Abstract
Long-term persistence of species characterized by a reduced effective population size is still a matter of debate that would benefit from the description of new relevant biological models. The island endemic specialist Drosophila sechellia has received considerable attention in evolutionary genetic studies. On the basis of the analysis of a limited number of strains, a handful of studies have reported a strikingly depleted level of genetic variation but little is known about its demographic history. We extended analyses of nucleotide polymorphism in D. sechellia to a species-wide level using 10 nuclear genes sequenced in 10 populations. We confirmed that D. sechellia exhibits little nucleotide-sequence variation. It is characterized by a low effective population size, >10-fold lower than that of D. simulans, which ranks D. sechellia as the least genetically diverse Drosophila species. No obvious population subdivision was detected despite its fragmented geographic distribution on different islands. We used approximate Bayesian computation (ABC) to test for demographic scenarios compatible with the geological history of the Seychelles and the ecology of D. sechellia. We found that while bottlenecks cannot account for the pattern of molecular evolution observed in this species, scenarios close to the null hypothesis of a constant population size are well supported. We discuss these findings with regard to adaptive features specific to D. sechellia and its life-history strategy.
PATTERNS of nucleotide variation are shaped by the evolutionary and demographic history of species. Considerable research has focused on detecting adaptive evolution from polymorphism data in a large number of organisms, including Drosophila (Baines et al. 2004; Tenaillon et al. 2004; Schmid et al. 2005; Heuertz et al. 2006). Among Drosophila, most evolutionary studies aiming at understanding species demography and selective history have been undertaken in the model species Drosophila melanogaster and its close relative D. simulans (Wall et al. 2002; Orengo and Aguade 2004; Dumont and Aquadro 2005; Li and Stephan 2006). The latter species and its endemic siblings, D. sechellia and D. mauritiana, have already proven a useful and major model system for evolutionary genetic studies related to speciation (Sawamura et al. 1993; Coyne and Orr 1998; Gleason et al. 2005; Haerty and Singh 2006). However, demographic studies on large samples are simply missing in the two endemic species, although there is an increasing number of examples revealing complex demographic histories in many Drosophila species (Haddrill et al. 2005; Bachtrog and Andolfatto 2006; Baudry et al. 2006; Pool et al. 2006).
D. sechellia, endemic to the Seychelles archipelago, has drawn much attention since its discovery in 1980 (Tsacas and Bächli 1981). The species, among the youngest of the melanogaster subgroup, is estimated to have originated ∼250,000–500,000 years ago (Kliman et al. 2000; Lachaise et al. 2004; McDermott and Kliman 2008). This species is one of the few Drosophila species to have a well-defined ecology. It breeds exclusively on the ripe fruits of the Rubiaceae Morinda citrifolia (Lachaise and Silvain 2004; Cariou et al. 2009) and is the only one among the four species of the melanogaster complex to resist the volatile lethal components of Morinda fruit (R'kha et al. 1991; Farine et al. 1996). Genomic regions involved in the genetic determinism of this adaptation have been localized (Jones 1998; Jones 2005); they include the olfactory receptors (Stensmyr et al. 2003; Dekker et al. 2006) and two odorant binding proteins responsible for the loss of avoidance to Morinda (Matsuo et al. 2007).
D. sechellia is characterized by an overall depleted level of genetic diversity (Cariou et al. 1990; Hey and Kliman 1993; Kliman and Hey 1993; Kliman et al. 2000; Morton et al. 2004), which was first attributed to a strong founder effect at the origin of the species with a persistent small population size (Cariou et al. 1990; Kliman and Hey 1993). However those previous studies suffer from two major limitations. First they have relied on a too-restricted number of Drosophila strains to be conclusive. A species-wide sample (multiple strains from multiple populations) may reveal a strong population structure resulting in biases when the sample is limited to a single population. Second, a number of alternative scenarios may account for the observed pattern of depleted genetic variation including (i) a host specialization to M. citrifolia at the time of speciation; (ii) a recent shift from a primary endemic host to M. citrifolia, following its introduction to Seychelles with the establishment of human settlements as recently as 300 years ago (Lachaise and Silvain 2004); and (iii) a bottleneck accompanying the Holocene marine transgression 10,000 years ago resulting in a 600-fold reduction of the land masses of the Seychelles.
To get new insight into the pattern of variation, structure, and demography of D. sechellia, we collected a set of 10 populations in 9 Seychelles islands thereby covering the presently described species range. We sequenced 10 loci (3 autosomal and 7 X linked) with an average of 7.5 individuals per population and genotyped an mtDNA marker. On the basis of a data set of 758 sequences, we estimated genetic diversity, investigated underlying genetic structure, and used approximate Bayesian computation (ABC) methods to test for demographic scenarios compatible with the biogeographical history of the species.
MATERIALS AND METHODS
Sample collection:
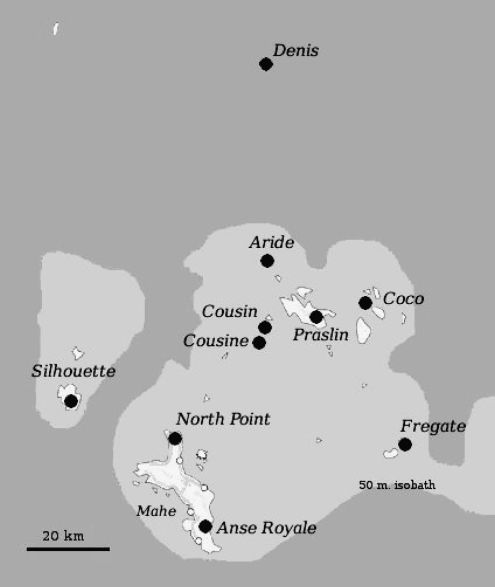
Since its discovery on Cousin island, D. sechellia was further identified on the Praslin, Frégate, and Mahé islands of the Seychelles archipelago. We recently found D. sechellia on five additional granitic islands, namely, Silhouette, Cousine, Coco, Aride, La Digue, and Denis, a coralline island located 100 km north of Mahé. We collected 92 D. sechellia from 10 locations on nine islands (Figure 1). Upon collection, all flies were immediately preserved in absolute ethanol. Molecular analyses were performed on males only.
Figure 1.—
Geographic distribution of D. sechellia sampling sites.
Molecular markers and DNA sequencing:
Mitochondrial DNA:
Among the D. melanogaster complex, mitochondrial DNA carries an extremely low diversity and is highly structured (Solignac 2004). Only one mitochondrial haplotype is known for D. sechellia. To further confirm this and to test for population subdivision, we selected a target sequence (part of NADH dehydrogenase) that allows the discrimination of all the mitochondrial haplotypes among species of the melanogaster complex.
