Abstract
A novel peptide, pal9a, was purified from the venom duct extract of the turrid snail, Polystira albida (superfamily Conoidea, family Turridae), collected in the Gulf of Mexico. Its primary structure was determined by automated Edman degradation and confirmed by mass spectrometry. Turritoxin pal9a contains 34 amino acid residues, including 6 Cys residues arranged in the pattern C-C-C-C-C-C (framework IX, where “-“ represents one or more non-Cys amino acids), which characterizes the P-conotoxins. Peptide pal9a is the first P-conotoxin-like turritoxin characterized from a member of family Turridae of the Western Atlantic. The primary structure of turritoxin pal9a, NVCDGDACPDGVCRSGCTCDFNVAQRKDTCFYPQ-nh2 (-nh2, amidated C-terminus; calculated monoisotopic mass, 3679.48 Da; experimental monoisotopic mass, 3678.84 Da), shows variable degrees of low sequence similarity with framework IX-toxins from turrid (three species of Lophiotoma, and four species of Gemmula), terebrid (Hastula hectica), and Conus species of the Indo-Pacific (C. textile, C. gloriamaris, C. amadis, and C. litteratus) and of the Western Atlantic (C. regius). During the comparison of peptide pal9a with the other framework IX-toxins known to date, we realized that, in general, these peptides are hydrophilic, acidic compounds that have not been found in the fish-hunting Conus species studied thus far; we also found support for the notion that they may belong to several distinct gene superfamilies, even those from the same species. Given the broad distribution of framework IX-toxins within superfamily Conoidea, it will be interesting to identify the still-unknown molecular targets of P-conotoxins, P-conotoxin-like turritoxins, and P-conotoxin-like augertoxins.
Keywords: Conoidea, Turridae, Polystira albida, turritoxin, P-conotoxin, framework IX
1. Introduction
Superfamily Conoidea (also known as suborder Toxoglossa) is a group of marine predatory snails that inhabit tropical and subtropical waters worldwide. This superfamily includes, among others, the families Conidae (cone snails or cones), Terebridae (terebrids), and Turridae (turrids), whose members, with some exceptions belonging to Terebridae and Turridae, are equipped with a highly specialized venom apparatus that is primarily used to paralyze and capture their prey; distinct species of cones prey on fish, mollusks, and worms, whereas terebrids and turrids feed on worms (mainly polychaetes) [39].
The venoms of the cone snails (500–700 species) have been extensively studied, and have been shown to contain 50–200 different, physiologically active venom peptides [31] that are injected into the prey through a disposable, hollow radular tooth. The peptide toxins in Conus (the only genus of family Conidae) venoms generally have 7–40 amino acid residues and 2 to 4 disulfide bonds, and they often contain diverse post-translational modifications [5]. Conus toxins (conotoxins) bind voltage-gated ion channels, ligand-gated ion channels, G-protein-coupled receptors, or neurotransmitter transporters, generally with high affinity and specificity; this causes disruption of neuromuscular transmission and consequently paralysis in the prey [29]. Conus venoms are extremely potent, and some species have caused human fatalities [10]. Due to these properties some conotoxins have been used as molecular tools to study ion channels and receptors [2] and as potential pharmaceuticals [9, 22, 42]; indeed, a peptide from Conus magus that blocks N-type calcium channels(ω-conotoxin MVIIA) was approved in 2004 for the treatment of chronic pain [44]. Conotoxins are produced by proteolytic processing of precursors with the canonical structure: signal peptide - “pro” region - mature toxin. Based on the sequence of the signal peptide, conotoxins are classified into gene superfamilies, whereas their biological activities and molecular targets define pharmacological families [29]. So far, thirteen superfamilies of conotoxins (T, A, J, O, M, P, I, S, L, D, C, V, G) have been identified [1, 14, 16, 18, 24, 29, 32, 33], and a correlation has been found between the superfamily and the arrangement of Cys residues within the sequences of the mature conotoxins belonging to it. Each gene superfamily includes one to two Cys patterns, and one to five pharmacological families. Based on the same criterion, five superfamilies of conopeptides have been proposed: two of them (the conopressins and the contryphans) contain one disulfide bridge, whereas the other three (the contulakins, the conantokins, and the conorfamides) do not have Cys residues [29].
Despite the numerous species of terebrids (~300) [4] and turrids (~9,000) [36], venoms of these species have been studied only recently.
The first study of a terebrid venom was conducted on Terebra subulata, from which one I-conotoxin-like and two O-conotoxin-like peptides (“augertoxins”) were purified and sequenced; however, the precursors of the latter peptides do not show conserved signal peptide sequences, which are characteristic of conotoxin superfamilies and, except for the disulfide bridges, none of the three peptides contains post-translational modifications [13]. From the terebrid Hastula hectica eight toxins were purified, and the same number of toxin sequences were deduced from cDNA clones, one of which corresponds to one of the purified peptides; Cys arrangements similar to those of the A-, O-, P-, G-, J/L-, and V-conotoxins were found, but also two Cys frameworks not described in conotoxins [15].
The first study of turrid venoms, those of Polystyra albida and Gemmula periscelida captured in the Gulf of Mexico, revealed the presence of long (~93 residues), Met-rich peptides that also contain numerous Tyr and Arg residues but few, if any, Cys residues (“turritoxins”); interestingly, the two peptides (one from each species) seem to be homologous [23]. From L. olangoensis, the sequences of 23 toxins were determined by cDNA cloning, and they were classified into 16 gene superfamilies; Cys arrangements similar to those of I-, O-, P-, and S-conotoxins were found, as well as six Cys patterns that have not been described in conotoxins [43]; the authors concluded that the I-conotoxin-like peptides of Lophiotoma are related to the I2-conotoxin superfamily. In addition, these authors found two precursors that were predicted to yield: 1) a mature toxin similar to the turritoxins first characterized from P. albida and G. periscelida [23], and 2) four short peptides, one with four Cys residues. From Gemmula speciosa, P-conotoxin-like peptides were recently purified and sequenced; in this case, the cloned precursors of two additional peptides had identical signal sequences, which demonstrates the existence of a toxin gene superfamily in this and other Gemmula species that were also studied [12].
