of SoxS, MarA, and Rob (average = 50%), because it recognizes the same degenerate binding site, and because it activates a subset of the genes of the SoxS/MarA/Rob regulon2. The gene encoding TetD resides on transposon Tn1035,36 and its transcription is negatively regulated by the TetC repressor37. The inducer that inactivates TetC and thus the physiological role of the TetC/TetD system is unknown. With TetD also being destabilized by Lon protease, the system resets by the same process as that of SoxS and MarA.
In summary, the regulation of the SoxR/S, MarR/A and TetC/D systems follow similar off-on pathways carried out in two stages by two genes: sensor proteins SoxR, MarR and TetC respond to their respective inducers by turning on the synthesis of the respective response regulators SoxS, MarA and TetD. These response regulators bind to the same degenerate sequence and activate transcription of an overlapping set of genes, although to different degrees. The respective systems are also turned off by similar processes: once the inducing stress has been relieved, de novo synthesis of the response regulator ceases, residual regulator is degraded by Lon protease, and expression of the regulon returns to the basal level.
Rob differs in several significant ways from other members of the subset. First, instead of being synthesized de novo in response to an inducing stress, Rob is expressed constitutively at about 5,000–10,000 molecules per cell38,39. Second, the constitutively expressed Rob molecules are inactive, as evidenced by the fact that a null mutation of Rob has little or no effect on the expression level of the regulon’s genes it has the potential to activate14. Curiously, when overexpressed from a plasmid, full-length Rob activates target gene expression as does its N-terminal domain, which contains the dual helix-turn-helix domains characteristic of AraC/XylS proteins1,40,41. Adding to the enigma is the observation that purified Rob is able to bind DNA and efficiently activate transcription of target genes in vitro5. A third difference between Rob and its paralogous proteins is its distinct cellular localization: Rob is sequestered into 3–4 immunostainable foci42. Fourth, unlike SoxS, MarA, and TetD, which are single domain proteins, Rob contains a second, C-terminal domain (CTD) of about 180 amino acids12,38.
Recently, a role for Rob’s CTD has been revealed. Rosner et al.43 reported that two forms of dipyridyl, 2, 2′-dipyridyl, an iron chelator, and 4, 4′-dipyridyl (hereafter called “DIP”), which does not chelate iron, are inducers of Rob-dependent transcription in vivo. And, Rosenberg et al. determined that unconjugated bile salts and medium-chain fatty acids like decanoate (DEC) also enhance Rob’s in vivo activity44. Spectroscopic methods showed that the inducers interact directly with Rob’s CTD43,44. However, as mentioned above, mobility shift and in vitro transcription assays showed that purified Rob is fully active in DNA binding and Rob-dependent transcription activation of target genes, i.e., neither activity requires or is enhanced by the presence of an inducer43,44. Thus, since inducers are required for the activity of full-length Rob in vivo but not in vitro, the inducers must confer some property on Rob that is also achieved during its purification. Moreover, this regulatory property almost certainly requires Rob’s CTD because (i) Rob’s N-terminal domain alone is able to activate in vivo transcription of target genes in the absence of inducers40, (ii) the inducers of Rob activity in vivo interact with Rob’s CTD in vitro but have no effect on Rob activity43,44, and (iii) the activity of native SoxS, which lacks a domain equivalent to Rob’s CTD, is also not enhanced by DIP or DEC43,44 (K. L. Griffith and R. E. Wolf, Jr., unpublished results).
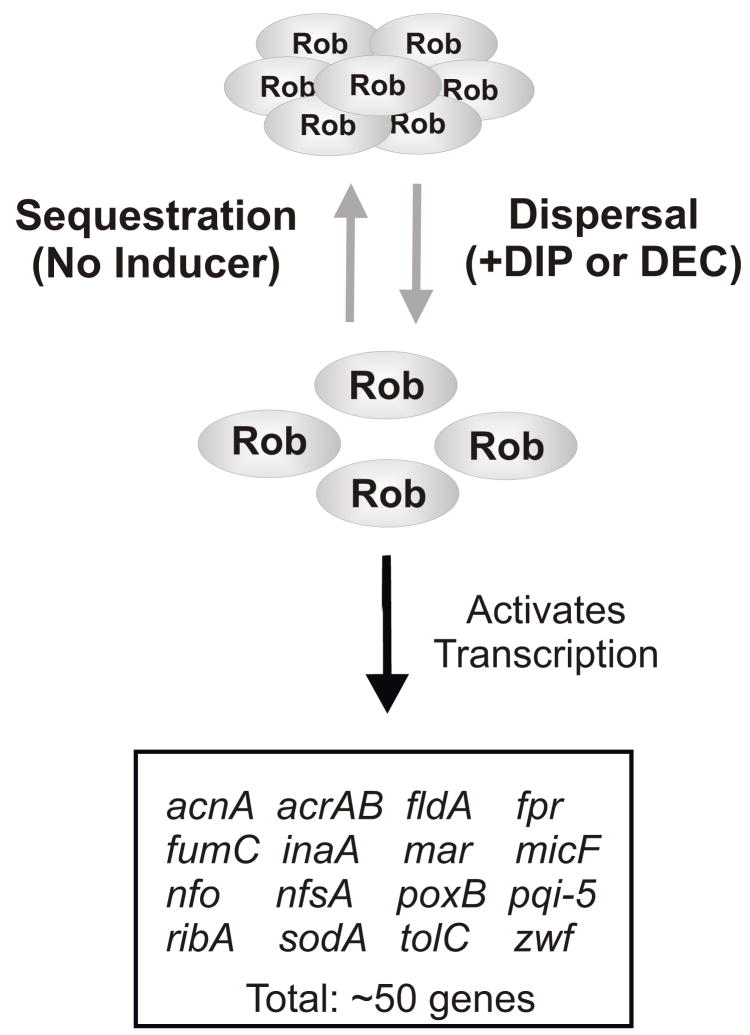
Here, we describe experiments demonstrating that Rob’s cellular localization provides a novel mechanism for regulating its activity. We call this mechanism “sequestration-dispersal” (Fig. 1). Thus, under non-inducing conditions, Rob is inactive because its CTD mediates its sequestration into intracellular foci that prevent Rob from interacting with the transcriptional machinery. Then, upon the addition of inducer, Rob is rapidly released from its sequestered state and dispersed Rob is free to activate transcription of the regulon’s genes. In addition, we show that full-length Rob is stable because its CTD blocks proteolytic degradation from the N-terminus by Lon protease. Moreover, since fusing Rob’s CTD to the C-terminus of SoxS forms a stable chimera whose activity is regulated by sequestration-dispersal, Rob’s CTD has novel properties that can be conferred on another protein to which it is fused. In particular, Rob’s CTD functions as an anti-sigma factor in regulating the co-sigma factor-like activity of SoxS when SoxS resides in the SoxS-Rob chimera27.
Figure 1.
Sequestration-dispersal as the mechanism that regulates the activity of Rob as a transcriptional activator. In the absence of an inducer, Rob is sequestered into immunostainable foci, which prevents its access to DNA; the mechanism of sequestration is unknown. Upon treatment of cells with an inducer, e.g., DIP or DEC, Rob becomes dispersed and transcription activation of target genes ensues.
Results
Rationale for using the SoxS-Rob chimera as a Rob surrogate for the study of sequestration-dispersal
According to the hypothesis that sequestration-dispersal regulates the activity of Rob, the role of the inducers is to disperse sequestered Rob so that Rob is free to activate transcription of its target genes. As a first step in testing the hypothesis, we needed to visualize Rob within immunostainable foci like those originally reported by Azam et al.42. Accordingly, we grew strain GC4468 in broth to A600 ~ 0.5 and subjected the cells to indirect immunofluorescence microscopy by the same method as described by Azam et al.42 and using the same anti-Rob serum. Initially, we were able to visualize very faint immunofluorescent foci within individual cells; however, over time the activity of the anti-serum decreased until it was no longer useful. We then purified N-his6-Rob and prepared rabbit polyclonal antiserum against it. Unfortunately, repeated attempts to visualize Rob with the IgG fraction of this antiserum were unsuccessful because only a weak fluorescence signal was produced.
In another attempt to visualize the intracellular form of Rob under non-inducing conditions, we constructed a SoxS-Rob chimera in which Rob’s CTD, comprised of 183 amino acids, was fused to the C-terminus of SoxS. Thus, the SoxS moiety should confer on the chimera the DNA binding and transcription activation properties of SoxS. Moreover, if our hypothesis is correct, Rob’s CTD should regulate the activity of SoxS by mediating sequestration of the chimera and by rendering it inducible and dispersible by DIP and DEC. Accordingly, we made the SoxS-Rob chimera and were confident it would serve our purpose because we had previously prepared a high-titer, polyclonal rabbit antiserum directed against N-his6-SoxS and had affinity-purified the anti-N-his6-SoxS antibodies25.
Rob’s CTD mediates sequestration of SoxS in the SoxS-Rob chimera
The SoxS-Rob chimera was cloned into the medium copy plasmid pBAD33 so that it could be expressed under control of the arabinose-inducible promoter, PBAD. Experiments described below demonstrated that the SoxS-Rob chimera binds DNA and activates transcription of target genes; in particular, its activity is enhanced by treatment of growing cells with DIP and DEC. Thus, the SoxS-Rob chimera has the potential to serve as a Rob surrogate in evaluation of the sequestration-dispersal hypothesis.