Nuclear DNA:
We intended to get variation from loci combining intergenic regions, introns, and exons. We therefore sequenced 10 genic regions: 3 autosomal (amyrel, cecropin psiII, and janus-ocnus regions) and 7 X-linked loci (period, sqh, vermilion, white, otu, pgd, and zeste). The number of sequences for each locus and each population is recorded in Table 1.
TABLE 1.
Number of sequences per gene and per population studied in D. sechellia
| Population | amyrel | period | cecropin | janusB-ocnus | sqh | vermilion | white | otu | pgd | zeste | NADH Dase II |
|---|---|---|---|---|---|---|---|---|---|---|---|
| North Point | 10 | 10 | 4 | 6 | 9 | 9 | 8 | 8 | 8 | 7 | 2 |
| Anse Royale | 10 | 9 | 3 | 6 | 8 | 8 | 9 | 9 | 8 | 9 | 2 |
| Praslin | 9 | 10 | 3 | 6 | 8 | 8 | 8 | 8 | 6 | 8 | 2 |
| Aride | 10 | 10 | 5 | 6 | 8 | 8 | 8 | 8 | 8 | 8 | 2 |
| Coco | 6 | 10 | 4 | 6 | 8 | 8 | 8 | 8 | 7 | 8 | 2 |
| Cousine | 10 | 10 | 3 | 6 | 8 | 8 | 8 | 8 | 8 | 8 | 2 |
| Cousin | 8 | 8 | 2 | 6 | 8 | 8 | 8 | 8 | 8 | 8 | 2 |
| Frégate | 7 | 9 | 3 | 6 | 8 | 8 | 8 | 8 | 8 | 8 | 2 |
| Silhouette | 10 | 7 | 4 | 6 | 9 | 9 | 8 | 8 | 8 | 8 | 2 |
| Denis | 8 | 9 | 3 | 6 | 8 | 9 | 8 | 8 | 8 | 8 | 2 |
| Total | 88 | 92 | 34 | 60 | 82 | 83 | 81 | 81 | 77 | 80 | 20 |
We extracted DNA from single flies using the DNAeasy tissue kit (QIAGEN). The sequences of all the primers used and detailed PCR conditions for each marker are available upon request. PCR products were purified with a QIAquick PCR purification kit (QIAGEN). In case of multiple amplifications, the fragment of expected length was isolated and purified using a Millipore DNA gel extraction column. All PCR products were directly sequenced in both directions. Particular attention was given to avoid mis-scoring singleton heterozygotes in sequence analysis. Hence heterozygotes and all the cecropin fragments were cloned using the Dual Promoter TA cloning kit with pCR II vector (Invitrogen). Several clones were subsequently sequenced to recover the different alleles and to assess true single nucleotide polymorphism. Because cloning is time consuming, we limited the number of flies (half) for autosomal loci, which, however, resulted in the same number of sequences as for X-linked loci. All of the sequencing was carried out using an ABI-3130-30 sequencer.
Population structure analyses:
Genetic differentiation between pairs of populations was evaluated by pairwise fixation index FST using Arlequin v.3.01 (Schneider et al. 2000). Significance of FST values was ascertained by 10,000 random permutations, and P-values were adjusted with the Bonferroni correction.
Neighbor-joining trees were performed for each marker with Mega v.3.1 (Kumar et al. 2004) with the pairwise deletion option and the Tamura 3-parameters distance. For each tree, 1000 bootstrap resampling was done.
To test for clustering of individuals, we used Structure v.2.2 (Pritchard et al. 2000). The autosomal loci were excluded because we were not able to determine their gametic phases easily and because only half of the individuals were sequenced, resulting in a number of missing data. We tested for linkage disequilibrium (LD) among and within the seven X-linked loci using Fisher exact tests (data not shown). Because no significant LD was detected among loci, they were considered independent and their sequences were concatenated. Because only 1.5% of the total pairwise comparisons reveal significant intralocus LD we decided not to account for linkage following the guidelines of Structure user manual v.2.2 (Pritchard et al. 2007).
We used the admixture model that allows mixed ancestries of individuals, and the correlated allele frequency model, given that this model is the most appropriate for closely related populations. We performed 5 independent runs for each value of K, the number of clusters, ranging from 1 to 10 with 9.105 iterations and a burn-in period of 50,000. To detect the number of populations that best fit our data we first looked at the log probabilities [Pr(X|K)] and associated variances for each K. Second, we used the method of Evanno et al. (2005). Briefly, this method estimates ΔK, the rate of change in the log probability of data between successive K and the corresponding variance of log probabilities.
DNA polymorphism and statistical tests of neutrality:
Sequences were aligned using BioEdit, v.7.0.5 (Hall 1999). We used DNAsp v.4 (Rozas et al. 2003) to estimate standard population genetic diversity parameters: the number of polymorphic sites S, the number of mutations in external branches (singletons), the number of insertion–deletion sites (indel), the number of haplotypes K, the haplotypic diversity Hd, the average number of pairwise differences π (Tajima 1983), and the Watterson estimator θw (Watterson 1975) (π and θw are estimators of the population mutation parameter θ). We also calculated the last two parameters on synonymous, nonsynonymous, and silent sites. In the standard neutral model θ = 4Neμ for autosomal loci, θ = 3Neμ for X-linked loci and θ = Neμ for mitochondrial loci, where Ne is the effective population size and μ the neutral mutation rate. To test for the neutral equilibrium model, we calculated Tajima's D (Tajima 1989) called thereafter Dt, Fu and Li's F (Fu and Li 1993), and Fay and Wu's H (Fay and Wu 2000) statistics. For H, substitutions were polarized using sequences from D. simulans (whole genome data, NCBI). We also tested by coalescent simulations the confidence interval of Hd with 1000 replicates, S being fixed (fixing θ produced similar results). Finally, we estimated the population recombination parameter, C (Hudson 1987), which estimates 4Nc and 3Nc, respectively, for autosomal and X-linked loci, where c is the rate of recombination per generation per base pair. All of the analyses described above excluded insertion/deletion polymorphisms.