Here we report the biochemical characterization of a P-conotoxin-like peptide (Cys framework IX), pal9a, from a turrid snail of the Western Atlantic, Polystira albida. This peptide shows low sequence similarity with P-conotoxin-like turritoxins and augertoxins, and P-conotoxins from species of the Indo-Pacific and the Western Atlantic. In comparing peptide pal9a with the other framework IX-toxins, we found that, in general, these peptides are hydrophilic, acidic compounds that have not been found in piscivorous Conus species, and that even those from the same species may belong to several distinct gene superfamilies.
2. Materials and methods
2.1. Collection and storage of specimens
The specimens of Polystira albida were collected by dredging at depths of ~65 m along the coasts of the Bay of Campeche, Mexico. The captured organisms were frozen at −20°C and, upon arrival at the laboratory, they were stored at −70°C until used.
2.2. Crude venom extraction
The shells of 10 specimens were thawed and then centrifuged at 1,693 × g for 5 min at 4°C to release the organisms. The venom ducts were dissected with clamps and scissors while keeping the bodies of the snails on a Petri dish on ice. The venom ducts were homogenized in 500 μl of extraction solution (2% (v/v) trifluoroacetic acid (TFA) in 40% (v/v) aqueous acetonitrile (MeCN)) with a mechanically driven Potter-Elvehjem tissue grinder with glass pestle. The mixture was centrifuged at 11,130 × g for 15 min at 4°C, the supernatant was saved, and the process was repeated four times. The supernatants (crude venom extract) were combined. The total protein content of the extract was determined by the Lowry method, using bovine serum albumin as standard (DC Protein Assay; Bio-Rad, Hercules CA). The crude venom (final volume, 1.8 ml) was stored at −20°C until purified.
2.3. Crude venom fractionation and peptide purification
The venom crude extract was fractionated at room temperature by Reversed Phase-High Performance Liquid Chromatography using a C18 analytical column (Vydac; 218TP54, 4.6 × 250 mm, 5 μm particle diameter; 300 Å pore size) equipped with a C18 guard column (Vydac; 218GK54, 4.6 × 10 mm, 5μm particle diameter, 300 Å pore size), filter (Alltech; 28689, 4 mm, 2 μm pore size), and a 5-ml sample-loading loop. The elution was conducted at 1 ml/min, employing solutions A (0.1% (v/v) aqueous TFA) and B (0.1% (v/v) TFA in 90% (v/v) aqueous MeCN). A linear gradient from 5 to 95% solution B was developed over 90 min. The absorbance of the eluate was monitored at 220 nm. Before the injection, the crude venom (1.8 ml) was diluted to 5 ml with solution A.
Further purification was achieved by means of the same analytical C18 column, C18 precolumn, and sample-loading loop as above. The selected fraction was lyophilized, dissolved in 1 ml of extraction solution and the volume was adjusted to 5 ml with solution A. The sample was divided into two 2.5-ml aliquots; before the injection, each aliquot was diluted to 5 ml with solution A and then centrifuged at 11,130 × g for 15 min at room temperature. The elution was performed at 1 ml/min, employing the same solutions as above. A linear gradient from 27 to 41% solution B was developed over 56 min.
Final purification was carried out on an analytical C8 column (Vydac; 208TP54, 4.6 × 250 mm, 5 μm particle diameter, 300 Å pore size) with a C8 precolumn (Varian, 0120-MG, 4.6 × 15 mm, 5 μm particle size, 100 Å pore size) and a 500-μl sample-loading loop. The selected fraction was lyophilized and dissolved in 100 μl of extraction solution and then divided into two 50-μl aliquots; before the injection, each aliquot was diluted to 500 μl with solution A and then centrifuged at 11,130 × g for 15 min at room temperature. The elution was conducted at 1 ml/min, employing the same solutions as above. A linear gradient from 10 to 32% solution B over 88 min was employed.
2.4. Primary structure analysis
Sequence analysis and peptide quantitation were performed by automated Edman degradation in a Procise 491 Protein Sequencing System (Applied Biosystems, Foster City CA), using the Pulsed-Liquid Method and the 20 Amino Acid PTH Standard (Perkin Elmer Applied Biosystems, 400879). Peptide pa19a was sequenced without reduction and alkylation of potential disulfide bridges and Cys residues, respectively.
2.5. Molecular mass determination
A sample of the native peptide (~160 pmol) was subjected to matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry on a Voyager DE Mass Spectrometer (Applied Biosystems) equipped with delayed ion extraction. The spectra were obtained in positive reflector mode using sinapinic acid as matrix.
2.6. Sequence comparison
The sequence of the turritoxin characterized in this work was compared with the reported mature sequences of P-conotoxins (framework-IX) [18] and with framework-IX toxins from turrid [12, 43] and terebrid [15] species. The sequences were aligned with CLUSTALW2, employing the default settings [20, 40]. The alignment was entered into MEGA 4.0 [38], and the phylogenetic analysis was performed with the neighbor-joining method [37] using the Jones-Taylor-Thornton (JTT) matrix [17] and the Poisson correction [45], with pairwise deletion of gaps. Bootstrap values [7] were estimated from 2000 replicates (95% confidence) [11] with a random seed.
3. Results
3.1. Peptide purification
In order to continue the characterization of the toxins of Polystira albida, five minor hydrophilic peaks, eluting from ~23 to ~31 min during the fractionation of the crude venom (total protein, 3.8 mg) on an analytical C18 column were chosen for study. The chemical characterization of the first four components has not been completed and will be reported elsewhere. The fifth fraction (arrow in Fig. 1A) displayed one major component and several minor components after rechromatography on the same column; the major peak (arrow in Fig. 1B) was further purified on an analytical C8 column, yielding one major peak (arrow in Fig. 1C).
Figure 1.
