Abstract
Purpose
We determined the effects of selenomethionine, the major organic selenium containing compound found in the diet and the form of selenium being used in the Selenium and Vitamin E Cancer Prevention Trial, on prostate cancer cells.
Materials and Methods
We assessed global transcript profiles of selenomethionine treated LNCaP using cDNA microarrays and compared them to those of cells treated with methylselenic acid, a direct precursor of methylselenol, which is the active form of selenium in vivo.
Results
After treatment with selenomethionine 2,336 unique genes showed expression changes of at least 1.5-fold in at least 3 time points during 48 hours and 366 unique transcripts differed significantly between selenomethionine and methylselenic acid treated LNCaP. Approximately half of the 76 cell cycle regulated genes affected by selenomethionine were down-regulated and enriched for genes associated with the G2/M phase. Flow cytometry analysis showed that selenomethionine induced G2/M arrest in LNCaP at low concentrations. Selenomethionine also affected expression levels of 35 known androgen responsive genes and 18 of these transcripts showed changes that were the inverse of those seen after androgen stimulation. At high concentrations selenomethionine decreased prostate specific antigen promoter driven luciferase expression.
Conclusions
Selenomethionine modulates transcript levels of genes involved in a number of biological processes, including cell cycle/apoptosis androgen signaling, signal transduction and transcriptional regulation. Although the pathways affected paralleled in many ways those that are modulated by methylselenic acid, distinct differences in transcript patterns and effects on cell cycle regulation suggest that different selenium compounds could exert unique effects in prostate cells.
Keywords: prostate, prostatic neoplasms, selenomethionine, microarray analysis, gene expression
Several epidemiological studies demonstrated an inverse association with plasma, serum or tissue selenium levels and the subsequent risk of PC1. The NPC trial showed a decrease in overall cancer death rates by 50% in the selenium treated group and a 66% decrease in PC diagnosis rates at 6.4 years of followup.2 The ongoing SELECT will test definitively whether selenium, vitamin E or the combination will prevent PC.3 Whereas the NPC trial used a selenized yeast preparation containing a mixture of organic selenium compounds including SM, SELECT tests whether pure SM will prevent PC.
Although the cancer protective activities of all selenium compounds have been proposed to be mediated through methylselenol, a key metabolite of selenium in vivo, different forms of selenium seem to vary in their effectiveness for producing biological changes.4,5 For example, SM and MSA, which is a direct precursor of methylselenol, reportedly arrest human LNCaP PC cells at different phases of the cell cycle.6,7 We reported that MSA produced striking changes of the transcriptional programs in LNCaP in vitro.7 To better understand the effects of SM on prostate cells we determined the global gene expression changes induced by SM in LNCaP using cDNA microarrays. We compared these changes to those elicited by MSA to test the hypothesis that different organic selenium compounds could affect prostate cells differently.
MATERIALS AND METHODS
Cell Culture and Treatment, Total RNA Isolation and cDNA Microarray Hybridizations
LNCaP cells were cultured in RPMI 1640 with 5% defined bovine serum (HyClone®), which contributed 13 nM selenium to the medium. Cells were treated with 10 μM SM or vehicle and were then lysed 1, 2, 4, 6, 9, 12, 15, 18, 24 and 48 hours after treatment. Total RNA was isolated and hybridized to cDNA microarray containing 24,164 unique genes, as described previously.7 Each sample from a single time point after SM treatment was analyzed on 1 array and no replicates were generated.
Data Processing and Analysis
Fluorescence intensities for each spot on the microarrays were acquired, analyzed and stored, as described previously.7 Only spots with a signal intensity of greater than 50% above background in Cy5 and Cy3 channels in at least 70% of microarray experiments were used for subsequent analysis. We arbitrarily selected transcripts whose expression level varied at least 1.5-fold after treatment compared to controls in at least 3 experiments examined. Genes in the resulting data table were ordered by their patterns of expression using hierarchical clustering analysis8 and visualized using Treeview software (http://rana.lbl.gov/EisenSoftware.htm).
Expression data on SM treated cells were compared to data on MSA treated cells, which we reported previously.7 Transcripts differentially expressed in response to SM and MSA were identified using the SAM procedure (http://www-stat.stanford.edu/~tibs/SAM/).9
QRT-PCR
Total RNA from untreated and treated cells was reverse transcribed. The cDNA product was mixed with iQ™ SYBR® Green Supermix and primers of choice. Gene products were amplified in triplicate using an iCycler® iQ Real-Time PCR Detection System according to manufacturer instructions. GAPDH transcript levels were assayed simultaneously with each of 8 genes and used to normalize transcript levels in treated and untreated cells. Table 1 lists the primer sequences used.
Table 1.
