Abstract
Insulin stimulates GLUT4 (glucose transporter 4) translocation in adipocytes and muscles. An emerging picture is that Rab10 could bridge the gap between the insulin signalling cascade and GLUT4 translocation in adipocytes. In the present study, two potential effectors of Rab10, GDI (guanine-nucleotide-dissociation inhibitor)-1 and GDI-2, are characterized in respect to their roles in insulin-stimulated GLUT4 translocation. It is shown that both GDI-1 and GDI-2 exhibit similar distribution to GLUT4 and Rab10 at the TGN (trans-Golgi network) and periphery structures. Meanwhile, GDI-1 clearly interacts with Rab10 with higher affinity, as shown by both immunoprecipitation and in vivo FRET (fluorescence resonance energy transfer). In addition, the participation of GDIs in GLUT4 translocation is illustrated when overexpression of either GDI inhibits insulin-stimulated GLUT4 translocation in 3T3-L1 adipocytes. Taken together, we propose that GDI-1 is preferentially involved in insulin-stimulated GLUT4 translocation through facilitating Rab10 recycling.
Keywords: adipocytes, fluorescence resonance energy transfer (FRET), guanine-nucleotide-dissociation inhibitor (GDI), glucose transporter 4 (GLUT4), Rab10, total internal reflection microscopy (TIRFM)
Abbreviations: BFA, brefeldin A; EGFP, enhanced green fluorescent protein; FRET, fluorescence resonance energy transfer; GAP, GTPase-activating protein; GDI, guanine-nucleotide-dissociation inhibitor; GLUT4, glucose transporter 4; GSV, GLUT4 storage vesicle; HA, haemagglutinin; HEK-293 cell, human embryonic kidney 293 cell; mKO, monomeric Kushibara Orange; PM, plasma membrane; TIRFM, total internal reflection microscopy; TGN, trans-Golgi network
INTRODUCTION
Insulin regulates glucose uptake into adipocytes and muscles by controlling the amount of GLUT4 (glucose transporter 4) on the PM (plasma membrane) [1,2]. The main signalling pathway downstream of the insulin receptor in GLUT4 translocation involves sequential activation of PI3K (phosphoinositide 3-kinase) and Akt, and subsequent inactivation of AS160 by Akt-mediated phosphorylation. AS160 is one of the substrates of Akt and possesses a Rab GAP (GTPase-activating protein) domain which could stimulate the intrinsic GTPase activity of its cognate Rab protein and increase the portion of GDP-bound Rab protein [3–5]. Rab protein functions as a molecular switch oscillating between GTP- and GDP-bound forms. GTP-bound Rab protein is considered as the active form and recruits various effectors to initiate vesicle transport [6,7]. Inactivation of the GAP domain of AS160 upon phosphorylation by insulin-stimulated Akt renders its cognate Rab protein to the GTP-bound form, which then initiates GLUT4 translocation. The emergence of AS160 as a downstream molecule of Akt and the fact that it contains a functional GAP domain has shed valuable light on the linkage between the insulin signalling pathway and GLUT4 translocation and has inspired fruitful studies in the search of Rab proteins downstream of AS160. This has led to identification of several Rab proteins that probably participate in insulin-stimulated GLUT4 translocation, including Rab10 in adipocytes [8,9] and Rab8A and Rab14 in muscle cells [10].
Rab protein regulates intracellular vesicle transport with the orchestration of a variety of effectors. On the donor membrane, a GEF (guanine-nucleotide-exchange factor) activates Rab protein by promoting the exchange of GDP for GTP, and a GAP inactivates Rab protein by triggering GTP hydrolysis. Activated Rab protein initiates vesicle transport and is transferred to the acceptor membrane along with the vesicle. When converted into the GDP-bound form on the acceptor membrane, Rab protein is removed from the membrane into the cytosol by GDIs (guanine-nucleotide-dissociation inhibitors) [11,12]. Cytosolic complexes of GDP-bound Rab and GDI carry all the information that is needed for returning Rab protein back to the donor membrane, allowing multiple rounds of delivery [6,7].
Although Rab10 has been implicated in insulin-stimulated GLUT4 translocation in adipocytes, the role of GDI in this process has not been addressed yet. In the present study, we investigated the involvement of GDI-1 and GDI-2 in insulin-stimulated GLUT4 translocation and provide evidence that GDI-1 interacts preferentially with Rab10.
EXPERIMENTAL
Plasmid construction, cell culture and transfection
Construction of GLUT4–EGFP (where EGFP is enhanced green fluorescent protein) has been described previously in [13]. GLUT4–mKO (where mKO is monomeric Kushibara Orange) was constructed with EGFP substituted by mKO. Human GDI-1 and GDI-2 cDNA were purchased from SANYING Biotechnology (Wuhan, China). EGFP-, mKO- and HA (haemagglutinin)-tagged GDIs were constructed by inserting GDIs to the C-terminal of EGFP, mKO and HA tag respectively. mKO and 3×FLAG epitope sequence synthesized by AuGCT Biotechnology (Beijing, China) were fused to the N-terminal of Rab4A and Rab10 to generate mKO–Rab4A, mKO–Rab10, FLAG–Rab4A and FLAG–Rab10. Rab4A(S27N) and Rab10(T23N) were generated using the QuikChange Site-Directed Mutagenesis kit from Stratagene. mKO–Syntaxin 6 was constructed by placing Syntaxin 6 at the C-terminal end of mKO [14]. Golgi–EGFP (No. 632464; Clontech) contains the N-terminal 81 amino acids of the human β-1,4-galactosyltransferase and labels the transmedial region of the Golgi apparatus [15–17]. mKO–EGFP employed in FRET (fluorescence resonance energy transfer) experiments was constructed by placing EGFP adjacent to the C-terminal of mKO.