Demographic inferences using ABC methods:
To investigate further if bottlenecks associated with the geographical history of the Seychelles or with a possible man-linked introduction of Morinda have to be considered in a comprehensive evolutionary history of D. sechellia (see Introduction), we used an approximate Bayesian computation approach. The bases of the ABC method are described in Beaumont et al. (2002). Briefly, it compares summary statistics at one locus computed on the observed data (summary statistics) to those computed at the same locus on simulated data. To account for differences in effective population size between autosomal loci (4N) and X-linked loci (3N), the observed summary statistics were multiplied by 4/3 for X-linked loci. Simulated data were obtained for each locus from coalescent simulations that include various demographic scenarios. The likelihood of a demographic scenario depends on the difference between the summary statistics computed from the simulated data in this scenario and the summary statistics computed on the observed data (weighted for X-linked loci as described above). The closer these estimates are, the higher the likelihood of a given demographic scenario. We followed the protocol described by Excoffier et al. (2005) (Figure 2). Coalescent simulations were performed independently for 10 loci using ms (Hudson 2002). Input parameters (locus length, sample size) for the coalescent simulations were estimated by considering the whole data set at each locus, given that our phylogeographical analyses find no clear and consistent evidence of subdivision (see results).
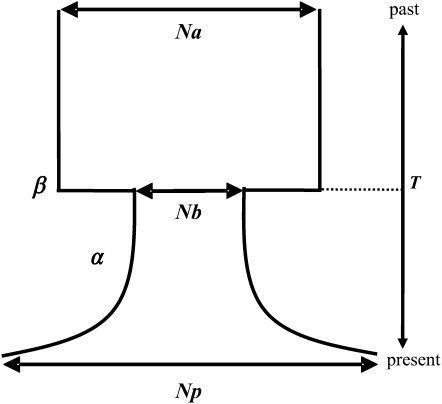
Figure 2.—
Bottleneck tested in the simulations. Na, Nb, and Np, respectively, are the ancestral, bottleneck, and present effective population sizes; T is the time of the bottleneck in unit of 4Np generations; β measures the ratio of Na over Nb and therefore the reduction of the effective population size during the bottleneck; and α is the exponential growth rate.
We simulated bottlenecks as shown in Figure 2. At time T = t/4Np, where t is the time to the bottleneck in generations, and Np is the present effective population size (population sizes were weighted for X-linked loci), the ancestral effective population size (Na) is reduced to Nb (bottle neck population size) with β = Na/Nb. Each bottleneck is followed by an exponential growth of rate, α = (−1/T)log(Nb/Np) until present. On the basis of ecological observations during our field studies, the generation time was set to 20 generations per year. We simulated two types of bottlenecks, one occurring 300 years ago and corresponding to a putative introduction of M. citrifolia on the Seychelles, and the other occurring during the last marine transgression which ended 10,000 years ago. Within each type of bottleneck, a wide grid of scenarios was tested. Na ranged between 50 and 4 × 106, Nb between 50 and 2 × 105 and Np between 50 and 2 × 105. The mutation rate per site was set to 1.5 × 10−9 following the estimation on D. simulans (Wall et al. 2002). This resulted in the following prior distributions for the demographic parameters: θ uniform[0.018;2.4], β uniform[0.00025;800], α300 uniform[−492;1106] for bottlenecks occurring 300 years ago and α10,000 uniform[−15;34] for bottlenecks occurring 10,000 years ago. Because reliable estimates of the population recombination rate are difficult to obtain and may vary from one locus to another, we chose to consider it as a nuisance parameter drawn from a uniform distribution ranging from 1 to 10. Within each bottleneck type, 1 million simulations corresponding to independent drawing of parameter values in the prior distributions were performed. Note that scenarios explored include the null hypothesis of a constant population size, where Na = Nb = Np (α = 0 and β = 1). We defined strong bottlenecks as those resulting in a reduction from Na to Nb at least equal to 10. We also tested for both exponential growth (α > 0) or decline (α < 0) of population size after the bottleneck.
We computed five summary statistics on our simulated scenarios for each of the 10 loci: S, K, π, H, and Dt. We then used the abcEst2 program (Excoffier, http://cmpg.unibe.ch/people/Excoffier-perso.htm), which computes Euclidian distances between the values of the observed summary statistics and the values of the simulated summary statistics. Parameters (α, β, and θ) were estimated on the 1000 best scenarios by weighted linear regression.
RESULTS
Population structure analyses:
We computed pairwise FST values between the 10 populations for the 10 loci. Of these 450 pairwise comparisons (supporting information, Table S1), 19 were significant after Bonferroni corrections (6 in pgd and 13 in sqh; Table 2). Significant values involve two of the largest islands for sqh, Mahé (North Point + Anse Royale) and Silhouette and are more difficult to interpret for pgd. Although significant values before Bonferroni corrections often concern the North Point population, no clear pattern emerges from the FST analysis. In addition, a lack of resolution from neighbor-joining trees was obtained both when considering concatenated sequences or individual genes (data not shown).
TABLE 2.
Pairwise FST measures of population differentiation for pgd (above diagonal) and sqh (below diagonal) after Bonferroni correction
| North Point | Anse Royale | Praslin | Aride | Denis | Frégate | Coco | Cousine | Cousin | Silhouette | |
|---|---|---|---|---|---|---|---|---|---|---|
| North Point | — | 0.00 | 0.49 | 0.67 | 0.45 | 0.42 | 0.43 | 0.48 | 0.55 | 0.45 |
| Anse Royale | 0.38 | — | 0.57 | 0.71 | 0.5 | 0.48 | 0.49 | 0.46 | 0.61 | 0.39 |
| Praslin | 0.51 | 0.15 | — | 0.06 | 0.003 | 0.00 | 0.00 | 0.42 | 0.00 | 0.59 |
| Aride | 0.81 | 0.36 | 0.09 | — | 0.06 | 0.21 | 0.15 | 0.49 | 0.00 | 0.68 |
| Denis | 0.74 | 0.23 | 0.07 | 0.00 | — | 0 | 0.00 | 0.14 | 0.00 | 0.37 |
| Frégate | 0.76 | 0.33 | 0.10 | 0.01 | 0.006 | — | 0.00 | 0.27 | 0.02 | 0.42 |
| Coco | 0.57 | 0.39 | 0.20 | 0.40 | 0.37 | 0.39 | — | 0.16 | 0.00 | 0.37 |
| Cousine | 0.55 | 0.24 | 0.02 | 0.14 | 0.12 | 0.14 | 0 | — | 0.38 | 0.02 |
| Cousin | 0.64 | 0.14 | 0.00 | 0.05 | 0.00 | 0.02 | 0.36 | 0.14 | — | 0.57 |
| Silhouette | 0.10 | 0.00 | 0.28 | 0.51 | 0.41 | 0.47 | 0.45 | 0.34 | 0.34 | — |
Significant values are indicated in boldface type.