Accordingly, an overnight culture of strain RA4468 (Δrob::kan) containing pBAD33-SoxS-Rob was grown at 37°C in LB medium containing chloramphenicol (25μg/ml). The culture was diluted 1:100 and grown under the same conditions to A600 = 0.1 at which time arabinose was added to a concentration of 0.02%. After allowing expression of the SoxS-Rob chimera by growth under these conditions for 1 h, samples were taken, fixed, and treated with affinity-purified anti-SoxS antibodies and then with Alexa 488-conjugated secondary antibodies. The fixed preparations were then subjected to indirect immunofluorescence microscopy.
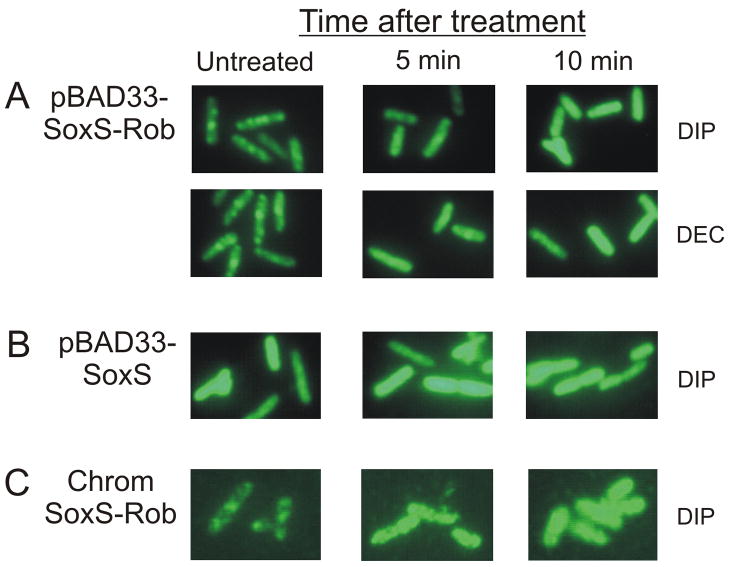
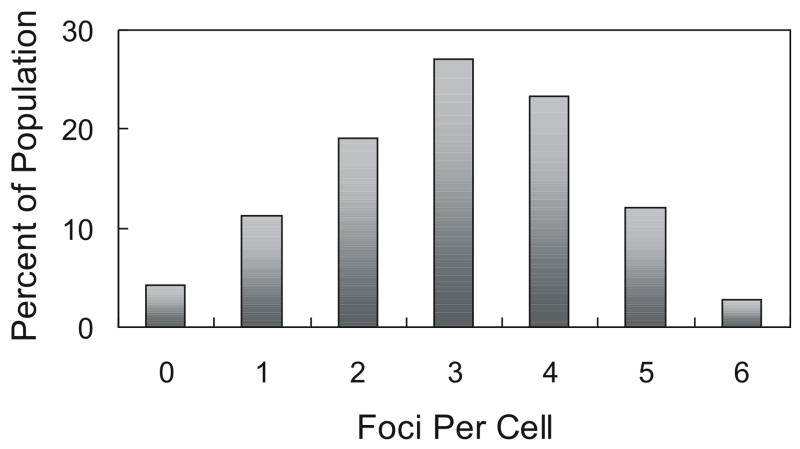
The SoxS-Rob chimera was localized into several immunostainable, punctate foci that were randomly distributed throughout individual cells (Fig. 2A), a localization pattern similar to that observed previously with native Rob42. The number of SoxS-Rob foci varied from zero to six per cell: ~70% of the cells had two, three, or four foci, while the remaining 30% had zero, one, five or six foci (Fig. 3). The size of the foci tended to vary between cells (Fig. 2A and data not shown). This suggests that the observed number of foci per cell might under represent the true number because the limitations of the resolution of the microscopy might have prevented us from determining whether a given focus actually contains multiple foci. Nonetheless, the addition of Rob’s CTD to the C-terminus of SoxS appears to confer on the chimera a localization pattern similar to that of native, full-length Rob, forming multiple punctate foci within individual cells.
Figure 2.
Cellular localization of SoxS-Rob and native SoxS. Strain RA4468 carrying pBAD33-SoxS-Rob or pBAD33-SoxS was grown to A600 ~0.1, induced with 0.02% arabinose for 1 hr, and treated with the following: 0.02% arabinose only; 0.02% arabinose with 3 mM DIP; or 0.02% arabinose with 8 mM DEC. Cells were fixed and subjected to indirect immunofluorescence microscopy as described in Materials and Methods. A. SoxS-Rob. B. Native SoxS.
Figure 3.
Distribution of SoxS-Rob foci within individual cells. The distribution of SoxS-Rob foci present in individual cells of a representative experiment of SoxS-Rob expressed from pBAD33 is shown. The number of foci per cell is expressed as a percentage of the 214 cells analyzed.
An alternative interpretation is that SoxS drives the sequestration of the SoxS-Rob chimera into the punctate foci. To test this possibility, we carried out indirect immunofluorescence microscopy on cells expressing native SoxS from plasmid pBAD33-SoxS. No immunostainable foci were observed; instead, fluorescence was uniformly distributed throughout the cell (Fig. 2B). Thus, Rob’s CTD is responsible for sequestration of the SoxS-Rob chimera. Moreover, we infer that the sequestration of full-length, native Rob observed by Azam et al.42 also depends on the CTD.
Rob’s CTD mediates the ability of DIP and DEC to induce the dispersal of the SoxS-Rob chimera from the sequestered state
Cells of strain RA4468 carrying pBAD33-SoxS-Rob growing exponentially at 37°C in LB medium were treated with 0.02% arabinose to induce expression of the chimera. After 1 hr, the culture was divided into three parts, with one part being untreated, one being treated with DIP, and one being treated with DEC. Incubation was continued and samples were taken at several times thereafter. Fig. 2A shows that following treatment with the inducers, the immunostainable foci disappeared and were replaced by a uniform, diffuse staining throughout the cells. Dispersal was rapid, as the foci began to disappear within 5 min after an inducing treatment and were almost completely gone after 10 min of induction (Fig. 2A).
The intensity of the fluorescence signal was greater in cells treated with inducer than in untreated cells (Fig. 2A). This enhanced signal is not due to an intrinsic fluorescent property of the inducers since no fluorescence was observed when cultures of strain RA4468 carrying vector plasmid pBAD33 were treated with DIP or DEC (data not shown). We speculate that the observed increase in fluorescence during DIP or DEC treatment of the strain expressing the SoxS-Rob chimera is due to the release of the chimera from its sequestered state; i.e., when SoxS-Rob is sequestered, only molecules on the surface of the bundles can react with the primary antibody whereas by causing dispersal of the chimera from the bundles, an inducing treatment significantly increases the number of reactive SoxS-Rob molecules.
Just as Rob’s CTD is responsible for sequestration of SoxS in the SoxS-Rob chimera, so too is it required for its dispersal. Thus, treatment of cells expressing SoxS from pBAD33-SoxS with inducers DIP or DEC had no effect on the diffuse staining pattern or fluorescence intensity compared to untreated cells (Fig. 2B) In summary, Rob’s CTD mediates both sequestration and dispersal of the SoxS-Rob chimera encoded by plasmid pBAD33-SoxS-Rob; by inference, it also mediates sequestration and dispersal of full-length, native Rob.
DIP induces dispersal of chromosomally encoded SoxS-Rob
Conceivably, the dispersal of SoxS-Rob produced from plasmid pBAD33-SoxS-Rob could be effected by its elevated abundance. Accordingly, we used “Recombinant Enhancement by Selection for Survival” (RESS), our scarless method of recombineering (R. Toughiri and R.E.W. Jr., unpublished results), to replace the N-terminal 107 amino acids of chromosomally encoded Rob with the 107 amino acids of SoxS. The preparation of strain MF100 carrying the chimeric gene in the chromosome of strain N7840 is described in Materials and Methods.
A culture of strain MF100 carrying chromosomally encoded SoxS-Rob was grown in LB medium at 37°C to A600 = ~0.2. A negative control sample was taken and fixed for indirect IFM. Then, the culture was treated with 5 mM DIP and samples were taken and fixed after 5 min and 10 min of inducing treatment.
As expected from the work of Azam et al.42 and that described above (Fig. 2A), chromosomally encoded SoxS-Rob produced in the absence of DIP was sequestered into ~3 immunostainable clusters per cell and very little fluorescence was observed in the remaining intracellular milieu (Fig. 2C). More importantly, as with SoxS-Rob expressed from pBAD33-SoxS-Rob (Fig. 2A), the immunostainable foci in strain MF100 began to disappear within 5 min of inducing treatment and were almost completely gone by 10 min (Fig. 2C). As above, after induction, the fluorescent foci in the cells were replaced by a uniform, diffuse staining whose overall fluorescence was considerably higher than that in the uninduced cells containing aggregated SoxS-Rob. Thus, this experiment and those above demonstrate that the relative abundance of SoxS-Rob does not affect its sequestration under non-inducing conditions or its dispersal upon induction.
Regulation of the activity of the SoxS-Rob chimera: enhancement of transcription activation by DIP and DEC
The experiments described above clearly demonstrate that the SoxS-Rob chimera is a excellent surrogate of Rob for studies of cellular localization and the effects of inducers on it. Nonetheless it is important to determine whether the chimera accurately reflects the regulatory properties of native Rob, in particular the ability of the chimera to activate transcription of target genes and the regulation of that activity by the inducers DIP and DEC. Accordingly, we compared the regulatory properties of the chimera to those of native Rob with two functional tests. In one, we determined the effect of the inducers on the ability of the two proteins to activate in vivo transcription from the Rob-dependent promoter, inaA; in the other, we conducted a plate test that reports the ability of native Rob and the chimera to access chromosomal DNA.