Culture and electroporation of 3T3-L1 cells have been described previously in [18]. HEK-293 (human embryonic kidney 293) cells were transfected with Lipofectamine™ 2000 according to the manufacturer's instructions (Invitrogen).
Antibodies and reagents
Anti-FLAG M2 resin (A2220) and antibody (F1804) were purchased from Sigma. Anti-HA antibody (ab9110) was purchased from Abcam. BFA (brefeldin A; B6542) was purchased from Sigma. Unless otherwise stated, all other reagents were purchased from Sigma.
Immunoprecipitation and Western blotting
At 36 h post-transfection, HEK-293 cells (on a 10-cm-diameter plate) were washed twice with PBS and then lysed in 1 ml of lysis buffer (50 mM Tris/HCl, pH 8.0, 500 mM NaCl, 0.1% Nonidet P40, 1 mM dithiothreitol and protease inhibitor tablets from Roche Applied Science). Protein complexes were immunoprecipitated with anti-FLAG–agarose resin for 2 h at 4 °C. Then resins were washed four times with the lysis buffer and eluted in SDS sample buffer. The eluted proteins were separated by SDS/PAGE, followed by Western blotting with corresponding antibodies according to standard procedures.
TIRFM (total internal reflection microscopy) imaging
The TIRFM setup was constructed based on through-the-lens configuration as described previously in [13]. Experiments were performed 2 days after electroporation in KRBB solution (129 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 5 mM NaHCO3, 10 mM Hepes, 3 mM glucose, 2.5 mM CaCl2, 1.2 mM MgCl2 and 0.1% BSA; pH adjusted to 7.2). Prior to experiments, transfected adipocytes were serum starved for 2 h and transferred to a closed perfusion chamber. All experiments were performed at 30 °C. Insulin was applied at a final concentration of 100 nM throughout the study and was perfused at 0 min. For quantification of the time course of insulin-stimulated GLUT4–EGFP translocation, acquired images were processed as follows. First, the cell boundary was detected by a program developed by Matlab (The Math Works) [19]. Then, mean fluorescence intensity within the cell boundary was measured. Finally, mean fluorescence intensities from different time points were normalized with the intensity of the image captured at 0 min immediately prior to insulin perfusion. Intensity profiles from different cells were averaged and the S.E.M. was calculated.
Acceptor photobleaching FRET
Experiments were carried out as described by Bossuyt et al. [20]. Briefly, HEK-293 cells were transfected with EGFP–GDIs and mKO–Rabs, EGFP fused with mKO (mKO–EGFP) as a positive control, and mKO–Rab10(T23N) and EGFP empty vector as a negative control. All images were acquired using a confocal microscope (FV500; Olympus) equipped with 488 nm and 532 nm lasers. First, pre-acceptor photobleaching images were acquired using sequence mode to avoid fluorescence leakage. Then the acceptor (mKO) was bleached by repetitive scanning of the cell with a 532 nm laser tuned to full power for 3 min. Finally, post-acceptor photobleaching images were acquired with all acquisition parameters and laser powers identical with those applied to pre-acceptor photobleaching image acquisition.
Statistics
For normally distributed data, population averages are given as the means±S.E.M., and statistical significance was tested using the Student's t test.
RESULTS
Both GDI-1 and GDI-2 colocalize with GLUT4 at the TGN (trans-Golgi network) and cell periphery
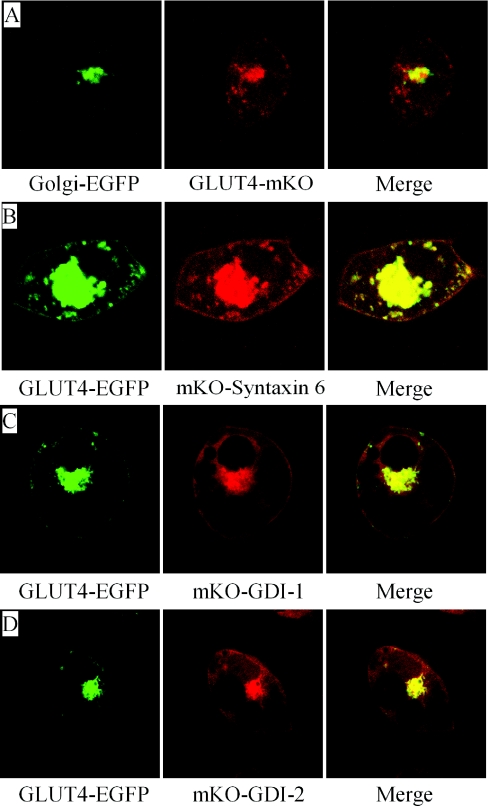
There are two GDIs expressed in 3T3-L1 adipocytes, namely GDI-1 and GDI-2 [21,22]. To investigate their potential roles in insulin-stimulated GLUT4 translocation, their subcellular localization was examined. GLUT4 was distributed in distinct small vesicles throughout the cytoplasm and in tubulo-vesicular structures in the perinuclear region (Figures 1A and 1B). Colocalization with TGN markers, including Golgi–EGFP and mKO–Syntaxin 6, shows that the tubulo-vesicular structures where GLUT4 aggregated in 3T3-L1 adipocytes should be the TGN (Figures 1A and 1B, and Supplementary Figure S1 at http://www.BiochemJ.org/bj/422/bj4220229add.htm). Clearly, both GDI-1 and GDI-2 exhibited similar distribution to GLUT4 at the TGN and periphery structures near the cell membrane (Figures 1C and 1D).
