Abstract
Gene expression is a complex trait determined by various genetic and nongenetic factors. Among the genetic factors, allelic difference may play a critical role in gene regulation. In this study we globally dissected cis (allelic) and trans sources of genetic variation in F1 hybrids between two Arabidopsis thaliana wild accessions, Columbia (Col) and Vancouver (Van), using a new high-density SNP-tiling array. This array tiles the whole genome with 35-bp resolution and interrogates 250,000 SNPs identified from resequencing of 20 diverse A. thaliana strains. Quantitative assessment of 12,311 genes identified 3811 genes differentially expressed between parents, 1665 genes with allele-specific expression, and 1688 genes controlled by composite trans-regulatory variation. Loci with cis- or trans-regulatory variation were mapped onto sequence polymorphisms, epigenetic modifications, and transcriptional specificity. Genes regulated in cis tend to be located in polymorphic chromosomal regions, are preferentially associated with repressive epigenetic marks, and exhibit high tissue expression specificity. Genes that vary due to trans regulation reside in relatively conserved chromosome regions, show activating epigenetic marks and generally constitutive gene expression. Our findings demonstrate a method of global functional characterization of allele-specific expression and highlight that chromatin structure is intertwined with evolution of cis- and trans-regulatory variation.
GENETIC variation leads to phenotypic variation through changes in gene expression (King and Wilson 1975). At the same time differential gene expression represents a molecular profile of phenotypic differentiation. Sequence polymorphisms within gene regulatory elements can affect transcription rate or transcript stability of the associated allele, while trans-genetic polymorphisms cause variation in transcript abundance of both target alleles. Dissection of local (potentially cis) and distant (trans) sources of variation can be accomplished by the mapping of expression quantitative trait loci (eQTL) in a segregating population (Brem et al. 2002; Schadt et al. 2003; Yvert et al. 2003; Morley et al. 2004; Ronald et al. 2005b; Keurentjes et al. 2007; West et al. 2007). Using this approach, the effects of causal genetic variations can be tested individually, and their additive effects or nonadditive interactions could be assessed in that mapping population (Brem and Kruglyak 2005; Brem et al. 2005). In eQTL mapping studies the characterization of cis- and trans-genetic variation depends on positional cutoff and an unambiguous separation of true cis and trans effect is impossible (Ronald et al. 2005b). An alternative approach detects cis-regulatory variation by direct testing of allele-specific expression (ASE) in a heterozygous system (Cowles et al. 2002; Yan et al. 2002; Lo et al. 2003; Wittkopp et al. 2004; de Meaux et al. 2006; Kiekens et al. 2006; Springer and Stupar 2007; Guo et al. 2008). The existence of trans-genetic variation is revealed by testing for a deviation between hybrid allelic expression and a mix of the parental expression profiles (Wittkopp et al. 2004; Springer and Stupar 2007; Guo et al. 2008). Since parental genotypes vary for both cis and trans effect, comparison of parental expression variation with the hybrid cis effect highlights loci regulated by “composite” trans difference, e.g., multiple trans eQTL, developmental and environmental dependency, and cis × trans interaction. In this design we are not mapping the controlling trans loci but only categorizing targets controlled in trans.
In early studies, ASE was detected by single-base extension of a primer adjacent to the variant single nucleotide polymorphism (SNP) (Cowles et al. 2002; Wittkopp et al. 2004; Carrel and Willard 2005). Several recent studies applied a variety of technologies to scale up the tested genes (Jeong et al. 2007; Bjornsson et al. 2008; Guo et al. 2008; Serre et al. 2008). The use of a microarray-based genomics approach to globally test ASE takes advantage of transcribed SNPs or single-feature polymorphisms (SFPs) (Ronald et al. 2005a). In the present study we used a very high-density SNP-tiling array, which interrogates 250,000 SNPs identified from resequencing of 20 diverse Arabidopsis thaliana accessions (Clark et al. 2007). Quantitative assessment of allelic variation in RNA samples was achieved by comparison with genomic DNA 1:1 mixture. The DNA signal provides an empirical heterozygous genotype to account for probe hybridization effect. It thus serves as the reference for equal allelic expression. This simple approach allows a true high-throughput identification of ASE in RNA samples. This study presents the first genome scale dissection of composite cis- and trans-regulatory effects in a complex eukaryotic organism. By examining the genome patterns of genes controlled in cis or by trans, our study suggested that chromatin structure may have profound effects on the evolution of cis- and trans-regulatory variation.
MATERIALS AND METHODS
Plant materials:
Seeds of A. thaliana accessions Col-0 (accession no. CS22625) and Van-0 (accession no. CS22627) were obtained from Arabidopsis Biological Resource Center and propagated for one generation. Seeds were stratified at 4° for 8 days in water containing 10 mg/liter gibberellic acid-3 (Sigma). Seeds were then planted in soil and grown in a greenhouse with 16 hr light (cool white light supplemented with incandescent) and 8 hr dark at 20°. After growing for 28 days, plants were crossed using the main stem flower buds. Four replicate crosses for each of Col × Col, Van × Van, Van (♀) × Col (♂), and Col (♀) × Van (♂) were generated by pairing different paternal and maternal plants (16 Col and 16 Van plants in total). The cross experiment was repeated on the same pair of parents. For each replicate cross, the seeds from the two experiments were combined and used as one maternal seed batch. About 250 seeds from each maternal seed batch were grown on a single Petri dish. After gas sterilization for 4 hr, seeds were plated on a total of 16, 0.7% agar (Sigma) plates supplemented with 0.5× Murashige and Skoog salts (Sigma). Seed plates were placed horizontally in a growth chamber (Percival Scientific, model E361) after stratification at 4° for 5 days. Seedlings were grown for 7 days under a diurnal mode with 12 hr light (cool white light supplemental with red light) and 12 hr dark at 20°.
Sample preparation and microarray hybridization:
Seedlings grown on each plate were split for genomic DNA and RNA preparation. Genomic DNA was isolated from 100 seedlings per plate using DNeasy plant mini kit (QIAGEN). DNA concentration was measured by NanoDrop (Thermo Scientific). For genomic DNA 1:1 mixtures, the four Col and four Van genomic DNA samples were randomly paired without replacement. For each pairing, 100 ng Col gDNA and 100 ng Van gDNA were mixed and labeled using BioPrime DNA labeling system (Invitrogen) with conditions modified as previously described (Borevitz et al. 2003). For the genomic DNA mixture series, one Col and one Van genomic DNA sample were selected and mixed at 1:0, 5:1, 3:1, 2:1, 1:1, 1:2, 1:3, 1:5, and 0:1. A total of 200 ng from each of the mixtures were then labeled using the BioPrime labeling system (Invitrogen). Total RNA was isolated from an additional 120 seedlings per plate using RNeasy plant mini kit (QIAGEN). For RNA 1:1 mixtures, the four Col and four Van RNA samples were randomly paired without replacement. A total of 40 μg from each of the parental RNA mixtures and F1 hybrid RNA samples were enriched for poly-(A) RNA using Oligotex mRNA mini kit (QIAGEN). For each sample, 11 μl out of 20 μl poly-(A) RNA were mixed with 166 ng random hexamer (Invitrogen) and subjected to first-strand cDNA synthesis (Invitrogen) in a total volume of 20 μl at 42° for 1 hr. The 20-μl first-strand reaction was used in second-strand cDNA synthesis (Invitrogen) in a total volume of 150 μl at 16° for 2 hr. Samples were then subjected to RNase treatment at 37° for 20 min with 20 units RNaseH (Epicentre), 1 unit RnaseA, and 40 units RNaseT (Ambion). Double-stranded cDNA was cleaned using Qiaquick PCR purification kit (QIAGEN) and labeled using the BioPrime labeling system (Invitrogen). About 16 μg of labeled product from genomic DNA or from double-stranded cDNA were subjected to hybridization to AtSNPtile1 array (Affymetrix) using standard gene expression array washing/staining protocol. For the genomic DNA concentration series, labeled products of Col or Van genomic DNA were hybridized to the array at total amounts of 6 μg, 9 μg, 12 μg, 15 μg, and 18 μg. It should be noted that as our labeling protocol generates double-stranded targets the antisense transcription cannot be tested.
