Abstract
Pmel17 is a melanocyte protein necessary for eumelanin deposition 1 in mammals and found in melanosomes in a filamentous form. The luminal part of human Pmel17 includes a region (RPT) with 10 copies of a partial repeat sequence, pt.e.gttp.qv., known to be essential in vivo for filament formation. We show that this RPT region readily forms amyloid in vitro, but only under the mildly acidic conditions typical of the lysosome-like melanosome lumen, and the filaments quickly become soluble at neutral pH. Under the same mildly acidic conditions, the Pmel filaments promote eumelanin formation. Electron diffraction, circular dichroism, and solid-state NMR studies of Pmel17 filaments show that the structure is rich in beta sheet. We suggest that RPT is the amyloid core domain of the Pmel17 filaments so critical for melanin formation.
While amyloids are usually associated with disease processes, there are several amyloids that clearly are functional. The curli fibers on the surface of E. coli (and other Enterobacteria) have a role in host cell adhesion and biofilm formation (1), hydrophobins protect the surface of fungi (2), as do amyloids on the surface of eggs of fish (3) or silkmoths (4). There is even an amyloid-based prion (infectious protein), [Het-s] of Podospora anserina, whose properties suggest it may be functional for the host, rather than a disease (5).
The melanosome is an organelle, related to both endosomes and lysosomes, in which melanin is synthesized in cells in the skin and eye (reviewed in ref. 6). As it is synthesized, the melanin is deposited on fibrils, which are found in parallel arrays in the melanosome lumen. In addition to enzymes such as tyrosinase and DOPAchrome tautomerase that prepare the tyrosine-derived substrates for polymerization into melanin, a structural protein, called variously Pmel17/gp100/SILV is critical for melanin formation. Pmel17 was first identified genetically as the silver gene of mice whose mutation produces hypopigmentation (7). A cDNA clone whose transcript was preferentially expressed in melanocytes was named Pmel17 and mapped to the mouse silver locus (8).
Pmel17 is a 668-residue protein with a single transmembrane domain, a short C-terminal cytoplasmic domain, and a large luminal domain with several distinct regions [Fig. 1; reviewed in (9)]. Pmel17 is the major component of the melanosome fibrils as shown by absence of the fibers in melanocytes from silver mice (10), immunostaining of fibers by anti-Pmel monoclonal antibodies (11), and the appearance of fiber-containing melanosome-like structures in nonpigment cells induced by overexpression of Pmel17 (12). Cleavage by a Kex2-group protease after the dibasic residues 468 and 9, producing a C-terminal fragment Μβ including the transmembrane and cytoplasmic domains and the N-terminal fragment Μα with most of the luminal domain, is necessary for fibrillogenesis (13). Further cleavages of Μα produce ΜαΝ and ΜαC fragments, the latter including the filament-forming region (14, 15). The parallel with gelsolin and amylin amyloid formation following their cleavage from precursors by similar proteases was noted (13). Indeed, Fowler et al. found that isolated melanosomes from bovine eyes stain with thioflavin S and Congo red, 2 dyes relatively specific for amyloids (16). Moreover, they reported that purified Mα, when diluted from guanidine, formed amyloid within 2 s, as measured by thioflavin T fluorescence, in a pH-independent reaction. This Pmel17 amyloid, as well as amyloids of Aβ and α-synuclein, nearly doubled the rate of melanin formation in vitro (16).
Fig. 1.