Figure 1. GDI-1 and GDI-2 colocalize with GLUT4 at the TGN and periphery structures.
Adipocytes were co-transfected with Golgi–EGFP and GLUT4–mKO (A), GLUT4–EGFP and mKO–Syntaxin 6 (B), GLUT4–EGFP and mKO–GDI-1 (C) or GLUT4–EGFP and mKO–GDI-2 (D), and were observed under confocal microscopy.
Both GDI-1 and GDI-2 exhibit similar distribution to Rab10
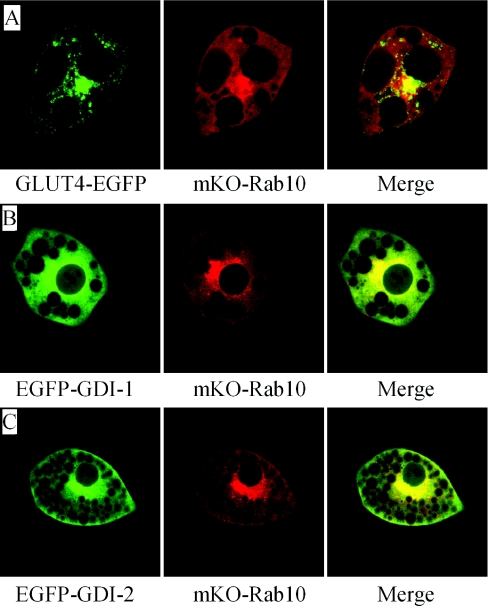
From previous reports that Rab10 plays important roles in insulin-stimulated GLUT4 translocation, it is plausible that the GDI involved in insulin-stimulated GLUT4 translocation should function in concert with Rab10 [8,9]. Rab10 aggregated at the TGN and was also distributed throughout the cytosol, which partially overlaps with GLUT4 in the same cell (Figure 2A). Meanwhile, both GDI-1 and GDI-2 shared a similar distribution pattern to Rab10 (Figures 2B and 2C), with aggregation at the TGN.
Figure 2. GDI-1 and GDI-2 colocalize mostly with Rab10.
Adipocytes were co-transfected with GLUT4–EGFP and mKO–Rab10 (A), EGFP–GDI-1 and mKO–Rab10 (B) or EGFP–GDI-2 and mKO–Rab10 (C), and were observed under confocal microscopy.
Aggregation of GLUT4, Rab10 and both GDIs at the TGN implies that the TGN might be the donor membrane of insulin-stimulated GLUT4 translocation, where Rab10 could initiate GSV (GLUT4 storage vesicle) formation and GDIs could return recycled Rab10.
BFA treatment inhibits insulin-stimulated GLUT4 translocation
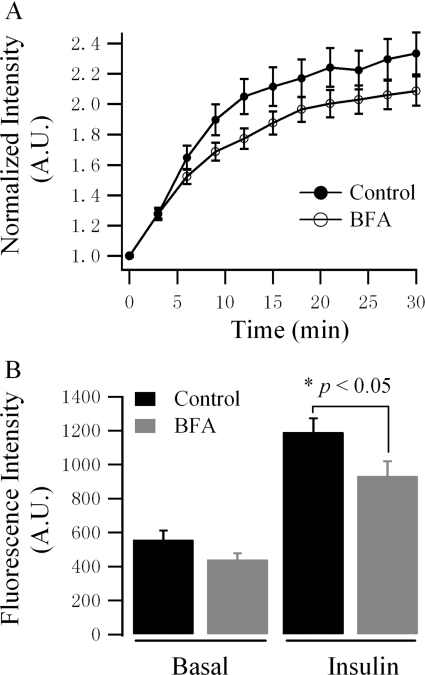
To estimate the role of the TGN in GLUT4 translocation, BFA was employed to disrupt the function of the TGN [23–26]. Pre-incubation of 3T3-L1 adipocytes with 10 μM BFA for 20 min partially inhibited insulin-stimulated GLUT4 translocation (Figure 3A). Quantification of GLUT4 distribution revealed that BFA treatment reduced GLUT4–EGFP translocated to the cell surface in response to insulin stimulation, whereas GLUT4–EGFP distribution in the basal condition was not significantly changed (Figure 3B). Thus it is probably through reducing GSV pool size rather than modifying GLUT4 distribution that disruption of the TGN function inhibits insulin-stimulated GLUT4 translocation.
Figure 3. BFA treatment inhibits insulin-stimulated GLUT4 translocation.