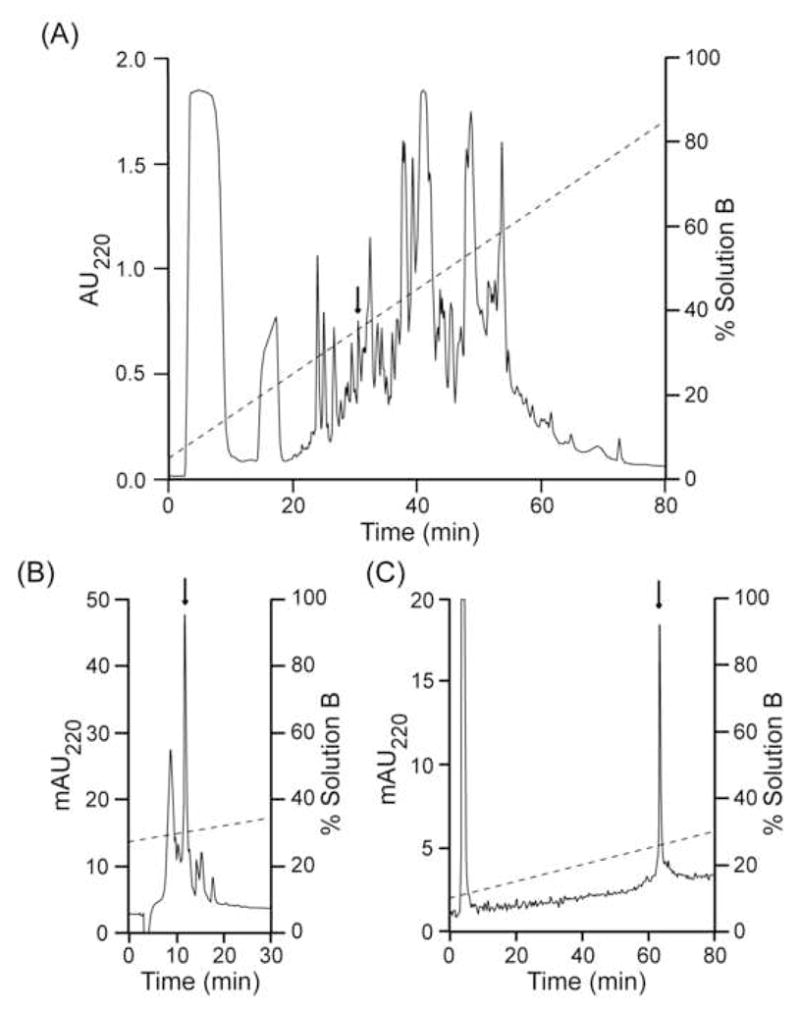
Purification of peptide pal9a by RP-HPLC. All chromatographies were conducted at room temperature, and the eluates were monitored for absorbance at 220 nm (AU220, absorbance units; mAU220, absorbance milliunits). Panel A, fractionation of the venom extract from 10 P. albida specimens by an analytical C18 column eluted with a linear gradient from 5 to 95% solution B over 90 min at 1 ml/min. HPLC solutions were 0.1% (v/v) aqueous TFA (sol. A) and 0.1% (v/v) TFA in 90% (v/v) aqueous MeCN (sol. B). Panel B, the peak indicated by the arrow in panel A was further purified on the same column using a linear gradient from 27 to 41% solution B over 56 min at 1 ml/min, employing the same solutions as above. Panel C, the peak indicated by the arrow in panel B was further purified on an analytical C8 column using a linear gradient from 10 to 32% solution B over 88 min at 1 ml/min, employing the same solutions as above.
3.2. Molecular mass determination
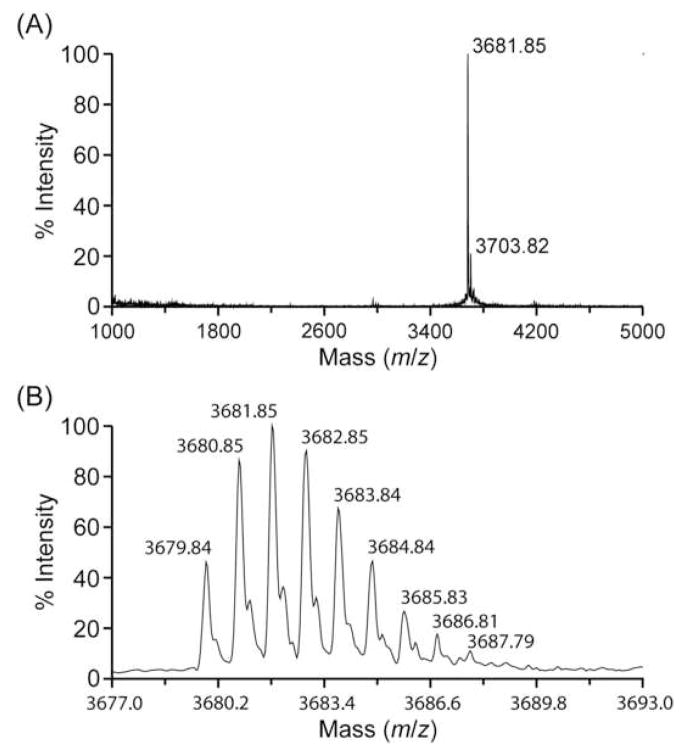
The MALDI-TOF mass spectra were obtained using 20% of the purified peptide and are shown in Figure 2. In panel A, the m/z signal at 3,681.85 corresponds to an average mass of 3,680.85 Da; the minor m/z signal at 3,703.82, which is ~22 units higher than the major m/z signal, is probably derived from a small fraction of sodium adducts. In panel B, the deconvoluted spectrum from panel A shows a m/z signal at 3,679.84, which corresponds to a monoisotopic mass of 3,678.84 Da.
Figure 2.
Molecular mass of peptide pal9a. Panel A, MALDI-TOF mass spectrum of the purified peptide; the m/z signal at 3,681.85 corresponds to an average mass of 3,680.85 Da; the minor m/z signal at 3,703.82, which is ~22 units higher than the major m/z signal, is probably derived from a small fraction of sodium adducts. Panel B, the deconvoluted spectrum from panel A shows a m/z signal at 3,679.84 that corresponds to a monoisotopic mass of 3,678.84 Da.
3.2. Peptide sequencing
Automated Edman sequencing (39 cycles) using 25% of the purified peptide (~160 pmol) yielded an unambiguous sequence up to position 34 (Figure 3). At positions 3, 8, 13, 17, 19, and 30, no increase of any PTH-derivative was observed; given that the peptide was sequenced without alkylation of Cys residues, we tentatively assigned these positions as Cys. At position 32, two residues increased by similar, very small amounts (Ser, 0.4 pmol; Tyr, 0.3 pmol), which might be due to sequence microheterogeneity.
