Abstract
Horizontal gene transfer (HGT) describes the transmission of genetic material across species boundaries and is an important evolutionary phenomenon in the ancestry of many microbes. The role of HGT in plant evolutionary history is, however, largely unexplored. Here, we compare the genomes of six plant species with those of 159 prokaryotic and eukaryotic species and identify 1689 genes that show the highest similarity to corresponding genes from fungi. We constructed a phylogeny for all 1689 genes identified and all homolog groups available from the rice (Oryza sativa) genome (3177 gene families) and used these to define 14 candidate plant-fungi HGT events. Comprehensive phylogenetic analyses of these 14 data sets, using methods that account for site rate heterogeneity, demonstrated support for nine HGT events, demonstrating an infrequent pattern of HGT between plants and fungi. Five HGTs were fungi-to-plant transfers and four were plant-to-fungi HGTs. None of the fungal-to-plant HGTs involved angiosperm recipients. These results alter the current view of organismal barriers to HGT, suggesting that phagotrophy, the consumption of a whole cell by another, is not necessarily a prerequisite for HGT between eukaryotes. Putative functional annotation of the HGT candidate genes suggests that two fungi-to-plant transfers have added phenotypes important for life in a soil environment. Our study suggests that genetic exchange between plants and fungi is exceedingly rare, particularly among the angiosperms, but has occurred during their evolutionary history and added important metabolic traits to plant lineages.
INTRODUCTION
Horizontal gene transfer involves the transmission of genetic material between distinct evolutionary lineages (Ragan, 2001b; Doolittle et al., 2003; Richards et al., 2003; Andersson, 2005; Keeling and Palmer, 2008). It is now well established that horizontal gene transfer (HGT) is an important process in prokaryotes (Jain et al., 1999; Doolittle et al., 2003), providing a source for genomic innovation in many microbial lineages (Jain et al., 2003). Analysis of protozoan genomes has also demonstrated that HGT is a factor in some eukaryotic genomes (Richards et al., 2003; Andersson, 2005; Eichinger et al., 2005; Loftus et al., 2005; Keeling and Palmer, 2008), suggesting that universal eukaryotic cellular features, such as the possession of linear chromatin-based chromosomes, intron-exon gene structures, and the nuclear envelope, are not barriers to HGT.
It has been argued that the phagotrophic predatory lifestyle of many eukaryote cells may effectively establish a gene transfer ratchet between the predator and prey, increasing the frequency of opportunity for prey-to-predator gene transfer events (Doolittle, 1998). Several studies of phagotrophic eukaryotes appear to be consistent with the principle of the “you are what you eat hypothesis” because several phagotrophic single-celled protozoa possess genes of recent prokaryotic ancestry (Andersson et al., 2003; Archibald et al., 2003; Eichinger et al., 2005; Loftus et al., 2005). There is, however, also some evidence for HGT involving nonphagotrophic fungi and oomycetes (Garcia-Vallve et al., 2000; Friesen et al., 2006; Richards et al., 2006a; Belbahri et al., 2008).
Evidence for HGTs involving plants that have cell walls and are nonphagotrophic is currently very limited and generally only of a specific type and origin, involving transmission of transposable elements (Diao et al., 2006), plastid-derived endosymbiotic gene transfers (Martin et al., 2002), prokaryote-derived gene transfers (the majority of which appear to be anciently derived; Richards et al., 2006b; Moustafa et al., 2008), Agrobacterium tumefaciens–mediated DNA transfer (Aoki and Syno, 1999; Intrieri and Buiatti, 2001), cross-species hybridization events that lead to very recent gene transfer events and occur between relatively recently derived evolutionary lineages (Kim et al., 2008), or gene transfer between mitochondrial genomes of plants (Cho and Palmer, 1999; Bergthorsson et al., 2003, 2004; Richardson and Palmer, 2007). The latter type of HGT, involving cross-species mitochondrial transfer events, represents one of the few forms of gene transfer into the plant kingdom that occurred after the evolution of multicellularity, loss of phagotrophy, and colonization of terrestrial environments. The majority of other forms of HGT into plants appear to predate the rise of land plants. A recent study of the Vitis vinifera organelle genomes, for instance, demonstrated additional cases of transfer between the chloroplast and mitochondria and from closterovirus to the V. vinifera mitochondrial genome (Goremykin et al., 2008). The study furthermore suggested that many reported cases of plant mitochondrial gene transfer may not stand up to rigorous phylogenetic analyses, which include appropriate models of site rate heterogeneity. Meanwhile, other studies have highlighted the need to account for the full genomic inventory of the gene family and an adequate taxon sampling (Cusimano et al., 2008) when evaluating cases of HGT in plant genomes. Taken together, these data have suggested a less diverse web of transfer events in land plants when compared with phagotrophic-microbial eukaryotes.
Increasingly, phylogenomic analysis suggests that HGT has occurred between microorganisms with overlapping ecological niches or that share close associations, such as host and parasite, microbial prey and predator, and partners within a symbiosis (Andersson et al., 2003; Archibald et al., 2003; Beiko et al., 2005; Loftus et al., 2005; Richardson and Palmer, 2007). Furthermore, it appears that such gene transfers are often important for defining the habitat range of recipient lineages (Garcia-Vallve et al., 2000; Friesen et al., 2006). The land plants and fungi are very distant relatives (Rodriguez-Ezpeleta et al., 2005; Burki et al., 2008), with numerous metabolic and developmental differences, but they do share a multitude of ecological associations, including mycorrhizal, endophytic, and commensal interactions, plant diseases, and the widespread saprotrophic degradation of plant material by fungi. Indeed, it is widely suggested that diversification of both the plant and fungal kingdoms in terrestrial environments has, in part, been dependent on their shared (Wang and Qiu, 2006) and ancient (Pirozynski and Malloch, 1975) ecological associations, raising the possibility that plant-fungi HGT may be a hitherto unexplored factor in their collective evolutionary history.
The detection of interspecies gene transfers is best achieved by generation of a strongly supported phylogenetic tree that contradicts the known species phylogeny, placing the gene of one taxonomic group within a phylogenetic cluster of an unrelated taxa (Richards et al., 2003; Andersson, 2005; Keeling and Palmer, 2008). Additional methods have also been applied, including surrogate sequence composition and codon usage methods, but have been demonstrated to be unreliable without phylogenetic analyses (Ragan, 2001a). Alternative methods, such as taxon distribution and nearest neighbor analyses, have also been used to identify peculiar gene ancestries that could be symptomatic of HGT (Keeling and Inagaki, 2004; Friesen et al., 2006).