GLUT4–EGFP-transfected 3T3-L1 adipocytes were pre-incubated without (Control/Basal) or with 10 μM BFA for 20 min and then GLUT4 distribution and insulin-stimulated GLUT4 translocation were measured using TIRFM. (A) Serum-starved adipocytes were stimulated with 100 nM insulin, and insulin-stimulated GLUT4–EGFP translocation was observed over 30 min. Insulin was perfused at 0 min, immediately after the first image was acquired. Intensities of images from one single cell over time were normalized with the intensity of the first image and then intensity profiles from different cells were averaged. Results are represented as the means±S.E.M. (Control, n=37 cells; BFA, n=47 cells). (B) The amount of GLUT4 on the cell surface in basal and insulin-stimulated conditions. Adipocytes were randomly selected and the intensities of the cell surface were measured. For basal conditions, measurement was carried out prior to insulin perfusion; for insulin conditions, measurement was carried out 30 min post-insulin perfusion. Results are represented as the means±S.E.M. (Control, n=37 cells; BFA, n=47 cells).
GDI-1 binds to Rab10 with higher affinity
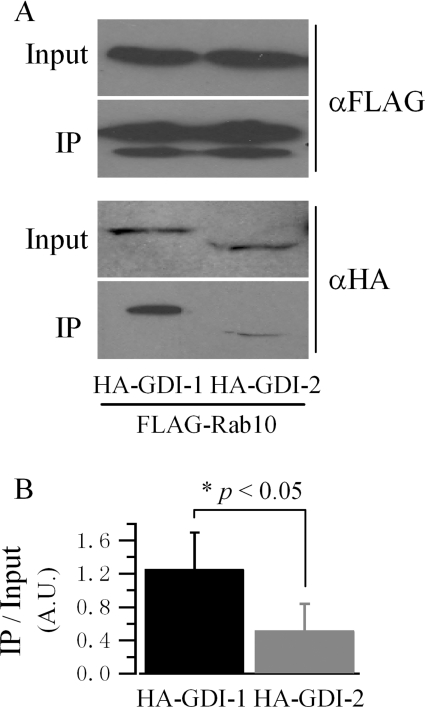
To explore the potential selectivity of the two GDIs towards Rab10, immunoprecipitation was used. FLAG–Rab10(T23N), a mutation which favours the GDP-bound form [27], was co-transfected with HA–GDI-1 or HA–GDI-2 in HEK-293 cells. FLAG–Rab10 was immunoprecipitated with anti-FLAG resin, and co-immunoprecipitated GDIs were detected with anti-HA antibody. An increased amount of protein was detected in the GDI-1 lane compared with the GDI-2 lane, indicating that GDI-1 may interact with Rab10 with higher affinity (Figure 4).
Figure 4. GDI-1 binds to Rab10 with higher affinity.
HEK-293 cells were co-transfected with FLAG–Rab10(T23N) and HA–GDI-1 or HA–GDI-2 as indicated. Protein complexes were immunoprecipitated (IP) with anti-FLAG resin (αFLAG) and detected with anti-HA (αHA) antibody for the presence of GDIs. (A) Representative result of the immunoprecipitation of GDIs with Rab10 as the bait protein. (B) Quantification of the interactions between GDIs and Rab10. Results are represented as the means±S.E.M. of three independent experiments. The IP GDI band intensity is normalized with the input one (A.U., absorbance units).
GDI-1 possesses higher affinity towards Rab10
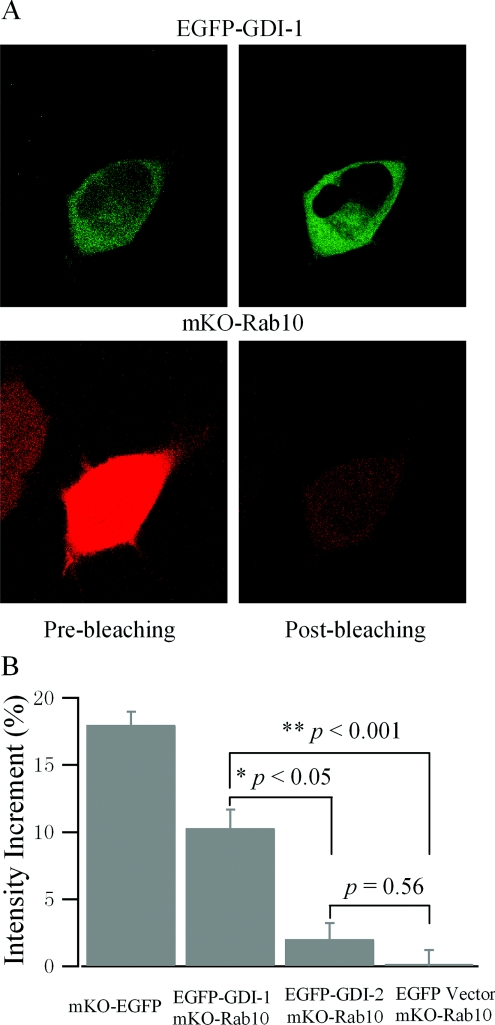
To confirm the results from the immunoprecipitation, interactions between GDIs and Rab10 were further characterized using in vivo FRET. As shown in Figure 5(A), HEK-293 cells were co-transfected with EGFP–GDI-1 and mKO–Rab10(T23N). The intensity of EGFP–GDI-1 was determined by both pre- and post- photobleaching of mKO–Rab10. Clearly, photobleaching of mKO–Rab10 significantly increased the fluorescence intensity of EGFP–GDI-1, indicating robust FRET between these two proteins. In contrast, the fluorescence intensity of EGFP–GDI-2 was not significantly increased after photobleaching of mKO-Rab10 (Figure 5B). Quantification of the interactions between GDI-1 or GDI-2 and Rab10 using in vivo FRET further confirms that GDI-1 does possess higher affinity towards Rab10.