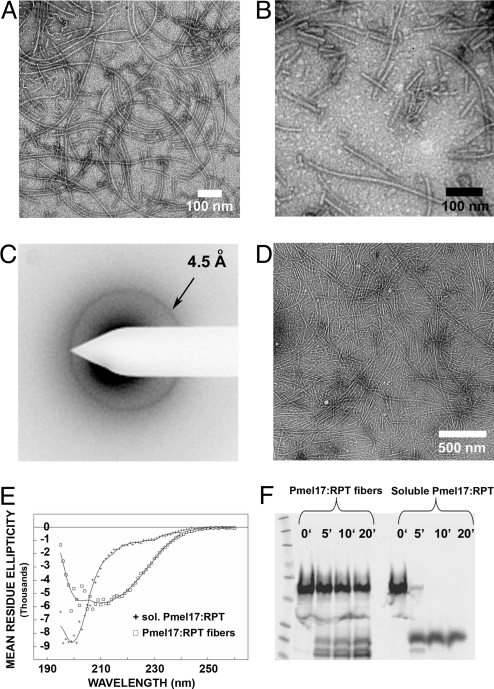
The Pmel RPT domain forms fibers under nondenaturing conditions. (A) Full-length Pmel17 is composed of 10 domains. SIG, signal peptide; NTD, N-terminal domain; PKD, polycystic kidney disease-like domain; RPT, proline/serine/threonine-rich repeat domain; KRG, kringle-like domain; TM, transmembrane domain; CTD, C-terminal domain; GAP1, GAP2, and GAP3, undefined domains. Known N-glycosylation sites are indicated above the NTD, RPT, and GAP3 domains by a “Y”. The RPT domain is O-glycosylated. The fragments examined for amyloid formation are shown with endpoints and His6 tag, if any. (B) SDS/PAGE (10%) analysis of RPT purified under denaturing conditions. (C) Sequence of the RPT domain, showing the imperfect repeats rich in proline, serine and threonine residues. (D) Transmission electron micrographs of Pmel17:RPT fibers negatively stained with uranyl acetate.
The Pmel17 protein includes (N-terminal to C-terminal) a signal sequence, an N-terminal region (NTR) lacking any revealing homology, a polycystic kidney disease domain (PKD), a repeat (RPT) region with 10 imperfect copies of the sequence “pt.e.gttp.qv.,” a kringle-like domain (KRG), the transmembrane domain, and the cytoplasmic domain (9). Several of these domains are necessary for proper sorting of Pmel17 into the melanosome compartment. The RPT region of Pmel17 is not essential for the protein's proper inclusion in melanosome precursor organelles but is crucial for the formation of the melanosome filaments (14, 17). There are 4 splice variants of human Pmel17, lacking either 7 amino acids near the transmembrane domain or 42 residues inside the repeat domain (18, 19). The most abundant form lacks only the 7 residues near the transmembrane domain and was used in our study.
Amyloid is a filamentous protein form characterized by a cross β-sheet structure (β-strands aligned perpendicular to the filament axis), protease-resistance and special staining properties. Using solid-state NMR, amyloids of the Alzheimer's Aβ peptide and the diabetes-associated amyloid peptide amylin are in-register parallel β-sheets (identical residues of different molecules aligned along the fiber long axis) (20–22), as were shown to be the infectious amyloids of the prion domains of Sup35p, Ure2p, and Rnq1p that are the basis of the yeast prions [PSI+], [URE3], and [PIN+] (23–25). Electron spin resonance studies showed in-register parallel β-sheet structures of amylin (26), α-synuclein (Parkinson's disease) (27), and Tau (Alzheimer's disease and other ‘tauopathies’) (28). Some fragments of pathogenic peptides and crystal structures of peptides capable of forming amyloid filaments are found to have antiparallel or parallel structures (29–33), and recently the D23N mutant of Aβ has been found to adopt an antiparallel structure (34). In contrast, the Podospora prion [Het-s] is based on amyloid of the HET-s protein with a β-helical structure (35, 36).
Aβ, amylin, and many other human amyloids are composed of a fragment of a precursor protein, and the yeast and fungal prion proteins Ure2p, Sup35p, Rnq1p, and HET-s have subdomains capable of propagating the prion in vivo and forming amyloid in vitro. Similarly, we sought to determine the domain of Pmel17 responsible for fiber formation, and found that of the fragments examined, only the RPT region formed long, unbranched filaments with the characteristics of amyloid. These filaments formed only at the mildly acid pH of melanosomes, but quickly dissolved at neutral pH. We show that this amyloid of the RPT region promotes eumelanin formation in vitro and has other properties in common with Pmel filaments of melanosomes.
Results
The Pmel17 Repeat Domain (RPT) Forms Filaments.