Primer sequences used in QRT-PCR
| Gene Symbol (primer sequence) |
|---|
| STK17A: |
| Forward-5′ TTTGTCTGAGTCGGCTGTTG 3′ |
| Reverse-5′ TGCCTTTTCCATCCTGAAAG 3′ |
| RAB39B: |
| Forward-5′ CATCACTCGCGCCTACTACA 3′ |
| Reverse-5′ GGGCTGAACGTGTACTTTGG 3′ |
| AKT1-1: |
| Forward-5′ AGAAGCAGGAGGAGGAGGAG 3′ |
| Reverse-5′ CCCAGCAGCTTCAGGTACTC 3′ |
| GIP2: |
| Forward-5′ AATGCGACGAACCTCTGAAC 3′ |
| Reverse-5′ TCAGCCAGAACAGGTCGTC 3′ |
| NFKB2: |
| Forward-5′ TACCTGGTGATCGTGGAACA 3′ |
| Reverse-5′ GATAGGTCTTTCGGCCCTTC 3′ |
| GUCY1A3: |
| Forward-5′ TTCTTCCCGGCATCATAAAG 3′ |
| Reverse-5′ TTCACAAACTCGCTGCAATC 3′ |
| NKX3-1: |
| Forward-5′ GGACTGAGTGAGCCTTTTGC 3′ |
| Reverse-5′ AGGTTACCCATTTTGGGGAC 3′ |
| CYP1B1: |
| Forward-5′CCCAAGGACACTGTGGTTTT3′ |
| Reverse-5′ TCATCACTCTGCTGGTCAGG3′ |
| GAPDH: |
| Forward-5′CGACCACTTTGTCAAGCTCA3′ |
| Reverse-5′ GGGTCTTACTCCTTGGAGGC3′ |
Cell Cycle Distribution Assay
Cells treated with SM for 48 hours and control cells cultured in parallel were collected by trypsinization. Cell cycle distribution was determined after propidium iodide (Sigma®) staining, as described previously.7 We chose this time point because previous studies in LNCaP showed significant changes in cell cycle distribution 48 hours after SM treatment. Also, it allowed us to compare our data to cell cycle distribution changes induced at an identical time point after MSA treatment, as we reported previously.7 All experiments were done in triplicate.
Transient Transfection and Luciferase Assay
A PSA 7 kb promoter-luciferase (PSA-Luc) reporter construct was co-transfected into LNCaP with a β-galactosidase reporter construct using Lipofectamine 2000 (Invitrogen™). Cells were treated with MSA at 5 or 10 μM or SM at 10 or 50 μM 24 hours after transfection. After another 24 hours treated and control cells were lysed with lysis buffer (BD™ Biosciences) and luciferase activity was measured using a BD Luciferase Reporter Assay Kit. Corresponding β-galactosidase activity was measured using a β-Galactosidase Assay System (Promega Biosciences, San Luis Obispo, California). The β-galactosidase activity in each cell lysate was used to normalize luciferase activity. Reported values represent the average of 3 independent transfections.
RESULTS
SM Induced Gene Expression Changes in LNCaP Cells
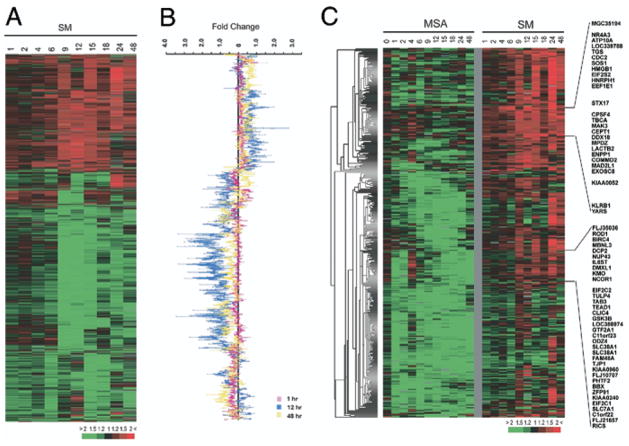
We identified 2,336 unique genes whose expression changed at least 1.5-fold in response to 10 μM SM in at least 3 samples during 48 hours and we used hierarchical clustering analysis to group genes based on their similarities in expression across the time course (fig. 1, A). We chose a concentration of 10 μM SM because it would inhibit LNCaP cell growth without significant cytotoxicity6,10 and allow us to compare our data to gene expression changes induced by an identical concentration of MSA, which we reported previously.7 The most significant changes in gene expression occurred at 12 hours. This could be appreciated in a plot of the fold change in gene expression for all 2,336 genes 1, 12 and 48 hours after SM treatment (fig. 1, B). Genes functioning in a number of biological processes, such as cell cycle and cell death, were enriched, as shown by GOTM (http://genereg.ornl.gov/gotm/) (fig. 2). A complete list of the gene names and the raw data are available at http://www.stanford.edu/~hongjuan/SM. Only 62 of the 2,336 genes changed expression at least 2-fold in at least 3 time points in response to SM. These transcripts are involved in various biological processes, including transcriptional regulation, cell adhesion/cytoskeleton organization, cell growth/apoptosis, signal transduction and transport (table 2).
Fig. 1.
A, hierarchical clustering analysis of gene expression changes induced by SM in LNCaP. Data are arranged in time ascending order of 1, 2, 4, 6, 9, 12, 15, 18, 24 and 48 hours. Columns represent data from single time point after SM treatment. Rows represent single gene expression across time course (top). Red areas indicate up-regulated transcripts. Green areas indicate down-regulated transcripts. Color saturation degree corresponds to gene expression ratio (bottom). Full transcript identities and raw data are available at http://www.Stanford.edu/~hongjuan/SM. B, expression fold changes induced by SM at 1, 12 and 48 hours. Genes are in same order as in part A. C, hierarchical clustering analysis of genes with significantly higher expression in SM vs MSA treated cells, as identified by SAM analysis. Data on each treatment condition are arranged in time ascending order (top).