Validation of allele-specific expression:
For each of 16 selected genes, a SNP within the largest exon was selected. PCR primers and extension primers were designed using Assay Design 3.1 (Sequenom). Primer sequences were listed in Table S6. For each sample poly-(A) RNA from ∼2 μg total RNA was reverse transcribed using 40 units of Superscript III (Invitrogen) and 0.5 μg Oligo(dT)18-20 primer (Invitrogen) in a total volume of 20 μl at 48° for 1 hr. First strand cDNA was amplified using gene-specific primer pairs with the following PCR conditions: denature at 94° for 3 min, 42 cycles of 94° for 15 sec, 65° for 15 sec, 72° for 20 sec, and extension at 72° for 5 min. PCR products were cleaned with Qiaquick PCR purification kit (QIAGEN), and submitted to the DNA sequencing facility of University of Chicago Cancer Research Center for extension reaction and Mass spectrometry assay (Sequenom).
Data analysis:
Probe intensities were background corrected as described previously (Borevitz et al. 2003). The log intensity for the ∼0.9 m SNP probes were then quantile normalized. For genomic DNA concentration series, the probe intensity was background corrected but without quantile normalization. We found that when Col genomic DNA hybridized to the array ∼7% SNPs having Col allele intensity < Van allele intensity, while when Van genomic DNA hybridized to the array ∼5% SNPs having Van allele intensity < Col allele intensity. Unknown thermodynamic property might contribute to these abnormal probe bindings. When comparing the Col genomic hybridization with Van genomic hybridization however, LARs from Col sample > LARs from Van sample for >99% SNPs. This holds for the SNP probes located within the significant cis-variation genes and cis-variation introns. Thus our significant calls were not biased to the SNP probes that bind abnormally.
For each annotated gene, the SNPs polymorphic between Col and Van (Clark et al. 2007) were mapped to promoter, gene, and downstream regions of the A. thaliana genome (TAIR version 7). Here the promoter was defined as the transcriptional start of the tested gene to the transcriptional stop of the upstream gene on the same strand. Correspondingly, downstream region was defined as the transcriptional stop of the tested gene to the transcriptional start of the downstream gene on the same strand. Genes containing one or more SNPs within the transcribed region were each tested by three separate linear models (Figure S2b), using LARs across SNPs and strands. As the correlation coefficient of LARs between sense and antisense strands was moderate (ρ = 0.54, P < 2.2E-16, n = 220,044) on the basis of genomic DNA 1:1 mixture hybridization, we treated the data points from sense and antisense strands as independent data points. For each tested effect, genewise d scores were calculated as d = coefficient / (standard deviation + s0), where s0 is the median of permutation d scores across genes and across 1000 permutations (Tusher et al. 2001). The d score is a modified t statistic that includes a constant in the denominator as a minimum probe variance. This statistic ensures that a minimum magnitude of allelic variation must be obtained to be called significant (Tusher et al. 2001). FDRs were determined by 1000 permutations as described previously (Zhang et al. 2008a).
For the sequence polymorphism analysis, positional information for SFPs and indels were obtained from a previous study (Zhang et al. 2008a). Genes were separated to cis, trans, and background groups. As only a small number of genes overlap between cis and trans groups, we included these overlapped genes in both cis and trans groups in the analysis. For absolute gene expression level, the gene expression values for each gene were obtained by median polish across three replicates and across gene probes. For expression specificity analysis, the gene probe intensities were divided by sample median for normalization. The gene expression values were obtained by median polish across three replicates and across gene probes. For each gene, the median expression value across samples was subtracted. Gene expression entropy E was calculated using the remaining positive gene expression value I as follows: pj = Ij/Σ Ij, Ei = −Σ (pj log pj), where i represents gene and j represents sample.
The analysis scripts were written in R (File S1).
RESULTS
Array-based detection of allelic variation in RNA samples:
Our custom-designed array, AtSNPtile1, interrogates ∼250,000 SNPs with Col as reference genotype, among which ∼55,000 SNPs are polymorphic between Col and Van. Each SNP is interrogated by four 25-mer probes with two alleles, Col and non-Col, on each of the two strands (supporting information, Figure S1). As the probe sequences for the two alleles on the same strand differ at the middle base, the target allele will preferentially bind to the perfect match over the mismatch SNP probe. Thus for a given target allele the mismatch binding provides a good estimation of the background probe hybridization effect. As such, the allele intensity ratio (Col allele/non-Col allele) estimates the relative amount of two target alleles with probe effect largely normalized across strands and SNPs. To stabilize the variance, log-transformed allele intensity ratio (LAR) was used for analysis. To determine the effectiveness of the LAR for detecting allelic variation, we mixed the genomic DNA of Col and Van at a range of ratios prior to random labeling and hybridization. A strong correlation (ρ = 0.69, P < 2.2E-16, n = 770,154) was observed between the LAR and the log allele ratio of template mixture (Figure 1A). We further tested whether LAR is robust to the difference in target concentration, i.e., variation of overall gene expression level. Labeled targets from Col and Van genomic templates were each hybridized to the array at a range of total amounts. Little correlation between LAR and the overall target amount was observed (ρ = 0.0033, P < 0.00053, n = 1,100,220) (Figure 1B). Indeed, for the majority of SNP probes, target amount has negligible effect on LAR in comparison with allelic composition of template mixture (Figure 1C). Because of this, we decided to use genomic DNA 1:1 mixture as reference of equal allele expression.
Figure 1.—
Detection of ASE using LAR as a measurement of allelic composition. Col allele is denoted as allele A and non-Col allele (Van allele) is denoted as allele B. The LARs across polymorphic SNPs were linear regressed against (A) the log allele ratios of template DNA mixture, and (B) the amount of target derived from Col DNA template (left 5 lanes) or Van DNA template (right 5 lanes). Data from antisense strand is colored orange and sense strand colored blue. Genomic hybridizations of Col and Van were added in A with template allele A/allele B set as 10/1 and 1/10, respectively. Note that the scanner setting for the two experiments was different so the variance of LARs of the two experiments was different, which does not affect the conclusion. (C) For each SNP on each strand, the LARs were linear regressed against the log allele ratio of template DNA mixture (blue) or the amount of target (orange). The regression coefficients (x-axis) were plotted against the corresponding r-squares (y-axis).