Figure 3.
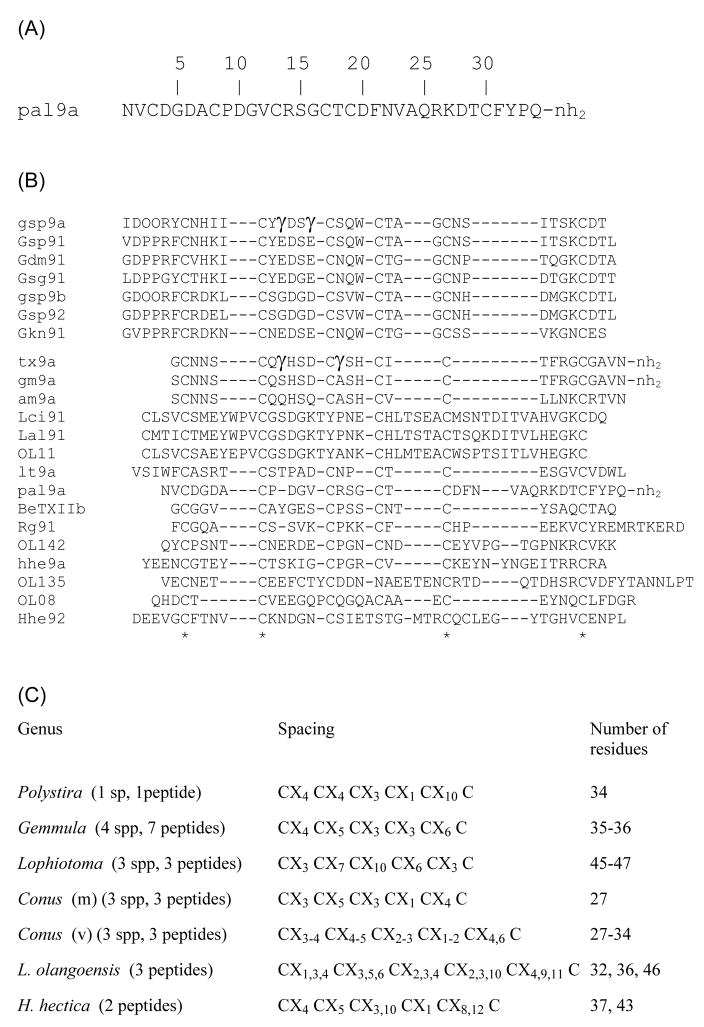
Primary structure of turritoxin pal9a and comparison with other framework IX-conopeptides. Panel A, Sequence of peptide pal9a from P. albida. Panel B, CLUSTALW2 multiple alignment of peptide pal9a with other framework IX-conopeptides (either purified or cloned) from Gemmula speciosa (gsp9a, Gsp91, gsp9b, Gsp92), G. diomedea (Gdm91), G. sogodensis (Gsg91), G. kieneri (Gkn91), C. textile (tx9a), C. gloriamaris (gm9a), C. amadis (am9a), C. litteratus (lt9a), C. betulinus (BeTXIIb), C. regius (Rg91), Lophiotoma olangoensis (OL11, OL142, OL135, OL08), L. cingulifera (Lci91), L. albina (Lal91), and Hastula hectica (hhe9a, Hhe92). The one-letter code is employed for standard amino acids. For posttranslational modifications: O, 4-hydroxy-Pro;γ, γ-carboxyglutamate;-nh2, amidated C-terminus. Identical residues (*) are shown for all the peptides (bottom line). Panel C, comparison of intercysteine spacings of all the known framework IX-conopeptides; the feeding type of the Conus species is indicated in parentheses: m, molluscivorous; v, vermivorous.
Assuming that the six Cys residues form three disulfide bonds, the theoretical monoisotopic masses of the putative species containing Ser at position 32 are 3,604.43 Da (for a free C-terminus) and 3,603.45 (for an amidated C-terminus); the average of these values is 74.90 Da lower than the experimental monoisotopic mass (3,678.84 Da). With the same assumption for the Cys residues, the theoretical monoisotopic masses of the putative species containing Tyr at position 32 are 3,680.46 Da (for a free C-terminus) and 3,679.48 Da (for an amidated C-terminus); the average of these values is 1.13 Da higher than the experimental monoisotopic mass (3,678.84 Da). Therefore, we concluded that the small increase of Ser at position 32 was an artifact, and that the purified turritoxin contains Tyr at this position. The theoretical monoisotopic mass of the 34-residue peptide sequence containing Tyr at position 32 are 3,680.46 Da for a free C-terminus and 3,679.48 Da for an amidated C-terminus; the latter value is in good agreement with the experimentally determined monoisotopic mass (3,678.84 Da). Thus, we concluded that this turritoxin is a 34-residue peptide containing 3 disulfide bridges and an amidated C-terminus (Figure 3A).
4. Discussion
The purification and biochemical and biological characterization of peptides contained in the venoms of turrid snails has been hampered by two factors: 1) to obtain enough biological material is very difficult because these species tend to inhabit deep waters, and they do not live in concentrated populations; and 2) turrids generally are smaller than most of the cone snails that have been studied, and therefore they have short and very narrow venom ducts [12, 43].
Here we report the purification and primary structure determination of a minor peptide from the venom of the turrid snail Polystira albida, collected in the Gulf of Mexico. Following a recent proposal for the nomenclature of conoidean venom peptides [12], the turritoxin characterized in this work was named pal9a. This peptide contains 34 amino acid residues, including six Cys residues in the arrangement C-C-C-C-C-C (where “-“ represents one or more non-Cys residues). This peptide does not contain post-translational modifications except the disulfide bridges and the amidated C-terminus. Due to the small number of collected organisms, which have very short, narrow venom ducts, the amount of the purified peptide was not enough for even a preliminary biological characterization, and for determining the connectivity of the Cys residues.