We examined filament formation by various purified fragments of Pmel17 diagrammed in Fig. 1A. Mα (p1216) extends from the N terminus (except for the signal peptide) to the Kex2 cleavage site inside GAP2, and is reported to form amyloid within seconds when diluted from guanidine into buffer, independent of pH (16). Our attempts to observe this rapid or even gradual filament formation with Mα using the reported conditions (or others) were unsuccessful. We did find filament formation with the RPT domain (Pmel17:RPT) over a period of weeks when incubated without agitation or seeding (Fig. 1D and Fig. S1A). For the first few days, no filaments were observed. At intermediate times, sharply curved narrow filaments formed (squiggles), mixed with amorphous aggregates and soluble material (Fig. S1A). From 28 to 53 days, straighter, gently curved filaments were observed to form, eventually becoming the major species, but not entirely replacing the squiggles (Fig. S1A). Other purified Pmel fragments, P1208, P1204, P1192, and P1194 (see Fig. 1A) failed to form the straight filaments with or without agitation, under the same conditions (Fig. S1B). P1192 and P1194, which differ from the RPT domain only by short added segments at the N- and C-terminal extremities, did form the squiggles (Fig. S1B).
pH Instability of RPT Filaments.
The melanosome is a lysosome-related organelle, and, like lysosomes, is known to be acidic with a pH as low as 4.0 (37, 38). Furthermore, acidic pH is critical for pigment formation (39). In fact, we only observed filament formation by RPT when monomer solutions were incubated at pH 5.0 (Mα did not form filaments under these conditions). At higher pH, RPT remained monomeric and could not form filaments. Moreover, when filaments formed at pH 5.0 were exposed to neutral pH conditions, they rapidly dissociated (Fig. 2C). Static light scattering supported this observation, showing the dissociated fibers were composed of monomer and small oligomers with an average mass of 24 ± 0.2 kDa (see SI Text). Electron microscopic examination of fibril preparations exposed to various pHs (Fig. 2A) show that fibers remained at pH 4 to 6, but were nearly completely dissolved at pH 7 or above. A time course of thioflavin T fluorescence likewise showed loss of fluorescence within seconds of adjusting the pH to 7 or above (Fig. 2C). Fluorescence of the lone Trp residue in fibers formed at pH 5.0 shifts to longer wavelengths at pH 6.0 or 8.0, indicating greater exposure to the solvent under those conditions (Fig. S2).
Fig. 2.
pH stability of RPT fibers. (A) Transmission electron micrograph images showing pH stability between pH 4.0–9.0 of RPT fibers formed at pH 5.0. (B) Comparison of ThT fluorescence of RPT with that of Ure2p, Sup35NM, and HET-s at pH 5.0. (C) RPT fibers monitored by thioflavin T over time at pH 5.0–8.0. (D) RPT fibers monitored by thioflavin T over time at pH 5.0–8.0.
Seeding or Agitation Produces a More Uniform Filament Population.
A portion of filaments formed from RPT without agitation over weeks (as in Fig. 1D) was sonicated and used to seed filament formation by a fresh batch of RPT protein. Under these conditions, almost exclusively straighter filaments were formed within 7 days (Fig. 3A). These filaments are approximately 6–10 nm in diameter and have a uniform appearance. Attempts to seed other Pmel17 fragments (including P1216 (Mα), P1208, P1204, P1194, and P1192) with filaments of the RPT domain were unsuccessful. Freshly prepared RPT was gently agitated over the course of 1 week without seeding. The resulting preparation showed exclusively straight filaments which were, as expected, shorter than the straight filaments present in preparations incubated without agitation (Fig. 3B). These filaments had a uniform appearance and were used for all further experiments.
Fig. 3.