Fig. 2.
Bar charts show biological process categories of genes affected by SM. Red lettering indicates statistically significant enrichment, as identified by GOTM. Black lettering indicates no statistically significant enrichment, as identified by GOTM. A and a, biological processes of genes up-regulated by SM. B and b, and C and c, biological processes of genes down-regulated by SM, respectively.
Table 2.
Genes whose expression changed at least 2-fold in at least 3 time points during 48 hours in response to SM
| Biological Process | Gene Symbol |
|---|---|
| Transcriptional regulation | ATF2, CBX6, CNNM3, IKBKB, L3MBTL4, TBX1, TEAD3, THG-1 |
| Cell adhesion/cytoskeleton | CLSTN3, COL6A1, DST, LASP1, PARVB, SCARB1,* SEMA5A,* TAOK2* |
| Cell growth/apoptosis | LTBR,* PLG, PRKCZ,* SHC1,* TAOK2* |
| Signal transduction | ECE2, GRK6, LTBR,* PRKCZ,* RAP2A, RGS19IPI, SEMA5A,* SHC1* |
| Transport | ATP6V0A1, EHD1, NARF, NDUFV1, SCARB1,* SLC25A1, STX17, TXNRD2, VPS4A |
| Other | ACO2, ARS2, C16orf40, CDC91L1, CRMP1, DUSP3, HPN, KLHDC3, MAP3K11, PCK2GPX1, PMVK, POLL, PPM1B, TREX1, UBE2S, WWP2 |
| Unknown | ACD, AKTIS1, C11orf24, C19orf29, CYHR1, DKFZP564I1171, EEIG1, FLJ14466, ITM2C, LOC134121, LOC283400, MRPL54, PCNXL3, SITPEC |
Gene included in more than 1 category.
Comparison of Expression Changes Induced by SM and MSA
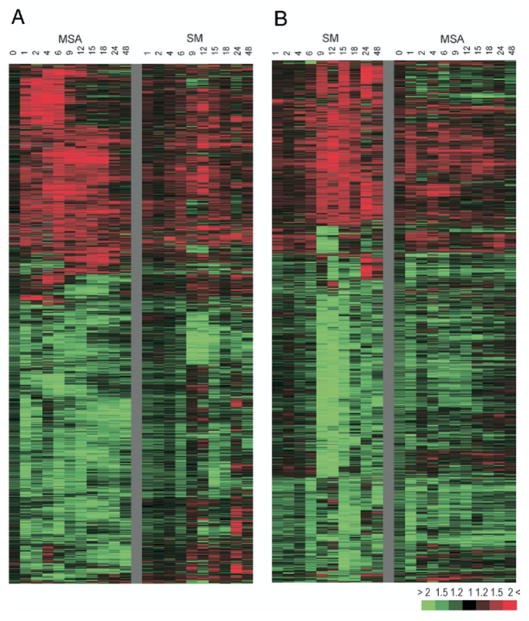
Of the transcripts 2,336 and 2,165 showed that expression changed at least 1.5-fold compared to controls in at least 3 experiments during 48 hours in response to SM and MSA, respectively, at a concentration of 10 μM. The overlap between the data sets was limited. Tables 3 and 4 list the top 100 genes modulated by MSA but not by SM or vice versa. Only 6% of the transcripts up-regulated by MSA were also up-regulated by SM, accounting for 14% of genes whose expression was increased by SM. Of the transcripts down-regulated by MSA 21% showed decreased expression in response to SM, accounting for 23% of the total genes suppressed by SM. However, genes selected as up-regulated or down-regulated by 1 compound using the current data selection criteria usually showed similar changes in expression levels in response to the other compound, although they were often not as strong or as consistent across time points and, therefore, they were not selected (fig. 3). In addition, several pathways, such as cell cycle/apoptosis and signal transduction, were affected similarly by the 2 compounds (table 5).
Table 3.
Top 100 genes with known function modulated by MSA but not by SM
| 50 Genes up-regulated by MSA but not SM: | |
| Cell cycle/apoptosis | JUN,* TNFRSF12A, MAD, ING1,* CDKN1A, FTH1 |
| Cell-cell signaling/signal transduction | DPYSL3,* FN1, RAB31, TXNRD1,* PITPNC1 |
| Metabolism | DPYSL3,* GCLM, KIAA1363, AKAP6,* NAT2, UGDH,* OAT |
| Regulation of transcription | EGR1, ATF3, JUN,* HIF0, SNAI2, HIST1H1C, MAD, PERI, CRSP7, SOX9, AF5Q3, 1IRF6, CALR, ELF3, SMARCB1 |
| Other functions | ERP70, AKR1C2, GADD45B, TNFRSF12, ATUFT1, DNAJB9, DNAJC3, MS4A8B, FLJ12587, ING1,* FN1, SPTAN1, CARF, AKAP6,* MANIA1, GABARAPL1, GSN, ULBP2, UGDH,* H3F3B, PJA2, TXNRD1,* DMN, DUSP10, TMSB4Y, PIM2, DNAJB11, FYN, MAP1B |
| 50 Genes down-regulated by MSA but not SM: | |
| Cell cycle/apoptosis | CDK2, CENPF, ANLN, TTK, STK6,* CHAF1A, CCNH |
| Signal transduction | ITGA4,* GUCY1A3, ERBB2IP, PIK4CA,* AMH |
| Transcription regulation | UHRF1, HIPK2, NCORI1, BRCA1, EMX2, KIAA1041, TULP4, ZBTB1, ADNP, MYST4, TRIP8, E2F1, FOXA1, NKX3–1 |
| Other functions | SCD, ITGA4,* KLK2, PRPF8, BRIP1, EIF2C2, RDH11, HMMR, USP34, BIRC6, EPB41, TYMS, TSN, TJP1, ABCA7, CKLFSF4, KIF11, PIK4CA, STK6,* TNFRSF10D, FLJ22329, KIS, KCTD15, EXO1, LANCL2, HPS4, STEAP2, RPL26L1 |
Gene appears in more than 1 category.