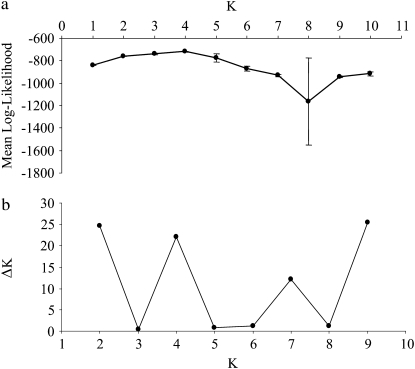
To further test for clustering of individuals, we performed assignment tests using Structure after concatenation of the 7 X-linked loci (Figure 3a). Because several values of K lead to similar likelihood values, we followed the method of Evanno et al. (2005). Values of ΔK as a function of K distinguished similarly K = 2, K = 4, and K = 9 (Figure 3b). For K = 9, the proportion of the sample assigned to each cluster is roughly symmetric, an indication that this partition is meaningless. Table S2 indicates the membership proportion of each predefined population considering K = 2 or K = 4. Six out of the 10 predefined populations had membership coefficients >30% for at least 2 clusters for K = 4. When the number of clusters was set to 2, only 6 out of the 10 predefined populations were clearly assigned to 1 cluster (membership coefficient >80%). Note that in our study, ΔK values were at most ∼25, while usually correct K values are attributed for ΔK values comprised between 50 and 100 (Evanno et al. 2005; Martien et al. 2007). In summary, Structure did not allow to infer precisely a number of clusters, none of these partitions were biologically meaningful and individuals from the same island could have several assignations. These results are consistent with low FST values and poorly resolved NJ trees and suggest that instead of considering an erroneous population structure, it is more parsimonious to consider the species as a whole for subsequent analyzes.
Figure 3.—
Attempt using Structure analyses to identify the number of clusters best fitting the 7 X-linked loci. (a) Mean log-likelihood for K = 1–10 obtained from 5 runs of 9.105 iterations and a burn-in period of 50,000. (b) Values of ΔK as a function of K following Evanno et al. (2005).
Nucleotide diversity and polymorphism pattern in D. sechellia:
We sequenced 446 bp of the NADH gene in 20 individuals (two per population, Table 1). We did not detect any variation indicating that all D. sechellia individuals share the same mtDNA type, namely the se type as described in Solignac (2004).
A total of 15 kb were aligned over the 10 genomic regions, of which 7.8 kb were coding sequences, corresponding to a total of 758 aligned sequences (Table 3). Our sequencing panel included 92 flies with an average of 7.5 individuals per population. Note that because a duplication of the entire region for the cecropin marker was suspected, we retained for analysis only sequences that were unambiguously assigned to the same orthologous region (a total of 34 sequences, Table 1). Three indels were recorded: at the end of the second exon of amyrel, generating a stop codon, in the intergenic region of the cecropin psiII and in the upstream region of sqh. They occurred in several individuals from different islands.
TABLE 3.
Summary statistics of the 10 loci for all the samples
| Locus | Location | n | L bp | indel | S | ηe | K | πtotal | θtotal | πs | θs | πns | θns | πsilent | θsilent | Dt | F | H | C | Hd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| amyrel | 2R | 88 | 1640 (1482-57-101) | 19 | 12 | 2 | 15 | 1.14 | 1.47 | 3.3 | 2.5 | 0.6 | 0.9 | 2.1 | 2.4 | −0.59 | −0.07 | 1.45 | 0.002 | 0.7 (0.68–0.93) |
| cecropin | 3R | 34 | 1522 (353-61-1108) | 1 | 5 | 2 | 6 | 0.66 | 0.96 | 1.4 | 3 | 0.2 | 0.9 | 0.8 | 1 | −0.86 | −0.83 | 0.67 | ND | 0.72 (0.06–0.84) |
| janus-ocnus | 3R | 60 | 685 (240-62-383) | 0 | 1 | 0 | 2 | 0.14 | 0.31 | 0 | 0 | 0.5 | 1.1 | 0 | 0 | −0.70 | 0.19 | 0.09 | ND | 0.1 |
| Average automal | 2R–3R | 0.65 | 0.91 | 1.6 | 1.8 | 0.4 | 1.0 | 1.0 | 1.1 | −0.72 | −0.24 | 0.74 | ||||||||
| period | X | 92 | 757 (499-258-0) | 0 | 7 | 1 | 10 | 1.12 | 1.82 | 3.1 | 4.3 | 0.6 | 0.5 | 2.1 | 3 | −0.92 | −0.11 | 0.78 | 0.048 | 0.59 (0.33–0.86) |
| sqh | X | 82 | 1833 (525-64-1244) | 10 | 8 | 1 | 11 | 0.91 | 0.88 | 3.9 | 1.8 | 0 | 0 | 1.2 | 1.1 | 0.08 | 0.42 | 1.23 | 0.006 | 0.75 (0.31–0.83) |
| vermilion | X | 83 | 1498 (1053-355-90) | 0 | 3 | 0 | 4 | 0.22 | 0.4 | 1. | 1.6 | 0 | 0 | 0.5 | 0.9 | −0.83 | 0.38 | 0.30 | ND | 0.32 (0.09–0.69) |
| white | X | 81 | 1216 (889-327-0) | 0 | 6 | 0 | 7 | 1.18 | 0.99 | 1.9 | 0.9 | 0.5 | 0.6 | 2 | 1.5 | 0.44 | 1.07 | 1.07 | ND | 0.82 (0.25–0.79) |
| otu | X | 81 | 1388 (1047-341-0) | 0 | 4 | 0 | 6 | 0.68 | 0.58 | 2.2 | 2.5 | 0.5 | 0.25 | 0.9 | 1 | 0.34 | 0.89 | 0.58 | 0.005 | 0.52 (0.14–0.73) |
| pgd | X | 77 | 2191 (568-1498-125) | 0 | 7 | 2 | 10 | 0.82 | 0.65 | 6.3 | 4.5 | 0 | 0 | 1 | 0.8 | 0.65 | −0.11 | 0.69 | 0.019 | 0.77 (0.33–0.81) |
| zeste | X | 80 | 2154 (1164-182-808) | 0 | 3 | 1 | 4 | 0.15 | 0.28 | 0 | 0 | 0 | 0 | 0.3 | 0.5 | −0.88 | −0.75 | 0.29 | ND | 0.31 (0.07–0.69) |
| Average X linked | X | 0.73 | 0.80 | 2.7 | 2.2 | 0.2 | 0.2 | 1.1 | 1.3 | −0.16 | 0.25 | 0.71 | ||||||||
| All average | 2R–3R–X | 0.70 | 0.83 | 2.3 | 2.1 | 0.3 | 0.4 | 1.1 | 1.2 | −0.33 | 0.11 | 0.72 |
n, number of sequences; L, total length of the sequences in base pair (length of exon, intron, and intergenic sequences); S, number of polymorphic sites; ηe, number of unique mutation (singletons); K, number of haplotypes; π (× 103), nucleotidic diversity; θ (× 103), Watterson estimator (total, s, ns, and silent refer to all, synonymous, nonsynonymous, and silent sites); Dt, Tajima's D statistic; F, Fu and Li's F statistic; H, Fay and Wu's statistic; C, estimate of the population recombination rate per base pair (4Nc for autosomal loci; 3Nc for X-linked loci; ND, not determined); Hd, haplotypic diversity (95% confidence interval).