Here, we have investigated the incidence and ancestry of HGT between plants and fungi. We compared sequences of the entire predicted proteomes from six plant species, representing a wide taxonomic diversity, with 84 eukaryotic and 69 prokaryote genome sequences (see Supplemental Table 1 online), and generated individual phylogenetic trees using optimized models that account for site rate heterogeneity for every plant gene family that showed a higher similarity to fungal genes than those of any other genome. This process identified nine plant-fungi gene transfers. The results of these analyses suggest a pattern of rare and ancient transfers between plants and fungi.
RESULTS
In this study, we set out to investigate the incidence of HGT between plants and fungi. To do this, we first generated automated Perl scripts to allow BLAST searches of the full gene inventories of all available plant genome sequences, Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Selaginella moellendorffii, Sorghum bicolor, and Physcomitrella patens, against a database of 1,566,625 gene sequences. These included 46 fungal species as well as five green algae. A total of 5866 proteins were clustered using OrthoMCL to group together potential orthologs (e-value cutoff 1e−20; inflation value 1.5). Every predicted plant protein sequence that showed a higher BLAST similarity score to a gene from a fungus rather than an algal, protist, prokaryote, or animal species was retained for subsequent analysis (Table 1 shows results of these comparisons). This initial screen resulted in 943 clusters and 746 singletons and a final list of 1689 sequence data sets that, based on BLAST profiles, were considered to be candidate plant-fungi HGTs and therefore retained for phylogenetic analyses. We were aware, however, that comparisons of BLAST profiles can be misleading, and the closest BLAST hit is often not the nearest neighbor on a phylogenetic tree (Koski and Golding, 2001). It is therefore possible that such a BLAST-based genome comparison may miss a significant number of candidate plant-fungi HGTs. To test for such a possibility, we clustered the predicted proteome of the rice genome into ortholog groups by Markov Chain clustering (Li et al., 2003) and then independently generated a phylogeny for every rice cluster group (3177 phylogenies). Together, these two protocols identified 4866 gene families for phylogenetic analyses.
Table 1.
Identification of Putative HGTs between Plants and Fungi for Phylogenetic Analyses
| Arabidopsis | O. sativa | P. patens | P. trichocarpa | S. moellendorffii | S. bicolor | |
|---|---|---|---|---|---|---|
| Total number of proteins | 31,913 | 26,777 | 35,938 | 45,555 | 22,285 | 35,899 |
| Top hit versus fungi | 1,206 | 1,045 | 776 | 1,426 | 802 | 1,043 |
| Top hit versus fungi, number of transposable elements | 1,160 | 852 | 764 | 1,331 | 794 | 965 |
BLASTp (e-value cutoff 1e−20) was used to compare predicted proteins from Arabidopsis, O. sativa, P. patens, P. trichocarpa, and S. moellendorffii with sets of predicted proteins from 159 species, as described in Methods.
Testing for the Incidence of Plant-Fungal HGT Using Phylogenetic Analysis
To test for HGT, we established an automated gene-by-gene phylogeny pipeline to generate a PhyML tree (Guindon and Gascuel, 2003) for each of the 4866 genes identified in the analyses of the Oryza genome and the BLAST comparisons (see Methods) and then manually inspected the results of each of the 4866 phylogenies. To account for both taxon distribution and phylogenetic methods of detecting HGT, we identified plant-fungi HGT phylogenies based on two criteria: a gene phylogeny demonstrating a plant gene sequence branching within a cluster of sequences from fungal taxa (or vice versa) or a phylogeny that demonstrated a diverse plant-specific gene family absent from all other taxa except a narrow taxonomic group of fungi (or vice versa). Using this approach, we detected 38 putative plant-fungi HGT candidates, of which two were detected using only the whole rice genome approach, 35 were detected using the BLAST-based survey, and one was detected using both search protocols.
To test the reliability of the pipeline phylogenetic data for the candidate plant-fungi HGTs, we manually checked each multiple amino acid sequence alignment by adding, where required, additional sequence sampling available in GenBank that was not initially included in our phylogenetic analysis pipeline database. Additional sequence data were identified and added to our alignments using manual BLASTp and PSI-BLAST sequence similarity searches of GenBank (nonredundant database). This process demonstrated that 24 of the data sets were not candidate HGTs because we could identify third party taxa not present in our pipeline database, which possessed high similarity to either the plant or fungal gene. We also compared each data set to the taxonomically broad EST eukaryotic database (TBestDB; O'Brien et al., 2007) and the GenBank EST database to improve taxon sampling where possible and to check for gene/taxon distributions that might be inconsistent with a HGT hypothesis. This resulted in 14 candidate HGT data sets, which were selected for manual phylogenetic analyses.
Testing the Plant-Fungi HGT Hypothesis
The 14 manually masked alignments were analyzed using a range of phylogenetic methods, including Bayesian topology searches (Ronquist and Huelsenbeck, 2003), fastML bootstrap analysis using heuristic searches that begin with parsimony-derived starting trees (Stamatakis, 2006), and neighbor joining–derived starting trees (Guindon and Gascuel, 2003). In addition, we also used a branch-by-branch SH topology test to investigate the resolution of each node within the tree topology (Anisimova and Gascuel, 2006). Analysis of HGTs in plant mitochondrial genomes has suggested that adequate assessment and correction for site rate heterogeneity (variation in substitution rates across the alignment) is an important factor for investigating HGT in plant genomes (Goremykin et al., 2008). When using all four of these methods, we implemented the optimal substitution matrices and models of site rate heterogeneity, including a gamma distribution, and in some cases a proportion of invariant sites, as assessed by Modelgenerator analysis (Keane et al., 2004) (see Supplemental Figures 1 to 9 online for details of the models used). After this additional round of analyses, five further data sets did not show phylogenetic tree topologies consistent with our HGT criteria, and we therefore excluded them from subsequent analyses. Table 2 lists the remaining nine putative HGTs, which we selected for further study.
Table 2.