Table 4.
Top 100 genes with known function modulated by SM but not MSA
| 50 Genes up-regulated by SM but not MSA: | |
| Cell growth/apoptosis | CASP8AP2,* CENPE, RASA1, CDC2, BNIP2, TGFA,* NBS1, SYK* |
| Signal transduction | CASP8AP2,* PIK3C2A, RASA1, FZD4, RAB30, HSPC121, SMARCE1, TGFA,* SYK,* KLRB1 |
| Transcription regulation | PRPF4B,* CBF2, ZF, ATRX, RLF, SCML1, SMARCE1, L3MBTL |
| Cell adhesion/matrix remodeling | PPFIA2, ADAM9, FOLH1, IGSF1 |
| Other functions | PRPF4B,* LUC7A, ABCA5, CPN1, SNRPN, TPR, ADH4, ITCH, NKTR, SGCB, SSB, FLJ10618, SCN2A2, PRLR, PCLO, EIF5, SLC26A4, USP15, CALD1, WDR31SFRS12, EIF3S7, MGST1, KIAA0674, ENPP1, LOC317671 |
| 50 Genes down-regulated by SM but not MSA: | |
| Cell growth/apoptosis | IGFBP2, SHC1,* CCT4, BCL2L12, PLG* |
| Signal transduction | SHC1,* MAP3K11, SNX15, GIP2,* GPRK6, LIMK1,* CSNK1D |
| Transcription regulation | PSMC3,* NFKB2, CBX6, SMARCA2, FOXA2, FLJ25045, PPP5C, RPC8, GRLF1, PLG,* DOT1L, FXR2 |
| Other functions | NDUFV1, PSMC3,* SLC6A8, HMBS, RLUCL, SHFM3, SMTN, MVK, G1P2,* TBCD, NT5C, PTK9L, CD2BP2, TNIP1, 1KBKB, U5-200KD, EPHX1, ATP6V0A1, CDC37, NDUFB7, CKB, ALAD, SLC25A11, NARF, MAP3K14, PMVK, ARFGAP1, COL6A1, BPAG1, CD151, LIMK1 |
Gene appears in more than 1 category.
Fig. 3.
Overview of 4,145 transcripts whose expression changed at least 1.5-fold compared to control in at least 3 experiments in response to MSA (A) or SM (B). Columns represent data from 1 time point after treatment. Rows represent single gene expression across time course. Red areas indicate up-regulated transcripts. Green areas represent down-regulated transcripts. Color saturation degree corresponds to gene expression ratio of treated cells vs controls (color bar, bottom right).
Table 5.
Common genes modulated by MSA as well as SM
| Common genes up-regulated by MSA + SM: | |
| Cell cycle/apoptosis | BIRC2, STK17A, ICEBERG,* CDC14A, TGFB2* |
| Cell-cell signaling/signal transduction | FGF12,* PPFIA1,* PRKAB2, RAB39B,* TGFB2* |
| Protein targeting/transport | SRP54, COPB2, SNX2, RAB39B* |
| Transcription regulation | TCF21, PEG3, ZNF154, CREB3, FBXO7* |
| Other functions | TRA1, NUCB2, GCA, ICEBERG,* MRCL3, MT2A, FGF12,* SEC63, EM1LIN3, PPFIAI,* GCC2, HLA-E, FLJ21934, IFRD1, NQO2, ORC41, SSR3, SV2B, FBXL6, RPL12, UBB, FBXO7* |
| Common genes down-regulated by MSA + SM: | |
| Cell cycle/apoptosis | DAPK3,* PLK,* LOC283431, C20orfI50, CDKN2D, CCND1, TPX2, INHBB,* IGF1R,* PCAF,* MDM4, ERBB2,* RBBP6, LIMD1,* AURKB,* CDC25B, CDC25A, AKT1,* RBM5, LMNB2, MXD4,* ATF5,* LTBR* |
| Transcription regulation | CTDSPL, ZFD25, FLJ20259, TOX, POLR2A, LMO4,* RENT1, INHBB,* PCAF,* HTATSF1, ZNF481, GTF2F1, MADH2, LIMD1,* NCOA5, ELL2, TBX1, NR2F6,* TRAP95, MXD4,* ATF5,* SF1, MYBL2, NFIC, HOXB13, HEYL, ZNF287, HCFC1, KIAA1333 |
| Metabolism | GCAT, NAGLU, CA14, MUS81, APOA4, MAN2A1, FLJ10851, PGAM2, TK1 |
| Signal transduction | PRKCI,* FAT,* DAP4, EGFR,* GNAQ, DVL3, IGFIR,* ILIRI,* ERBB2,* LIMD1,* NMU, AKT1,* ARHGAP1, ARL5, STRN4, NR2F6,* LTBR* |
| Immune response | IGLL1, INHBB,* TRGV9, IL1R1,* IL6ST, TAPBP, LTBR* |
| Amino acid phosphorelation/dephosphorelation | DAPK3,* PLK,* DKFZP434C131, PRKC1,* STYX, AURKB,* KPI2, PTPN4, AKT1* |
| Electron transport | TXN2, LMO4,* EGFR,* CYP1B1, TXNRD2 |
| Cell adhesion | FAT,* PCDHB5, CNTNAP2, PKP3, LAMC1 |
| Other functions | SMC5L1, VARS2, VARS2L, SLC25A1, PGGT1B, KIAA0676, SLC25A13, TTS–2.2, SLC30A7, DLAT, DPP9, POLS, LIG3, DDT, AP4E1, FUT11, COPA, AP1GBP1, ENSA, SMCIL1, FNBP2, LRRN4, CPEB3, RBM14, FKBP14, GALNT2, KCNAB1, GEMIN7, SFRS10, C20orfl4, LRP6, FKBP8, PPP2R1A, SEC6, RNASEH2A, RNPC1, CLONE24922, MGC2408, LASP1, NUMAI, GPT2, HS2ST1, EIF4G1, IF2, NUP153, SLCO4A1, MDH2, THOP1 |
Gene appears in more than 1 category.