The first peptide with the Cys pattern C-C-C-C-C-C characterized from the venom of a marine snail was toxin BeTXIIa, from the vermivorous species Conus betulinus, from the South China Sea [6]. Soon thereafter, this Cys arrangement was found in the spasmodic peptide tx9a from the molluscivorous species C. textile from the Indo-Pacific, and in a cDNA clone from the piscivorous species C. geographus from the same region [21], which when synthesized (gm9a) elicited the same symptoms in mice [26]; these peptides, known as “the spasmodic peptides”, defined the P-superfamily of conotoxins and the Cys framework number IX [21]. Toxin tx9a contains two gamma-carboxy-Glu residues and an amidated C-terminus, whereas gm9a is predicted to have an amidated C-terminus. Recently, this pattern was also found in a peptide from C. amadis, a mollusk-hunting species from the Indo-Pacific, and in clones from the worm-hunting species C. regius (one clone), from the Western Atlantic [18] and C. litteratus (nine clones corresponding to three variants, one of which, lt9a, was identified in the venom of this species) [34].
Framework-IX venom peptides have also been found in members of families Turridae and Terebridae. In the turrid Lophiotoma olangoensis from the Philippines, the sequences of four P-type peptides were deduced from a venom duct cDNA library, and one sequence was also cloned from the related species L. cingulifera and L. albina; in these cases, no post-translational modifications other than the disulfide bonds were predicted. Except for the Cys pattern, these peptides are heterogeneous (in sequence and Cys spacings), and the authors concluded that these peptides probably do not belong to a single superfamily [43]. Recently, two post-translationally modified (with 4-hydroxy-Pro and γ-carboxy-Glu residues) P-conotoxin-like peptides were characterized from the turrid Gemmula speciosa from the Philippines; in addition, the sequences of two cDNA clones related to these peptides were reported, as well as one cloned sequence each from G. sogodensis, G. diomedea, and G. kieneri. In contrast to the clones from Lophiotoma, the signal sequences of the five cloned precursors were reported, and they are almost identical, which demonstrates the existence in these Gemmula species of a toxin gene superfamily designated the Pg-superfamily; however, although the Cys spacings are totally conserved among the seven peptides or clones, the purified or predicted mature toxins are very divergent in sequence [12]. Regarding the terebrid snails, one P-conotoxin-like peptide, hhe9a, and two related cDNA clones (one of which corresponded to the purified peptide) were characterized from Hastula hectica from the Philippines. With the exception of the disulfide bridges, the purified peptide has no post-translational modifications, and the clones were not predicted to contain any other post-translational modification; the purified or predicted mature toxins have distinct Cys spacings and a low degree of sequence similarity [15].
It was proposed recently that the term “conopeptide” be used to refer to peptides contained in the venoms of all conoidean species, not just to those from Conus venoms [12]; in the rest of this discussion we have followed this recommendation.
Given that a low degree of sequence similarity was found between the signal peptide and the mature regions of framework IX-conopeptides expressed in the terebrid H. hectica (clone Hhe9.1, peptide hhe9a) and in the turrid L. olangoensis (clone Lol142) [15], we compared the sequence of the peptide characterized in this work, pal9a, not only with the known framework IX-turritoxins, but also with the other conopeptides mentioned above. The CLUSTALW2 multiple sequence alignment (Figure 3B) shows that only four out of six Cys residues can be considered as conserved among the 22 sequences, due to different Cys spacings in distinct groups of peptides or individual peptides (even within the same genus or species). However, several toxins share Cys spacings and a similar number of residues: the seven peptides from the four species of Gemmula, the three peptides from the three molluscivorous Conus species, and three peptides from three species of Lophiotoma. Despite these similarities, the sequence heterogeneity noticed by other authors for some peptides from the species of Lophiotoma [43] and Gemmula [12] is apparent. The peptides from the three vermivorous Conus species are slightly heterogeneous in size and Cys spacings, whereas the remaining peptides from L. olangoensis and H. hectica are very heterogeneous in size and Cys spacings. Peptide pal9a has a size similar to that of the toxins from the turrid Gemmula species, but only two of its Cys spacings are identical to those of these toxins (Figure 3C); in addition, turritoxin pal9a has little sequence (11–22%) identity to any framework IX-conopeptide (Table 1).
Table 1.
| gsp9a | Gsp91 | Gdm91 | Gsg91 | gsp9b | Gsp92 | Gkn91 | tx9a | gm9a | am9a | Lci91 | Lal91 | OL11 | lt9a | pal9a | BeTXIIb | Rg91 | OL142 | hhe9a | OL135 | OL08 | Hhe92 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gsp9a | 100 | 91 | 71 | 74 | 57 | 57 | 51 | 25 | 22 | 22 | 11 | 8 | 11 | 9 | 14 | 22 | 8 | 20 | 11 | 11 | 15 | 17 |
| Gsp91 | 100 | 75 | 72 | 63 | 61 | 57 | 25 | 22 | 22 | 11 | 8 | 11 | 12 | 14 | 14 | 8 | 16 | 11 | 11 | 18 | 16 | |
| Gdm91 | 100 | 75 | 61 | 58 | 65 | 18 | 14 | 22 | 8 | 8 | 11 | 25 | 14 | 14 | 14 | 16 | 11 | 13 | 18 | 16 | ||
| Gsg91 | 100 | 61 | 58 | 51 | 14 | 11 | 22 | 22 | 13 | 13 | 22 | 17 | 14 | 14 | 19 | 22 | 13 | 18 | 16 | |||
| gsp9b | 100 | 97 | 54 | 14 | 14 | 22 | 11 | 11 | 8 | 29 | 20 | 18 | 14 | 11 | 11 | 8 | 9 | 19 | ||||
| Gsp92 | 100 | 51 | 14 | 14 | 22 | 13 | 11 | 8 | 29 | 20 | 18 | 14 | 11 | 13 | 8 | 9 | 19 | |||||
| Gkn91 | 100 | 29 | 18 | 18 | 8 | 8 | 11 | 25 | 14 | 11 | 17 | 20 | 14 | 14 | 18 | 14 | ||||||
| tx9a | 100 | 88 | 59 | 11 | 7 | 14 | 22 | 22 | 22 | 18 | 18 | 14 | 14 | 14 | 14 | |||||||
| gm9a | 100 | 66 | 11 | 7 | 11 | 22 | 22 | 18 | 22 | 14 | 18 | 11 | 18 | 14 | ||||||||
| am9a | 100 | 14 | 18 | 22 | 14 | 22 | 25 | 18 | 14 | 18 | 25 | 22 | 14 | |||||||||
| Lci91 | 100 | 75 | 73 | 12 | 17 | 18 | 8 | 13 | 13 | 10 | 9 | 16 | ||||||||||
| Lal91 | 100 | 64 | 22 | 17 | 14 | 8 | 11 | 13 | 11 | 12 | 9 | |||||||||||
| OL11 | 100 | 9 | 14 | 18 | 8 | 11 | 16 | 6 | 6 | 13 | ||||||||||||
| lt9a | 100 | 19 | 18 | 16 | 29 | 16 | 16 | 12 | 19 | |||||||||||||
| pal9a | 100 | 18 | 17 | 11 | 14 | 17 | 15 | 14 | ||||||||||||||
| BeTXIIb | 100 | 22 | 22 | 14 | 14 | 14 | 7 | |||||||||||||||
| Rg91 | 100 | 17 | 14 | 20 | 15 | 14 | ||||||||||||||||
| OL142 | 100 | 27 | 16 | 12 | 11 | |||||||||||||||||
| hhe9a | 100 | 16 | 15 | 24 | ||||||||||||||||||
| OL135 | 100 | 9 | 16 | |||||||||||||||||||
| OL08 | 100 | 9 | ||||||||||||||||||||
| Hhe92 | 100 |
The percent identity scores were taken from CLUSTALW2 pairwise alignments (data not shown), calculated using the default settings (Matrix, Gonnet 250; Gap open penalty, 10.0; Gap extension penalty, 0.2; Gap separation penalty, 4.0; with exclusion of end gaps).