Nine Putative Plant-Fungi Gene Transfers Identified Following Detailed Phylogenetic Study
| HGT | Putative Functional Protein Annotation | Direction of Transfer | GenBank Accession No. |
|---|---|---|---|
| 1a | l-Fucose permease, sugar transporter | Fungi > Plant | EDQ83581 |
| 1b | Zinc binding alcohol dehydrogenase | Plant > Fungi | EDQ61440* |
| 1c | Major facilitator superfamily, membrane transporter | Fungi > Plant | EAU93280* |
| 2 | Phospholipase/carboxylesterase family protein | Fungi > Plant | XP_389330* |
| 3a | iucA/iucC family protein, siderophore biosynthesis | Fungi > Plant | EDR08700* |
| 3b | Unknown/conserved hypothetical protein | Fungi > Plant | EDQ68642 |
| 4a | DUF239 domain protein | Plant > Fungi | EDR02747 |
| 4b | Phosphate-responsive 1 family protein | Plant > Fungi | ABK92591* |
| 4c | Unknown/conserved hypothetical protein with similarity to zinc finger (C2H2-type) protein | Plant > Fungi | EDN23584 |
GenBank accession numbers of transferred genes are given, and the asterisks indicate accession numbers of the closest BLAST hit in GenBank to the HGT, rather than that of the recipient gene.
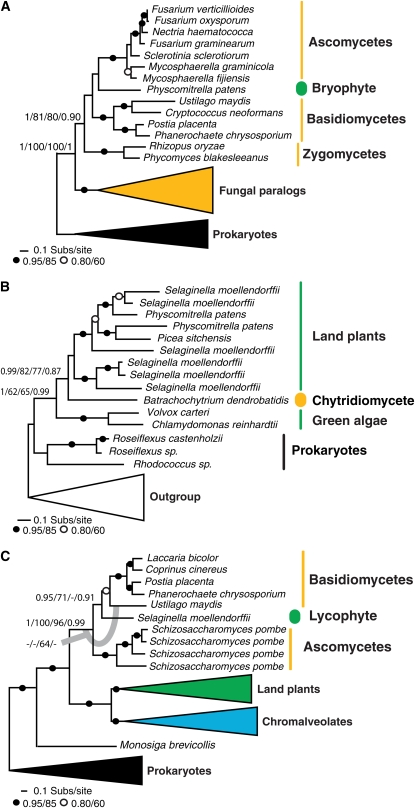
Three of the HGT data sets demonstrated a single plant or fungal gene (the recipient) clustering with, and crucially, within a phylogenetic cluster of fungi or plants, respectively (the putative donor) (Figure 1). We assessed the tree topologies by four separate methods. In all three phylogenies, the nodes that placed the recipient taxa with the donor taxa and within the donor taxon cluster were supported by a Bayesian posterior P value of 0.95 or greater. Furthermore, bootstrap analyses using two different fastML heuristic search methods demonstrated support for the HGT-like tree topology with 62% support or greater. In one case, the RAxML analyses showed a slightly different tree topology, shown in Figure 1C, but importantly the support for the recipient taxa branching with, and within, the donor cluster was still in excess of 64% bootstrap support.
Figure 1.
Phylogenetic Evidence for Plant-Fungi HGT.
Results of phylogenetic analyses of three candidates for which the tree topology results support a case for HGT between plants and fungi. All phylogenies are reduced versions of the full tree topologies (see corresponding Supplemental Figures 1 to 3 online for full details). Black dots are used when the relevant node is supported with 85% or more bootstrap support by both bootstrap methods. Open circles show cases when both bootstrap results are between 60 and 84%. For key nodes, actual support values are marked in the order Bayesian posterior probability/PhyML bootstrap/RAxML bootstrap/PhyML node-by-node SH test. Fungi groups are marked in yellow and plants in green. The transferred genes are marked by thick and colored ovals. Nonformal and nonequivalent higher taxonomic names are labeled on each phylogeny.
(A) Phylogeny of the putative l-fucose permease sugar transporter.
(B) Phylogeny of the putative zinc binding alcohol dehydrogenase.
(C) Phylogeny of the putative major facilitator superfamily membrane transporter.
The fourth topology assessment method involved a process whereby each topological branch was systematically collapsed and then each altered version of the tree topology was compared with the highest scoring tree using the SH topology comparison test (Anisimova and Gascuel, 2006). Generally, these branches are considered to be significantly supported when a value of 0.95 or greater is returned. In all three cases, the topology node that clustered the recipient sequences with the donor taxa was significantly supported by this topology test, suggesting that the recipient does not branch elsewhere in the phylogeny. However, in all three cases, the SH branch-by-branch analyses method did not significantly support the grouping of the recipient taxa within the donor phylogenetic cluster, although in all three cases, the SH value approached significance at the 0.95 level (0.9, 0.87, and 0.91; Figures 1A to 1C, respectively). However, these topology alterations and comparison tests do not represent the full HGT topology, but instead test only the viability of one specific node within the tree topology.
Because the results of the branch-by-branch topology search were ambiguous, and bootstrap values of 62 to 95% can be interpreted as weak, we further tested these three phylogenetic relationships using specific topology comparison tests. In all three cases, we generated alternative topologies for which we forced the putative donor taxa to form a monophyletic group to the exclusion of the putative recipient taxa and calculated the optimal phylogenetic tree under this constraint using a range of methods. These protocols specifically test support for the recipient taxa branching within the donor clade. In all three data sets, we could exclude the alternative non-HGT topology at the 5% significance level using either the SH test, the AU test, or both (Shimodaira and Hasegawa, 2001; Shimodaira, 2002) (see Supplemental Figures 1 to 3 online). When considered together, these data demonstrate three tree topologies where the leading explanation for the branching relationships is a plant-fungus HGT event.