Global cis- and trans-regulatory variation:
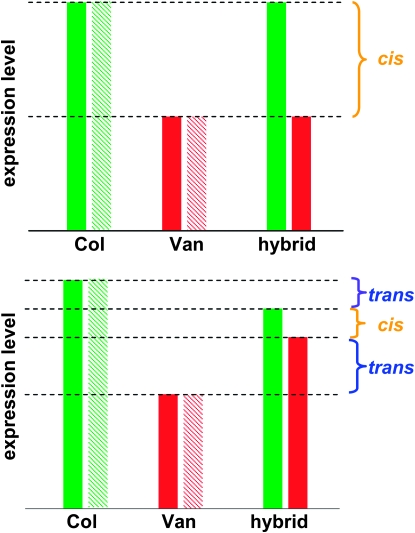
In a hybrid, gene expression difference between two alleles indicates regulatory polymorphism acting in cis, because both alleles are exposed to the same pool of trans factors. If, for a given gene, cis-genetic variation is the only regulatory variation, allele expression differences in the hybrid should be equal to half of the gene expression difference between diploid parents, i.e., expression difference measured between the two alleles in the parental RNA 1:1 mixture (Figure 2). Deviation between allelic expression in hybrid and allelic expression in 1:1 RNA mixture could attribute to trans-regulatory variation and genetic interactions involving trans variation (Figure 2). To test such composite cis and trans effects, we included in our study four biological replicates of parental RNA 1:1 mixture and reciprocal F1 hybrids. Using the LAR of genomic 1:1 mixture as a reference, parental expression difference was detected in parental RNA 1:1 mixture, ASE was detected across reciprocal F1 hybrids, trans variation was detected as difference between parental RNA 1:1 mixture and F1 hybrids, and imprinting effect was detected as difference between reciprocal F1 hybrids. We tested these effects in three separate linear models, each of which contains three orthogonal contrasts to fully partition the variance among four sample groups (Figure S2a and Figure S2b). For each effect, we applied a permutation-based false discover rate (FDR) (Tusher et al. 2001) to select significant calls (Figure S2c).
Figure 2.—
The experimental design. The diagram illustrates examples of regulatory variation acting only in cis (upper panel) and acting both in cis and in trans (lower panel). Gene expression levels of diploid parents and their hybrid were shown. If there is only cis genetic variation, half of the parental expression difference, i.e., the difference between the Col (green) and Van (red) solid bars, should be equal to the allele expression difference in the hybrid. If there are both cis and trans variation, this half of parental expression difference can be explained by cis effect (the allele expression difference in the hybrid), plus composite trans effect.
Among 12,311 genes analyzed, 3811 (FDR 0.1%) were differentially expressed between Col and Van parents as parental RNA 1:1 mixture differed in allelic ratio compared with the DNA 1:1 mixture (Table 1). Among these 3811 genes, 3558 were upregulated in Col and 253 upregulated in Van, suggesting a major trans factor. The overall upregulation of Col expression was unlikely due to systematic bias, i.e., Col target alleles do not preferentially hybridize to the probes, as demonstrated by the LAR distribution of genomic hybridizations (Figure S2d). ASE was detected for 1665 genes (FDR 0.3%) as significant deviation between genomic 1:1 DNA mixture and hybrid RNA allelic ratio (Table 1). Cis variation upregulates Col allele for 1075 genes and Van allele for 590 genes. A total of 1688 genes (FDR 0.2%) exhibited trans-regulatory variation where the parental RNA 1:1 mixture deviated in allelic ratio from the hybrid RNA (Table 1). Nearly all of the trans variation upregulates Col (1685). No imprinting effect was detected at the selected threshold (Table 1). It should be noted that allelic variation detected here may be caused by splicing variation, as a large portion of analyzed genes contain only one or two SNPs (Figure S2e). We found that SNP-containing exons, which are located within cis-variation genes, were enriched in differential splicing exons (Zhang et al. 2008a) by 1.7-fold with marginal significance (χ2 = 3.88, P < 0.049).
TABLE 1.
The number of genes significant for parental expression difference, cis effect, trans effect, or imprinting effect at different thresholds
| Difference between parents
|
cis effect
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Deltaa | Sig+b | Sig−b | Total | False | FDR (%) | Sig+b | Sig−b | Total | False | FDR (%) |
| 0.15 | 8962 | 1286 | 10248 | 4330 | 42 | 5098 | 3196 | 8294 | 3577 | 43 |
| 0.25 | 8066 | 1004 | 9070 | 1394 | 15 | 3839 | 2274 | 6113 | 543 | 8.9 |
| 0.35 | 7046 | 695 | 7741 | 256 | 3.3 | 2800 | 1618 | 4418 | 81 | 1.8 |
| 0.45 | 5799 | 501 | 6300 | 48 | 0.77 | 2002 | 1183 | 3185 | 25 | 0.80 |
| 0.55 | 4644 | 360 | 5004 | 13 | 0.25 | 1500 | 825 | 2325 | 11 | 0.46 |
| 0.65 | 3558 | 253 | 3811 | 4 | 0.11 | 1075 | 590 | 1665 | 5 | 0.30 |
| 0.75 | 2606 | 200 | 2806 | 1 | 0.048 | 773 | 453 | 1226 | 3 | 0.20 |
| 0.85 | 1657 | 165 | 1822 | 1 | 0.032 | 558 | 344 | 902 | 1 | 0.15 |
| 0.95 | 1077 | 127 | 1204 | 0 | 0.024 | 408 | 265 | 673 | 1 | 0.094 |
|
trans effect
|
Imprinting effect
|
|||||||||
| Deltaa | Sig+b | Sig−b | Total | False | FDR (%) | Sig+b | Sig−b | Total | False | FDR |
| 0.15 | 45 | 10018 | 10063 | 4627 | 46 | 11 | 0 | 11 | 2895 | 26317 |
| 0.25 | 12 | 8121 | 8133 | 1542 | 19 | 4 | 0 | 4 | 295 | 7379 |
| 0.35 | 6 | 6326 | 6332 | 302 | 4.8 | 3 | 0 | 3 | 8 | 282 |
| 0.45 | 5 | 4422 | 4427 | 47 | 1.1 | 3 | 0 | 3 | 1 | 37 |
| 0.55 | 3 | 2907 | 2910 | 11 | 0.37 | 0 | 0 | 0 | 0 | NA |
| 0.65 | 3 | 1685 | 1688 | 2 | 0.15 | 0 | 0 | 0 | 0 | NA |
| 0.75 | 3 | 729 | 732 | 1 | 0.080 | 0 | 0 | 0 | 0 | NA |
| 0.85 | 3 | 248 | 251 | 0 | 0.085 | 0 | 0 | 0 | 0 | NA |
| 0.95 | 2 | 91 | 93 | 0 | 0.080 | 0 | 0 | 0 | 0 | NA |
FDR was determined on the basis of 1000 permutations for 12,311 analyzed genes. Note that some FDR was >100%. This was due to the d score distribution of nonsignificant genes in real data being tighter than the null (Tusher et al. 2001), meaning no significant enrichment over background. Boldface highlights the number of significant genes described in the main text.
Delta, the thresholds.
Sig+ and Sig−, the number of genes significant for each of the two directions of comparison. For parental expression difference, + indicates that expression level is greater in Col; for cis effect, + indicates that cis variation upregulates Col; for trans effect, + indicates that trans variation upregulates Van; for imprinting effect, + indicates that Col allele is upregulated in Col-mother F1 hybrids.