Names were taken from the original references, except for the clones from C. amadis, L. cingulifera, and L. albina; in these cases we assigned the names according to the nomenclature recently proposed for conoidean venom peptides [12].
The identity scores of turritoxin pal9a with the other conopeptides are shaded.
The identity scores among the conopeptides that are grouped within the three branches of the consensus phylogenetic tree are in bold face and underlined.
The phylogenetic analysis supports the similarities revealed initially by the multiple sequence alignment. The two analyses (JTT matrix and Poisson correction) yielded essentially similar results when unrooted consensus trees were calculated with a cut-off value of 50% (data not shown). In Figure 4, three branches are clearly defined with 87%, 98%, and 73% probability, corresponding to the seven toxins from the Gemmula species, the three toxins from three Lophiotoma species, and the three toxins from the three molluscivorous Conus species, respectively. Thus, this analysis is consistent with the demonstrated existence of gene superfamilies of framework-IX conopeptides in Gemmula species [12] and molluscivorous Conus species [21] from the Indo-Pacific. However, this analysis did not clarify the relationship of peptide pal9a with any individual framework IX-toxin, or the relationships of the other eight peptides or clones (lt9a, BeTXIIb, Rg91, OL142, hhe9a, OL135, OL08, and Hhe92). In retrospect, these latter results are not unexpected, given the low sequence similarities between peptide pal9a and every other framework IX-conopeptide, and among the eight peptides or clones mentioned above and the other peptides (Table 1); this sequence heterogeneity may be related to differences in the origin of the peptide (taxonomical family, genus, species, and geographical region). In fact, it has been pointed out that it is likely that the four framework IX-conopeptides from L. olangoensis do not belong to a single gene superfamily [43].
Figure 4.

Evolutionary relationships of mature framework IX-conopeptides. The CLUSTALW2 multiple sequence alignment (Figure 3B) was entered into MEGA4 [38]. The probable evolutionary history was inferred using the Neighbor-Joining method [37]. The bootstrap consensus tree from 2000 replicates is taken to represent the evolutionary history of the sequences analyzed [7]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentages of replicate trees in which the associated sequences clustered together in the bootstrap test are shown next to the branches.
Despite the striking differences in sequence among some of the 22 framework IX-conopeptides (Table 1), we realized that, in general, they share two physicochemical properties: 1) with two exceptions (BeTXIIb and lt9a, both from vermivorous cone snails), they are hydrophilic molecules; and 2) with four exceptions (two from molluscivorous cones, one from a vermivorous cone snail, and one from a terebrid snail), they are acidic compounds predicted to have negative net charge at physiological pH (Table 2).
Table 2.
Physicochemical properties of framework IX-conopeptidesa
| Peptide or cloneb |
Grand average of hydropathicity (GRAVY)c |
Theoretical pIc | Net charged |
|---|---|---|---|
| gsp9a | −0.491e | 4.43e | −3 |
| Gsp91 | −0.500 | 4.83 | −2 |
| Gsg91 | −0.856 | 4.23 | −4 |
| Gdm91 | −0.839 | 4.83 | −2 |
| gsp9b | −0.564e | 4.75 | −2 |
| Gsp92 | −0.553 | 4.27 | −4 |
| Gkn91 | −1.006 | 4.94 | −1 |
| tx9a | −0.422e | 5.27e | −3 |
| gm9a | −0.141e | 6.64e | +1 |
| am9a | −0.385 | 8.32 | +2 |
| Lci91 | −0.194 | 4.57 | −4 |
| Lal91 | −0.280 | 6.87 | 0 |
| OL11 | −0.067 | 5.42 | −2 |
| pal9a | −0.456e | 4.28e | −1 |
| BeTXIIb | +0.007 | 4.00 | −1 |
| hhe9a | −0.949 | 7.76 | +1 |
| OL142 | −1.339 | 6.24 | 0 |
| Rg91 | −0.932 | 8.64 | +3 |
| OL135 | −1.135 | 3.85 | −10 |
| OL08 | −0.728 | 4.08 | −5 |
| Hhe92 | −0.447 | 4.25 | −5 |
| lt9a | +0.423 | 4.03 | −2 |
Values that are distinct from the majority are shaded.
Names were taken from the original references, except for the clones from C. amadis, L. cingulifera, and L. albina; in these cases we assigned the names according to the nomenclature recently proposed for conoidean venom peptides [12]. With the exception of BeTXIIb, all names with an initial capital letter correspond to clones.