RPT fibrils prepared by seeding or agitation. (A) Fibrils made by seeding with 5% RPT seeds for 2 weeks. (B) Fibrils made by 1 week incubation with gentle agitation. (C) Electron diffraction of RPT fibers formed in vitro exhibits a reflection at 4.5 ± 0.2 Å indicative of a β sheet structure. (D) Electron micrograph of sample used in C. (E) Circular dichroism spectra of soluble and fibrous RPT support a β-sheet rich structure of the fibrils. Based on curve-fitting, RPT is 6 ± 1% α-helix, 47 ± 2% β-sheet, 19 ± 1% β-turn, and 28 ± 1% remainder. (F) Equal concentrations of soluble and fibrous RPT exposed to Proteinase K support a protease-resistant core in the fiber form.
Characterization of Pmel RPT Filaments.
Amyloid is defined by its filamentous character, enhanced fluorescence on binding thioflavin T (or other dyes), elevated protease - resistance and a cross-beta sheet structure (40). Circular dichroism studies showed that the soluble RPT was largely unstructured, but the fibers showed a β sheet pattern (Fig. 3E). Electron diffraction with unoriented fibers showed the band at 4.5 ± 0.2 angstroms typical of β sheets (Fig. 3C). Limited digestion of RPT fibers with proteinase K showed them to be far more resistant to digestion than the soluble form of the protein (Fig. 3F). The fact that the monomers in fibers do not change in size on protease digestion suggests that most of the sequence is in amyloid form. Fibers of RPT show birefringence on staining with Congo Red (Fig. S3) and are stable in 2% SDS (Fig. S4).
On binding to RPT filaments at pH 5.0, thioflavin T displayed a dramatic increase of fluorescence compared to the soluble form, or even compared to other amyloid filaments, such as those of Sup35NM, full-length Ure2p, or the HET-s prion domain (Fig. 2B). However, this fluorescence is comparable to that of thioflavin T bound to the other amyloids at neutral pH. Thus, the RPT fibers have the characteristics of amyloid fibers.
RPT Fibers Promote Melanin Formation in Vitro.
Pmel17 is known to be necessary for melanin biogenesis. We find that RPT fibers increase the rate and yield of melanin formation in vitro by ≈3-fold compared to a control reaction containing only soluble RPT or no RPT, and continued incubation beyond 74 h showed up to 6-fold stimulation compared to these controls (Fig. 4A). Other amyloids, such as Sup35NM and HETs increase the efficiency of melanin synthesis (Fig. 4B) at rates comparable to RPT.
Fig. 4.
Amyloid of recombinant RPT accelerates melanin synthesis. (Upper) A time course of melanin synthesis in vitro shows that RPT fibers enhance melanin formation per unit time when compared to the soluble form. (Lower) Comparison of melanin synthesis facilitated by amyloid of RPT (q) with that of Sup35NM (p) and HET-s (l). Reactions were performed in duplicate.
Preliminary Solid-State NMR Studies of RPT Fibers Confirm β-Sheet Structure.
We labeled RPT with Met-1-13C at 5 sites or with Leu-1-13C at 3 sites, none close to each other in the linear sequence (Fig. 1C). The 1-dimensional spectrum of Met-1-13C-labeled filaments showed a minor peak (13% of signal) at a chemical shift typical of random coil, and a major peak (87%) at a lower frequency, indicative of β-sheet structure (Table 1). Leu-1-13C-labeled fibers showed 2 peaks, but both were shifted substantially below typical random coil values (Table 1). As a further check, we prepared RPT labeled with Val-1-13C at the 15 valine residues scattered through repeats 1–9 of the sequence. The 1-dimensional solid-state NMR spectrum shows a single peak at a chemical shift indicating β-sheet structure (Table 1). Thus, chemical shifts of Met, Leu, and Val labels indicate a high β-sheet structure of the fibers, consistent with CD data and the results of electron fiber diffraction mentioned above.
Table 1.