To determine whether the activity of the SoxS-Rob chimera as a transcription activator can be enhanced by DIP and DEC, we introduced plasmid pBAD18-SoxS-Rob into strain RA4468 carrying an inaA-lacZ transcriptional fusion on a single-copy prophage and measured β-galactosidase activity in the absence and presence of the inducers. For comparison, we also carried out the same experiment with the fusion strain carrying pBAD18-Rob.
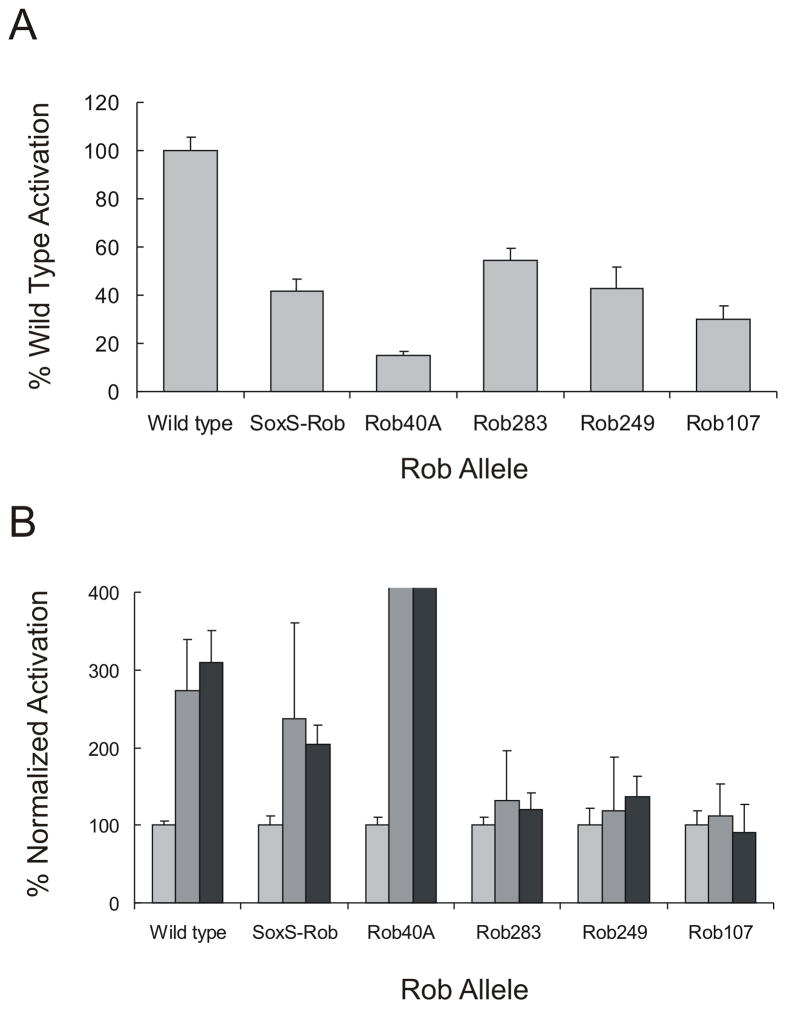
The data in Fig. 4B show that compared to the uninduced control cultures (arabinose treatment only), the addition of DIP and DEC increased SoxS-Rob-dependent activation of inaA transcription by 2.4- and 2.1-fold, respectively, enhancements similar to the 2.7-fold and 3.1-fold increase in activation obtained when cultures expressing native Rob were treated with the respective inducers. We also note that DIP and DEC have no inducing effects on SoxS43,44. Thus, in accord with the ability of Rob’s CTD to confer sequestration-dispersal on SoxS’s cellular localization, so too does it endow SoxS with the ability to respond to DIP and DEC as inducers of its activity in transcription activation. We also note that the activity of SoxS-Rob in the presence of DIP or DEC (Fig. 4) is approximately the same as that of native SoxS when expressed under similar conditions43,44.
Figure 4.
Transcription activation of inaA by variants of Rob and the effects of inducers DIP and DEC. Strain RA4468 containing an inaA-lacZ fusion and pBAD18-SoxS-Rob or variants of pBAD18-Rob were grown to A600 ~0.2 at which time they were treated with the following: 0.02% arabinose only (light grey bars); 0.02% arabinose with 3 mM DIP (grey bars); and 0.02% arabinose with 8 mM DEC (black bars). Cultures were harvested after treatment for 1 hr and β-galactosidase specific activity was determined as described in Materials and Methods. A, Transcription activation of the inaA-lacZ fusion by wild type Rob, SoxS-Rob, and Rob mutants. The β-galactosidase specific activity of wild type Rob was set to 100% and the other values are expressed relative to it. B, Effects of the inducers DIP and DEC on activation of transcription of the inaA-lacZ fusion. The β-galactosidase specific activity of each strain grown in the absence of an inducer (light grey bars) was set to 100% and the respective values for the cultures treated with DIP (grey bars) and DEC (black bars) are expressed relative to the uninduced values. The values for R40A induced by DIP (960%) and DEC (780%) have been truncated for reasons of scale and space.
Lastly, we compared the induction of the activity of chromosomally encoded SoxS-Rob to that of chromosomally encoded native Rob. To do so, we introduced plasmid pfumC, which carries a fumC-lac fusion9, into strain MF100 with the chromosomally encoded SoxS-Rob and into strain N7840, the rob+ parent of MF100. Assay of β-galactosidase expression showed that the extent of induction by DIP of chromosomally encoded SoxS-Rob, 3.5-fold, was approximately the same as the extent of DIP induction of chromosomally encoded Rob, 3.9-fold. And, the chromosomally encoded genes are as active and respond as well to the inducer as do the genes carried on plasmid pBAD33 (see Fig 4A). Thus, the plasmid encoded chimera mimics well the properties of chromosomally encoded Rob and SoxS-Rob.
Regulation of the activity of the SoxS-Rob chimera: toxicity induced by overexpression in the presence of DIP and DEC
A toxicity plate test was also used to compare the effect of inducers on the activity of SoxS-Rob to that of native Rob. Previously, we designed a plate test to aid in the characterization of the effects of single alanine substitutions of SoxS on its activity45. The plate test was based on the observation that overexpression of SoxS is toxic45: specifically, growth is severely inhibited when cells carrying plasmid pBAD18-his6-SoxS are plated on lactose tetrazolium medium supplemented with arabinose, which induces SoxS expression to a high level. With the DNA binding site for SoxS being highly degenerate10,11,46, we inferred45 that overexpression of SoxS is toxic because binding of SoxS to certain soxboxes in the chromosome interferes with function(s) essential to growth; i.e., these binding sites are not embedded in SoxS-dependent promoters but are just some of the many degenerate soxboxes scattered throughout the chromosome. This inference is consistent with the fact that toxicity is relieved by alanine substitutions that confer a defect in specific DNA binding45. Relevantly, Rosner et al. showed that overexpression of Rob in the presence of DIP is toxic, while growth is not inhibited when Rob is overexpressed in the absence of the inducer43. Accordingly, we determined whether overexpression of the SoxS-Rob chimera is toxic only when an inducer is present.
Before doing so, it was important to determine whether the toxicity conferred by overexpression of Rob in the presence of inducer requires DNA binding. Therefore, we introduced a single alanine substitution at position 40 in the recognition helix of Rob’s N-terminal helix-turn-helix DNA-binding motif and determined its effect on toxicity. As shown in the co-crystal structure of Rob in complex with micF DNA, this conserved arginine residue is surface-exposed and makes base-specific contacts with robbox DNA12. The analogous substitution in SoxS (R40A) is defective in DNA binding in vitro45 but has no effect on the interaction between SoxS and the α subunit of RNA polymerase in the yeast two-hybrid system27. Table 1 shows that the R40A substitution of Rob completely relieves the toxicity imposed by overexpression in the presence of DIP or DEC. Moreover, the relief of toxicity is not due to a reduction in protein abundance since RobR40A is as stable as wild type Rob (see Table 1 and below).
Table 1.
Effects of DIP and DEC on colony size and determination of the half-life of wild type and variant Rob proteins
| Addition to lactose tetrazolium plates containing 0.2% or 2% arabinose |
||||||||
|---|---|---|---|---|---|---|---|---|
| No treatment |
DIP (1 mM) |
DEC (3 mM) |
||||||
| Rob Allele | Representative Western Blot | Half-life | 0.2% | 2% | 0.2% | 2% | 0.2% | 2% |
| None | normal | normal | normal | normal | normal | normal | ||
| Wild type |
|
>20 hr | normal | normal | SMALL | NO GROWTH | SMALL | NO GROWTH |
| RobR40A |
|
>20 hr | normal | normal | normal | normal | normal | normal |
| Rob107 |
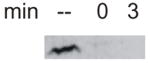
|
<1 min | normal | normal | normal | normal | normal | normal |
| Rob249 |

|
20 min | normal | SMALL | normal | SMALL | normal | SMALL |
| SoxS-Rob |
|
>20 hr | normal | normal | NO GROWTH | NO GROWTH | NO GROWTH | NO GROWTH |
| SoxS | 2 min | normal | normal | normal | normal | normal | normal | |
Overnight cultures of strain RA4468 [pBAD18-Rob] carrying different rob alleles were diluted10−6 and 0.1 ml plated on lactose tetrazolium plates with 0.2% or 2% arabinose along with DIP (1 mM), or DEC (3 mM). Growth phenotypes were determined after incubating the plates at 37°C for 15 hr. In separate experiments, the half-life of the respective proteins was determined by western blotting. The half-life of native SoxS27 was determined previously and the value is included here.