Figure 5. Rab10 preferably interacts with GDI-1 in vivo.
(A) Interaction of GDI-1 with Rab10 demonstrated by acceptor photobleaching FRET. HEK-293 cells were co-transfected with EGFP–GDI-1 and mKO–Rab10(T23N), and images were collected 36 h post-transfection. Pre-bleaching images of EGFP–GDI-1 and mKO–Rab10 were acquired first (left-hand column), and then cells were exposed to maximal intensity laser at 532 nm for 3 min to bleach mKO–Rab10 (acceptor). After mKO–Rab10 was bleached, post-bleaching images of EGFP–GDI-1 and mKO–Rab10 were acquired (right-hand column). (B) Quantification of the interaction of GDI-1 and GDI-2 with Rab10. Intensities of post-bleaching EGFP–GDI images are normalized with the corresponding pre-bleaching images, and mean intensity increments are displayed. An mKO–EGFP construction in which EGFP is directly fused to the C-terminal of mKO was employed as a positive control, and co-transfection of mKO–Rab10(T23N) with the EGFP empty vector was taken as a negative control. Results are represented as the means±S.E.M. (GDI-1, n=31 cells; GDI-2, n=35 cells).
Overexpression of either GDI protein inhibits insulin-stimulated GLUT4 translocation
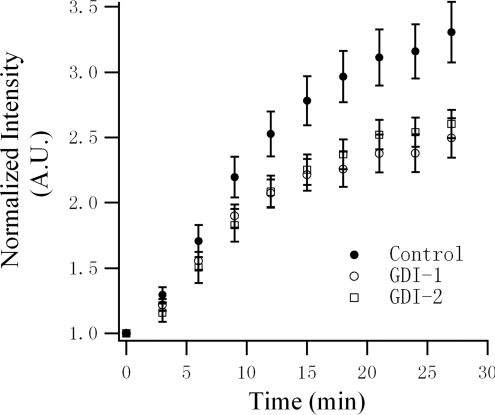
To verify their role in insulin-stimulated GLUT4 translocation, GDI-1 or GDI-2 was overexpressed in 3T3-L1 adipocytes. Insulin-stimulated GLUT4 translocation was measured with the GLUT4–EGFP reporter using TIRFM. Overexpression of either GDI protein partially, but significantly, inhibited insulin-stimulated GLUT4 translocation in 3T3-L1 adipocytes (Figure 6). Apparently, GDIs are involved in insulin-stimulated GLUT4 translocation, probably through interaction with Rab10.
Figure 6. Overexpression of either GDI-1 or GDI-2 inhibits insulin-stimulated GLUT4 translocation in 3T3-L1 adipocytes.
3T3-L1 adipocytes were co-transfected with GLUT4–EGFP and mKO–GDI-1 or mKO–GDI-2, and insulin-stimulated GLUT4 translocation was compared with GLUT4–EGFP-transfected adipocytes using TIRFM. Experiments and data preparation were carried out in the same way as described in Figure 3(A). Results are represented as the means±S.E.M. (Control, n=32 cells; GDI-1, n=43 cells; and GDI-2, n=41 cells). A.U., absorbance units.
Rab4A prefers binding to GDI-2
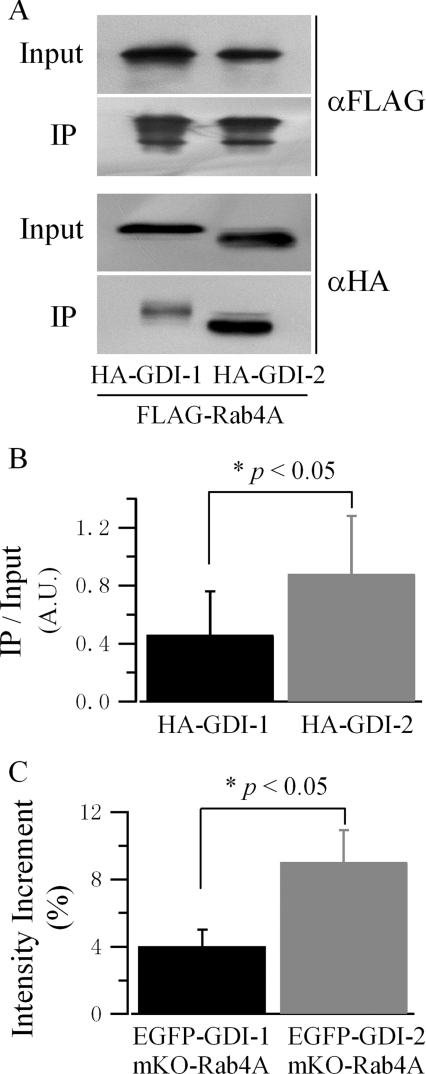
To establish the selectivity of GDIs towards Rab proteins further, the interaction of Rab4A, which has been reported to be involved in GLUT4 recycling [28,29], with both GDIs was also tested using both immunoprecipitation and in vivo FRET. With similar expression levels of FLAG–Rab4A(S27N), the mutation favouring the GDP-bound form [30], and HA–GDIs, more GDI-2 was immunoprecipitated with Rab4A (Figures 7A and 7B). Quantification of their interaction using in vivo FRET also revealed that GDI-2 interacted with Rab4A with higher affinity (Figure 7C).