Chemical shifts from one-dimensional solid-state NMR experiments
| Label | Chem. shift, ppm | Line width, ppm | Fraction of total peak, % | Random coil, ppm |
|---|---|---|---|---|
| Met-1-13C fibrils | 172.77 | 3.3 | 87 | 175.6 |
| 175.2 | 0.5 | 13 | ||
| Leu-1-13C fibrils | 172.24 | 2.3 | 45 | 175.9 |
| 173.77 | 1.7 | 55 | ||
| Val-1-13C fibrils | 172.65 | 2.6 | 100 | 174.6 |
Discussion
We have found that the RPT domain of the melanosome protein Pmel17 forms straight amyloid fibers characterized by filamentous morphology, protease-resistance, birefringence on staining with Congo Red, fluorescence on staining with thioflavin T, and high β-sheet content. Under our conditions, only this fragment (of those we have tried) does so. Several lines of evidence suggest that this amyloid corresponds to the core of the melanosome fibers.
First, the RPT domain is necessary for fiber formation in vivo (14, 17). Other domains are needed for proper sorting of Pmel17 into premelanosomes, but none but RPT have yet been found needed for fiber formation. Furthermore, antibodies directed against epitopes in RPT decorate the filaments (14).
Second, the striking pH dependence that we find for fiber formation and stability by the RPT domain corresponds to the known low pH of melanosomes and the pH dependence for melanin formation (39). The pH dependence of fiber formation is also understandable from the abundance of acidic residues in the RPT domain. This extraordinary pH instability of RPT fibers may explain why no ex vivo fibers have been isolated. Extreme care would be needed to isolate these filaments, an unusual property for the usually extremely robust amyloids.
Third, it is generally true that the prion domain alone or amyloid core domain forms amyloid far more readily when free than when bound to the rest of the protein. Prion proteins have distinct prion domains which comprise the core of their amyloid structures and are necessary and sufficient for prion propagation (35, 41–47). In general, the prion domain is stabilized in the nonprion form by being attached to the rest of the molecule (41, 43, 46, 48). Similarly, only when released from the precursor protein does the Aβ peptide form amyloid. This is consistent with the enhanced ability of RPT to form amyloid. It is possible that in vivo chaperones or other factors facilitate amyloid formation by larger fragments of Pmel17, such as Mα or MαC.
Fourth, the morphology of the straight filaments is consistent with the very straight appearance observed in vivo. The RPT fibers are approximately 6–10 nm in diameter, identical to those in melanocytes estimated to be 6–10 nm in diameter (49). Our RPT (13,914 Da) is unglycosylated and may be only the core of the amyloid structure while the filaments in vivo are composed of fragments >25 kDa [O-glycosylated (50), but probably not N-glycosylated (9, 51)] as estimated by SDS/PAGE (14). However, we find that our RPT, whose size (13,914 Da) we confirmed by light scattering (see SI Text) and by mass spectrometry migrates at approximately 26 kDa by SDS/PAGE using un-stained markers and >40 kDa using prestained markers. The in vivo fragments could be in fact smaller than predicted because of this anomalous migration on a 10% SDS/PAGE, and may prove to consist of no more than RPT.
Finally, the RPT fibers promote eumelanin formation in vitro as do Pmel17 fibers in vivo. However, this is not a strong argument because other unrelated amyloids do so as well. Taken together, our results indicate that amyloid of the RPT region corresponds to the core of the Pmel17 melanosome fibers, but it remains important to isolate such fibers and directly examine their structural properties.
We cannot explain our failure to reproduce the results of Fowler et al. (16). Mα purified as described or otherwise did not form amyloid under the described conditions (or other conditions). We believe it is unlikely that amyloid can form within 2 s as described, and the pH - independence of amyloid formation reported by Fowler et al. is at variance with our results and the pH requirement for filament formation in vivo.
The repeats in RPT are reminiscent of the repeat sequences in HETs which are central in determining the structure of HETs amyloid (35, 36). On the other hand, the repeats of the Sup35 prion domain are not essential for prion formation (52) and, unlike the β-helix structure of HETs, Sup35NM has an in-register β-sheet architecture (23). Thus one cannot infer structure from the presence of repeats, and further studies will be needed.