Intron retention is a common alternative splicing form in plants. Allele-specific intron expression in F1 hybrids could imply an intronic splicing difference. We tested the parental difference, cis effect, trans effect, and imprinting effect for a total of 6707 introns, using linear models and a permutation approach described above (Figure S2a, Figure S2b, and Figure S2c). A total of 1202 introns (FDR 0.1%) showed differential expression between parents, 803 enriched in Col and 399 in Van (Table 2). A total of 1584 introns (FDR 0.4%) were identified in F1 hybrids exhibiting cis variation, 995 enriched in Col allele, and 589 in Van allele (Table 2). No trans effect and imprinting effect were detected at the selected threshold (Table 2). Here cis effect detected in F1 hybrids is more than the detected parental difference by a number of 382. This is likely due to larger RNA sample variance than DNA sample variance for intron LAR (Figure S2F). This implies that for intron splicing there is less power to detect trans variation, which relies on the comparison of two RNA sample groups. Allele-specific intron expression could be due to ASE when two intron alleles are both not spliced. We found, however, no significant enrichment of allelic introns in ASE genes (χ2 = 0.049, P < 0.82).
TABLE 2.
The number of introns significant for parental splicing difference, cis effect, trans effect, or differential splicing between reciprocal F1 hybrids at different thresholds
| Difference between parents
|
cis effect
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Deltaa | Sig+b | Sig−b | Total | False | FDR (%) | Sig+b | Sig−b | Total | False | FDR (%) |
| 0.15 | 3238 | 1446 | 4684 | 1251 | 27 | 2997 | 1933 | 4930 | 1592 | 32 |
| 0.25 | 2430 | 1014 | 3444 | 150 | 4.4 | 2327 | 1448 | 3775 | 192 | 5.1 |
| 0.35 | 1806 | 731 | 2537 | 21 | 0.83 | 1783 | 1101 | 2884 | 42 | 1.5 |
| 0.45 | 1283 | 504 | 1787 | 5 | 0.27 | 1367 | 777 | 2144 | 15 | 0.69 |
| 0.55 | 803 | 399 | 1202 | 1 | 0.12 | 995 | 589 | 1584 | 6 | 0.39 |
| 0.65 | 502 | 289 | 791 | 1 | 0.065 | 675 | 449 | 1124 | 3 | 0.25 |
| 0.75 | 341 | 205 | 546 | 0 | 0.039 | 488 | 335 | 823 | 1 | 0.16 |
| 0.85 | 212 | 159 | 371 | 0 | 0.032 | 335 | 265 | 600 | 1 | 0.11 |
| 0.95 | 139 | 118 | 257 | 0 | 0.023 | 231 | 211 | 442 | 0 | 0.070 |
|
trans effect
|
Difference between reciprocal hybrids
|
|||||||||
| Deltaa | Sig+b | Sig−b | Total | False | FDR (%) | Sig+b | Sig−b | Total | False | FDR (%) |
| 0.15 | 14 | 690 | 704 | 1422 | 202 | 20 | 99 | 119 | 1094 | 919 |
| 0.25 | 4 | 151 | 155 | 59 | 38 | 13 | 3 | 16 | 43 | 267 |
| 0.35 | 1 | 41 | 42 | 7 | 17 | 0 | 0 | 0 | 3 | NA |
| 0.45 | 0 | 7 | 7 | 2 | 27 | 0 | 0 | 0 | 1 | NA |
| 0.55 | 0 | 0 | 0 | 1 | NA | 0 | 0 | 0 | 0 | NA |
| 0.65 | 0 | 0 | 0 | 0 | NA | 0 | 0 | 0 | 0 | NA |
| 0.75 | 0 | 0 | 0 | 0 | NA | 0 | 0 | 0 | 0 | NA |
| 0.85 | 0 | 0 | 0 | 0 | NA | 0 | 0 | 0 | 0 | NA |
| 0.95 | 0 | 0 | 0 | 0 | NA | 0 | 0 | 0 | 0 | NA |
FDR was determined on the basis of 1000 permutations for 6707 analyzed introns. Boldface highlights the number of significant introns described in the main text.
Delta, the thresholds.
Sig+ and Sig−, the number of genes significant for each of the two directions of comparison. See footnotes in Table 1.
Effect of cis- and trans-regulatory variation:
We examined the composite cis and trans effects of gene expression using the estimates obtained from linear modeling described above. We first examined the direction of cis and trans effects (Figure S3a). Here positive cis effect indicates that cis variation upregulates Col (or Col allele > Van allele in F1 hybrids); positive trans effect indicates that trans variation upregulates Col (or Col allele decreases and Van allele increases in F1 hybrids relative to parents); positive total effect indicates that expression level in Col is greater than that in Van. The Venn diagram shows that 27% (1047) of the parental differential genes were caused by only cis variation and 29% (1113) by only trans variation. The cis or trans effects of these genes were generally in the same direction as the total effects, as expected. About 2.2% (85) of the parental differential genes were caused by both cis and trans variation, mostly acting in the same direction. This holds for the remaining 41% (1566) of the parental differential genes for which neither cis nor trans effect was detected, as here cis and trans effects were each too small to be called significant at the selected threshold. About 32% (533) of the cis-variation genes and 29% (487) trans-variation genes did not overlap parental differential genes. For these genes, the detected cis or trans effects were masked in the parental lines by small trans or cis effects, respectively, which act in the opposite direction. This is consistent with extensive transgression of eQTL reported for A. thaliana accessions (Keurentjes et al. 2007; West et al. 2007). Between Col and Van the majority of trans-effect genes were upregulated in Col. This is probably because Van harbors a natural null mutation at ERECTA (Torii et al. 1996), a trans eQTL hot spot (Keurentjes et al. 2007). Genetic variation at this locus may cause many growth-related genes being upregulated in Col (Zhang et al. 2008a). Due to this large bias of trans effects, small positive cis effects (Col allele > Van allele in hybrids) tend to be revealed in parents, while small negative cis effects (Van allele > Col allele in hybrids) tend to be masked in parents.
We also examined the size of cis and trans effects relative to parental expression difference (Figure S3b). Parental expression difference showed strong correlation with cis effects (ρ = 0.81, P < 2.2E-16, n = 12,311) and moderate correlation with trans effects (ρ = 0.48, P < 2.2E-16, n = 12,311). A weak inverse correlation was observed between the magnitude of cis effects and trans effects (ρ = −0.13, P < 2.2E-16, n = 12,311). This is in line with the observation that only a small number of genes (144) were significant for both cis and trans effects (Figure S3a). Even for these genes the cis and trans regulatory variation could be separated between parental lines. An example was FLOWERING LOCUS C (FLC), where trans variation downregulated Col FLC while cis variation upregulated the Col allele in F1 hybrids (Figure 3A). This is because Col harbors a null mutation at FRIGIDA, a transcription activator of FLC, while Van contains a nonsense mutation at FLC (Werner et al. 2005) that leads to nonsense mediated decay of that allele. The composite trans effects detected in F1 hybrid system are the sum of additive and epistatic effects (trans, trans × trans and cis × trans). In a single hybrid pair, cis variation is not tested across segregating trans backgrounds, and thus, the cis × trans effect cannot be dissected. A recent study in Drosophila suggests however that cis × trans dependent regulation may not be common (Wittkopp et al. 2008a).