Since the possibility remains that the overexpression of Rob R40A in the presence of DIP or DEC is not toxic because the mutant protein has lost the ability to bind and respond to inducer, we transformed plasmid pBAD-RobR40A into the strain carrying the inaA-lacZ fusion and determined whether the residual transcription activation by the mutant protein can be enhanced by treatment of cells with DIP and DEC. Although the R40A substitution reduced Rob-dependent transcription activation to 15% of wild type (Fig 4A), activation was still enhanced seven- to ninefold by DIP and DEC (Fig 4B). Thus, despite being defective in DNA binding, RobR40A still retains enough structure and function to partially activate target gene transcription and to respond to DIP and DEC.
We then carried out the second functional test comparing the activities of SoxS-Rob and native Rob. An overnight culture of RA4468 containing pBAD-SoxS-Rob or pBAD-Rob was diluted and spread on lactose tetrazolium plates supplemented with 0.2% or 2% arabinose. Table 1 shows that after incubation at 37°C for ~15 hr the growth of both strains was inhibited when the medium contained DIP or DEC. Thus, cells carrying pBAD-Rob formed small colonies on plates with 0.2% arabinose and did not grow when the plates contained 2% arabinose while cells carrying pBAD-SoxS-Rob did not grow on either medium. Overexpression of SoxS-Rob or native Rob is not sufficient for toxicity, since both strains formed normal-sized colonies on arabinose plates when inducer was absent (Table 1). Moreover, DIP and DEC alone are not toxic because RA4468 containing pBAD18 grew normally on plates containing arabinose and DIP or DEC (Table 1).
Thus, the results from these two functional tests demonstrate that the SoxS-Rob chimera, formed by replacing the 107 N-terminal amino acids of Rob with full-length SoxS, retains the regulatory properties of native Rob, viz., the ability to activate transcription of target genes and the toxicity imposed by overexpression in the presence of an inducer. In turn, we can infer that the observed sequestration of the SoxS-Rob chimera that occurs in the absence of inducer renders the protein unable to activate transcription and non-toxic when overexpressed because it does not have access to chromosomal DNA. Moreover, we also infer that the addition of an inducer allows the chimera to gain access to chromosomal DNA and thereby activate transcription of target genes because the inducer releases it from the immunostainable foci. Lastly, the results of these functional tests with SoxS-Rob allow us to infer that what is true of the chimera is almost certainly true of native Rob. In particular, just as sequestration-dispersal regulates the activity of SoxS-Rob, so too must it regulate the activity of native Rob.
Identification of functional regions within Rob’s CTD
As described above, Rob’s CTD is required for regulating Rob’s activity; in particular, the addition of the CTD to the C-terminus of SoxS endows SoxS with the sequestration-dispersal properties of native Rob (Fig. 2). As a step toward elucidating the roles of various regions of the CTD in regulating the activity of Rob, we introduced into pBAD-Rob three deletions which remove increasing amounts of the CTD and we determined their effects on the regulation of Rob activity. Deletions Rob283 and Rob249 remove the distal 6 and 40 amino acids, respectively, from the C-terminus, while Rob107 removes the entire CTD, leaving only the DNA binding and transcription activation domain that is paralogous to that of SoxS. In preparing the deletions, we were careful not to create at the C-terminus the leucine-alanine-alanine tripeptide recognition sequence for ClpXP that allows efficient unfolding and degradation by this protease.
We first tested the effects of the deletions on transcription activation of the inaA-lac fusion by measuring the β-galactosidase activity produced by each strain. Compared to full-length Rob, all three deletions reduced transcription activation of inaA-lacZ and the larger the deletion, the greater the defect in transcription activation, i.e., activation was reduced by 46%, 57% and 70% when the strains expressed Rob283, Rob249, and Rob107 respectively (Fig. 4A). Thus, removing only six amino acids from Rob’s C-terminus caused about a twofold reduction in its ability to activate transcription, which shows that the full-length CTD is essential for maximum activity even though the CTD is not directly involved in DNA binding or transcription activation40. Moreover, Rob activity in the truncation mutants was not enhanced by treating the cells with the inducers DIP or DEC (Fig. 4B). This shows that all or virtually all of Rob’s CTD is required for regulation by inducers of its activity as a transcription activator. A likely interpretation of this result is based on the observation presented above that the CTD is required for sequestration (Fig. 2); as such, deletions removing even a small part of the CTD prevent sequestration, thereby allowing the dispersed protein to access chromosomal DNA and activate transcription in the absence of inducer. Thus, the role of the inducer is to antagonize the inactivating sequestration of Rob that is promoted by its CTD.
We note that Rosner et al. also prepared C-terminal truncations of Rob43. Although these deletions did not reduce Rob-dependent transcription of inaA-lacZ in the absence of DIP, their deletions, like ours, prevented induction of activity by DIP43. They concluded that elements of the CTD are required for induction of Rob activity by DIP and they suggested that DIP might reverse the sequestration previously observed by Azam et al.42.
Next, we used the plate test to determine whether the C-terminal truncations Rob249 and Rob107 affect the ability of Rob to access chromosomal DNA. Removing the entire CTD, i.e., Rob107, completely eliminated the effect of overexpression on colony size even in the presence of inducers (Table 1). According to the sequestration-dispersal hypothesis, removal of Rob’s CTD should prevent sequestration, with the result that overexpression of Rob107 should be highly toxic, because all molecules would have ready access to the chromosome and be available to bind to sites essential for viability. As discussed below, the absence of toxic effects of overexpression of Rob107 in the presence of the inducers is because the protein is very unstable (Table 1), such that few molecules are available for binding to sites in the chromosome that are essential for growth.
The effect on toxicity of removing only part of Rob’s CTD was determined in plate tests with Rob249 (Table 1). Strain RA4468 carrying pBAD-Rob249 grew normally on plates containing 0.2% arabinose, but colony size was reduced on plates with 2% arabinose. This intermediate level of toxicity in the absence of inducer is probably a combination of two effects of removing the 40 amino acids at Rob’s C-terminus: a reduction in or the absence of sequestration, i.e., partial or complete dispersal, and a lower abundance because of a protein half-life significantly shorter than that of wild type Rob (Table 1). In addition to conferring intermediate toxicity in the absence of inducer, Rob249 was refractory to the inducing effects on toxicity of DIP and DEC (Table 1). Accordingly, either the 40 amino acid truncation causes complete dispersal so that the inducers cannot further disperse Rob249 or Rob249 is only partially dispersed and the binding site for the inducers is non-functional or absent. Experiments described below (Table 2) show that increasing the abundance of Rob249 increases its toxicity and thus support the first explanation.
Table 2.
Effect of stabilizing Rob249 on the toxicity imposed by overexpression of wild type and variant Rob proteins and determination of the half-life of the respective proteins
| Arabinose concentration in lactose tetrazolium |
||||||
|---|---|---|---|---|---|---|
| Rob allele | Protease mutant | Representative western blot | Half-life | 0% | 0.2% | 2% |
| Rob107 | None | normal | normal | normal | ||
| Nhis6-Rob107 | None |

|
3 min | normal | normal | normal |
| Rob249 | None |
|
20 min | normal | normal | SMALL |
| Nhis6-Rob249 | None |

|
2.5 hr | normal | SMALL | NO GROWTH |
| Rob249 | Lon |

|
3.5 hr | normal | SMALL | NO GROWTH |
| Rob249 | Lon ClpQ |
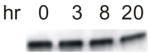
|
>20 hr | normal | NO GROWTH | NO GROWTH |
Overnight cultures of strain RA4468 containing various rob alleles in pBAD18-Rob were diluted10−6 and 100 μl spread on lactose tetrazolium plates with the appropriate amounts of arabinose (0%, 0.2% or 2%). Plates were incubated at 37°C for 15 hr prior to determining relative colony size. In separate experiments, the half-life of the respective proteins was determined by western blotting.
Rob is a stable protein and its stability depends on the CTD
SoxS, MarA, and TetD are very unstable proteins with half-lives ranging from 1–3 min2,28. As a consequence of this instability, these proteins do not accumulate to very high levels; for example, upon induction and de novo synthesis, the concentration of SoxS reaches a maximum of only 2500 molecules per cell25. In contrast to its paralogs, Rob is expressed constitutively38 and its abundance is maintained at about 10,000 molecules per cell47. Thus, with SoxS, MarA, and TetD being synthesized de novo in response to stress, accumulating to modest concentrations, and being unstable, while Rob is synthesized constitutively and is maintained at a higher concentration, we wanted to determine the half-life of Rob during steady-state growth.
Strain RA4468 containing pBAD18-Rob was treated with arabinose to induce Rob synthesis and after 1 hr of induction, chloramphenicol was added to block protein synthesis. Samples were taken at various times thereafter and the amount of Rob in each sample was determined by western blotting as described previously28. (Note that this experiment was carried out shortly after obtaining the anti-Rob serum from Dr. Ishihama, when its activity was still relatively high, as well as with antibodies we prepared against N-his6-Rob.) Unlike its sister proteins, Rob is very stable with >80% of the protein remaining 20 hr after protein synthesis was blocked (Table 1).