The RPT region of Pmel17 is the most highly variable [reviewed in (9)]. The absence of an RPT in the otherwise closely related protein Nmb also suggests a melanin-specific function for this domain of Pmel17. Because Pmel17 has an integral role in pigmentation, the rapid variation of the RPT region may reflect selection for changes in pigmentation having to do with camouflage, resistance to UV light, or other functions. Other species also have repeats, but generally completely different from those of humans. For example, compared with the human repeat pt.e.gttp.qv. Xenopus laevis has a avtv(aea)vpnqeq repeat while the zebrafish Danio rerio has 5 exact repeats of aaaeaentatdalatpavieae and the green spotted pufferfish Tetraodon nigroviridis has partially conserved repeats of veaaad. The different repeats may select out certain melanin variants affecting outer appearance. It will be of interest to determine whether amyloid formation is conserved across this wide span of sequences.
Experimental Procedures
Pmel17 Protein Expression and Purification.
Pmel17 gene fragments (p1216, p1208, p1204, p1194, p1192, and Pmel17:RPT (Fig. 1) with C-terminal his-tags (except p1216), were subcloned into pET21a(+) (Novagen) (Table S1) and expressed in E.coli BL21(DE3) RIPL (Stratagene). Shaken cultures (1 L Luria Broth) were grown at 37°C to an A600 = 0.4–0.5 and then induced with 1.0 mM IPTG for 4 h. For isotope labeling, cells were grown in defined amino acid medium as previously described (23). P1216 was purified as described by Fowler et al. (16). For other constructs, cells were collected by centrifugation (9,000 rpm) for 20 min at 4°C and then re-suspended in denaturing buffer (8M guanidine, 100 mM NaCl, 100 mM K2HPO4, pH 7.5, and 10 mM imidazole) and incubated for 60 min at room temperature with gentle agitation. The generated lysate was spun at 30,000 rpm for 45 min. The pellet was discarded and the supernatant was mixed with nickel-nitrilotriacetic acid agarose (Ni-NTA from Qiagen; 5 mL per l culture) and incubated at 4°C for 60 min with gentle agitation. The mixed Ni-NTA lysate was poured into a column and washed with 10-column volumes of buffer (8M urea, 100 mM NaCl, 100 mM K2HPO4, pH 7.5, and 20 mM imidazole). The protein was eluted with buffer containing 8 M urea, 100 mM NaCl, 100 mM K2HPO4, pH 7.5 and 250 mM imidazole. Pmel17 aggregates of p1216, p1208, p1204, p1194, p1192, and RPT were generated by dialyzing purified protein into 125 mM K acetate buffer, pH 5.0, and incubating at room temperature with and without gentle agitation for varying lengths of time. Typical protein concentrations were approximately 2 mg/mL.
Electron Microscopy.
Diluted samples were adsorbed on carbon-coated copper grids, stained with 3% aqueous uranyl acetate, and visualized with an FEI Morgagni transmission electron microscope operating at 80 kV.
Electron Diffraction.
A suspension of RPT fibers (2 mg/mL monomer concentration) was applied to a carbon-coated copper grid and incubated for approximately 30 min. Excess liquid was blotted and the grid was quickly washed with 0.5 mM potassium acetate, pH 5, before air drying. Electron diffraction images were collected using an 80 kV electron beam with a 350-mm camera length. Diffraction distances and atomic spacing were calibrated using thallous chloride crystals under identical microscope conditions.
Circular Dichroism Secondary Structure Analysis.
RPT fibers (0.1 mg/mL) in 5.0 mM K acetate buffer, pH 5.0, were analyzed using a Jasco J-715 spectropolarimeter as previously described (53).
Synthetic Melanogenesis.
The ability of RPT to enhance melanin synthesis was evaluated using a modified protocol (16). Briefly, RPT (0.125 mg) was added to 5 mM DL-DOPA in 0.25 mL of 125 mM K acetate buffer, pH 5.0. tyrosinase (2 μg) was added to initiate the reaction and incubated at room temperature. The reaction was stopped by centrifugation (15,000 rpm, 20 min). The pellets, which contain essentially all of the melanin product, were re-suspended in 1 M NaOH and heated at 60°C for 5 min. Absorbance was recorded at 400 nm. Synthetic melanin (Sigma no. M8631) was used as a standard to calculate melanin formation.