Calculated by means of ProtParam [8]: GRAVY [19]; pI [3]. For the sequences deduced by cDNA cloning, the parameters have been calculated without taking into account potential post-translational modifications.
Manually calculated using standard values for the pKas of the NH2-, COOH, and the side chain moieties of the standard amino acids [25]. For the sequences deduced from cDNA clones, the net charges have been calculated without taking into account potential residues of gamma-carboxy-Glu.
Approximate values, because hydroxylation of Pro residues, amidation of the C-terminus and/or gamma-carboxylation of Glu residues are not taken into account by ProtParam.
Interestingly, we also realized that, apparently, framework IX-conopeptides have not been reported from piscivorous Conus species. In order to confirm this idea, we searched the ConoServer database [18] and all the databases of the Entrez cross-database and found no framework IX-conotoxins from fish-hunting species. In addition, a comprehensive analysis of expressed sequence tags (ESTs) from the venom duct of the fish-hunting species C. striatus revealed the expression of 221 toxin-encoding ESTs belonging to the gene superfamilies A (132 ESTs), O (80 ESTs), T (6 ESTs), M (1 EST), S (1 EST), and contryphans (1 EST), but no ESTs of P-conotoxins [35]. For the case of Conus species, it has been pointed out that: 1) the outstanding variation in peptide composition of the venoms can be rationalized in terms of the different behavior, biotic interactions, and evolutionary history of the distinct species, and 2) there are toxin families characteristic of clades of related species. For example, in piscivorous Conus, δ- and κ-conotoxins have not been found in species belonging to clade II (such as C. geographus), a group that employs the “net strategy” for capturing prey, but these types of toxins are present in species within clade III (such as C. striatus), which use the “hook-and-line” strategy. Furthermore, it has been shown that there are clades that, despite using the same capture strategy, produce distinct sets of toxins; for instance, C. striatus (clade I, Indo-Pacific) expresses α-, “long” κA-, and ω-conotoxins but not αA-, ψ-, or κ-conotoxins, whereas C. purpurascens (clade III, Eastern Pacific) produces αA-, ψ-, and κ-conotoxins but not α-, “long” κA- or ω-conotoxins [30]. Thus, it is possible that framework IX-conoidean peptides are characteristic of mollusk- (some clades of cone snails) and worm-hunting species (turrid and terebrid snails, and some clades of cone snails), but are not produced by fish-hunting species (some clades of cone snails).
Thus far, the molecular target of any framework IX-conopeptide has not been identified. In fact, only peptides tx9a, gm9a, and hhe9a have been tested by bioassays; peptides tx9a and gm9a (the “spasmodic peptides”), when injected intracranially into mice, produce sensory hypersensitivity similar to that observed in mice carrying the spasmodic or the spastic mutations [21, 26], whereas peptide hhe9a did not elicit any detectable change when injected into the pseudocoelomic space of the nematode C. elegans [15]. Given that the symptomatology of the spasmodic and spastic mutant mice is the result of a deficit in Gly receptors, it has been suggested that this receptor might be the target of the spasmodic peptides; however, although preliminary experiments with cloned Gly receptors did not yield positive results, they left open the possibilities that these peptides block a specific subtype of Gly receptors and/or that they act noncompetitively on this receptor [21].
It has been pointed out that structure-function analyses of peptide toxins have yielded few clues that help to predict molecular targets [28]; this is especially true of peptides such as the framework IX-conopeptides with no structural counterparts whose pharmacological targets are known. There is an obvious toxinological interest in uncovering the molecular target(s) of this type of toxins; furthermore, as has been pointed out by Miles et al., among the six Cys-containing conopeptides, the scaffold of framework IX-peptides has the highest potential for structural and functional diversity [26], which has implications for protein engineering and for mimicking interesting epitopes [27, 41].
Although the molecular target of the framework IX-conopeptides is unknown, they have several noteworthy features: they have not been found in piscivorous Conus species; in general they are hydrophilic, acidic compounds, and it is very likely that they belong to distinct gene superfamilies, even those from the same species.
Acknowledgments
This investigation was supported by Grants: IN206701 (to E.H.C. and M.B.A.), IX211904 (to M.B.A.), and IN-204403 (to E.H.C.) from the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México, and GM 48677 from the National Institute of General Medical Sciences (to B.M.O.). R.A.C.R. was supported by a contract as Level III Researcher (E.H.C.) Assistant from the Sistema Nacional de Investigadores del Consejo Nacional de Ciencia y Tecnología, México. We thank the officers and crew of the RV Justo Sierra for the trawling operations to obtain the specimens of P. albida during the campaign SMG-6 under the direction of Dr. Adolfo Gracia Gasca. We gratefully acknowledge Dr. Dorothy D. Pless for revising the manuscript. We also thank Lic. Pilar Galarza for retrieving bibliographic references.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aguilar MB, López-Vera E, Ortiz E, Becerril B, Possani LD, Olivera BM, et al. A novel conotoxin from Conus delessertii with posttranslationally modified lysine residues. Biochemistry. 2005;44:11130–6. doi: 10.1021/bi050518l. [DOI] [PubMed] [Google Scholar]
- 2.Armishaw CJ, Alewood PF. Conotoxins as research tools and drug leads. Curr Protein Pept Sci. 2005;6:221–40. doi: 10.2174/1389203054065437. [DOI] [PubMed] [Google Scholar]
- 3.Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–39. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- 4.Bratcher T, Cernohorsky WO. Living Terebras of the world. New York: American Malacologists, Inc; 1987. [Google Scholar]
- 5.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–79. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JS, Fan CX, Hu KP, Wei KH, Zhong MN. Studies on conotoxins of Conus betulinus. J Nat Toxins. 1999;8:341–9. [PubMed] [Google Scholar]