Of the 4,145 transcripts whose expression changed in response to SM or MSA 419 (10.1%), representing 366 unique genes, were identified by SAM analysis as showing a statistically significant difference in expression between SM and MSA treated LNCaP cells with a false detection rate of 0.5%, indicating that SM can induce gene expression changes distinct from MSA. Notably 351 unique genes showed significantly higher expression in SM vs MSA treated cells (fig. 1, C). Of them 73 genes were cell cycle regulated and 10 were androgen regulated (table 6). Cell cycle regulated genes were enriched 7.3-fold compared to their expected frequency based on their number represented on the microarray (chi-square test p <0.001). For androgen regulated genes 1.3-fold enrichment was observed, although it was not statistically significant (p = 0.36). However, several typical androgen regulated genes, including KLK3, NKX3-1 and GUCY1A3, were down-regulated consistently during part of the time course by MSA but not by SM. These results suggest that MSA and SM exert different effects on gene expression in LNCaP.
Table 6.
Genes expressed at significantly higher level during 48 hours in SM vs MSA treated cells by SAM
| Biological Process | Gene Symbol |
|---|---|
| Cell growth/apoptosis | ADH4, ANLN, BIRC6, BMI1, BRCA1, BRCA2, BRRN1, BUB1B, C6orf139, CANP, CCNA2, CCNT2, CDC2, CDC7, CENPE, CHEK1, CSE1L, CUL4B, DEK, DHFR, DKFZp451J0118, EWSR1, FLJ12892, FLJ13912, FLJ23311, FLJ25416, GNAS, GPC6, HMMR, IFIT1, IQG AP3, KIF11, KIF23, KIS, KNTC2, LDLR, LOC139886, LOC157570, LOC200895, MAD2L1, MCM3, MCM4, MGC24665, MK167, NBS1, NR4A3, NRP1, OPA1, PCNA, PLK4, POLS, PNN, POLD3, RAMP, RASA1, RBPSUH, RDH11, SFPQ, SGOL2, SIL, SLC38A2, SMC4L1, SPAG5, STK17A, TADA2L, TF DP1, TMPO, TOP2A, TOPBP1, TOPK, TSN, UHRF1, USP1 |
| Androgen regulated | ABCC4, ADAM9, ATP11A, EEF1E1, GUCY1A3, KLK3, MCCC2, NEDD4L, NKX3–1 |
| Cell adhesion | CNTNAP2, DSC2, EVA1, FAT, ITGA4, LAMC1, MLLT4, NRP1, PCDH7, PNN, PPFIA2, SSX2IP |
Validation of Microarray Data by QRT-PCR
QRT-PCR was used to confirm changes in transcript levels for 8 genes measured on the microarrays after treatment with SM or MSA (fig. 4). Expression changes measured on the DNA microarray were consistent with those measured by QRT-PCR in virtually all 16 cases with only minor discrepancies observed at the limits of detection by either method (fig. 4, B).
Fig. 4.
Real-time RT-PCR shows validation of gene expression changes using microarray. A, expression changes of 8 genes in response to MSA and SM determined by microarray and viewed in Treeview. B, expression changes of same genes in response to MSA and SM at 6 and 12 hours, respectively, determined by RT-PCR and compared with those determined by microarray analysis. Levels of transcripts determined by RT-PCR in triplicate were normalized against those of GAPDH in same sample and they are shown in same order as in part A.