We identified a total of 56 segregating sites of which 9 were singletons (Table 3). Due to the low number of polymorphic sites in some genes, we could not estimate the population recombination rate, C, for all of them (Table 3). Neutrality tests failed to reject the neutral equilibrium model using various statistics (Dt, F, and H). For example, the Dt values are all close to zero (average Dt among 10 loci = −0.33; Table 3), 4 are positive and 6 negative. We compared the average Dt value and its variance (0.39) among 10 loci to a simulated distribution using the program of J. Hey (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA) and found no significant departure from neutral expectation (P-value of 0.19 and 0.92 for the mean and variance, respectively). The H values are all positive and most are close to zero (average H = 0.72). However, some results should be viewed with caution because of the low level of variability in some markers (see for example janus-ocnus).
The overall nucleotidic diversity of D. sechellia as estimated by π is on average 0.00065 for autosomal loci and 0.00073 for X-linked loci. Corresponding silent diversity is 0.001 and 0.0011, respectively, for autosomal and X-linked loci. Diversity estimates provided by θ are similar (Table 3). Synonymous diversity (on average 0.0018 as estimated by θ and 0.0016 as estimated by π) is 1.8 to 4 times higher than nonsynonymous diversity (on average 0.001 and 0.0004 for θ and π, respectively) for autosomal loci and 11 to 13.5 times higher for X-linked loci (Table 3). Interestingly, X-linked loci exhibit both a depleted level of nonsynonymous diversity and an elevated level of synonymous diversity as compared with autosomal loci. Under the neutral model, the level of variation in autosomes is expected to be higher than in X-linked loci simply because of differences of effective population size between the two classes of loci. Note that contrasting patterns have been found among related species (Andolfatto 2001). While our results raise an interesting issue, they remain difficult to interpret in regard with the restricted number of autosomal loci sampled and the large heterogeneity in diversity estimates they exhibit (θ and π estimated at synonymous sites range from 0.000 to 0.0033 and from 0.000 to 0.003, respectively). Assuming a neutral mutation rate ranging from 10−8 to 1.5 × 10−9, the effective population size estimated from silent diversity would be ∼100,000 individuals and no more than 200,000. These are low values for a Drosophila species and corroborate our field observations of the scarce distribution of D. sechellia in some islets.
Considering sequence variation in D. sechellia for all the markers, the most striking feature was that almost all variants were present in individuals from several islands. For example, one of the amyrel variants occurred in 24 sequences from the eight islands of the Seychelles sampled (data not shown). The number of haplotypes and the haplotypic diversity for each marker do not differ from neutral expectations, suggesting that linkage disequilibrium is weak.
Demographic inferences:
Although classical analyzes of molecular evolution do not support deviation from neutral equilibrium, the effect of bottlenecks on polymorphism patterns can be complex. For instance, very recent bottlenecks generally result in a skew toward high-frequency-derived variants and a positive Tajima's D, whereas for older bottlenecks, Tajima's D will become more negative and Fay and Wu's H more positive, reflecting the occurrence of new mutations. We were interested on further investigating the demographic history of the species searching for scenarios compatible with the observed patterns of variation.
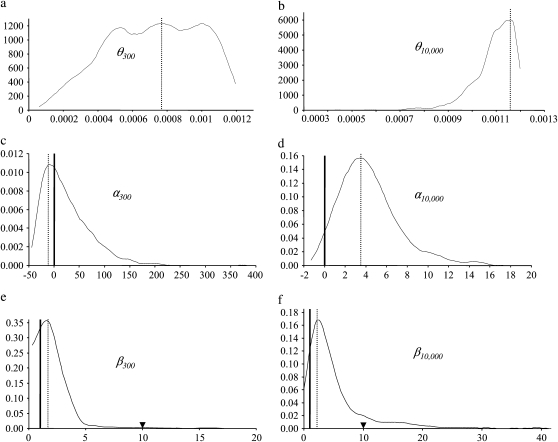
We performed coalescent simulations to test the influence of two types of bottleneck: a recent bottleneck occurring 300 years ago, mimicking a reduction of population size caused by a host shift at the time of the introduction of M. citrifolia and an old bottleneck occurring 10,000 years ago, corresponding to a reduction of population size caused by the drastic shrinkage of the Seychelles territory at the time of the sea level rise. For both bottleneck types, we performed 1 million simulations covering a broad range of scenarios. Euclidian distances between simulated values and observed values for five summary statistics obtained for the 1000 best scenarios were estimated. Overall, distances were lower for the scenarios occurring 300 years ago than for those occurring 10,000 years ago (data not shown). Posterior density curves of the three demographic parameters θ, α, and β corresponding to the 1000 best scenarios are presented in Figure 4. Values of α = 0 and β = 1 corresponding to the null hypothesis are indicated as well as the threshold that we defined to designate strong bottlenecks, i.e., β > 10. The modes, means, medians, and 95% confidence intervals for each distribution are summarized in Table 4.
Figure 4.—
Posterior density curves of demographic parameters (θ, α, and β) for two types of bottleneck occurring 300 or 10,000 years ago. A dotted vertical line indicates the mode of each distribution. Values for the null hypothesis corresponding to α = 0 and to β = 1 are indicated by a bold vertical line. Black arrows on Figure 4, e and f correspond to severe bottleneck threshold (β > 10).
TABLE 4.