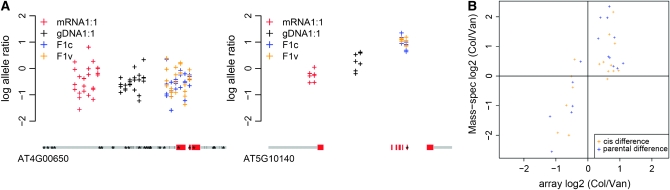
Figure 3.—
Validation of significant loci. (A) FLC is significant for both cis and trans effects. FRIGIDA, a transcriptional activator of FLC, shows no expression difference between Col and Van, although Col harbors a functional mutation at FRIGIDA (left panel). Col allele of FLC was upregulated in F1 hybrids due to a functional FRIGIDA brought by Van parent, while Van allele was downregulated due to a nonsense mutation at FLC in Van (right panel). (B) The validation of 16 randomly selected ASE genes. Each locus was tested for parental RNA 1:1 mixture (blue) and F1 hybrid RNA sample (orange). The log2 allele ratios of Mass-spectrometry peak heights (y-axis) were plotted against the log2 allele ratios of AtSNPtile1 hybridization intensity (x-axis).
To validate our method experimentally, 16 genes were randomly selected from the 1665 ASE gene list and tested by single-base extension coupled with Mass-spectrometry. Each locus was tested for both the parental RNA 1:1 mixture and F1 hybrid RNA samples. The peak-height allele signal was analyzed just as the probe allele intensity on the array, by using the genomic DNA 1:1 mixture as a reference. The log allele ratios from the Mass-spectrometry assay were linear regressed against the LARs from microarray hybridization. The correlation between the two approaches was high (ρ = 0.74, P < 1.08E -06, n = 32). The regression slope was 1.3, indicating that estimation from Mass-spectrometry assay tends to exceed that from microarray hybridization (Figure 3B). Other microarray studies have shown a downward bias of fold change magnitude with improved precision due to quantile normalization (Bolstad et al. 2003).
Sequence polymorphisms for cis- and trans-variation genes:
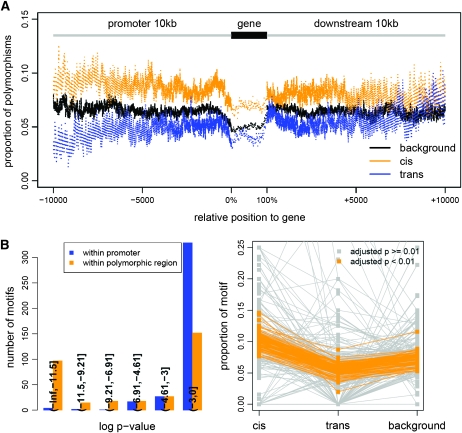
We examined the extent of local sequence polymorphism (deletions >200 bp, SFPs and SNPs in Van relative to Col) for genes regulated in cis, by trans, and genes without significant cis or trans effect (background). For each gene group, promoters were aligned at the transcriptional start, downstream regions were aligned at the transcriptional stop, and genes were divided into 100 percentiles on the basis of position for a scaled comparison. The proportion of sequence polymorphism was calculated for each percentile within a gene as well as for each position across up to 10-kb promoter and 10-kb downstream regions. Cis variation genes were consistently more polymorphic than background genes across analyzed regions (regression coefficient = 0.019, ρ = 0.81, P < 2.2E-16, n = 40,200), while trans-variation genes appeared to be more conserved than background genes (regression coefficient = −0.013, ρ = 0.63, P < 2.2E-16, n = 40,200). Such difference was largely due to the presence of SFPs and particularly indels (Table S1). Assuming that the proximal ends of promoters are enriched for functional cis elements, we would expect the divergence in level of sequence polymorphism among gene groups would decrease toward distal ends of promoters. On the contrary, such difference is throughout the promoter and downstream regions (Figure 4A) and extends to the neighboring genes (Figure S4a).
Figure 4.—
Sequence polymorphisms for cis- and trans-variation genes. (A) The proportion of sequence polymorphisms within promoter, gene, and downstream regions for cis (orange), trans (blue), and background (black) gene groups. For each gene group, promoters were aligned at the −1 position relative to gene start; downstream regions were aligned at the +1 position relative to gene stop, and genes were divided into 100 percentiles on the basis of the position within the gene. For each position from −1 to −10 kb and from +1 to +10 kb, and for each percentile, the proportion of polymorphic sites was calculated. SFPs were counted as 25-bp polymorphic sites. (B) Functional motifs were differentially affected by sequence polymorphisms for cis, trans, and background gene groups. Motifs were mapped to promoter regions. For each motif, its occurrence relative to the occurrence of all other motifs was counted for each gene group, and a test of equal proportion across the three gene groups was performed to obtain the P-value (blue in left panel). For each motif, its occurrence in polymorphic regions (overlapping with SNPs, SFPs, and indels) relative to its occurrence in nonpolymorphic regions was counted for each gene group, and a test of equal proportion across the three gene groups was performed to obtain the P-value (orange in left panel). The proportion of a given motif within the polymorphic region (the number of occurrences within promoter polymorphic regions/the number of occurrence within entire promoter regions) was calculated for each gene group (gray in right panel). Motifs significantly different (adjusted P-value < 0.01) across the three gene groups for the distribution in polymorphic vs. nonpolymorphic regions were colored orange.
We also examined the sequence polymorphism distribution for allelic introns. To test whether sequence polymorphisms relevant to intronic splicing are preferentially located at certain positions, analyzed introns and their upstream/downstream exons were each divided into 10 percentile bins on the basis of position, and the proportion of sequence polymorphism was calculated for each percentile. The proportion of sequence polymorphism was generally higher in cis-variation introns than in background introns, across all analyzed regions. The difference was most obvious however for the 3′ end of the upstream exon, the 3′ half of the intron, and the 5′ end of the downstream exon (Figure S4b). Taken together, the data fit with a model of cis-regulatory variation generally being detected in regions of high diversity between Col and van, which extends dozens of kilobases in both directions due to historical linkage disequilibrium.
Cis genetic variation likely affects functional motifs. To test this, we mapped the plant-specific motifs (Higo et al. 1999) to promoter regions of the analyzed genes and examined the motif distribution. The relative occurrence of functional motifs was very similar across cis, trans, and background gene groups. When restricted to a 1-kb promoter region, only two functional motifs were significantly enriched in trans variation genes (test of equal proportions, adjusted P-value < 0.01). These motifs are involved in ABA responses (ABRE-related sequence) and ABA/light responses (CACGTGG-box motif) (Higo et al. 1999). In comparison, the relative occurrence of functional motifs within polymorphic region was quite different among the three groups. For all of the 107/327 motifs showing significant difference (test of equal proportions, adjusted P-value < 0.01), they fell within polymorphic regions the most frequently in cis variation genes while the least frequently in trans-variation genes (Figure 4B). There is no significant difference in the number of total motifs among three gene groups (Table S2a). The motif density of the cis-variation genes, however, was slightly lower than that of background genes (Table S2a). TATA box motifs were shown to be enriched in genes with distinct expression variability (Landry et al. 2007; Choi and Kim 2008). Examination of the six TATA box motifs indicated that only the distribution of TATA box 1 sequence (CTATAAATAC) was significantly different among cis, trans, and background gene groups (Table S2b).