Phylogenetic Evidence for Plant-Fungi HGT Supported by Alternative Topology Tests
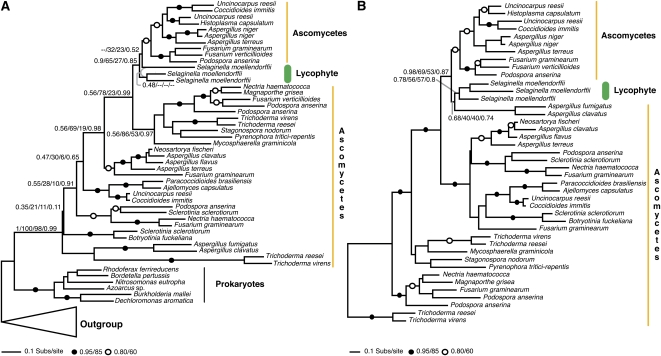
An additional putative case of fungi-to-plant gene transfer, which showed three genes from the lycophyte Selaginella spp branching within the fungal ascomycete clade, is shown in Figure 2A. This relationship was weakly supported in three of the topology assessment methods used, whereas the PhyML bootstrap method showed the Selaginella genes branching within the ascomycete cluster with three moderately supported nodes with bootstrap values of 69, 78, and 65%, respectively. To test the phylogenetic support for these three Selaginella sequences grouping within the ascomycete fungal clade (the key phylogenetic relationship for inferring a fungi-to-plant HGT event), we constrained a monophyletic branching order for the specific ascomycete clade within which Selaginella branches and calculated alternative tree topologies using distance and parsimony methods (Swofford, 2002). For each alternative topology, we recalculated branch lengths and site likelihood values using ML in P4 (Foster, 2004) for the alternative topologies and compared the resulting topologies with the MrBayes and PhyML topologies using the AU and SH tests in Consel (Shimodaira and Hasegawa, 2001). We found that we could reject all five alternative topologies with monophyly of the ascomycete clade enforced at <0.1% confidence level. In contrast with the weakly supported MrBayes, RAxML, and branch-by-branch SH test analysis, this provides strong evidence that the Selaginella sequence branches within the ascomycete clade. The most parsimonious explanation of this result is therefore a fungi-to-plant HGT.
Figure 2.
Phylogenetic Evidence for Plant-Fungi HGT with Weak Topology Support Values but Confirmed by Alternative Topology Tests.
(A) The phylogeny of the phospholipase/carboxylesterase family protein demonstrates moderate support for a fungi-to-plant HGT by PhyML bootstrap methods but weak support by all three other topology support assessment methods. Alternative topology tests gave strong support for the placement of the Selaginella sequences within the specific ascomycete cluster, suggesting an ascomycete-to-Selaginella HGT.
(B) The Selaginella sequences do not form a monophyletic cluster in the phylogeny. To test this, we performed a second analysis focusing the taxon sampling on the branches local to the Selaginella group and increasing character sampling. The phylogenetic trees shown are reduced versions of the full tree topologies (see Supplemental Figure 4 online for full details). The figures are labeled using the same conventions described for Figure 1.
The phylogeny also showed that the three Selaginella genes were not monophyletic (Figure 2A). To test the support for paraphyly of the lycophyte and ascomycete clades, we performed a second phylogenetic analysis (see Supplemental Figure 4 online) removing distantly related sequences and adjusting the alignment character sampling accordingly. Phylogenetic analyses based on this second alignment demonstrated weak support (56/57% bootstrap support) for a monophyletic clade of Selaginella sequences (Figure 2B), suggesting that the putative gene transfer was a single fungal-to-plant transfer event and that the paraphyly of the Selaginella sequences within the ascomycete clade (shown in Figure 2A) is a phylogenetic tree reconstruction artifact.
Plant-Fungi HGTs of Potential Prokaryotic Origin
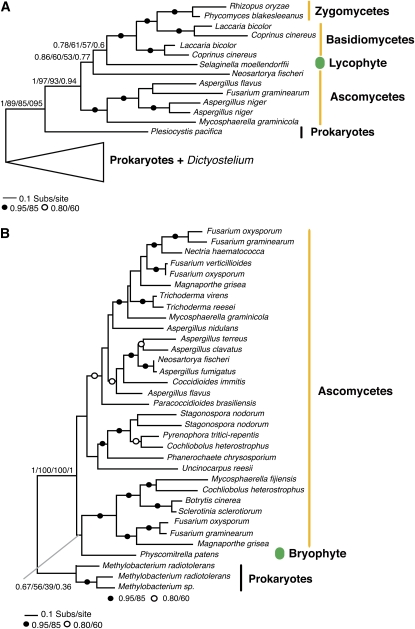
We detected two data sets, which, based upon a combination of the taxon distribution of the gene family and multiple phylogenetic analyses, were consistent with fungi-to-plant HGT. In two cases that are shown in Figures 3A and 3B, the gene family analyzed was present in a diverse group of fungi, a taxonomically broad group of prokaryotes, and a single plant genome. Together, this analysis shows that these gene families demonstrate a highly punctate taxonomic distribution that is suggestive of HGT. In both cases, the plant genes branch within the fungal cluster, suggesting fungi-to-plant HGT. However, in the first case (Figure 3A), the phylogenetic relationship of the fungal taxa is not consistent with the current consensus of the fungal phylogeny (Fitzpatrick et al., 2006; James et al., 2006) with two basidiomycete paralogs branching with the zygomycetes separately from the ascomycetes. In the second case (Figure 3B), topology support for the plant gene branching within the fungal clade is weak; therefore, in both cases, the HGT hypothesis relies primarily on the observed taxonomic distribution and only secondarily on the phylogenetic data. As these gene families are absent in all other eukaryotes sampled and therefore restricted to a relatively narrow taxonomic range of fungi, but are nevertheless present in a wide taxonomic diversity of prokaryotes, these data are consistent with an original HGT from prokaryotes-to-fungi and then two additional fungal-to-plant gene transfers.
Figure 3.
Evidence for Plant-Fungi HGT Based on a Prokaryote Tagged-Chain Transfer Hypothesis.
In the absence of strong phylogenetic tree topological support, it has been argued that a putative eukaryote-to-eukaryote HGT can be inferred when it is rooted by an uncontroversial case of prokaryote-to-eukaryote HGT. We detected two additional examples of HGT supported by this form of evidence.
(A) Phylogeny of the putative bifunctional iucA/iucC siderophore biosynthesis protein.
(B) Phylogeny of the unknown/conserved hypothetical protein. All phylogenies are reduced versions of the tree figure and the phylogenetic analysis implemented. See Supplemental Figures 5 and 6 online for full details. The figures are labeled using the same conventions described for Figure 1.