Statistics on the posterior distributions of the demographic parameters
| Parameter | Mode | Mean | Median | 95% C.I. | |
|---|---|---|---|---|---|
| α | −10.02 | 24.18 | 13.68 | −44.33–222.51 | |
| t = 300 YA | β | 1.71 | 2.38 | 1.75 | 0.38–4.7 |
| θ × 103 | 0.00077 | 0.00072 | 0.00073 | 0.00021–0.00119 | |
| α | 3.55 | 4.33 | 3.9 | −0.87–10.33 | |
| t = 10,000 YA | β | 2.2 | 5.29 | 3.62 | 0.029–16.75 |
| θ × 103 | 0.00116 | 0.00109 | 0.0011 | 0.00093–0.0012 |
The five summary statistics (S, K, π, H, and Dt) were computed from 1 million simulations at both times (bottleneck occurring 300 or 10,000 years ago) and then compared to the observed data. The 1000 scenarios that best fit the data were kept to estimate the posterior distributions.
The posterior density curve of θ for a bottleneck occurring 300 years ago rapidly reaches a plateau and steeply decreases for values >0.001, while for a bottleneck occurring 10,000 years ago, the best estimate obtained is 0.00116. Several observations can also be made regarding the demographic scenarios: first, confidence intervals for α and β include the values expected under the null hypothesis of α = 0 and β = 1 (Table 4). Second, most scenarios fall far below the threshold of strong bottlenecks (Figure 4, e–f). Third, the modes are consistent with a null-to-mild exponential growth (α300 = −10.02 and α10,000 = 3.55) and a weak decrease in population size during the bottleneck phase (β300 = 1.71 and β10,000 = 2.2). Taken together, results from our simulations tend to exclude severe bottlenecks as the cause of the low diversity of D. sechellia, and scenarios close to the null hypothesis of a constant population size receive the strongest support.
DISCUSSION
The present study provides a clear and realistic evaluation of the genetic diversity of D. sechellia at a species-wide level. On the basis of a unique data set generated from a collection of 92 individuals from nine islands, we highlighted the patterns of polymorphism observed throughout the whole species range. Previous studies showed that almost all nucleotide variations described were singletons simply because they were based on a limited number of strains (Kliman and Hey 1993; Kliman et al. 2000). Our work somewhat challenges the pessimistic view of McBride (2007) concerning the use of population genetic tools in D. sechellia. The pattern of frequency spectrum we observed suggests a long common history of the species as a whole. Hence, no obvious population structure was evidenced either between Silhouette and the rest of the former Seychelles Bank, which were isolated before its submersion ∼10,000 years ago or among the different islands of the Seychelles archipelago. In addition, our results bring new insights into D. sechellia demographic history and suggest the long-term persistence of a small effective population size.
A very low current effective population size:
The overall nuclear diversity of silent sites is on average ∼0.001 for D. sechellia. The species is 10 to 20 times less polymorphic than any other species of the melanogaster subgroup (D. orena excluded as only known from a unique isofemale line) (Kliman et al. 2000; Lachaise et al. 2000; Lachaise and Silvain 2004; Stephan and Li 2007; M.-L. Cariou, unpublished data). Such low genetic variation suggests a small population size. Indeed, we estimate the effective population size of D. sechellia to be close to 100,000, much less than the 2–6 millions estimated for its close mainland relative, D. simulans (Sawyer and Hartl 1992; Wall et al. 2002). Among Drosophila, several lineages comprise island endemics or rare species of which only a few have been studied. Of the obscura group, the endemic D. guanche, which is restricted to some isolated gorges of the relictual Laurisilva forest of Tenerife island, showed a nucleotide variation at silent sites twice that of D. sechellia (θ = 0.0022 in the RpII215 region, 6.9 kb) (Perez et al. 2003). The effective population size of the rare mainland species, D. miranda, is estimated to be in the order of 1 million (Yi et al. 2003) and roughly fivefold smaller than that of D. melanogaster (Bachtrog 2008). For both species the difficulty of collecting them in the wild is consistent with a current low population size (A. Gonzales, unpublished data; Yi et al. 2003). Estimates of a million have been given for two species of the virilis group, D. ezoana and D. littoralis, which however have higher levels of polymorphism, 1.2–1.3% (Vieira 2002). D. sechellia appears unique among Drosophila in the sense that it harbors the lowest genetic diversity and the smallest population size so far recorded.
Rates of substitutions at synonymous and nonsynonymous sites in D. sechellia are comparable to what is observed in D. melanogaster (Bierne and Eyre-Walker 2006; Bachtrog 2008). In that way, the pattern of variability in D. sechellia does not seem to agree with Woolfit and Bromham's (2005) conclusion that island lineages have significantly higher ratios of nonsynonymous-to-synonymous substitution rates than mainland lineages. A low effective population size would reduce the strength of purifying selection resulting in an increase in the rate of fixation of slightly deleterious amino acid mutations. Indeed Kliman et al. (2000) reported that genes from D. sechellia have accumulated mutations at a rate that is ∼50% higher than the same genes from D. simulans. They also described a significant excess of unpreferred codon substitutions at synonymous sites. The recent survey of 136 olfactory and gustative receptor genes in D. sechellia has revealed an increase in gene loss due to a high rate of lack-of-function mutations (Mcbride 2007). D. sechellia is also the only species showing deletions that cause genes that are highly conserved among species of the melanogaster subgroup and Drosophila in general to be nonfunctional, like Amyrel (see results; Da Lage et al. 2007) and Amy (Shibata and Yamazaki 1995). All these observations are consistent with a stronger effect of genetic drift in a historically small-sized population of D. sechellia.
The demographic history of D. sechellia:
On the basis of three nuclear genes polymorphism, Hey and Kliman (1993) pointed out that they were not able to distinguish between a scenario involving a recent population bottleneck and a scenario in which the population size of D. sechellia had been restricted since the species' formation. Later, Kliman et al. (2000) used a standard population genetic approach and proposed that D. sechellia has carried a low persistent population size since its origin. These authors, however, did not test alternative scenarios. In the present study, we combined a large population sample and the recent ABC methods to test for demographic scenarios compatible with the biogeographical history of the species and cover a wide range of bottleneck conditions. Our results exclude severe bottlenecks and support the idea that D. sechellia is an island-endemic species that has always carried a low effective population size. Simulation results are consistent with the low number of singletons and the pattern of polymorphism across the species range, as well as the elevated substitution rate described by Kliman et al. (2000) for this species compared to its close relatives, D. simulans (mainland) and D. mauritiana (island). Interestingly, the two island endemics, D. sechellia and D. mauritiana, have contrasted evolutionary histories. D. mauritiana is a generalist species (David et al. 1989), which has a large population size and has probably experienced a recent expansion (Kliman et al. 2000), while D. sechellia is highly specialized with a small population size.