Cis- and trans-variation genes are associated with distinct chromatin states:
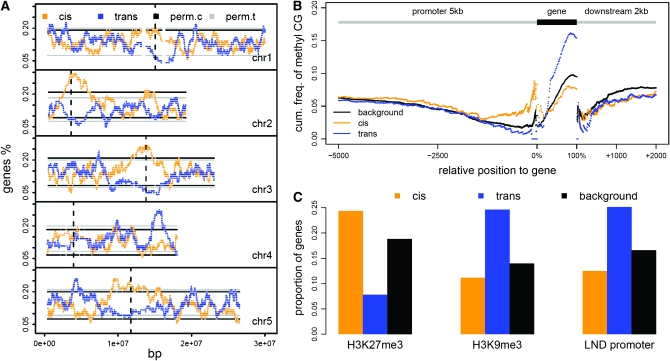
The frequency of sequence polymorphism varies along a chromosome (Borevitz et al. 2007; Clark et al. 2007). This could potentially affect the chromosomal distribution of cis- and trans-variation genes. Sliding windows by 120-gene bin size revealed a high proportion of cis- and low proportion of trans-variation genes around pericentromeric regions (Figure 5A). Within euchromatin arms, more trans- than cis-variation genes were observed in general (Figure 5A). The analysis also revealed two significant trans-variation gene clusters, one located at the right arm of chromosome 4 and the other at the left arm of chromosome 5 (Figure 5A), implying possible chromatin level regulation. Overall the chromosome distribution of cis genes showed a modest negative correlation (ρ = −0.28, P < 2.2E-16, n = 11,716) with that of trans genes. As the analyzed genes have to contain at least one SNP, sampling bias could increase toward euchromatin arms (Springer and Stupar 2007), implying an overestimation of cis- but underestimation of trans-variation genes toward euchromatin arms. We thus examined directly the correlation between chromosomal distribution of cis/trans effects with that of sequence polymorphism and gene distance (distance between two genes on the same strand), both of which exhibited clear chromosomal trends decreasing from pericentromeric regions toward euchromatin arms (Figure S5). Indeed, chromosomal distribution of cis-variation genes was positively correlated with that of sequence polymorphism (ρ = 0.48, P < 2.2E-16, n = 11,716) and gene distance (ρ =0.39, P < 2.2E-16, n = 11,716). In contrast, chromosomal distribution of trans-variation genes was negatively correlated with that of sequence polymorphism (ρ = −0.45, P < 2.2E-16, n = 11,716) and gene distance (ρ = −0.41, P < 2.2E-16, n = 11,716). Similar correlation patterns along chromosome were observed using quantitative measurements of cis/trans effects (Table S3). Although the proportion of sequence polymorphism was significantly different between cis- and trans-variation genes (two-sample t-test P < 2.9E-11, n = 3336), gene distance was not (two-sample t-test P < 0.94, n = 3349).
Figure 5.—
cis (orange), trans (blue), and background (black) genes are associated with distinct chromatin status. (A) Chromosome distribution of cis, trans, and background genes. The proportion of genes within sliding windows of 120 gene bin size was calculated for each gene group. To determine statistical significance, gene order was permutated within chromosome and the proportion of genes was calculated for each group. The 95% confidence lines on the basis of 1000 permutations were plotted for cis (black) and trans (gray) variation genes. (B) CG methylation of cis, trans, and background genes. For each gene group, promoters were aligned at the transcriptional start, genes were divided into 100 percentiles on the basis of position, and the downstream regions were aligned at the transcriptional stop. For each position within promoter 5 kb and within downstream region 2 kb, and for each gene percentile, the cumulative frequency of CG methylation was calculated as the number of constitutive CG methylation within the region from the aligned position to the tested position divided by the number of CCGG sites within that region. (C) Histone modification for cis, trans, and background genes. The proportion of genes containing H3K27me3, H3K9me3, or low nucleosome density (LND) promoters was calculated for each gene group. Note that as the proportions of H3K9me3 were generally high, they were divided by 3 to fit the plotting scale.
The chromosomal distribution of cis- and trans-variation genes implies that they could be associated with distinct chromatin structure (Gilbert et al. 2004) and epigenetic modifications (Zhang et al. 2007; Zilberman et al. 2007). Cytosine methylation and histone modification are two major epigenetic marks that reflect local chromatin activity. We first examined the CG methylation pattern for cis- and trans-gene groups, using data from a previous study that detects global CG methylation in Col and Van (Zhang et al. 2008b). The pattern of constitutive CG methylation (constitutive across Col and Van) was very different between cis- and trans-variation genes. cis genes showed a high level of CG methylation across the proximal promoter and 5′ portion of gene, while trans genes toward the 3′ portion of gene (Figure 5B). CG methylation within promoter often leads to gene repression while methylation within the gene body increases steady-state gene expression (Zhang et al. 2006, Zhang et al. 2008b; Zilberman et al. 2007). Distribution of polymorphic CG methylation (polymorphic between Col and Van) was quite similar between cis- and trans-variation genes, except for a very short region immediately downstream (Figure S6). In A. thaliana, genes associated with histone 3 Lys 27 trimethylation (H3K27me3) are expressed at a low level with tight regulation, while genes associated with low nucleosome density regions (LND) are constitutively expressed with low tissue specificity (Zhang et al. 2007). We found that the proportion of H3K27me3-associated genes was much higher in cis-variation genes (χ2 = 168, P < 2.2E-16, d.f.= 2) while the proportion of LND-associated genes was much higher in trans-variation genes (χ2 = 103, P < 2.2E-16, d.f. = 2) (Figure 5C). Interestingly, the proportion of histone 3 Lys 9 trimethylaiton (H3K9me3)-associated genes (Turck et al. 2007) was also much higher in trans-variation genes (χ2 = 111, P < 2.2E-16, d.f. = 2) (Figure 5C). H3K9me3 is previously thought to be a repressive epigenetic mark (Eissenberg and Shilatifard 2006). Recent studies suggest, however, that H3K9me3 could activate gene expression (Wiencke et al. 2008). In contrast to epigenetic marks, there was no significant difference in the proportion of small RNA or microRNA target genes among cis, trans, and background genes (Table S4).
Gene expression specificity for cis- and trans-variation genes:
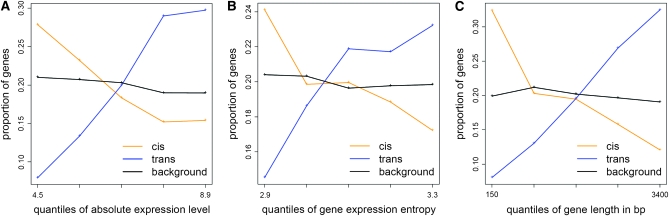
Distinct epigenetic properties among cis, trans, and background genes implied that the three groups might have different gene expression patterns. We thus examined the overall expression level and expression specificity for the three groups. The gene expression values were obtained for 63 diverse tissues in Col wild-type background (Schmid et al. 2005). The analyzed genes were divided into five percentile bins on the basis of absolute expression level. For each percentile bin, the proportion of cis, trans, and background genes was calculated. Genes regulated in cis were more common among the genes expressed at lower levels, while genes regulated in trans made up a greater proportion of genes expressed at higher levels (Figure 6A). Gene-expression specificity was measured by expression entropy (Zhang et al. 2006; Ritchie et al. 2008) (materials and methods). cis variation genes showed lower entropy (higher tissue specificity) while trans-variation genes higher entropy (lower tissue specificity) as compared to the background genes (Figure 6B). Gene length is correlated with both gene body CG methylation level and absolute expression level (Zhang et al. 2006, Zhang et al. 2008b; Zilberman et al. 2007). Indeed, cis-variation genes were generally shorter while trans-variation genes longer than background genes (Figure 6C).