Since among the four paralogous proteins only Rob has a domain in addition to that which renders it a member of the AraC/XylS family, we speculated that this region, the CTD, is important for stabilizing the protein. Thus, we used the above described deletions, Rob283, Rob249 and Rob107, to determine the role of Rob’s CTD on Rob stability by measuring the half-life of each truncated protein in comparison with that of native Rob.
Interestingly, removing only 6 amino acids from the CTD (Rob283) was sufficient to reduce the half-life from >20 hr to ~10 min (data not shown). Similarly, the deletion of 40 amino acids from Rob’s C-terminus, i.e., Rob249, also significantly destabilized the protein (Table 1). Moreover, Rob107, which retains only Rob’s NTD, the segment that is homologous to the 107 N-terminal amino acids of SoxS, is very unstable (Table 1). Indeed, Rob107 was barely detectable during steady-state growth and no protein was observed immediately following treatment with antibiotics, which prevented us from accurately determining its half-life (Table 1).
As mentioned above, native SoxS is also very unstable with a half-life of 2 min28. However, addition of Rob’s CTD onto the otherwise unstable SoxS resulted in a stable chimeric protein with a half-life of >20 hr (Table 1). Thus, we conclude from these experiments that Rob’s NTD is intrinsically unstable like its paralogs SoxS, MarA, and TetD and that the CTD functions to stabilize Rob (and SoxS-Rob), presumably by protecting the vulnerable NTD from proteolytic degradation.
Amino acids at or near the N-terminus are essential for the intrinsic instability of Rob’s NTD
Previous work showed that amino acids at or near the N-terminus of SoxS and MarA serve as substrates for Lon protease, since modifications to the N-terminus, e.g., by the addition of an N-terminal his6 tag, partially protect both proteins from degradation by Lon28. To determine whether the same is true of Rob, we appended a his6 tag to the N-terminus of Rob107 and measured the half-life of N-his6-Rob107. The modification increased the half-life from immeasurable to 3 min (Table 2). We also added a his6 tag to the N-terminus of Rob249, whose half-life is 20 min. The tag increased the stability by 7.5-fold to 2.5 hr. Therefore, just like SoxS and MarA, the N-terminus of Rob appears to contain a recognition site(s) for proteolytic degradation28,48.
In previous work, a series of mutants deficient in the 5 main AAA-ATP proteases (Lon, FtsH, ClpXP, ClpAP, and ClpYQ) was used to identify Lon as the major protease responsible for the degradation of SoxS and MarA28. We found that Lon is also responsible for destabilizing Rob249, as a Δlon mutation increased its half-life 10-fold (t1/2 = 3.5 hr) (Table 2). None of the other single protease mutants affected the stability of Rob249 (data not shown). Thus, Lon is the primary protease responsible for the degradation of Rob and a Lon recognition site(s) is present within the N-terminus of SoxS, MarA, and Rob.
Since SoxS was completely stable in a Lon FtsH double mutant (t1/2 >20 hr) 28, we determined whether multiple protease mutations could further enhance the stability of Rob249 (see Table 1 of Griffith et al., 2004 for a complete list of strains). Indeed, Rob249 was completely stable in a Lon ClpQ double mutant (Table 2). Thus, just as FtsH plays an accessory role in the degradation of SoxS, so too does ClpYQ function in combination with Lon in the degradation of Rob249.
Stabilizing Rob249 increases its toxicity upon overexpression even in the absence of an inducer
Increasing the stability of SoxS and MarA, by either modifying the N-terminus or by introducing a mutation in a key protease, enhances the overexpression toxicity observed with our plate test28. As described above, overexpression of Rob249 only confers an intermediate level of toxicity, which is not increased by the presence of inducers DIP and DEC (Table 1). We hypothesize that the absence of toxicity is because its short half-life prevents it from accumulating to the level necessary to bind to robboxes located within chromosomal sequences essential to viability. Accordingly, the absence of toxicity upon overexpression of Rob249 should be overcome if overexpression is conducted under conditions that increase its abundance.
Even when overexpressed on plates containing 2% arabinose, the size of Rob249 (t1/2 = 20 min) colonies was only slightly reduced (Table 2). However, increasing Rob249’s half-life by 7.5-fold through the addition of an N-his6 tag (t1/2 = 2.5 hr) or 10.5-fold by expression in the strain with the Δlon mutation (t1/2 = 3.5 hr) significantly enhanced toxicity, producing small colonies on plates with 0.2% arabinose and preventing growth on plates with 2% arabinose (Table 2). Moreover, toxicity was dramatically increased when Rob249 was expressed in the Lon ClpQ double mutant (Rob249 t1/2 >20 hr), i.e., no growth was observed on plates supplemented with as little as 0.02% arabinose (Table 2 and data not shown). The increased toxicity in the Lon ClpQ double mutant must be due to overexpression of Rob249 since cultures of the double mutant containing the pBAD18 vector grew normally at all arabinose concentrations (data not shown). Thus, unlike full-length Rob, whose overexpression is toxic only in the presence of an inducer (because it is sequestered in the absence of an inducer), overexpression of Rob249 with a truncation of the CTD leads to binding to robboxes in essential chromosomal sites and concomitant toxicity, even in the absence of DIP or DEC, provided that means are taken to increase the abundance of the otherwise unstable protein.
Discussion
In this work, we report that Rob’s CTD is endowed with two functions that regulate Rob’s activity as a transcriptional activator, with one acting directly to mediate sequestration-dispersal and the other acting indirectly by protecting from proteolysis the regions in the NTD that carry out DNA-binding and transcription activation. The sequestration-dispersal hypothesis for regulation of Rob’s activity was derived from several previous observations. First, the ~10,000 constitutively expressed molecules of Rob per cell are relatively inactive in their ability to activate transcription of the regulon’s genes14,40 whereas Rob133, which lacks most of the CTD, is considerably more active40. Second, and most importantly, Azam et al. showed that under normal growth conditions, when Rob is inactive, the protein is sequestered into 3–4 immunostainable foci42. Third, the in vivo activity of constitutively expressed Rob is induced by the iron-chelating and non-chelating forms of dipyridyl and by lipophilic agents present in the mammalian gut such as bile salts and DEC43,44. Fourth, the inducers interact directly with the CTD, but they have no effect on DNA binding or transcription activation of purified Rob in vitro43,44, which implies that solubilization of Rob during purification renders it fully active.
From this prior knowledge, we predicted that inducers activate Rob in vivo by interacting with the CTD of sequestered Rob, that this interaction causes Rob to be released from the immunostainable foci observed by Azam et al.42, and that dispersed Rob is active in transcription activation because it is free to bind to robboxes in Rob-dependent promoters. Thus, sequestration represents the inactive, “off” state of the regulator and dispersal represents the active, “on” state. As such, the key test of the model was to determine by indirect immunofluorescence microscopy whether the sequestered, inactive form of Rob is dispersed rapidly upon treatment of growing cells with an inducer.
As mentioned above, we were unable to test the hypothesis with Rob itself, because by the time these experiments were conducted the anti-Rob antibodies lacked sufficient activity. However, we were fortunate to have affinity-purified, high-titer anti-SoxS antibodies25 and to make use of them we prepared a SoxS-Rob chimera in which Rob’s CTD was fused to the C-terminus of native SoxS. The chimera proved to be an excellent surrogate for Rob because it too formed intracellular, immunostainable aggregates (Figs. 2A and 3) and both its activity as a transcription activator (Fig. 4B) and its toxicity upon overexpression (Table 1) were enhanced by inducers DIP and DEC, as is the case with native Rob. Moreover, because native SoxS does not form immunostainable foci (Fig. 2B), the inactivity of the anti-Rob antibodies proved to be serendipitous because the experiments with the chimera allowed us to determine that Rob’s CTD is sufficient to confer sequestration on another protein. It will be interesting to see whether the CTD can confer sequestration on a totally unrelated protein.
SoxS15–19, MarA29–33, and TetD2,37 are the direct transcription activators of the SoxS/MarA/TetD/Rob regulon. However, they are the second components of two-gene, two-stage systems in which the first components, SoxR, MarR and TetC, respectively, are sensor-regulators which receive an inducing environmental signal, e.g., superoxide or salicylate, respectively, (the signal for TetC is unknown), that modifies them such that de novo synthesis of the respective activators ensues. Rob, on the other hand, is synthesized constitutively38 and inactivated by sequestration; it becomes activated when the CTD receives an inducing signal43,44 that leads to dispersal and subsequent transcription activation of the regulon’s genes (see Figs 2–4).
The two fundamentally different mechanisms that lead to activation of transcription of the same regulon raise interesting evolutionary questions that may be based on the physiology of the systems. For example, why should the response to a lethal agent like reactive oxygen species require the de novo synthesis of the protein that turns on the defense mechanism? Even though SoxS, MarA, and TetD are relatively small proteins that function as monomers5, they still must achieve an intracellular concentration sufficient to rapidly mount the defense response. With the binding sites being highly degenerate, ~65,000 per fast-growing cell25,34, the regulators must also be able to identify activator-dependent promoters in the virtual sea of sequence-equivalent but non-functional binding sites scattered throughout the chromosome. We have presented evidence that activation by pre-recruitment solves this conundrum26 and appropriation of RNA polymerase by binding of the activator to the UP-element-binding determinant of the α subunit may enhance the efficiency of the process27. Still, the low information content of the activator binding sites in promoters11 is difficult to explain. One process that does make physiological sense is that SoxS, MarA, and TetD are highly unstable, such that when the inducing stress is relieved, de novo synthesis ceases and the residual activator is rapidly degraded2,28. Rapid degradation is most likely necessary because overexpression of the respective proteins is toxic45. Moreover, since the proteins appear to interact directly with RNA polymerase and divert it from UP-element containing rRNA genes to stress-response genes27, maximum growth in the absence of stress requires that all RNA polymerase molecules be available for transcription of genes for essential housekeeping functions. Thus, although it would be relatively simple to design a two-component system that would efficiently and rapidly regulate a 50-member regulon that provides a defense against a variety of unrelated, stress-inducing signals, the current systems work and hence need not be targets for evolutionary improvement. Moreover, simpler systems with constitutively synthesized activators that respond to these inducing signals may pose hazards because the equilibrium between the inactive and active forms in the absence of inducer may yield too much active activator for the good of the cell.