In these cases, the HGT hypothesis relies in part on taxon distribution of this gene family; therefore, it is important to confirm taxon distribution in a wide range of eukaryotes. In both cases, searches of TBestDB (O'Brien et al., 2007) and the GenBank EST database revealed no additional gene/taxon distributions that would be inconsistent with the HGT hypothesis. To test further that the taxon sampling was optimal, we ran PSI-BLAST analyses for five iterations, using the putative recipient gene as a BLAST seed. Apart from one case, we found only putative homologs from fungi or prokaryotes, consistent with the sampling shown in Figures 3A and 3B and Supplemental Figures 5 and 6 online, and highly divergent eukaryotic genes, which, based on sequence alignment, appear to be unrelated to the target gene families. However, we detected highly divergent forms of the gene family shown in Figure 3A, present in the social amoeba species Dictyostelium discoideum and Dictyostelium purpureum and therefore added these sequences to the phylogenetic analysis. Our analysis provided strong support for placing the Dictyostelium genes separately from the fungal and plant genes (see Supplemental Figure 5 online), suggesting they had a separate prokaryotic origin from the plant/fungal clade. The additional eukaryotic taxon sampling is therefore not likely to be relevant to the fungal-to-plant HGT event. Furthermore, the analysis showed some support for the hypothesis that these Dictyostelium genes represent a separate prokaryote-to-Dictyostelium HGT (Eichinger et al., 2005). However, our analysis did not provide strong support for the placement of the Dictyostelium genes and did not identify a strongly supported prokaryote sister group, preventing us from identifying a candidate donor taxonomic group (see Supplemental Figure 5 online).
Gene Family Taxon Distribution as Evidence for Plant-to-Fungi HGTs
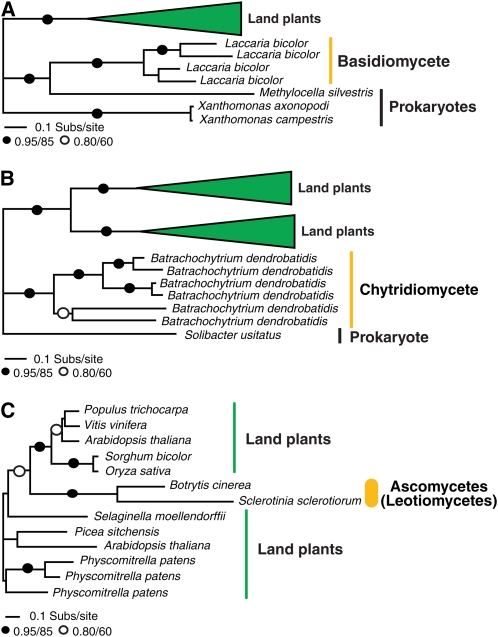
Two additional putative HGTs are shown in Figures 4A and 4B that demonstrate gene families that we observed to be restricted to a diverse collection of plant genomes, but also present in a single fungal species and a very low number of prokaryote genomes. This taxon distribution suggests plant-to-fungi HGT either directly or involving prokaryote intermediates. To test that the taxon sampling was optimal for these gene families, we ran a PSI-BLAST search using the putative fungal recipient (transferred) gene as a BLAST seed for five iterations. We found a range of sequences with very low sequence similarity in a range of prokaryote and metazoan taxa, which, based on sequence alignment, appear to be unrelated to the target gene families. We also used the putative recipient gene as a BLAST seed to search TBestDB (O'Brien et al., 2007) and the GenBank EST database for additional eukaryotic homologs not present in GenBank. In both cases, we found no further putative homologs, with one exception; we detected a putatively homologous gene to the phosphate-responsive 1 family protein (Figure 4B) in the Glaucocystis nostochinearum EST database. Only a partial sequence was available for this glaucocystophyte alga, so it was not included the phylogenetic analyses. However, the Glaucocystophyceae are an algal group closely related to the land plants (Rodriguez-Ezpeleta et al., 2005). The plant-to-fungal HGT hypothesis is based upon the proposition that this gene family is a Plantae-specific innovation that has then undergone transfer to the fungus Batrachochytrium dendrobatidis and the prokaryote Solibacter usitatus.
Figure 4.
Evidence for Plant-Fungi HGT Based on Gene Family Taxon Distribution.
Phylogenetic analyses of taxonomic distribution of HGT candidates among plants, a subsection of fungi, and (in two cases) a very restricted distribution of prokaryotes. These highly punctate taxon distributions suggest that these three genes have been horizontally transferred from plants to fungi. Phylogenies are shown to summarize the taxon distribution of this gene family.
(A) Phylogeny of the DUF239 domain protein.
(B) Phylogeny of the phosphate-responsive 1 family protein.
(C)Phylogeny of the unknown/conserved hypothetical protein with similarity to zinc finger (C2H2 type) protein. All phylogenies are reduced versions of the full tree topologies (see Supplemental Figures 7 to 9 online for full details). The figures are labeled using the same conventions described for Figure 1.
Figure 4C demonstrates an additional gene family only present in plants and two closely related leotiomycete (ascomycete) genomes Botrytis cinerea and Sclerotinia sclerotiorum (Fitzpatrick et al., 2006; James et al., 2006). Based on the taxon distribution of these gene families and the failure to detect additional homologs from different taxonomic groups using PSI-BLAST of GenBank and tBLASTn searches of TBestDB (O'Brien et al., 2007) and the GenBank EST database, the taxon distribution suggests a punctate pattern of taxonomic distribution consistent with HGT.
Together, these three data sets suggest plant-to-fungi HGT events either as direct transfer events, in the case of the data sets reported in Figure 4C, or possibly involving additional transfers to prokaryote genomes, either secondarily or as an intermediate event between the plant donor and fungal recipient (Figures 4A and 4B). The wide taxon distribution among the plants and presence of multiple plant paralogues in two of the gene families analyzed (see Supplemental Figures 7 and 8 online) suggests that this gene family may have arisen and diversified in the plant kingdom. Support for the HGT hypothesis in these three cases relies entirely, however, upon the taxonomic distribution of the gene family, and as such, it is important that these putative transfers should be treated with caution because they may require revision as more genomic data becomes available from a wider number of species.
Testing Alternative Hypotheses to HGT
For all nine candidate HGT events, unidentified ancestral gene duplication events and subsequent gene losses (hidden paralogy) represent an alternative hypothesis to HGT. For six of the candidate HGTs (Figures 1A to 3C), we investigated the possibility of hidden paralogy by identifying the backbone species tree (see Supplemental Figures 1 to 6 online) and identifying the number of gene duplication and gene losses required to generate the HGT-like tree topologies. In all cases, the hidden paralogy scenarios required extensive ancestral gene duplications followed by multiple, highly complex patterns of gene maintenance and differential gene loss across highly divergent evolutionary branches of the eukaryotes. This pattern of gene family evolution is unlikely because it requires each of the gene functions to be selected for maintenance over long periods of evolutionary divergence and then for the selection to cease independently on numerous different occasions, resulting in gene losses in multiple divergent eukaryotic lineages. The narrow taxonomic distribution of the gene family for three of the putative HGTs shown in Figures 4A to 4C indicates that the alternative hypothesis of hidden paralogy is even more unlikely because it requires the gene family to be anciently derived, maintained during the early diversification of the eukaryotes, and then for numerous independent gene loss events to have occurred in all other lineages sampled. We conclude that plant-fungi HGT is the most consistent hypothesis for the observed patterns of gene ancestry of the nine gene families presented here, given current genome sampling.