- 7.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 8.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 9.Grant MA, Morelli XJ, Rigby AC. Conotoxins and structural biology: a prospective paradigm for drug discovery. Curr Protein Pept Sci. 2004;5:235–48. doi: 10.2174/1389203043379710. [DOI] [PubMed] [Google Scholar]
- 10.Gray WR, Olivera BM, Cruz LJ. Peptide toxins from venomous Conus snails. Annu Rev Biochem. 1988;57:665–700. doi: 10.1146/annurev.bi.57.070188.003313. [DOI] [PubMed] [Google Scholar]
- 11.Hall BG. Phylogenetic trees made easy. Sunderland: Sinauer Associates, Inc; 2004. p. 50. [Google Scholar]
- 12.Heralde FM, III, Imperial J, Bandyopadhyay PK, Olivera BM, Concepcion GP, Santos AD. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon. 2008;51:890–7. doi: 10.1016/j.toxicon.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imperial JS, Watkins M, Chen P, Hillyard DR, Cruz LJ, Olivera BM. The augertoxins: biochemical characterization of venom components from the toxoglossate gastropod Terebra subulata. Toxicon. 2003;42:391–8. doi: 10.1016/s0041-0101(03)00169-7. [DOI] [PubMed] [Google Scholar]
- 14.Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, et al. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry. 2006;45:8331–40. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- 15.Imperial JS, Kantor Y, Watkins M, Heralde FM, III, Stevenson B, Chen P, et al. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. J Exp Zoolog B Mol Dev Evol. 2007;308:744–56. doi: 10.1002/jez.b.21195. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez EC, Olivera BM, Teichert RW. αC-conotoxin PrXA: a new family of nicotinic acetylcholine receptor antagonists. Biochemistry. 2007;46:8717–24. doi: 10.1021/bi700582m. [DOI] [PubMed] [Google Scholar]
- 17.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 18.Kaas Q, Westermann JC, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24:445–6. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- 19.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 21.Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay P, Cruz LJ, et al. The spasmodic peptide defines a new conotoxin superfamily. Biochemistry. 2000;39:1583–8. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- 22.Livett BG, Gayler KR, Khalil Z. Drugs from the sea: conopeptides as potential therapeutics. Curr Med Chem. 2004;11:1715–23. doi: 10.2174/0929867043364928. [DOI] [PubMed] [Google Scholar]
- 23.López-Vera E, Heimer de la Cotera EP, Maillo M, Riesgo-Escovar JR, Olivera BM, Aguilar MB. A novel structural class of toxins: the methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea) Toxicon. 2004;43:365–74. doi: 10.1016/j.toxicon.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Loughnan M, Nicke A, Jones A, Schroeder CI, Nevin ST, Adams DJ, et al. Identification of a novel class of nicotinic receptor antagonists: dimeric conotoxins VxXIIA, VxXIIB, and VxXIIC from Conus vexillum. J Biol Chem. 2006;281:24745–55. doi: 10.1074/jbc.M603703200. [DOI] [PubMed] [Google Scholar]
- 25.Mathews CK, van Holde KE, Ahern KG. Biochemistry. 3. San Francisco, CA: Benjamin/Cummings, an Imprint of Addison Wesley Longman; 1999. p. 128. [Google Scholar]
- 26.Miles LA, Dy CY, Nielsen J, Barnham KJ, Hinds MG, Olivera BM, et al. Structure of a novel P-superfamily spasmodic conotoxin reveals an inhibitory cystine knot motif. J Biol Chem. 2002;277:43033–40. doi: 10.1074/jbc.M206690200. [DOI] [PubMed] [Google Scholar]
- 27.Mourier G, Servent D, Zinn-Justin S, Ménez A. Chemical engineering of a three-fingered toxin with anti-α7 neuronal acetylcholine receptor activity. Protein Eng. 2000;13:217–25. doi: 10.1093/protein/13.3.217. [DOI] [PubMed] [Google Scholar]
- 28.Norton RS. Toxin Structure and Function: What Does Structural Genomics Have To Offer? In: Ménez A, editor. Perspectives in molecular toxinology. New York: John Wiley & Sons, Inc; pp. 159–73. [Google Scholar]
- 29.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–98. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Olivera BM. Conus venom peptides: Reflections from the biology of clades and species. Annu Rev Ecol Syst. 2002;33:25–47. [Google Scholar]
- 31.Olivera BM, Rivier J, Scott JK, Hillyard DR, Cruz LJ. Conotoxins. J Biol Chem. 1991;266:22067–79. [PubMed] [Google Scholar]
- 32.Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, et al. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27:2174–81. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 33.Peng C, Liu L, Shao X, Chi C, Wang C. Identification of a novel class of conotoxins defined as V-conotoxins with a unique cysteine pattern and signal peptide sequence. Peptides. 2008;29:985–91. doi: 10.1016/j.peptides.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Pi C, Liu J, Peng C, Liu Y, Jiang X, Zhao Y, et al. Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics. 2006;88:809–19. doi: 10.1016/j.ygeno.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Pi C, Liu Y, Peng C, Jiang X, Liu J, Xu B, et al. Analysis of expressed sequence tags from the venom ducts of Conus striatus: focusing on the expression profile of conotoxins. Biochimie. 2006;88:131–40. doi: 10.1016/j.biochi.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Powell AWB. The molluscan families Speightiidae and Turridae: An evaluation of the valid taxa, both recent and fossil, with lists of characteristic species. Auckland, New Zealand: Unity Press Limited; 1966. [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Taylor JD, Kantor YI, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (= Toxoglossa) (Gastropoda) Bull Nat Hist Mus Lond (Zool) 1993;59:125–70. [Google Scholar]
- 40.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vita C, Drakopoulou E, Vizzavona J, Rochette S, Martin L, Ménez A, et al. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc Natl Acad Sci U S A. 1999;96:13091–6. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang CZ, Chi CW. Conus peptides - a rich pharmaceutical treasure. Acta Biochim Biophys Sin (Shanghai) 2004;36:713–23. doi: 10.1093/abbs/36.11.713. [DOI] [PubMed] [Google Scholar]
- 43.Watkins M, Hillyard DR, Olivera BM. Genes expressed in a turrid venom duct: divergence and similarity to conotoxins. J Mol Evol. 2006;62:247–56. doi: 10.1007/s00239-005-0010-x. [DOI] [PubMed] [Google Scholar]
- 44.Wermeling DP. Ziconotide, an intrathecally administered N-type calcium channel antagonist for the treatment of chronic pain. Pharmacotherapy. 2005;25:1084–94. doi: 10.1592/phco.2005.25.8.1084. [DOI] [PubMed] [Google Scholar]
- 45.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York NY: Academic Press; 1965. pp. 97–166. [Google Scholar]