Similar questions may be asked of Rob. First, why is Rob made constitutively rather than being turned on by de novo synthesis in response to a stress-inducing signal and turned off by cessation of its synthesis and degradation of the remaining protein, as is sufficient for SoxS, MarA, and TetD? A simple answer might be that the survival threat posed by lipophilic agents in the mammalian gut might require such a rapid response, more rapid than the threats posed by superoxide and salicylate, that active Rob must be nearly instantaneously available and available at a concentration sufficient to turn on the defense response genes rapidly and efficiently. Note that as a one-stage, one-gene system, constitutively expressed Rob’s DNA-binding, transcription activation domain needed a means for its activity to be regulated. Hence, as is usually the case for members of the AraC/XylS family1, Rob is endowed with an inducer-binding domain.
A second question may be asked of Rob. Why is the activity of constitutively expressed Rob regulated by a novel mechanism, sequestration-dispersal? Would not simple regulation of its DNA-binding activity by binding of the inducing ligand to the CTD suffice, as is the case for the majority of bacterial transcription factors, e.g., Catabolite Activator Protein49,50. Again, the answer may be that two properties of the system are prohibitive. First, if Rob is not rapidly cleared from the cell, some RNA polymerase might continue to activate stress response genes at the expense of the expression of genes used for maximizing growth. While this is undoubtedly a worthwhile trade-off when survival to a stress is at stake, it is probably not worthwhile in the absence of stress. Second, binding of Rob to sites that are not in promoters would likely reduce viability. Again, from a population standpoint, this may be a worthwhile trade-off under stressful conditions but would not be worthwhile once the stress has been overcome.
Given that SoxS, MarA, and TetD are very unstable proteins2,28, that Rob is sequestered into immunostainable foci, and that Rob’s CTD is required for sequestration, we considered whether Rob was likely to be stable or unstable. On the one hand, Rob might have been intrinsically unstable but stabilized by sequestration. Then, upon induction and release from the foci, Rob would be available to activate transcription of the regulon’s genes while its abundance is regulated by auto-repression51 (K.L.G and R.E.W., Jr., unpublished results). Subsequently, as the stress subsides, Rob would return to its inactive, sequestered form, perhaps by the action of a sequestration factor. This instability despite the presence of the large CTD would be consistent with the fact that the instability of SoxS is unaffected by the addition of a his6 tag to the C-terminus, whereas stability is enhanced 13-fold by the addition of a his6 tag to its N-terminus 28. On the other hand, Rob might have been intrinsically stable, with the CTD being responsible for stabilizing the otherwise unstable NTD. Indeed, the crystal structure of Rob bound to micF DNA is consistent with this possibility12, since Rob’s C-terminal amino acids form a “ridge-like” projection which lies over the N-terminal amino acids and would thereby have the potential to protect them from proteolytic attack. The fact that previous genetic and biochemical experiments carried out in vivo and in vitro have shown that a site essential for proteolysis by Lon resides within the 20 amino acids at the N-terminus of SoxS is also consistent with the possible protective effect of Rob’s CTD48.
Indeed, Rob proved to be very stable, with a half-life greater than 20 hr (Table 1). Moreover, and importantly, the SoxS-Rob chimera was also very stable (Table 1). Therefore, since SoxS is intrinsically unstable with a half-life of ~ 2 min, Rob’s CTD appended to the C-terminus of SoxS is able to confer stability on it. As such, the CTD is not only sufficient to allow SoxS to form intracellular foci and induction by DIP and DEC, but it is also sufficient to stabilize this otherwise unstable protein.
Removing as few as 6 amino acids from Rob’s C-terminus (Rob283) reduced its half-life to 10–20 minutes (data not shown) and removing the entire CTD (Rob107) rendered it highly unstable (Table 1) like its paralogs SoxS, MarA, and TetD2,28. As with SoxS, adding an N-his6 tag to truncated Rob proteins enhanced their stability, e.g., the tag increased the half-life of Rob249 from 20 minutes to 2.5 hr (Table 2). The truncations also caused loss of inducer-dependence. In addition, just as Lon is the primary protease that degrades SoxS, so too are the protease-susceptible, C-terminally truncated Rob proteins also degraded by Lon. Moreover, the C-terminal truncations not only reduce transcription activation of an inaA-lacZ fusion but they also eliminate the ability of Rob activity to be induced by DIP and DEC (Fig. 4). These effects are likely the result of the combination of the destabilization and reduced abundance of the Rob fragments compared to full-length Rob, the destruction of the inducer binding site, and potentially the release of Rob from the foci. Thus, it will be interesting to determine which Rob fragments if any can be sequestered and which if any can bind inducer.
This work raises many interesting questions. For example, how does the system reset once the inducing stress is relieved? We imagine that the process is initially effected by the AcrAB efflux pump, which is induced by bile salts and which then eliminates them from the cell44. Thus, when Rob is no longer bound by inducer, does it contain the intrinsic capacity to become sequestered or is a “cellular sequestration factor” required? Another important question is whether the activity of other bacterial transcription factors is regulated by sequestration-dispersal through the action of a small molecule inducer. It seems highly likely that some inducers that enhance the activity of activators do so by causing their dispersal from a sequestered state. Thus, even though this work describes the first example of sequestration-dispersal as the mechanism that regulates the activity of a bacterial transcription activator, we expect that when additional experiments are done, e.g., indirect immunofluorescence microscopy with activator-specific antibodies or perhaps glycerol gradient centrifugation, other examples will be revealed.
Recently, we presented evidence that SoxS functions as a co-sigma factor27: by interacting with the DNA binding determinant of the α subunit of RNAP, SoxS diverts RNAP from “strong”, UP element-containing promoters, e.g., the promoters of the genes encoding rRNA, to the superoxide stress-inducible promoters of the SoxRS regulon. In addition, Dangi et al.52 showed that MarA also makes protein-protein contact with the same domain of the alpha subunit. And, in unpublished work, we have found that Rob functions as a co-sigma factor: just like SoxS and MarA, Rob interacts with the DNA binding determinant of the α subunit of RNAP and in so doing diverts RNAP from UP element-containing promoters to the promoters of the Rob regulon (E.F. Keen III and R.E.W., Jr, unpublished results).
The mechanism of sequestration-dispersal and the role of Rob’s CTD in it are particularly interesting in this context. Since Rob’s NTD functions as a co-sigma factor and since Rob’s CTD mediates sequestration of the NTD and thereby its inactivation, the CTD is playing the role of an anti-sigma factor. Thus, under non-inducing conditions, the ability of the co-sigma factor to divert RNAP to promoters of the Rob regulon is blocked by sequestration, just as the binding of an anti-sigma factor like RseA blocks the action of its cognate sigma factor, σE53. Then, when a stress-inducing signal, e.g., lipophilic agents like bile salts, is encountered, Rob becomes dispersed and gains access to the transcription machinery, just as misfolded proteins in the periplasm leads to release of σE from RseA so that σE can activate transcription of the stress-response genes54. What is particularly unique about the Rob system is that the co-sigma factor, the NTD, and its anti-sigma factor partner, the CTD, both reside in a single polypeptide chain. Moreover, as shown here, Rob’s CTD is also able to function as an anti-sigma factor when fused to another protein, e.g., SoxS, which has co-sigma factor-like activity27.
Materials and Methods
Bacterial strains
All experiments except those employing mutations of protease genes or the chromosomally encoded SoxS-Rob chimera (strain MF100) were carried out with strain RA446838, an isogenic derivative of strain GC4468 (F− ΔlacU169 rpsL)16 carrying a Δrob::kan mutation. The preparation of RA4468 carrying an inaA-lacZ fusion in single copy on a lambda prophage has been described10 Cloning was done with strain DH5α as the recipient. The set of strains containing mutant proteases is described elsewhere28. The preparation of strain MF100 is described below.
Plasmid constructions
Standard recombinant DNA techniques were used to clone the rob gene from strain GC4468 into plasmid pBAD18 in which its expression is under the control of the arabinose-inducible PBAD promoter55. The rob coding sequence was amplified by PCR from chromosomal DNA of strain GC4468 using oligonucleotide primers whose sequences are available upon request of the corresponding author. The conditions used for PCR were: 35 cycles at 95°C for 30 sec; 52°C for 30 sec, and 72°C for 90 sec. PCR products were digested with restriction enzymes XbaI and HindIII and ligated into plasmid pBAD18 (Novagen), which had been digested with the same two restriction enzymes. An aliquot of the ligation mixture was transformed into strain DH5α and the transformed cells were plated onto LB agar with ampicillin (50 μg/ml). DNA was isolated from the transformants by the alkaline lysis method and purified using Qiagen spin-columns as described by the manufacturer. The DNA sequence was determined at the UMBC core facility and the construct was designated pBAD-Rob. To prepare pBAD-N-his6-Rob, a his6 tag was added onto the N-terminus of rob by add-on PCR, essentially as described for N-his6-SoxS25. Termination codons were introduced into the rob coding sequence in pBAD-Rob by QuikChange mutagenesis according to the manufacturer’s instructions (Stratagene). The SoxS-Rob chimera was constructed by using PCR-SOEing56 to add the sequence encoding the last 182 amino acids of Rob, in frame, to the 3′-end of the soxS coding sequence; the chimera was then cloned into pBAD3355 and pBAD18 as described above.