Figure 7. Rab4A favours binding to GDI-2.
(A) A representative result of the immunoprecipitation of GDIs with Rab4A as the bait protein. HEK-293 cells were co-transfected with FLAG–Rab4A(S27N) and HA–GDI-1 or HA–GDI-2. Immunoprecipitation (IP) and Western blotting were carried out in the same way as described in Figure 4(A). (B) Quantification of the interactions between GDIs and Rab4A. Results are represented as the means±S.E.M. of two independent experiments. Band intensity is normalized as described in Figure 4(B). (C) Quantification of the interaction of GDI-1 and GDI-2 with Rab4A by acceptor photobleaching FRET. HEK-293 cells were co-transfected with EGFP–GDI-1 or EGFP–GDI-2, and mKO–Rab4A(S27N). Experiments and data preparation were carried out in the same way as described in Figure 5(B). Results are represented as means±S.E.M. (GDI-1, n=25 cells; GDI-2, n=25 cells).
DISCUSSION
Progress in the research of insulin-stimulated GLUT4 translocation has revealed a model in which AS160 and Rab10 play important roles in the regulation of GLUT4 translocation in adipocytes [3–5,8,9,31]. AS160 has a functional GAP domain, which remains active in the basal state [3,4,31]. Insulin-activated Akt phosphorylation of AS160 inhibits its GAP activity, thus shifting more Rab10 to the active GTP-bound form. GTP-bound Rab10 could recruit a variety of Rab effectors to facilitate the transport of GLUT4 storage vesicles towards the PM [8,9,32,33].
Apparent colocalization of GLUT4 and Rab10 revealed in the present study is consistent with the report by Sano et al. [8], where Rab10 was found at GSVs purified from 3T3-L1 adipocytes with anti-GLUT4 antibody. In combination with previous findings that the GAP domain of AS160 shows activity against Rab10 [5], that overexpression of Rab10(Q67L), which favours the GTP-binding form, increases GLUT4 on the cell surface in the absence of insulin stimulation [9], and that Rab10 knock-down inhibits insulin-stimulated GLUT4 translocation [8,9], the model centring AS160 and Rab10 in GLUT4 translocation in adipocytes is firmly favoured, although potential roles of other Rab proteins should also not be neglected [5,10].
Aggregation of GLUT4, Rab10 and both GDIs at the TGN reveals that the TGN might be the donor membrane where Rab10 initiates GSV formation. This hypothesis has previously been proposed in some of the models depicting GLUT4 translocation [2,34]. Supportive evidence includes that a large fraction of GLUT4 is distributed at the TGN [35], that internalized GLUT4 rapidly traverses the endosomal system en route to Syntaxin 6 and 16-positive TGN [36], that newly synthesized GLUT4 enters GSVs at the TGN in a GGA [Golgi-associated γ-adaptin ear homology domain Arf (ADP-ribosylation factor)-interacting protein]-dependant manner [37,38], and that disturbance of the function of Syntaxin 6, a TGN-localized protein, affects GLUT4 translocation [39]. Although previous studies using BFA to interfere with TGN function gave ambiguous results about the role of the TGN in GLUT4 translocation [23–26], in the present study it was found that BFA treatment partially inhibits insulin-stimulated GLUT4 translocation (Figure 3). Although the inhibitory effect is minor, it is noteworthy that a model assigning GSV formation at the TGN does not imply that the TGN-localized GLUT4 would be directly translocated to the cell surface in response to insulin stimulation. GSVs represent the standing insulin-sensitive pool of GLUT4 and have already been formed in the resting state. They are capable of redistributing to the cell surface in a short time when stimulated with insulin. In this way, it should not be expected that the disturbance of TGN function with BFA could produce a severe inhibition of insulin-stimulated GLUT4 translocation.
Although both GDI-1 and GDI-2 could bind to a wide range of Rab proteins [22,40], either GDI shows distinct functional roles in the context of living cells [21,41]. Specifically, the expression level of the two GDIs is different, with a higher expression level of GDI-1 in 3T3-L1 adipocytes [22], and their subcellular distribution is also distinct [21]. The demonstration that GDI-1 binds to Rab10 with higher affinity, whereas GDI-2 favours Rab4A, in the present study, reveals a certain selectivity of GDIs towards different Rab proteins and probably implies a general mechanism underlying the interaction between GDIs and Rab proteins in the cell.
Overexpression of GDI inhibits insulin-stimulated GLUT4 translocation in 3T3-L1 adipocytes. These results confirm the participation of GDIs in insulin-stimulated GLUT4 translocation. The defect probably lies in that overexpressed GDI could compete with downstream Rab effectors, e.g. GDFs (GDI-displacement factors) [6], in binding Rab10, and then sequester Rab10 in its GDP-bound form in the cytosol. Thus inefficient delivery of Rab10 back to the donor membrane could result in reduced GLUT4 translocation. Moreover, it is noteworthy that overexpression of GDI-2 inhibited insulin-stimulated GLUT4 translocation to a similar extent to GDI-1, although its affinity to Rab10 is lower. This may imply that some other Rab protein(s) (e.g. Rab4A) favoured by GDI-2 is also involved in GLUT4 translocation, and the inhibition of which by overexpressed GDI-2 also causes reduction in insulin-stimulated GLUT4 translocation.