SM Affected Transcript Levels of Cell Cycle Regulated Genes and Cell Cycle Progression
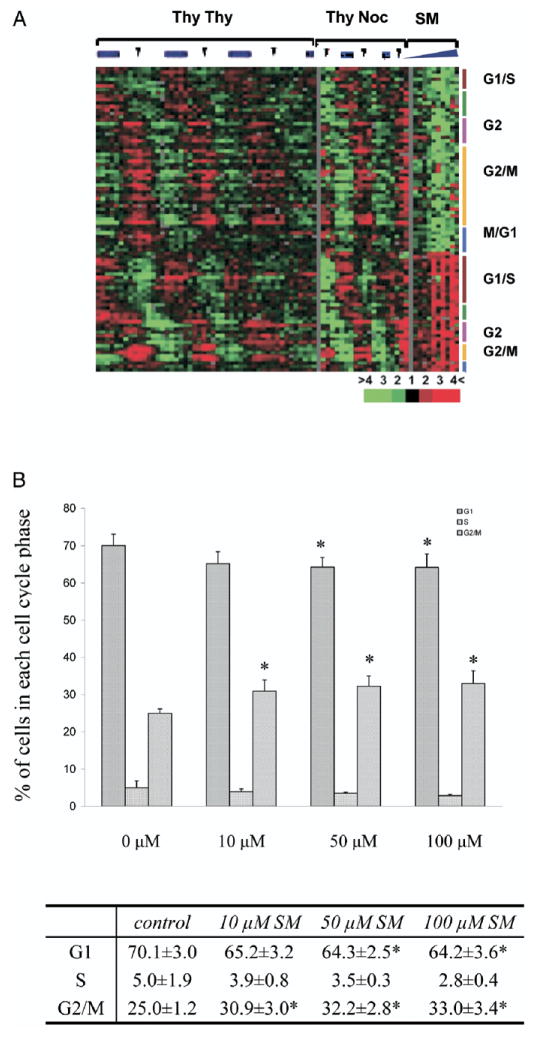
The 2,336 transcripts modulated by SM were compared to a set of 1,134 transcripts that vary periodically as synchronized cells progress through the cell cycle.11 A total of 89 transcripts, representing 76 unique genes, were found in common, representing a 1.3-fold enrichment that was statistically significant (chi-square test p <0.005, fig. 5, A). Approximately half of the transcripts were down-regulated and they were enriched for the G2/M phase of the cell cycle (47%). In the half of the transcripts that showed increased expression there was enrichment for the G1/S-phase (36%). In addition, flow cytometry analysis showed that cells treated with 10, 50 and 100 μM SM showed a significant increase in the percent at G2/M phase with a corresponding depletion of cells in G0/G1 and S phases (fig. 5, B). At higher concentrations (50 and 100 μM) SM treated cells demonstrated a significant increase in the sub-G0/G1 population compared to controls, suggesting apoptosis induction at these concentrations.
Fig. 5.
SM effects on cell cycle regulated genes and cell cycle progression. A, cell cycle regulated transcripts previously identified as being down-regulated or up-regulated by SM. Right, number of transcripts belonging to different cell cycle phases and SM effect on expression of these genes, organized as in figure 1. Left, pattern of these genes across multiple cell cycles in HeLa cells. Thy-Thy indicates double thymidine block to synchronize cells at S-phase before release. Thy-Noc indicates thymidinenocodazole block to synchronize cells at mitosis before release. Green bar represents S-phase. Red arrowheads indicate mitosis, as determined by flow cytometry or bromodeoxyuridine labeling. B, LNCaP cell cycle distribution after SM treatment by flow cytometry analysis. Asterisk indicates statistically significantly different.
Effects of SM on Androgen Signaling Pathways
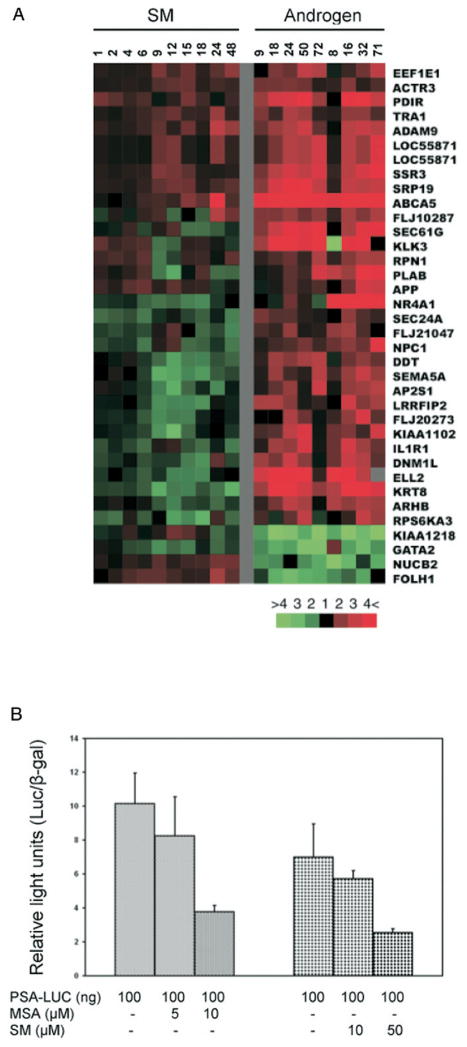
We compared the 2,336 transcripts affected by SM to a set of 567 androgen regulated transcripts, representing 398 unique genes that we identified previously.12 Half of the 35 genes found in common showed similar expression changes in response to SM and androgen, while the remainder showed changes that were opposite (fig. 6, A). The effects of SM on androgen regulated genes were relatively modest in the number of genes affected and the magnitude of the expression changes compared to what we observed previously in MSA treated LNCaP cells. Furthermore, after 24 hours of treatment 10 μM SM did not affect PSA promoter driven luciferase activity. However, 50 μM treatment resulted in a 60% decrease of PSA-luciferase activity, comparable to that observed with only 10 μM MSA (fig. 6, B). Therefore, SM can suppress androgen mediated transcription but it required much higher concentrations than MSA to achieve similar effects.