Replacement of the N-terminal 107 amino acids of chromosomally encoded Rob with SoxS
RESS (R. Toughiri and R.E.W., Jr., unpublished results), a variation of “gene gorging”57, was used to construct strain MF100, which carries the gene for the SoxS-Rob chimera in place of the chromosomal rob gene.
In the first step of RESS, a DNA cassette carrying the recognition sequence for the I-SceI meganuclease and a gene encoding kanamycin resistance (Kan-r) was prepared by PCR and then introduced by recombineering58 into rob by selection for Kan-r. The template for preparation of the DNA cassette was plasmid pKD4, which encodes Kan-r58. The upstream primer, 5′-upRSK-3′, contained 50 bases homologous to the DNA upstream of Rob’s start codon, followed by the 18 bp recognition sequence for the I-SceI meganuclease, followed by the 19 base sequence of priming site 1 for amplification of the kan gene58. The downstream primer, 5′-downRK-3′, contained 70 bases homologous to the DNA of rob downstream of codon 107 followed by the 19 base sequence of priming site 2 for amplification of the kan gene58. The sequences of the primers are available upon request.
The recombineering plasmid pKD4658 was introduced by electroporation into strain N7840 (ΔlacU169 Δmar rpsL). The Red recombinase enzymes, which effect homologous recombination of linear DNA, were induced by treatment with 2% arabinose. The cells were made electrocompetent and transformed with the DNA cassette. The transformed cells were plated on LB medium containing Kan (10 μg/ml) at 30°C. Kan-r transformants were selected and, after purification, the recombinants were cured of pKD46 by growth in LB medium at 37°C in the absence of ampicillin. To determine whether the constructs had the correct configuration, the cassette was amplified by colony PCR using primers homologous to rob sequences upstream of the start codon and downstream of codon 107. The sequence of the N-terminal Rob-I-SceI-Kan junction was verified by DNA sequencing with primer k158, which anneals to the template strand of kan.
In the second step of RESS, plasmid pACBSR57, which contains the Red genes and the gene for the I-SceI meganuclease, both expressed under control of the PBAD promoter, was introduced into a Kan-r recombinant by electroporation and selection for chloramphenicol (Cm) resistance on LB + Cm (25 μg/ml) plates at 37°C. A transformant was inoculated into Rich Defined Medium (RDM; TEKnova, Inc.) containing 0.2% arabinose and incubated at 37°C for 30 min before Cm was added. Incubation was continued until A600 = 0.4 at which time the cells were prepared for electroporation.
The donor DNA fragment contained the entire SoxS coding sequence flanked by sequences homologous to DNA upstream of the Rob start codon and downstream of Rob codon 107. It was prepared by PCR with plasmid pBAD33-SoxS-Rob as the template. The upstream primer, 5′-upRS-3′, had 50 bases of DNA homologous to rob sequences upstream of the Rob start codon followed by 18 bases homologous to the first six codons of SoxS. The downstream primer, 5′-downRS-3′, had 50 bases of DNA homologous to rob sequences downstream of Rob codon 107 followed by 18 bases homologous to the last six codons of SoxS.
The donor DNA fragment was electroporated into electrocompetent cells of the “selection” strain carrying pACBSR. The transformed cells were transferred to RDM containing 0.2% arabinose and Cm (25 μg/ml) and incubated at 37°C overnight. Cells surviving expression of the meganuclease were obtained by plating on LB plates + Cm followed by overnight incubation. Colonies that appeared were replica plated onto LB plates + Kan (10 μg/ml). Over 80% of the colonies had the desired kanamycin-sensitive (Kan-s) phenotype. Five Kan-s clones were subjected to colony PCR and DNA sequencing. All proved to carry the 107 amino acids of SoxS in place of the corresponding 107 amino acids at the N-terminus of chromosomally encoded Rob and thus encoded the SoxS-Rob chimera. One clone was named MF100.
After RESS was fully developed and implemented, we learned that Cox et al. had published a similar method of scarless recombineering59.
Indirect immunofluorescence microscopy
The method used for indirect immunofluorescence microscopy was adapted from that of Azam et al.42. Overnight cultures containing pBAD33-SoxS -Rob or pBAD33-SoxS were diluted 1:100 into LB medium supplemented with 25 μg/ml chloramphenicol and grown with vigorous aeration at 37°C until the A600 reached approximately 0.1, at which time synthesis of SoxS-Rob or SoxS was induced by adding arabinose to each culture to a final concentration of 0.02% and growth was continued at 37°C with vigorous aeration. After 1 hr of induction, each culture was divided into three portions: one was left untreated; a second was treated with 5 mM 4, 4′-dipyridyl (DIP); and the third was treated with 8 mM decanoate (DEC). Incubation was continued under the same conditions for various lengths of times. Then, 1 ml aliquots were removed and the cells were fixed with 10 ml of 80% methanol for 1 hr at room temperature. After fixation, the cells were collected by centrifugation at 2500 g for 5 min at 4°C and washed twice with PBST (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mm KH2PO4, and 0.05% Tween 20). After resuspending the washed cells in 0.5 ml of 80% methanol, 10 μl samples were spotted onto a Gold Seal Rite-On Fluorescent Antibody Microslide (Fisher Scientific). The cell suspension was allowed to completely dry on the slide, and then the cells were permeabilized by treatment with freshly prepared lysozyme (1 mg/ml) for 5 min at room temperature. After four washes with PBST, blocking buffer (2% BSA in PBST) was added to the permeabilized cells and the slides were incubated for 15 min at room temperature. Rabbit polyclonal anti-SoxS serum25, affinity-purified by the Pocono Rabbit Farm, was diluted 1:1000 in blocking buffer and added to the slide, which was incubated overnight at 4°C in a humidified chamber. To remove any unbound, primary antiserum, the slides were washed 4 times with PBST and 2 times with blocking buffer and then incubated for 15 min at room temperature with blocking buffer. Then, the buffer was removed and replaced with Alexa488-labeled, goat anti-rabbit secondary antibody (Molecular Probes) (diluted 1:500 in blocking buffer) and the slides were incubated for 1 hr at room temperature in a humidified chamber. Slides were washed 5 times with PBST and mounted with 90% glycerol in PBS. Images were collected using a Zeiss Axioplan2 fluorescence microscope with a 63X objective lens and a 2.5X ocular lens. Brightness and contrast were adjusted using Adobe Photoshop 7 and the same settings were used for all images.
The same method was used for indirect immunofluorescence microscopy of strain MF100 except the cells were induced with 5 mM DIP at A600 = 0.2 and the primary antibody was diluted 1:250. The brightness and contrast of these images were also adjusted with Adobe Photoshop 7 and the same settings were used for each image.
Assay of β-galactosidase activity
β-galactosidase activity was measured from strains containing transcriptional fusions to inaA-lacZ using our previously described high-throughput method60. Duplicate samples were taken and assayed twice in a given experiment; values with a standard error >12% were discarded. The values represented in Fig. 4 are an average of at least three independent experiments and the bars represent standard error expressed as the percent of the mean.
Determination of the effects of DIP and DEC on transcription activation
Plasmids pBAD18-Rob and pBAD18-SoxS-Rob were transformed into strain RA4468 carrying an inaA-lacZ transcriptional fusion on a single copy prophage. Triplicate cultures of the two strains were grown in LB medium at 37°C to A600 = 0.2 and each culture was divided into three equal portions. All three subcultures of the triplicate cultures of strains RA4468 [pBAD-Rob] and RA4468 [pBAD-SoxS-Rob] were treated with 0.02% arabinose to induce Rob or SoxS-Rob expression, respectively. In addition, one culture of each set received no further treatment, another received DIP and the third received DEC. After continuing the incubation at 37°C for 1 h, the cells were harvested from the 18 cultures and β-galactosidase activity was measured as above. The triplicate values for the control cultures treated with arabinose but not with DIP or DEC were averaged and set at 100% and the triplicate values for the DIP- and DEC-treated cultures of the strains expressing native Rob and chimeric SoxS-Rob were averaged and their mean values were expressed in relation to the 100% values for the respective control cultures. RA4468[pBAD-Rob] strains carrying Rob truncations Rob283, Rob249 and Rob107 or the substitution R40A were prepared and analyzed by the same methods.
Plate tests for toxicity imposed by overexpression
The toxicity of strains over-expressing SoxS was previously described45. The same approach was used to determine the toxicity of overexpressed Rob and its derivatives. Overnight cultures of the appropriate strains were diluted 10−6 and 0.1 ml was spread on lactose tetrazolium plates containing ampicillin (50 μg/ml) and 0%, 0.02%, 0.2%, or 2% arabinose. Where appropriate, a final concentration of 1 mM DIP or 3 mM DEC was added to the plates. Following incubation of the plates at 37°C for ~18 hr, colony size was determined relative to strains containing the pBAD18 vector.