Lachaise and Silvain (2004) proposed that D. sechellia could have shifted recently from a primary host to M. citrifolia, according to their hypothesis of an introduction of Morinda by man that may be as recent as 300 years ago. The present results tend to contradict this hypothesis. First, our fieldwork shows that D. sechellia is found exclusively on Morinda all over its species range and all over the year. Second, our simulations are not consistent with demographic variations due to a recent shift but support an ancient specialization putatively associated with D. sechellia speciation.
How to explain such a low persistent population size?
Because of their small geographical range, island endemics are generally thought to have small effective population size. However, among the melanogaster subgroup, the island endemic species exhibit a large range of genetic diversity, D. sechellia being consistently the least variable for autosomal and X-linked loci. Thus, being confined to a restricted area may not be a factor that fully explains the extremely low polymorphism level found in D. sechellia. In this study, we also excluded demographic factors.
Life-history traits may result in a small effective population size with a loss of genetic variability over a long evolutionary period (Frankham et al. 2002). D. sechellia has a reduced number of ovarioles, lower female egg production, and low reproductive capacity compared to its close relatives D. mauritiana and D. simulans (R'kha et al. 1997). Such features may account for the reduced population size observed and the resulting depleted level of variability in D. sechellia. Moreover, oogenesis is stimulated by Morinda in D. sechellia, but inhibited in its mainland relative D. simulans. Morinda is clearly an oviposition attractant for D. sechellia but a repellent for D. simulans (R'kha et al. 1991; Amlou et al. 1998) indicating the specificity and the importance of this trait. We suggest that the small effective population size of D. sechellia results from a trade-off between life-trait performances and the use of a highly predictable resource (M. citrifolia) at all stages of development, from eggs to adults. Not only does M. citrifolia bear fruit throughout the year, but the fruit is toxic for most, if not all other Drosophila species. M. citrifolia therefore constitutes a unique habitat for D. sechellia, which is resistant to the toxic effects of the fruit (Jones 1998, 2005). We suggest that low population size, the cost of low fecundity, may be balanced by the benefit of being less affected by demographic hazards. Interestingly, Milot et al. (2007) recently showed that albatrosses have not suffered greatly from their long impoverished diversity because of a conjunction of particular life-history traits and population history. We suggest that D. sechellia has evolved a similar survival strategy that may explain its historically low effective population size.
Acknowledgments
The authors thank M.J.L. Loustau-Lalanne, Secrétaire Général à l'Environnement, for giving us permission to carry out Drosophila research in the Seychelles and for encouragement. We thank S. Remie, the Director of Conservation at the Ministry of Environment, for his support and his collaborators for their help. We are grateful to M. A. Cedras, manager, and his team for providing facilities in the Vallée-de-Mai. We thank M. and S. Betts, managers, for giving us access to facilities on Aride Nature Reserve. We thank the Island Development Company for their support on Silhouette and Ron and Gill Gerlach from the Nature Protection Trust of Seychelles for their friendly help. We are grateful to the Marine Park Authority for support and assistance on Curieuse and Coco islands. We thank N. Shah and Nature Seychelles for giving us access to Cousin Island. We thank Masons Travel for the facilities offered on Denis Island and S. Hill and B. Sachse for their help on Fregate Island. We are grateful to E. Robillard for technical assistance. We thank F. Depaulis, F. Austerlitz, and D. Casane for helpful discussions and L. Excoffier for making available a new version of the software. O. Tenaillon greatly helped in the ABC modelling, providing us with C programs and advice on the procedure and interpretation of the simulated data. We thank two reviewers and the editor for thoughtful comments that greatly improved the manuscript. This work was funded by the French Ministry of Environment and Sustainable Development through the program “Invasions Biologiques” (CV 02000216), by the Centre National de la Recherche Scientifique and by the ANR Biodiversity program (ANR-06-BDIV-003-01).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.108.092080/DC1.
References
- Amlou, M., B. Moreteau and J. R. David, 1998. Larval tolerance in the Drosophila melanogaster species complex toward the two toxic acids of the D. sechellia host plant. Hereditas 129 7–14. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., 2001. Contrasting patterns of X–linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18 279–290. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., and P. Andolfatto, 2006. Selection, recombination and demographic history in Drosophila miranda. Genetics 174 2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog, D., 2008. Similar rates of protein adaptation in Drosophila miranda and D. melanogaster, two species with different current effective population sizes. BMC Evol. Biol. 8 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines, J. F., A. Das, S. Mousset and W. Stephan, 2004. The role of natural selection in genetic differentiation of worldwide populations of Drosophila ananassae. Genetics 168 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, E., N. Derome, M. Huet and M. Veuille, 2006. Contrasted polymorphism patterns in a large sample of populations from the evolutionary genetics model Drosophila simulans. Genetics 173 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont, M. A., W. Zhang and D. J. Balding, 2002. Approximate Bayesian computation in population genetics. Genetics 162 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, N, and A. Eyre-Walker, 2006. Variation in synonymous codon use and DNA polymorphism within the Drosophila genome. J. Evol. Biol. 19 1–11. [DOI] [PubMed] [Google Scholar]
- Cariou, M.-L., D. Lachaise, P. Matyot, J. Gerlach, C. Montchamp, D. Legrand and S.F. McEvey 2009. Drosophilidae of Seychelles, pp. 355–385 in The Diptera of the Seychelles Islands, edited by J. Gerlach. Pensoft, Sofia, Bulgaria/Moscow.
- Cariou, M. L., M. Solignac, M. Monnerot and J. R. David, 1990. Low allozyme and mtDNA variability in the island endemic species Drosophila sechellia (D. melanogaster complex). Experientia 46 101–104. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1998. The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353 287–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Lage, J. L., G. Kergoat, F. Maczkowiak, J.-F. Silvain, M.-L. Cariou and D. Lachaise, 2007. A phylogeny of Drosophilidae using the Amyrel gene: questioning the Drosophila melanogaster species group boundaries. J. Zool. Syst. Evol. Res. 45 47–63. [Google Scholar]
- David, J. R., S. F. McEvey, M. Solignac and L. Tsacas, 1989. Drosophila communities on Mauritius and the ecological niche of D. mauritiana (Diptera, Drosophilidae). Revue Zool afr-J. Afr. Zool. 103 107–116. [Google Scholar]
- Dekker, T., I. Ibba, K. P. Siju, M. C. Stensmyr and B. S. Hansson, 2006. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr. Biol. 16 101–109. [DOI] [PubMed] [Google Scholar]
- Dumont, V. B., and C. F. Aquadro, 2005. Multiple signatures of positive selection downstream of notch on the X chromosome in Drosophila melanogaster. Genetics 171 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno, G., G. Regnaut and J. Goudet, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., A. Estoup and J. M. Cornuet, 2005. Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics 169 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine, J.-P., L. Legal, B. Moreteau and J.-L. Le Quere, 1996. Volatile components of ripe fruits of Morinda citrifolia and their effects on Drosophila. Phytochemistry 41 433–438. [Google Scholar]
- Frankham, R., J. D. Ballou and D. A. Briscoe, 2002. Introduction to Conservation Genetics. Cambridge University Press, Cambridge, UK.