Fig. 6.
Androgen responsive genes modulated by SM. Genes that occur more than once are represented by multiple clones on arrays. A, SM affected transcripts present in list of androgen responsive transcripts identified by DePrimo et al.12 Left, gene expression patterns from 2 time courses induced by treatment of LNCaP with synthetic androgen R1881. Red arrowheads indicate well characterized androgen regulated genes. B, effects of SM and MSA on luciferase activity driven by PSA promoter after 24 hours of treatment normalized against luciferase activity in control cells. Asterisks indicate significantly different effects on reporter construct by selenium compounds.
DISCUSSION
Previous studies in several mammalian systems showed that the choice of chemical form and dose of selenium can strongly influence the observed biological effects.4,5 For example, p-XSC (1,4-phenylenebis(methylene)selenocyanate) significantly decreases the lung tumor incidence in mice exposed to NNK (4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone), whereas SM does not.13 MSA and methylselnocysteine are equally effective for preventing DMBA (7,12-dimethylbenz(a)anthracene) induced rat mammary tumors, while SM is only half as effective.14,15 In LNCaP cells in vitro 10 μM MSA produces 50% inhibition in cell growth after 48 hours of treatment, while SM requires doses of 130 μM to achieve similar growth inhibition.6,7 Differences in the biological effects between SM and other selenium compounds have been attributed to differences in their conversion to methylselenol4,16 or to depletion of SM by its nonspecific incorporation into proteins.4 However, our finding of differences in global gene expression patterns induced by SM and MSA suggests that SM and other selenium compounds target different molecular and biological pathways in the cell.
Several studies, including ours, demonstrate that MSA and SM cause cell cycle arrest at different phases and our data show that they also change expression levels of cell cycle regulated genes differently. MSA decreases expression of transcripts from all phases of the cell cycle, suggesting that it causes LNCaP to exit the cell cycle rather than produce arrest at a specific phase.7 On the other hand, SM produces enrichment for up-regulated transcripts in the G1/S-phase and down-regulated transcripts in the G2/M phase, including several that are critical to G2/M progression, such as CDC25B and CDC25A. Consistent with gene expression changes, SM induced G2/M arrest of the cell cycle, whereas MSA resulted in an accumulation of cells at the G0/G1 phase. The modest increase in LNCaP in G2 (5% to 6%) after treatment with 10 to 100 μM SM was consistent with a previous study showing a 13% increase in cells in G2/M after treatment with 500 μM SM.6
Whether different selenium compounds can influence the androgen signaling pathway differently is less clear. Our studies showed that MSA as well as SM produced mixed effects on the transcript levels of androgen responsive genes since about half of the changes in expression suggested suppressed androgen signaling, while the remaining changes were consistent with stimulation. In addition, MSA affected the expression of many more AR and androgen responsive genes to a much greater degree than SM. In part it might have been due to the different effects of SM and MSA on AR levels. We previously reported that 10 μM MSA decreases transcript and protein levels of AR and PSA,7 possibly through suppressing AR binding to the androgen responsive element,17 while 10 μM SM has no effect on AR and PSA expression levels.18,19 However, 50 μM SM or longer exposure of LNCaP to SM at low concentrations (5 and 10 μM) also decreased AR mediated reporter gene expression.20 To our knowledge whether high SM levels could act on androgen responsive elements has not been tested.
The different effects of SM and MSA on the transcriptional programs of PC cells might have important implications in the interpretation and outcome of PC intervention trials using selenium. It is possible that the mixture of organic selenium compounds found in the diet and in selenized yeast, as in the NPC intervention trial, could show different biological effects than pure SM. If SELECT is a negative trial, possible interpretations could be that selenium compounds do not protect against PC or selenium compounds other than SM are important for cancer prevention. Improved understanding of the mechanisms of action of selenium for PC prevention and the development of bio-markers germane to the pathways involved could clarify whether the differences in action observed in vitro are relevant in vivo.
CONCLUSIONS
SM exerts diverse effects on the transcriptional program of LNCaP, influencing transcript levels of genes involved in a number of biological processes, including cell cycle/apoptosis, signal transduction and transcriptional regulation. We observed similar, broadly based effects after treatment of LNCaP with MSA, suggesting that selenium compounds could influence prostate carcinogenesis through several pathways. Although the biological pathways affected by SM and MSA are similar, many important expression features differ. Our results suggest that SM can elicit gene expression changes distinct from MSA in LNCaP and they substantiate the hypothesis that different forms of selenium produce different effects on prostate cells. Further experiments are necessary to determine whether these distinct transcript patterns between SM and MSA represent secondary effects or distinct primary responses.
Acknowledgments
Supported by United States Army MRMC Prostate Cancer Research Program Postdoctoral Traineeship Award W81XWH-04-1-0080 (HZ) and the Oxnard Foundation.