Mass Spectrometry.
Incorporation of methionine-1-13C with no apparent leakage of label to other amino acids was shown by analyzing a Glu-C digested sample by LC/MS/MS as described in detail in Fig. S5.
NMR.
Solid-state NMR experiments were performed on selectively 13C-labeled samples at 9.39 T (100.4 MHz 13C NMR frequency) using an InfinityPlus NMR spectrometer (Varian, Inc.) and a double-channel magic-angle spinning (MAS) probe from Varian. One-dimensional 13C NMR spectra of lyophilized samples in thick-walled 3.2-mm rotors (Varian) at room temperature were recorded at an MAS frequency of 20 kHz with 1H-13C cross-polarization (54) and using 2-pulse phase-modulated decoupling (55).
Supplementary Material
Acknowledgments.
We thank Mickey Marks (University of Pennsylvania, Philadelphia) for clones of Pmel17, Eric Anderson for mass spectrometry work, Rob Tycko and Vince Hearing for fruitful discussions, and our colleagues for critical reading of the manuscript. This work was supported by the Intramural Program of the National Institute of Diabetes, Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906509106/DCSupplemental.
References
- 1.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sunde M, Kwan AH, Templeton MD, Beever RE, MacKay JP. Structural analysis of hydrophobins. Micron. 2008;39:773–784. doi: 10.1016/j.micron.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Podrabsky JE, Carpenter JF, Hand SC. Survival of water stress in annual fish embryos: Dehydration avoidance and egg amyloid fibers. Am J Physiol Regulatory Integrative Comp Physiol. 2001;280:R123–R131. doi: 10.1152/ajpregu.2001.280.1.R123. [DOI] [PubMed] [Google Scholar]
- 4.Iconomidou VA, Vriend G, Hamodrakas SJ. Amyloids protect the sildmoth oocyte and embryo. FEBS Lett. 2000;479:141–145. doi: 10.1016/s0014-5793(00)01888-3. [DOI] [PubMed] [Google Scholar]
- 5.Benkemoun L, Saupe SJ. Prion proteins as genetic material in fungi. Fungal Genet Biol. 2006;43:789–803. doi: 10.1016/j.fgb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Raposo G, Marks MS. Melanosomes - dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn LC, Thigpen LW. The silver mouse: A recessive color variation. J Hered. 1930;21:495–498. [Google Scholar]
- 8.Kwon BS, et al. A melanocyte-specific gene, Pmel17, maps near the silver coat color locus on mouse chromosome 10 and is in a syntenic region on human chromosome 12. Proc Natl Acad Sci USA. 1991;88:9228–9232. doi: 10.1073/pnas.88.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theos AC, Truschel ST, Raposo G, Marks MS. The Silver locus product Pmel17/gp100/Silv/Me20: Controversial in name and function. Pigment Cell Res. 2005;18:322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou BK, et al. Identification of a melanosomal matrix protein encoded by the murine si (silver) locus using “organelle-scanning”. Proc Natl Acad Sci USA. 1994;91:7076–7080. doi: 10.1073/pnas.91.15.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. Distinct protein sorting and localization to premelanosomes, melanosomes and lysosomes in pigmented melanocytic cells. J Cell Biol. 2001;152:809–823. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell. 2001;12:3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berson JF, et al. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol. 2003;161:521–533. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoashi T, et al. The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers. J Biol Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 15.Harper DC, et al. Premelanosome amyloid-like fibrils are composed of only Golgi-processed forms of Pmel17 that have been proteolytically processed in endosomes. J Biol Chem. 2008;283:2307–2322. doi: 10.1074/jbc.M708007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler DM, et al. Functional amyloid formation within mammalian tissue. Plos Biology. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theos AC, et al. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicularendosomes involved in organelle morphogenesis. Dev Cell. 2006;10:343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adema GJ, de Boer AJ, Vogel AM, Loenen WAM, Fegdor CG. Molecular characterization of the melanocyte lineage-specific antigen gp100. J Biol Chem. 1994;269:20126–20133. [PubMed] [Google Scholar]