Taken together, our results propose that the TGN could be the donor membrane of insulin-stimulated GLUT4 translocation, and GDI-1 is preferably involved in insulin-stimulated GLUT4 translocation through interaction with Rab10.
Online data
AUTHOR CONTRIBUTION
Yu Chen and Tao Xu designed the research and wrote the paper. Yu Chen, Yongqiang Deng and Jinzhong Zhang performed the research. Lu Yang and Xiangyang Xie contributed some of the reagents.
ACKNOWLEDGEMENTS
We thank Professor David E. James (Garvan Institute of Medical Research, Sydney, Australia) for providing human Rab4A and murine Rab10 cDNA templates, and our colleague Dr Liangyi Chen for valuable discussions.
FUNDING
This work is supported by the National Science Foundation of China [grant number 30630020]; the National Basic Research Program of China [grant number 2004CB720000]; the CAS Project [grant number KSCX1-YW-02-01]; and the Beijing Natural Science Foundation [grant number 5092017].
References
- 1.Watson R. T., Kanzaki M., Pessin J. E. Regulated membrane trafficking of the insulin-responsive glucose transporter 4 in adipocytes. Endocr. Rev. 2004;25:177–204. doi: 10.1210/er.2003-0011. [DOI] [PubMed] [Google Scholar]
- 2.Bryant N. J., Govers R., James D. E. Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 2002;3:267–277. doi: 10.1038/nrm782. [DOI] [PubMed] [Google Scholar]
- 3.Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J. Biol. Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 4.Sano H., Kane S., Sano E., Miinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 5.Miinea C. P., Sano H., Kane S., Sano E., Fukuda M., Peranen J., Lane W. S., Lienhard G. E. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem. J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer S., Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 7.Seabra M. C., Wasmeier C. Controlling the location and activation of Rab GTPases. Curr. Opin. Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Sano H., Roach W. G., Peck G. R., Fukuda M., Lienhard G. E. Rab10 in insulin-stimulated GLUT4 translocation. Biochem. J. 2008;411:89–95. doi: 10.1042/BJ20071318. [DOI] [PubMed] [Google Scholar]
- 9.Sano H., Eguez L., Teruel M. N., Fukuda M., Chuang T. D., Chavez J. A., Lienhard G. E., McGraw T. E. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Ishikura S., Bilan P. J., Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem. Biophys. Res. Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- 11.Ullrich O., Stenmark H., Alexandrov K., Huber L. A., Kaibuchi K., Sasaki T., Takai Y., Zerial M. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J. Biol. Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 12.Sasaki T., Kikuchi A., Araki S., Hata Y., Isomura M., Kuroda S., Takai Y. Purification and characterization from bovine brain cytosol of a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to smg p25A, a ras p21-like GTP-binding protein. J. Biol. Chem. 1990;265:2333–2337. [PubMed] [Google Scholar]
- 13.Li C. H., Bai L., Li D. D., Xia S., Xu T. Dynamic tracking and mobility analysis of a single GLUT4 storage vesicle in live 3T3-L1 cells. Cell Res. 2004;14:480–486. doi: 10.1038/sj.cr.7290251. [DOI] [PubMed] [Google Scholar]
- 14.Watson R. T., Pessin J. E. Functional co-operation of two independent targeting domains in syntaxin 6 is required for its efficient localization in the trans-Golgi network of 3T3L1 adipocytes. J. Biol. Chem. 2000;275:1261–1268. doi: 10.1074/jbc.275.2.1261. [DOI] [PubMed] [Google Scholar]
- 15.Roth J., Berger E. G. Immunocytochemical localization of galactosyltransferase in HeLa cells: codistribution with thiamine pyrophosphatase in trans-Golgi cisternae. J. Cell Biol. 1982;93:223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbero P., Bittova L., Pfeffer S. R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izumikawa T., Koike T., Shiozawa S., Sugahara K., Tamura J., Kitagawa H. Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. J. Biol. Chem. 2008;283:11396–11406. doi: 10.1074/jbc.M707549200. [DOI] [PubMed] [Google Scholar]
- 18.Bai L., Wang Y., Fan J., Chen Y., Ji W., Qu A., Xu P., James D. E., Xu T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab. 2007;5:47–57. doi: 10.1016/j.cmet.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y., Wang Y., Ji W., Xu P., Xu T. A pre-docking role for microtubules in insulin-stimulated glucose transporter 4 translocation. FEBS J. 2008;275:705–712. doi: 10.1111/j.1742-4658.2007.06232.x. [DOI] [PubMed] [Google Scholar]
- 20.Bossuyt J., Despa S., Martin J. L., Bers D. M. Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J. Biol. Chem. 2006;281:32765–32773. doi: 10.1074/jbc.M606254200. [DOI] [PubMed] [Google Scholar]
- 21.Shisheva A., Buxton J., Czech M. P. Differential intracellular localizations of GDP dissociation inhibitor isoforms. Insulin-dependent redistribution of GDP dissociation inhibitor-2 in 3T3-L1 adipocytes. J. Biol. Chem. 1994;269:23865–23868. [PubMed] [Google Scholar]