DNA Contamination as an Alternative Hypothesis to HGT
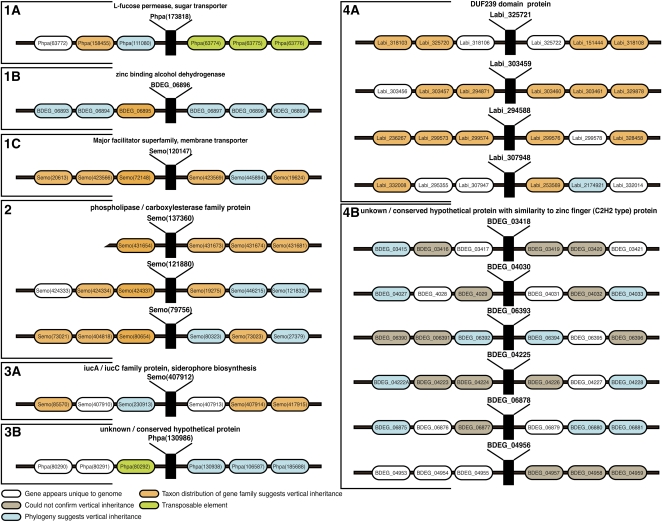
Fungi and plants share many intimate and longstanding ecological associations. It is therefore possible that the nine putative HGTs that we detected based on comparative genome analysis could be the result of contamination during DNA extraction in individual genome sequencing projects. To test for this possibility for each putative HGT, we identified and characterized three predicted protein-encoding genes that were located immediately 5′ and 3′ of each putative HGT candidate locus in each of the recipient genome sequences. HGTs that we found to be present in two or more recipient genomes (Figure 4C) were judged to be independent samples and therefore unlikely to be identical gene contamination events. We performed comparative genomics and phylogenetic analyses for all 106 of these tightly linked genes. This demonstrated that the HGT candidate loci were physically contiguous with neighboring genes, which showed a tree topology consistent with vertical inheritance generally congruent with the species phylogeny, as shown in Figure 5. We conclude that all nine putative transferred genes correspond to chromosomal loci located within a portion of native genome sequence and that they are not the result of DNA contamination.
Figure 5.
Results of Analyses of the Genome Contigs of Recipient Taxa Demonstrating the Evolutionary Ancestry of Genes Linked to the Plant-Fungi HGT.
This analysis demonstrates the location of the HGTs with respect to transposable elements and vertically inherited genes. In all nine cases, the analyses shows that the HGT is linked to a gene showing vertical phylogenetic inheritance or representing a gene family for which the taxon distribution suggests vertical inheritance. Because all nine HGTs are nested within a portion of native genome, DNA contamination can be ruled out as a possible source of artifact among the plant-fungus HGTs. Furthermore, in both cases that involve HGT from a fungus to the bryophyte moss P. patens, the HGT is positioned next to a putative transposable element. The alphanumeric label given in to the top right of each genome contig corresponds to Figures 1A to 4C. These data are reported in more detail in Supplemental Table 2 online.
Putative Functional Assignment of Plant-Fungi HGTs
The potential identities and biological functions of the plant-fungi HGT candidates were investigated based on the similarity of their putatively encoded products to known proteins.
Of the nine putative HGTs identified, four were identified as conserved hypothetical proteins or proteins with domains of unknown function (DUF); therefore, putative functional annotation was not possible. One HGT data set (Figure 1A) included protein sequences putatively encoding a l-fucose permease transporter family protein (FucP), which facilitates the uptake of l-fucose, also known as 6-deoxy-l-galactose (Gunn et al., 1994). A putative two-domain zinc binding alcohol dehydrogenase was encoded by an HGT candidate from the Plantae clade to the chytrid B. dendrobatidis (Figure 1B). Analysis of other forms of alcohol dehydrogenase-encoding genes demonstrated multiple prokaryote-to-eukaryote HGT events, suggesting that other alcohol dehydrogenase enzymes have replaceable functional roles and have undergone numerous replacement by HGT (Andersson et al., 2006). The third candidate HGT was a gene transfer from fungi to S. moellendorffii (Figure 1C). PFAM and CDD analyses suggest that this gene belongs to a large and highly diverse family of membrane transporter genes (Major Facilitator Superfamily [MFS_1]; Marchler-Bauer et al., 2005). The fourth HGT candidate identified (Figure 2) showed sequence similarity to the phospholipase/carboxylesterase protein family, members of which are reported to have the capability of hydrolyzing carboxylic ester bonds and have been suggested to have broad substrate specificity across the protein family (Kim et al., 1997; Marchler-Bauer et al., 2005). The fifth HGT (Figure 3A) family has two discrete protein domains, iucA and iucC, responsible for the two sequential steps in conversion of N epsilon-acetyl-N epsilon-hydroxylysine to the siderophore aerobactin (de Lorenzo and Neilands, 1986). Siderophores are low molecular mass iron chelators employed by both fungi and bacteria for iron uptake and storage (Haas et al., 2008). The bacterial enzymes that show some similarity to the Selaginella iucA and iucC bifunctional genes synthesize siderophores independently of nonribosomal peptide synthetases, suggesting that this transfer is possible without the establishment of a nonribosomal peptide synthetase pathway.
The only other candidate HGT gene for which we could infer a potential function was a putative phosphate-responsive 1 family protein-encoding gene, shown in Figure 4B. This gene family had numerous plant-specific paralogs (see Supplemental Figure 8 online) and was present in a wide diversity of plant taxa, the chytrid fungus B. dendrobatidis, and one bacterium, S. usitatus. A putative plant homolog in tobacco (Nicotiana tabacum) BY-2 cells shows increased transcription and accumulation of the protein in the cytoplasm prior to the start of plastid and nuclear DNA synthesis, when exposed to phosphate, suggesting that this gene plays a role during phosphate-induced plant cell division (Sano et al., 1999). The functional role of this HGT derived gene in either the Solibacter or Batrachochytrium is difficult to predict at present.