Abbreviations and Acronyms
- AR
androgen receptor
- GAPDH
glyceraldehyde-3-dehydrogenase
- GOTM
Gene Ontology Tree Machine
- MSA
methylselenic acid
- NPC
Nutritional Prevention of Cancer
- PC
prostate cancer
- PCR
polymerase chain reaction
- PSA
prostate specific antigen
- QRT-PCR
quantitative reverse transcriptase-PCR
- SAM
significance analysis of microarrays
- SELECT
Selenium and Vitamin E Cancer Prevention Trial
- SM
selenomethionine
References
- 1.Combs GF., Jr Status of selenium in prostate cancer prevention. Br J Cancer. 2004;91:195. doi: 10.1038/sj.bjc.6601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffield-Lillico AJ, Dalkin BL, Reid ME, Turnbull BW, Slate EH, Jacobs ET, et al. Selenium supplementation, baseline plasma selenium status and incidence of prostate cancer: an analysis of the complete treatment period of the Nutritional Prevention of Cancer Trial. BJU Int. 2003;91:608. doi: 10.1046/j.1464-410x.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- 3.Klein EA, Thompson IM, Lippman SM, Goodman PJ, Albanes D, Taylor PR, et al. SELECT: the selenium and vitamin E cancer prevention trial. Urol Oncol. 2003;21:59. doi: 10.1016/s1078-1439(02)00301-0. [DOI] [PubMed] [Google Scholar]
- 4.Ip C. Lessons from basic research in selenium and cancer prevention. J Nutr. 1998;128:1845. doi: 10.1093/jn/128.11.1845. [DOI] [PubMed] [Google Scholar]
- 5.Combs GF., Jr Considering the mechanisms of cancer prevention by selenium. Adv Exp Med Biol. 2001;492:107. doi: 10.1007/978-1-4615-1283-7_9. [DOI] [PubMed] [Google Scholar]
- 6.Menter DG, Sabichi AL, Lippman SM. Selenium effects on prostate cell growth. Cancer Epidemiol Biomarkers Prev. 2000;9:1171. [PubMed] [Google Scholar]
- 7.Zhao H, Whitfield ML, Xu T, Botstein D, Brooks JD. Diverse effects of methylseleninic Acid on the transcriptional program of human prostate cancer cells. Mol Biol Cell. 2004;15:506. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin-Kensicki EM, Eisen JS. Sclerotome development and peripheral nervous system segmentation in embryonic zebrafish. Development. 1997;124:159. doi: 10.1242/dev.124.1.159. [DOI] [PubMed] [Google Scholar]
- 9.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kajander EO, Harvima RJ, Kauppinen L, Akerman KK, Martikainen H, Pajula RL, et al. Effects of selenomethionine on cell growth and on S-adenosylmethionine metabolism in cultured malignant cells. Biochem J. 1990;267:767. doi: 10.1042/bj2670767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, et al. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3:RESEARCH0032. doi: 10.1186/gb-2002-3-7-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das A, Desai D, Pittman B, Amin S, El-Bayoumy K. Comparison of the chemopreventive efficacies of 1,4-phenylene-bis(methylene)selenocyanate and selenium-enriched yeast on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone induced lung tumorigenesis in A/J mouse. Nutr Cancer. 2003;46:179. doi: 10.1207/S15327914NC4602_11. [DOI] [PubMed] [Google Scholar]
- 14.Ip C, Thompson HJ, Ganther HE. Selenium modulation of cell proliferation and cell cycle biomarkers in normal and premalignant cells of the rat mammary gland. Cancer Epidemiol Biomarkers Prev. 2000;9:49. [PubMed] [Google Scholar]
- 15.Whanger PD. Selenocompounds in plants and animals and their biological significance. J Am Coll Nutr. 2002;21:223. doi: 10.1080/07315724.2002.10719214. [DOI] [PubMed] [Google Scholar]
- 16.Ip C, Thompson HJ, Zhu Z, Ganther HE. In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res. 2000;60:2882. [PubMed] [Google Scholar]
- 17.Dong Y, Zhang H, Gao AC, Marshall JR, Ip C. Androgen receptor signaling intensity is a key factor in determining the sensitivity of prostate cancer cells to selenium inhibition of growth and cancer-specific biomarkers. Mol Cancer Ther. 2005;4:1047. doi: 10.1158/1535-7163.MCT-05-0124. [DOI] [PubMed] [Google Scholar]
- 18.Bhamre S, Whitin JC, Cohen HJ. Selenomethionine does not affect PSA secretion independent of its effect on LNCaP cell growth. Prostate. 2003;54:315. doi: 10.1002/pros.10184. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Ni J, Messing EM, Chang E, Yang CR, Yeh S. Vitamin E succinate inhibits the function of androgen receptor and the expression of prostate-specific antigen in prostate cancer cells. Proc Natl Acad Sci U S A. 2002;99:7408. doi: 10.1073/pnas.102014399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JD, Pramanik R, Zhang X, Carey AM, Ragavan N, Martin FL, et al. Selenium- or quercetin-induced retardation of DNA synthesis in primary prostate cells occurs in the presence of a concomitant decrease in androgen receptor activity. Cancer Lett. 2005 doi: 10.1016/j.canlet.2005.07.037. [DOI] [PubMed] [Google Scholar]