- 22.Shisheva A., Sudhof T. C., Czech M. P. Cloning, characterization, and expression of a novel GDP dissociation inhibitor isoform from skeletal muscle. Mol. Cell. Biol. 1994;14:3459–3468. doi: 10.1128/mcb.14.5.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachaal M., Moronski C., Liu H., Jung C. Y. Brefeldin A inhibits insulin-induced glucose transport stimulation and GLUT4 recruitment in rat adipocytes. J. Biol. Chem. 1994;269:23689–23693. [PubMed] [Google Scholar]
- 24.Bao S., Smith R. M., Jarett L., Garvey W. T. The effects of brefeldin A on the glucose transport system in rat adipocytes. Implications regarding the intracellular locus of insulin-sensitive Glut4. J. Biol. Chem. 1995;270:30199–30204. doi: 10.1074/jbc.270.50.30199. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti R., Buxton J., Joly M., Corvera S. Insulin-sensitive association of GLUT-4 with endocytic clathrin-coated vesicles revealed with the use of brefeldin A. J. Biol. Chem. 1994;269:7926–7933. [PubMed] [Google Scholar]
- 26.Martin S., Ramm G., Lyttle C. T., Meerloo T., Stoorvogel W., James D. E. Biogenesis of insulin-responsive GLUT4 vesicles is independent of brefeldin A-sensitive trafficking. Traffic. 2000;1:652–660. doi: 10.1034/j.1600-0854.2000.010809.x. [DOI] [PubMed] [Google Scholar]
- 27.Schuck S., Gerl M. J., Ang A., Manninen A., Keller P., Mellman I., Simons K. Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic. 2007;8:47–60. doi: 10.1111/j.1600-0854.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 28.Imamura T., Huang J., Usui I., Satoh H., Bever J., Olefsky J. M. Insulin-induced GLUT4 translocation involves protein kinase C-λ-mediated functional coupling between Rab4 and the motor protein kinesin. Mol. Cell. Biol. 2003;23:4892–4900. doi: 10.1128/MCB.23.14.4892-4900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollenweider P., Martin S. S., Haruta T., Morris A. J., Nelson J. G., Cormont M., Le Marchand-Brustel Y., Rose D. W., Olefsky J. M. The small guanosine triphosphate-binding protein Rab4 is involved in insulin-induced GLUT4 translocation and actin filament rearrangement in 3T3-L1 cells. Endocrinology. 1997;138:4941–4949. doi: 10.1210/endo.138.11.5493. [DOI] [PubMed] [Google Scholar]
- 30.Odley A., Hahn H. S., Lynch R. A., Marreez Y., Osinska H., Robbins J., Dorn G. W., II Regulation of cardiac contractility by Rab4-modulated β2-adrenergic receptor recycling. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7082–7087. doi: 10.1073/pnas.0308335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larance M., Ramm G., Stockli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- 32.Pfeffer S. Membrane domains in the secretory and endocytic pathways. Cell. 2003;112:507–517. doi: 10.1016/s0092-8674(03)00118-1. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda M. Versatile role of Rab27 in membrane trafficking: focus on the Rab27 effector families. J. Biochem. 2005;137:9–16. doi: 10.1093/jb/mvi002. [DOI] [PubMed] [Google Scholar]
- 34.Dugani C. B., Klip A. Glucose transporter 4: cycling, compartments and controversies. EMBO Rep. 2005;6:1137–1142. doi: 10.1038/sj.embor.7400584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slot J. W., Geuze H. J., Gigengack S., Lienhard G. E., James D. E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shewan A. M., van Dam E. M., Martin S., Luen T. B., Hong W., Bryant N. J., James D. E. GLUT4 recycles via a trans-Golgi network (TGN) subdomain enriched in Syntaxins 6 and 16 but not TGN38: involvement of an acidic targeting motif. Mol. Biol. Cell. 2003;14:973–986. doi: 10.1091/mbc.E02-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson R. T., Khan A. H., Furukawa M., Hou J. C., Li L., Kanzaki M., Okada S., Kandror K. V., Pessin J. E. Entry of newly synthesized GLUT4 into the insulin-responsive storage compartment is GGA dependent. EMBO J. 2004;23:2059–2070. doi: 10.1038/sj.emboj.7600159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan A. H., Capilla E., Hou J. C., Watson R. T., Smith J. R., Pessin J. E. Entry of newly synthesized GLUT4 into the insulin-responsive storage compartment is dependent upon both the amino terminus and the large cytoplasmic loop. J. Biol. Chem. 2004;279:37505–37511. doi: 10.1074/jbc.M405694200. [DOI] [PubMed] [Google Scholar]
- 39.Perera H. K., Clarke M., Morris N. J., Hong W., Chamberlain L. H., Gould G. W. Syntaxin 6 regulates Glut4 trafficking in 3T3-L1 adipocytes. Mol. Biol. Cell. 2003;14:2946–2958. doi: 10.1091/mbc.E02-11-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shisheva A., Chinni S. R., DeMarco C. General role of GDP dissociation inhibitor 2 in membrane release of Rab proteins: modulations of its functional interactions by in vitro and in vivo structural modifications. Biochemistry. 1999;38:11711–11721. doi: 10.1021/bi990200r. [DOI] [PubMed] [Google Scholar]
- 41.Shisheva A., Czech M. P. Association of cytosolic Rab4 with GDI isoforms in insulin-sensitive 3T3-L1 adipocytes. Biochemistry. 1997;36:6564–6570. doi: 10.1021/bi970202g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.