Putative functional annotation of the nine gene transfers therefore did not suggest a strong functional trend among the plant-fungi HGTs (Table 2), but the group did include two membrane transporters and proteins that potentially function in obtaining iron. The acquisition of such genes by HGTs may therefore have been selected by a competitive advantage in the recipient to take up substrates with greater efficiency.
DISCUSSION
Phylogenomic comparison of the six plant genome sequences revealed 14 candidate HGTs between the plant and fungal kingdoms. Comparative genomics and phylogenetic analysis, which included models that account for site rate heterogeneity and, where appropriate, comparative topology tests, demonstrated nine cases where a plant-fungal HGT was the most consistent explanation for either the taxonomic distribution or the phylogeny of the gene family. This data included evidence for five transfers from fungi to plants and four from plants to fungi. Our analysis protocols, although not comprehensive, directly tested 1689 plant gene families that showed the highest similarity to sequences from fungi. Our protocol also tested 3177 rice gene families for HGT. Of the original 14 candidate HGT data sets, all were detected using the BLAST-based HGT identification survey, while only one was also detected using the rice genome-specific analyses. These comparisons suggest that the BLAST-based survey is a reliable means of initially identifying potential plant-fungi HGTs, prior to phylogenetic analyses.
To test that we had identified reliable incidences of HGT, we adopted a rigorous phylogenetic approach based on the identification of gene genealogies that were highly distinct from the expected species phylogeny, for example, showing topological support for a plant gene branching within a fungal phylogenetic cluster (for example, Figure 1a or 1c) or where the data sets showed a highly punctate taxon distribution consistent with a HGT hypothesis (Figures 3A and 3B). In the cases where the HGT hypothesis was based solely on taxon distribution, we also performed a five-iteration PSI-BLAST search of the GenBank nonredundant data set and then used the same sequence as a BLAST seed to search the taxonomically broad EST database, TBestDB (O'Brien et al., 2007), and the GenBank EST database for additional eukaryotic homologs. To remain in the selected HGT candidate list, no further putative homologs could be identified from any additional extant eukaryotic group as this would be inconsistent with our taxon distribution-based HGT hypotheses (Figures 3 and 4).
Within our candidate HGT data sets, we also found putative HGTs that appeared to have been the result of a two-step serial gene transfer involving a prokaryote-to-eukaryote transfer followed by a plant-fungi HGT (Figures 1A, 3A, and 3B). Interestingly, we have observed similar chain transfer events when investigating gene transfer between fungi and oomycetes (Phytophthora spp) (Richards et al., 2006a). Furthermore, Keeling and Palmer (2008) recently outlined such analytical results as an important alternative means of identifying HGTs between two eukaryotes. Their argument suggests that a prokaryote-to-eukaryote HGT can represent a tag, which can be used to identify a secondary eukaryotic-to-eukaryotic HGT in the absence of ideal sampling or strong phylogenetic support (Keeling and Palmer, 2008). These three data sets (Figures 1A, 3A, and 3B) are consistent with the scenario proposed by Keeling and Palmer (2008) and therefore suggest they represent fungi-to-plant gene transfer events. However, we recognize that such a transfer hypothesis should be treated with caution and may require revision when more widely distributed genomic sequence data become available.
Plant-Fungi HGTs Are Both Rare and Ancient
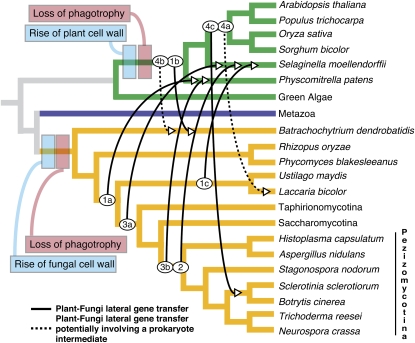
The recovery of only nine potential plant-fungal HGTs suggests that HGT between these two eukaryotic kingdoms has played only a very minor role in their evolution (0.53% in the BLAST defined analyses set). Interestingly, none of the HGT events involved donor or recipient lineages from the angiosperms (Figure 6). Instead, all of the plant-to-fungi HGTs involved transfer from lineages basal to the angiosperms or prior to the diversification of bryophytes, lycophytes, and angiosperms. Similarly, the five transfer events from fungi to plants were all acquired and/or retained by the lycophyte (three transfers) or the bryophyte (two transfers) lineage, while none of the detected transfers appear to have affected the angiosperm lineage. These data demonstrate the relative positioning of the HGT events and suggest either that barriers to fungal-to-plant HGT are more restrictive among the angiosperm taxa or that the two plant species that were analyzed that branch below the angiosperm radiation have ecological or cellular characteristics that make them more permissive to HGTs originating from fungi. It also seems clear, given that the transfers from fungi occurred prior to the radiation of the ascomycetes and in one case prior to the diversification of ascomycetes and basidiomycetes, and that plant transfers occurred from similarly basal lineages, that the transfers are all ancient HGT events. When considered together, the rare and ancient nature of plant-fungal HGT events has implications for our understanding of potential gene flow from transgenic plants, which is vital for evaluating the long-term propagation of genetically modified crops. The data reported here suggest that gene flow between plants and fungi appears to have been exceedingly rare over considerable evolutionary timescales and must be considered to be an extremely unlikely event from commercially cultivated crop species.
Figure 6.
A Model of the Plant-Fungi Transferome.
Schematic representation of how the nine candidate HGT events relate to the evolutionary history of plants and fungi. The point of origin and point of receipt of each HGT are marked by best approximation, based on available phylogenetic data. Candidates are labeled 1A to 4C and correspond to the trees shown in Figures 1A to 4C.
The rarity of HGT between plants and fungi is in stark contrast with studies of phagotrophic eukaryotes, which appear to show a regular pattern of gene transfer into eukaryotic genomes (Andersson et al., 2003; Archibald et al., 2003; Eichinger et al., 2005; Loftus et al., 2005). However, it is worth noting that the great majority of these transfers originate from prokaryotes. It remains to be seen whether prokaryote-derived HGT has played a major role during the diversification of the land plants and the fungi, although there are some initial reports to suggest this phenomenon is in play (Garcia-Vallve et al., 2000; Intrieri and Buiatti, 2001; Friesen et al., 2006; Richards et al., 2006a). However, if future comprehensive analysis proves that HGT from prokaryotes to plants or from prokaryotes to fungi is a relatively minor factor, as has been demonstrated here for gene transfer between plants and fungi, it would constitute further indirect evidence that phagotrophy is an important driving force for HGT.