Figure 6.—
The expression profiles are distinct for cis (orange), trans (blue), and background (black) genes. Genes were divided into five percentiles on the basis of (A) overall gene expression level, (B) gene expression entropy, or (C) gene length. For each percentile, the proportion of the genes falling within the percentile was calculated.
To test whether cis- and trans-variation genes are enriched in any functional category, we applied Fisher's exact test for gene ontology slim (GOslim) categories. cis-variation genes showed few enrichment in GOslim categories, while trans-variation genes were significantly enriched in several biological process, molecular function, and cellular component categories (Table S5). This implies that cis genes could be rather randomly distributed while certain biological functions are jointly regulated among those trans genes.
DISCUSSION
Our study suggests that genes regulated in cis are preferentially associated with closed chromatin marks while genes regulated in trans are associated with open chromatin. Chromatin structure affects DNA accessibility for a variety of nuclear processes including DNA repair and recombination (Surralles et al. 2002; Gilbert et al. 2004). Another observation is that genes regulated in cis tend to reside in genome regions (linkage disequilibrium blocks) that have been separated for longer evolutionary time and have accumulated structural changes. In line with this, a comparison of gene expression variation between intraspecific and interspecific Drosophila strains suggests that cis mutations seem to accumulate preferentially over time (Wittkopp et al. 2008b). A recent study introduced human chromosome 21 to mouse hepatocyte and demonstrated that human-specific transcriptional events are largely cis directed (Wilson et al. 2008). cis variations tend to affect genes with narrow expression regulation. Thus a cis mutation generally affects the expression regulation in a specific environment or developmental context. Accumulating evidence implicates that quantitative expression variation caused by cis mutation can lead to ecologically relevant phenotypic divergence (Wray 2007), for example pigment variation in Drosophila (Gompel et al. 2005) and skeletal reduction in sticklebacks (Shapiro et al. 2004). The extent to which cis-regulatory variation is adaptive and whether these are the first types of mutations or later compensatory changes requires further investigation (de Meaux et al. 2006).
A gene regulatory network is composed of regulatory genes and structural genes (Wittkopp et al. 2004). Genes regulated in cis would be distributed across both categories, while genes regulated in trans might preferentially include structural genes at terminal nodes of the network. Indeed we observed that trans-variation genes are significantly enriched in various enzyme activities and exhibit relatively low sequence polymorphism, consistent with a structural role. Studies in yeast (Landry et al. 2007) and Caenorhabditis elegans (Denver et al. 2005; Rifkin et al. 2005) suggest that trans mutations have relatively minor contribution in natural settings. trans mutations are potentially pleiotropic and tend to affect genes that express constitutively. Such large functional trade-offs, over time, would prevent their accumulation due to negative selection (Alonso and Wilkins 2005; Landry et al. 2007). Alternatively, recent mutations acting in trans could sweep through a population due to a large positive effect. One example includes chromatin regulator genes in yeast, which were identified as trans eQTL and which show signs of positive selection (Lee et al. 2006; Choi and Kim 2008). Another example of trans mutations at regulatory loci, that lead to a large selective advantage under specific environments, is FRIGIDA. This major regulatory gene determines the flowering time of A. thaliana winter annuals (Johanson et al. 2000). Natural accessions of A.thaliana accumulate independent functional sequence polymorphisms at this locus, which cause parallel phenotypic evolution of early flowering (Gazzani et al. 2003).
Distinct from gene expression variation that is contributed by both cis- and trans-regulatory difference, we found that intronic splicing variation is mostly controlled in cis. Splicing regulation is achieved by combinatorial control of splicing regulators and signaling pathways (Stamm 2002; Black 2003), which are generally not gene specific but affect many downstream splicing events (Stoilov et al. 2008). Unlike gene expression variation that largely results in quantitative difference among transcripts, splicing variation leads to structural difference with potentially severe consequences on cellular function. Thus selection could be much more extensive against a trans-splicing mutation than against a trans-expression mutation. On the other hand, alternative splicing is a major mechanism to generate novel protein function (Birzele et al. 2008). The splicing cis elements are often degenerate consensus sequences (Matlin et al. 2005) and are not pleiotropic. Thus cis-splicing variation could have a positive effect for biological processes that call for rapid evolution of molecular variety (Kazan 2003; Watson et al. 2005; Ule and Darnell 2006).
Our results confirm and extend the results from other ASE studies in plants (Springer and Stupar 2007; Guo et al. 2008). The approach of dissecting cis- and trans-regulatory variation using F1 hybrid system, however, has potential drawbacks. As mentioned above, the detection of trans effect relies on the subtraction of hybrid cis effect from parental expression difference. In the hybrid both additive trans effect and nonadditive trans × trans and cis × trans interaction could occur. The interpretation of the detected trans effect could be complicated when epistatic interaction is common. Another drawback of this approach is that epigenetic regulation could potentially interfere with the detection of cis-genetic variation.
Here we present the genomewide dissection of cis- and trans-regulatory variation among F1 hybrids and their parents using a newly released SNP-tiling array. We demonstrate this as a powerful platform to reveal allelic transcriptional variation in addition to transcript level differences. Our computational approach is a genotyping of RNA sample. This is accomplished by variance partitioning, among hybrids and parental RNA pools, and by including genomic hybridizations as reference, a method easily applied in other organisms. Given the intense interest in decoding cis elements, our approach is a powerful method to scan the genome for functional expression variation across genetic, developmental, and environmental perturbations.
Acknowledgments
We thank Han Xiao and Peter McCullagh (Department of Statistics, University of Chicago) for helpful discussion on the statistical analysis. We thank Juliette de Meaux (Max Planck Institute for Plant Breeding Research) and Andrew Cal (Department of Molecular Genetics and Cell Biology, the University of Chicago) for critical reading of the manuscript. We thank the greenhouse staff of the University of Chicago for taking care of the plants.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103499/DC1.
Microarray data included in this study have been deposited to Gene Expression Omnibus with accession no. GSE16520.