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, J. M., J. M. Jallon, J. D. Rouault and M. G. Ritchie, 2005. Quantitative trait loci for cuticular hydrocarbons associated with sexual isolation between Drosophila simulans and D. sechellia. Genetics 171 1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty, W., and R. S. Singh, 2006. Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol. Biol. Evol. 23 1707–1714. [DOI] [PubMed] [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41 95–98. [Google Scholar]
- Heuertz, M., E. De Paoli, T. Kallman, H. Larsson, I. Jurman et al., 2006. Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce. [Picea abies (L.) Karst] Genetics 174 2095–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and R. M. Kliman, 1993. Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Mol. Biol. Evol. 10 804–822. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50 245–250. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Jones, C. D., 1998. The genetic basis of Drosophila sechellia's resistance to a host plant toxin. Genetics 149 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C. D., 2005. The genetics of adaptation in Drosophila sechellia. Genetica 123 137–145. [DOI] [PubMed] [Google Scholar]
- Kliman, R. M., P. Andolfatto, J. A. Coyne, F. Depaulis, M. Kreitman et al., 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 1993. DNA sequence variation at the period locus within and among species of the Drosophila melanogaster complex. Genetics 133 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lachaise, D., P. Capy, M.-L. Cariou, D. Joly, F. Lemeunier et al., 2004. Nine relatives from one African ancestor: the population biology of the Drosophila melanogaster subgroup species, pp. 315–343 in The Evolution of Population Biology, edited by R. Singh and M. Uyenoyama. Cambridge University Press, Cambridge, UK.
- Lachaise, D., M. Harry, M. Solignac, F. Lemeunier, V. Benassi et al., 2000. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from Sao Tomé Island. Proc. Roy. Soc. Lond. 267 1487–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., and J. F. Silvain, 2004. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120 17–39. [DOI] [PubMed] [Google Scholar]
- Li, H., and W. Stephan, 2006. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2 e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, C. S., 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc. Natl. Acad. Sci. USA 104 4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott, S., and R. Kliman, 2008. Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus DNA sequence data. PLoS One 3 e2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martien, K. K., G. H. Givens, and E. Archer 2007. A note on the ability of STRUCTURE to correctly infer the number of populations for Bering-Chukchi-Beaufort Seas bowhead whales. International Whaling Commission. Paper SC/59/BRG34
- Matsuo, T., S. Sugaya, J. Yasukawa, T. Aigaki and Y. Fuyama, 2007. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milot, E., H. Weimerskirch, P. Duchesne and L. Bernatchez, 2007. Surviving with low genetic diversity: the case of albatrosses. Proc. Biol. Sci. 274 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, R. A., M. Choudhary, M. L. Cariou and R. S. Singh, 2004. A reanalysis of protein polymorphism in Drosophila melanogaster, D. simulans, D. sechellia and D. mauritiana: effects of population size and selection. Genetica 120 101–114. [DOI] [PubMed] [Google Scholar]
- Orengo, D. J., and M. Aguade, 2004. Detecting the footprint of positive selection in a european population of Drosophila melanogaster: multilocus pattern of variation and distance to coding regions. Genetics 167 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, J. A., A. Munte, J. Rozas, C. Segarra and M. Aguade, 2003. Nucleotide polymorphism in the RpII215 gene region of the insular species Drosophila guanche: reduced efficacy of weak selection on synonymous variation. Mol. Biol. Evol. 20 1867–1875. [DOI] [PubMed] [Google Scholar]
- Pool, J. E., V. Bauer Dumont, J. L. Mueller and C. F. Aquadro, 2006. A scan of molecular variation leads to the narrow localization of a selective sweep affecting both Afrotropical and cosmopolitan populations of Drosophila melanogaster. Genetics 172 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., X. Wen and D. Falush, 2007. Documentation for Structure software: version2.2.http://pritch.bsd.uchicago.edu/software/structure22/readme.pdf.
- R'kha, S., P. Capy and J. R. David, 1991. Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc. Natl. Acad. Sci. USA 88 1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R'kha, S., B. Moreteau, J. A. Coyne and J. R. David, 1997. Evolution of a lesser fitness trait: egg production in the specialist Drosophila sechellia. Genet. Res. 69 17–23. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-Delbarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Sawamura, K., T. Taira and T. K. Watanabe, 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. I. The maternal hybrid rescue (mhr) gene of Drosophila simulans. Genetics 133 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S. A., and D. L. Hartl, 1992. Population genetics of polymorphism and divergence. Genetics 132 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, B. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genomewide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000. ARLEQUIN, version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
- Shibata, H., and T. Yamazaki, 1995. Molecular evolution of the duplicated Amy locus in the Drosophila melanogaster species subgroup: concerted evolution only in the coding region and an excess of nonsynonymous substitutions in speciation. Genetics 141 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac, M., 2004. Mitochondrial DNA in the Drosophila melanogaster complex. Genetica 120 41–50. [DOI] [PubMed] [Google Scholar]
- Stephan, W., and H. Li, 2007. The recent demographic and adaptive history of Drosophila melanogaster. Heredity 98 65–68. [DOI] [PubMed] [Google Scholar]
- Stensmyr, M. C., T. Dekker and B. S. Hansson, 2003. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc. Biol. Sci. 270 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., J. U'ren, O. Tenaillon and B. S. Gaut, 2004. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21 1214–1225. [DOI] [PubMed] [Google Scholar]
- Tsacas, L., and G. Bächli, 1981. Drosophila sechellia, n.sp., huitième espèce du sous-groupe melanogaster des Iles Séchelles. [Diptera, Drosophilidae] Revue Fr. Ent. NS 3 146–150. [Google Scholar]
- Vieira, J., 2002. Two divergent species of the virilis group, Drosophila littoralis and Drosophila virilis, share a replacement polymorphism at the fused locus. Mol. Biol. Evol. 19 579–581. [DOI] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Woolfit, M., and L. Bromham, 2005. Population size and molecular evolution on islands. Proc. Biol. Sci. 272 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, S., D. Bachtrog and B. Charlesworth, 2003. A survey of chromosomal and nucleotide sequence variation in Drosophila miranda. Genetics 164 1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]