- 19.Nichols SE, Harper DC, Berson JF, Marks MS. A novel splice variant of Pmel17 expressed by human melanocytes and melanoma cells lacking some of the internal repeats. J Invest Dermatol. 2003;121:821–830. doi: 10.1046/j.1523-1747.2003.12474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antzutkin ON, et al. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of beta-sheets in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci USA. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balbach JJ, et al. Supramolecular structure in full-length Alzheimer's beta-amyloid fibrils: Evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luca S, Yau W-M, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baxa U, et al. Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 25.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci USA. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 27.Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of α-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 28.Margittai M, Langen R. Template-assisted filament growth by parallel stacking of tau. Proc Natl Acad Sci USA. 2004;101:10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benzinger TL, et al. Propagating structure of Alzheimer's beta-amyloid(10–35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkova AT, et al. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 31.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 33.Bu Z, Shi Y, Callaway DJ, Tycko R. Molecular alignment within beta-sheets in Abeta(14–23) fibrils: Solid-state NMR experiments and theoretical predictions. Biophys J. 2007;92:594–602. doi: 10.1529/biophysj.106.091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tycko R, Sciarretta KL, Orgel JPR, Meredith SC. Evidence for novel β-sheet structures in Iowa-mutant Aβ-amyloid fibrils. Biochemistry. 2009 doi: 10.1021/bi9002666. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasmer C, et al. Amyloid fibrils of the HET-s(218–279) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 37.Bhatnagar V, Anjaiah S, Burj N, Darshanam BN, Bramaiah A. pH of melanosomes of B 16 murine melanoma is acidic: Its physiological importance in the regulation of melanin biosynthesis. Arch Biochem Biophys. 1993;307:183–192. doi: 10.1006/abbi.1993.1577. [DOI] [PubMed] [Google Scholar]
- 38.Puri N, Gardner JM, Brilliant MH. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J Invest Dermatol. 2000;115:607–613. doi: 10.1046/j.1523-1747.2000.00108.x. [DOI] [PubMed] [Google Scholar]
- 39.Brilliant MH. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res. 2001;14:86–93. doi: 10.1034/j.1600-0749.2001.140203.x. [DOI] [PubMed] [Google Scholar]
- 40.Sipe JD, Cohen AS. Review: History of the amyloid fibril. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 41.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 42.TerAvanesyan A, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balguerie A, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitrenko YA, Pavon ME, Stone SI, Liebman SW. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet. 2007;51:309–319. doi: 10.1007/s00294-007-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King, et al. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 47.Baxa U, et al. Architecture of Ure2p prion filaments: The N-terminal domain forms a central core fiber. J Biol Chem. 2003;278:43717–43727. doi: 10.1074/jbc.M306004200. [DOI] [PubMed] [Google Scholar]
- 48.Kochneva-Pervukhova NV, Poznyakovski AI, Smirnov VN, Ter-Avanesyan MD. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharmyces cerevisiae. Curr Genet. 1998;34:146–151. doi: 10.1007/s002940050379. [DOI] [PubMed] [Google Scholar]
- 49.Hurbain I, et al. Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc Natl Acad Sci USA. 2008;105:19726–19731. doi: 10.1073/pnas.0803488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valencia JC, et al. Sialylated core 1 O-glycans influence the sorting of Pmel17/gp100 and determine its capacity to form fibrils. J Biol Chem. 2007;282:11266–11280. doi: 10.1074/jbc.M608449200. [DOI] [PubMed] [Google Scholar]
- 51.Maresh GA, et al. Differential processing and secretion of the melanoma-associated ME20 antigen. Arch Biochem Biophys. 1994;311:95–102. doi: 10.1006/abbi.1994.1213. [DOI] [PubMed] [Google Scholar]
- 52.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McPhie P. Circular dichroism studies on proteins in films and in solution: Estimation of secondary structure by g-factor analysis. Anal Biochem. 2001;293:109–119. doi: 10.1006/abio.2001.5113. [DOI] [PubMed] [Google Scholar]
- 54.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 55.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear Decoupling in Rotating Solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.