References
- Alonso, C. R., and A. S. Wilkins, 2005. Opinion: the molecular elements that underlie developmental evolution. Nat. Rev. Genet. 6 709–715. [DOI] [PubMed] [Google Scholar]
- Birzele, F., G. Csaba and R. Zimmer, 2008. Alternative splicing and protein structure evolution. Nucleic Acids Res. 36 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson, H. T., T. J. Albert, C. M. Ladd-Acosta, R. D. Green, M. A. Rongione et al., 2008. SNP-specific array-based allele-specific expression analysis. Genome Res. 18 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D. L., 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72 291–336. [DOI] [PubMed] [Google Scholar]
- Bolstad, B. M., R. A. Irizarry, M. Astrand and T. P. Speed, 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193. [DOI] [PubMed] [Google Scholar]
- Borevitz, J. O., S. P. Hazen, T. P. Michael, G. P. Morris, I. R. Baxter et al., 2007. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 12057–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J. O., D. Liang, D. Plouffe, H. S. Chang, T. Zhu et al., 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., and L. Kruglyak, 2005. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., J. D. Storey, J. Whittle and L. Kruglyak, 2005. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436 701–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296 752–755. [DOI] [PubMed] [Google Scholar]
- Carrel, L., and H. F. Willard, 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434 400–404. [DOI] [PubMed] [Google Scholar]
- Choi, J. K., and Y. J. Kim, 2008. Epigenetic regulation and the variability of gene expression. Nat. Genet. 40 141–147. [DOI] [PubMed] [Google Scholar]
- Clark, R. M., G. Schweikert, C. Toomajian, S. Ossowski, G. Zeller et al., 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342. [DOI] [PubMed] [Google Scholar]
- Cowles, C. R., J. N. Hirschhorn, D. Altshuler and E. S. Lander, 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32 432–437. [DOI] [PubMed] [Google Scholar]
- de Meaux, J., A. Pop and T. Mitchell-Olds, 2006. Cis-regulatory evolution of chalcone-synthase expression in the genus Arabidopsis. Genetics 174 2181–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, J. T. Streelman, S. K. Kim, M. Lynch et al., 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 37 544–548. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J. C., and A. Shilatifard, 2006. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 16 184–190. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., A. R. Gendall, C. Lister and C. Dean, 2003. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N. P. Carter et al., 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118 555–566. [DOI] [PubMed] [Google Scholar]
- Gompel, N., B. Prud'homme, P. J. Wittkopp, V. A. Kassner and S. B. Carroll, 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433 481–487. [DOI] [PubMed] [Google Scholar]
- Guo, M., S. Yang, M. Rupe, B. Hu, D. R. Bickel et al., 2008. Genome-wide allele-specific expression analysis using Massively Parallel Signature Sequencing (MPSS) reveals cis- and trans-effects on gene expression in maize hybrid meristem tissue. Plant Mol. Biol. 66 551–563. [DOI] [PubMed] [Google Scholar]
- Higo, K., Y. Ugawa, M. Iwamoto and T. Korenaga, 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S., Y. Hahn, Q. Rong and K. Pfeifer, 2007. Accurate quantitation of allele-specific expression patterns by analysis of DNA melting. Genome Res. 17 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., J. West, C. Lister, S. Michaels, R. Amasino et al., 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347. [DOI] [PubMed] [Google Scholar]
- Kazan, K., 2003. Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci. 8 468–471. [DOI] [PubMed] [Google Scholar]
- Keurentjes, J. J., J. Fu, I. R. Terpstra, J. M. Garcia, G. van den Ackerveken et al., 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 104 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekens, R., A. Vercauteren, B. Moerkerke, E. Goetghebeur, H. Van Den Daele et al., 2006. Genome-wide screening for cis-regulatory variation using a classical diallel crossing scheme. Nucleic Acids Res. 34 3677–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, M. C., and A. C. Wilson, 1975. Evolution at two levels in humans and chimpanzees. Science 188 107–116. [DOI] [PubMed] [Google Scholar]
- Landry, C. R., B. Lemos, S. A. Rifkin, W. J. Dickinson and D. L. Hartl, 2007. Genetic properties influencing the evolvability of gene expression. Science 317 118–121. [DOI] [PubMed] [Google Scholar]
- Lee, S. I., D. Pe'er, A. M. Dudley, G. M. Church and D. Koller, 2006. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc. Natl. Acad. Sci. USA 103 14062–14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, H. S., Z. Wang, Y. Hu, H. H. Yang, S. Gere et al., 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13 1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin, A. J., F. Clark and C. W. Smith, 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6 386–398. [DOI] [PubMed] [Google Scholar]
- Morley, M., C. M. Molony, T. M. Weber, J. L. Devlin, K. G. Ewens et al., 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin, S. A., D. Houle, J. Kim and K. P. White, 2005. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438 220–223. [DOI] [PubMed] [Google Scholar]
- Ritchie, W., S. Granjeaud, D. Puthier and D. Gautheret, 2008. Entropy measures quantify global splicing disorders in cancer. PLoS Comput. Biol. 4 e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, J., J. M. Akey, J. Whittle, E. N. Smith, G. Yvert et al., 2005. a Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 15 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, J., R. B. Brem, J. Whittle and L. Kruglyak, 2005. b Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 1 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E. E., S. A. Monks, T. A. Drake, A. J. Lusis, N. Che et al., 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422 297–302. [DOI] [PubMed] [Google Scholar]
- Schmid, M., T. S. Davison, S. R. Henz, U. J. Pape, M. Demar et al., 2005. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Serre, D., S. Gurd, B. Ge, R. Sladek, D. Sinnett et al., 2008. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 4 e1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, M. D., M. E. Marks, C. L. Peichel, B. K. Blackman, K. S. Nereng et al., 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428 717–723. [DOI] [PubMed] [Google Scholar]
- Springer, N. M., and R. M. Stupar, 2007. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm, S., 2002. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 11 2409–2416. [DOI] [PubMed] [Google Scholar]
- Stoilov, P., C. H. Lin, R. Damoiseaux, J. Nikolic and D. L. Black, 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl. Acad. Sci. USA 105 11218–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surralles, J., M. J. Ramirez, R. Marcos, A. T. Natarajan and L. H. Mullenders, 2002. Clusters of transcription-coupled repair in the human genome. Proc. Natl. Acad. Sci. USA 99 10571–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K. U., N. Mitsukawa, T. Oosumi, Y. Matsuura, R. Yokoyama et al., 1996. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., F. Roudier, S. Farrona, M. L. Martin-Magniette, E. Guillaume et al., 2007. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3 e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., R. Tibshirani and G. Chu, 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J., and R. B. Darnell, 2006. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 16 102–110. [DOI] [PubMed] [Google Scholar]
- Watson, F. L., R. Puttmann-Holgado, F. Thomas, D. L. Lamar, M. Hughes et al., 2005. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309 1874–1878. [DOI] [PubMed] [Google Scholar]
- Werner, J. D., J. O. Borevitz, N. H. Uhlenhaut, J. R. Ecker, J. Chory et al., 2005. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M. A., K. Kim, D. J. Kliebenstein, H. van Leeuwen, R. W. Michelmore et al., 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiencke, J. K., S. Zheng, Z. Morrison and R. F. Yeh, 2008. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene 27 2412–2421. [DOI] [PubMed] [Google Scholar]
- Wilson, M. D., N. L. Barbosa-Morais, D. Schmidt, C. M. Conboy, L. Vanes et al., 2008. Species-specific transcription in mice carrying human chromosome 21. Science 322 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430 85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2008. a Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics 178 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2008. b Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40 346–350. [DOI] [PubMed] [Google Scholar]
- Wray, G. A., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8 206–216. [DOI] [PubMed] [Google Scholar]
- Yan, H., W. Yuan, V. E. Velculescu, B. Vogelstein and K. W. Kinzler, 2002. Allelic variation in human gene expression. Science 297 1143. [DOI] [PubMed] [Google Scholar]
- Yvert, G., R. B. Brem, J. Whittle, J. M. Akey, E. Foss et al., 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35 57–64. [DOI] [PubMed] [Google Scholar]
- Zhang, X., J. Yazaki, A. Sundaresan, S. Cokus, S. W. Chan et al., 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhang, X., J. K. Byrnes, T. S. Gal, W. H. Li and J. O. Borevitz, 2008. a Whole genome transcriptome polymorphisms in Arabidopsis thaliana. Genome Biol. 9 R165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., O. Clarenz, S. Cokus, Y. V. Bernatavichute, M. Pellegrini et al., 2007. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., S. H. Shiu, A. Cal and J. O. Borevitz, 2008. b Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 4 e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., M. Gehring, R. K. Tran, T. Ballinger and S. Henikoff, 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat.Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]