Growth conditions for determining the half-life of Rob
Overnight cultures containing Rob and derivatives of Rob expressed from plasmid pBAD18 were diluted 1:100 into LB medium supplemented with ampicillin (100 μg/ml) and incubated at 37°C with vigorous aeration until the culture density reached A600 ~0.2. Cultures were treated with 0.02% arabinose to induce Rob expression and incubated at 37°C on a rotating shaker. After 1 hr of induction, protein synthesis was arrested by the addition of chloramphenicol (100 μg/ml). Aliquots were taken at specified times following the treatment with antibiotic and harvested by centrifugation at 6000 g for 5 min at 4°C. Cell pellets were resuspended in ice cold sonication buffer (50 mM Tris buffer, pH 7.9, 3 mM DTT, 1 mM EDTA). Extracts were prepared by sonication on ice with a Branson sonifier, applying 2 pulses of 40 sec with a 20 sec pause between each pulse. The samples were centrifuged at 13000 g for 20 min to pellet the insoluble cell material and the supernatant fluid from each sample was transferred to a sterile microfuge tube containing Laemmli gel loading dye. Proteins were separated by SDS-PAGE using a 12% or 18% Tris-glycine gel (Invitrogen).
Western blotting and determination of protein half-lives
The procedure for immunoblotting has been previously described25 Briefly, proteins were transferred from the Tris-glycine gels to polyvinylidene difluoride (PVDF) membranes (New England Nuclear) by electroblotting (Invitrogen). Western blots were performed using the appropriate polyclonal antiserum from the following three sources: original anti-serum against Rob kindly provided by A. Ishihama; a second preparation of anti-serum against N-his6-Rob (Pocono Rabbit Farm); and affinity-purified anti-serum directed against N-his6-SoxS25. The Enhanced Chemifluorescence (ECF) detection system (Amersham) was used according to the manufacturer’s instructions. Western blot signals were detected with a Storm PhosphorImager (Molecular Dynamics) and quantified using IMAGEQUANT software (Molecular Dynamics). The half-life values are an average of at least three independent experiments and were determined as described previously28.
Acknowledgments
We are grateful to A. Ishihama for the generous gift of anti-Rob antiserum, S. Gottesman for the collection of protease mutants and F. R. Blattner for providing plasmid pACBSR. We thank R.G. Martin and J.L. Rosner for providing strains, sharing unpublished data, and for many stimulating discussions. We also thank E. F. Keen III for his initial work on a SoxS-Rob chimera and R. Toughiri for developing the RESS method. This work was supported by Public Health Service grant GM27113 from the National Institutes of Health awarded to R.E.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol Molec Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith KL, Becker SM, Wolf RE., Jr Characterization of TetD as a transcriptional activator of a subset of genes of the Escherichia coli SoxS/MarA/Rob regulon. Mol Microbiol. 2005;56:1103–1117. doi: 10.1111/j.1365-2958.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- 3.Jair KW, Fawcett WP, Fujita N, Ishihama A, Wolf RE., Jr Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 4.Jair KW, Martin RG, Rosner JL, Fujita N, Ishihama A, Wolf RE., Jr Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jair KW, Yu X, Skarstad K, Thöny B, Fujita N, Ishihama A, Wolf RE., Jr Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa TM, Levy SB. Activation of the Escherichia coli nfnB gene by MarA through a highly divergent marbox in a class II promoter. Mol Microbiol. 2002;45:191–202. doi: 10.1046/j.1365-2958.2002.03006.x. [DOI] [PubMed] [Google Scholar]
- 7.Martin RG, Rosner JL. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol Microbiol. 2002;44:1611–1624. doi: 10.1046/j.1365-2958.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- 8.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fawcett WP, Wolf RE., Jr Purification of a MalE-SoxS fusion protein and identification of the control sites of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1994;14:669–679. doi: 10.1111/j.1365-2958.1994.tb01305.x. [DOI] [PubMed] [Google Scholar]
- 10.Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 11.Wood TI, Griffith KL, Fawcett WP, Jair KW, Schneider TD, Wolf RE., Jr Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol Microbiol. 1999;34:414–430. doi: 10.1046/j.1365-2958.1999.01598.x. [DOI] [PubMed] [Google Scholar]
- 12.Kwon HJ, Bennik MHJ, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nature Struct Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 13.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin RG, Gillette WK, Rosner JL. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- 15.Amábile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg JT, Monach P, Chou JT, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunoshiba T, Hidalgo E, Amábile Cuevas CF, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc Natl Acad Sci USA. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 22.Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J Biol Chem. 1997;272:5082–5086. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- 23.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo MS, Lee JH, Rah SY, Yeo WS, Lee JW, Lee KL, Koh YS, Kang SO, Roe JH. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith KL, Shah IM, Myers TE, O’Neill MC, Wolf RE., Jr Evidence for “pre-recruitment” as a new mechanism for transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem Biophys Res Commun. 2002;291:979–986. doi: 10.1006/bbrc.2002.6559. Erratum Biochem Biophys Res Commun 294, 1191. [DOI] [PubMed] [Google Scholar]
- 26.Griffith KL, Wolf RE., Jr Genetic evidence for pre-recruitment as the mechanism of transcription activation by SoxS of Escherichia coli: the dominance of DNA binding mutations of SoxS. J Mol Biol. 2004;344:1–10. doi: 10.1016/j.jmb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Shah IM, Wolf RE., Jr Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase α subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J Mol Biol. 2004;343:513–532. doi: 10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- 28.Griffith KL, Shah IM, Wolf RE., Jr Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol. 2004;51:1801–1816. doi: 10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 29.Alekshun MN, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–72. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ariza RR, Cohen SP, Bachhawat N, Levy SB, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SP, Levy SB, Foulds J, Rosner JL. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcription activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- 35.Braus G, Argast M, Beck CF. Identification of additional genes on transposon Tn10: tetC and tetD. J Bacteriol. 1984;160:504–509. doi: 10.1128/jb.160.2.504-509.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schollmeier K, Hillen W. Transposon Tn10 contains two structural genes with opposite polarity between tetA and IS10R. J Bacteriol. 1984;160:499–503. doi: 10.1128/jb.160.2.499-503.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe CM, Suzuki C, Laurie C, Simons RW. Regulation of the “tetCD” genes of transposon Tn10. J Mol Biol. 1997;270:14–25. doi: 10.1006/jmbi.1997.1094. [DOI] [PubMed] [Google Scholar]
- 38.Skarstad K, Thöny B, Hwang D, Kornberg A. A novel binding protein of the origin of the Escherichia coli chromosome. J Biol Chem. 1993;268:5365–5370. [PubMed] [Google Scholar]
- 39.Azam TA, AI Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity . J Biol Chem. 1999;274:33105–13. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 40.Ariza RR, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl Environ Microbiol. 1995;61:2302–2307. doi: 10.1128/aem.61.6.2302-2307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins with the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 43.Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J Bacteriol. 2002;184:1407–1416. doi: 10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 45.Griffith KL, Wolf RE., Jr A comprehensive alanine scanning mutagenesis of the Escherichia coli transcriptional activator SoxS: identifying amino acids important for DNA binding and transcription activation. J Mol Biol. 2002;322:237–257. doi: 10.1016/s0022-2836(02)00782-9. [DOI] [PubMed] [Google Scholar]
- 46.Griffith KL, Wolf RE., Jr Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol Microbiol. 2001;40:1141–1154. doi: 10.1046/j.1365-2958.2001.02456.x. Erratum Mol Microbiol 42, 571. [DOI] [PubMed] [Google Scholar]
- 47.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah IM, Wolf RE., Jr Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J Mol Biol. 2006;357:718–731. doi: 10.1016/j.jmb.2005.12.088. [DOI] [PubMed] [Google Scholar]
- 49.de Crombrugghe B, Chen B, Anderson WB, Gottesman ME, Perlman RL, Pastan I. Role of cyclic 3′, 5′-monophosphate and the cyclic adenosine 3′, 5′-monophosphate receptor protein in the initiation of lac transcription. J Biol Chem. 1971;246:7242–7248. [PubMed] [Google Scholar]
- 50.Majors J. Specific binding of CAP factor to lac promoter DNA. Nature. 1975;256:672–674. doi: 10.1038/256672a0. [DOI] [PubMed] [Google Scholar]
- 51.Schneiders T, Levy SB. MarA-mediated transcriptional repression of the rob promoter. J Biol Chem. 2006;281:10049–100585. doi: 10.1074/jbc.M512097200. [DOI] [PubMed] [Google Scholar]
- 52.Dangi B, Gronenborn AM, Rosner JL, Martin RG. Versatility of the carboxy-terminal domain of the α subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA. Mol Microbiol. 2004;54:45–59. doi: 10.1111/j.1365-2958.2004.04250.x. [DOI] [PubMed] [Google Scholar]
- 53.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-σ RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 54.Ades SE. Control of the alternative sigma factor σE in Escherichia coli. Curr Opin Microbiol. 2007;7:156–162. doi: 10.1016/j.mib.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 55.Guzman LM, Belin D, Carson MJ, Beckwith JR. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton RM, Cai Z, Ho SN, Pease L. Gene splicing by overlap extension: tailor made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 57.Herring CD, Glasner JD, Blattner FR. Gene replacement without selection: the introduction and regulatable suppression of amber mutations in E. coli. Gene. 2003;311:153–163. doi: 10.1016/s0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- 58.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PRC products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox MM, Layton SL, Jiiang T, Cole K, Hargis BM, Berghman LR, Bottje WG, Kwon YM. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotech. 2007;7:59–69. doi: 10.1186/1472-6750-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffith KL, Wolf RE., Jr Measuring β-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]