Putative Biological Functions of Plant-Fungi HGTs
Among the nine putative HGTs, we identified several gene acquisitions that would theoretically, based on putative functional annotation, aid colonization of a soil environment. The l-fucose permease sugar transporter from the bryophyte P. patens represents a distinct family of sugar transporters when compared with other sugar transporter families (Gunn et al., 1994). The methylpentose l-fucose is a constituent of many glycoproteins and glycolipids synthesized by microorganisms (Gunn et al., 1994) and is also present in appreciable quantities in soil environments (Cheshire, 1979; Gunn et al., 1994). A second putative transporter protein gene was also revealed as a HGT candidate (Figure 1C). This protein class contains single-polypeptide secondary carriers capable of transporting a range of small solutes across plasma membranes, including simple sugars, oligosaccharides, inositols, drugs, amino acids, nucleosides, organophosphate esters, Krebs cycle metabolites, and a large variety of organic and inorganic anions and cations (Marchler-Bauer et al., 2005). The solute transported is determined by minor changes in the amino acid sequence characteristics (Marchler-Bauer et al., 2005), and as such it is difficult to pinpoint the likely biological role of this horizontally transferred transporter.
The siderophore biosynthetic protein containing iucA/iucC domains (Figure 3A) also represents a putatively transferred function that bestows a clear selective advantage as a means of sequestering iron from the environment. Analyses of prokaryote-derived HGTs in the genome of the soil-dwelling D. discoideum have demonstrated 18 potential instances of prokaryote-to-Dictyostelium HGT (Eichinger et al., 2005). Interestingly, among the HGTs identified were gene families, which would potentially assist colonization of soil environments. One gene family included in the Dictyostelium HGT list is a very distant relative of the prokaryote-to-fungi-to-plant iucA-iucC siderophore synthesis gene reported here. Our phylogenetic analysis also suggests a prokaryote-to-Dictyostelium HGT and indicates a serial gene transfer from a prokaryote to the fungi followed by a fungi-to-plant HGT. In total, this analysis demonstrates three separate HGT events of a bifunctional siderophore biosynthesis enzyme into eukaryotic organisms. All three eukaryotic groups inhabit soil environments and would presumably benefit from enhanced iron appropriation afforded by the gain of the bifunctional iucA and iucC siderophore synthesis gene. Furthermore, analysis of interdomain HGT has demonstrated additional cases of gene transfer involving iron and siderophore transport proteins between Eubacteria and Archaea (Almeida et al., 2008).
Recent reports have suggested that siderophore production and homeostasis are important for fungal pathogens of both animals and plants and important also in plant-fungal symbioses (Haas et al., 2008). At least two of the HGTs might therefore be considered to have potentially aided colonization of a soil environment, facilitating the uptake of l-fucose and iron for instance, important metabolic resources available in many soil environments (Cheshire, 1979; Gunn et al., 1994; Haas et al., 2008), providing the selection for an HGT event to become maintained within a plant or fungal ancestor species.
In summary, we have used large-scale genome sequence data analysis to study the incidence of plant-fungi HGT. Our analyses used a rigorous and highly conservative set of phylogenetic methods for inferring HGT, which could be confidently predicted only for nine gene families. We conclude that HGT is both rare and ancient between fungi and plants, but it has clearly occurred and may have provided advantageous gene functions that could have conceivably widened substrate use and habitat spread in both plant and fungal species.
METHODS
HGT Detection Pipeline
Our analysis pipeline consists of a series of PERL scripts and was used to identify sequences with specific BLAST similarity patterns to sequences from specific taxonomic groups (all scripts used in this study are available on request). The pipeline then automatically constructed phylogenetic trees for each sequence identified with the target taxon/similarity profile. The phylogenies were calculated from taxon sampling using a bespoke MySQL database (www.mysql.com) containing predicted protein sequences (1,566,625 sequences in total) from 159 species representing a wide diversity of eukaryotes and representative prokaryote taxa (see Supplemental Table 1 online). Each candidate sequence was compared against sequences in the database using BLASTp (Altschul et al., 1997) and the best similarity hits from each species extracted (e-value cutoff 10−20). These sequences were aligned using MUSCLE (Edgar, 2004), highly conserved regions from this alignment were sampled using GBLOCKS (Castresana, 2000), and phylogenetic trees constructed using PhyML (Guindon and Gascuel, 2003) with a WAG + Γ + I substitution model (Γ + I parameters estimated by PHYML).
We used this pipeline first to identify all plant genes that, when compared against all other genomes except other plant genomes, showed a higher BLASTp similarity score to fungi than to any other group (Table 1). All sequences that had a high sequence similarity to the transposon data set (by comparison to Repbase [Jurka et al., 2005], a database of eukaryotic repetitive elements, using tBLASTn e-value cutoff of 10−20) were excluded because we aimed to study the effect of HGT on the putative functional proteome of fungi and plants. These 5866 proteins were clustered using OrthoMCL (Li et al., 2003) to group together potential orthologs (e-value cutoff of 1e−20; inflation value 1.5) producing 943 clusters and 746 singletons and 1689 gene homolog groups in total. We repeated this clustering step on the entire predicted proteome of rice (Oryza sativa), producing 3177 cluster groups. We then used the pipeline to generate PhyML trees for each of the 4866 sequence groups and manually inspected the resulting phylogenies. This process generated numerous phylogenies that were unresolved, either because sequence sampling was too narrow or because highly divergent sequences prevented resolution of the tree topology among the key plant and fungal branches. In these cases, we adjusted the sampling threshold to exclude highly divergent branches or increased the threshold to sample additional members of the gene family, depending on the results of the first phylogenetic tree, and reran the phylogenetic analyses pipeline for these data sets. We manually inspected all 4866 phylogenetic trees and identified any tree topology potentially suggesting a plant-fungus HGT. These data sets were analyzed in greater detail using a range of phylogenetic methods.
Phylogenetic Analysis
To test the phylogenetic results for all candidate HGT data sets, we manually refined the alignment taxon sampling by performing BLASTp searches of the GenBank nonredundant database, tBLASTn searches of TBest database (O'Brien et al., 2007) and GenBank EST database, and five-iteration PSI-BLAST searches of the GenBank nonredundant database. We then added complete putatively homologous protein sequences to our alignments, not initially included in our pipeline database. We masked each alignment to remove gaps and ambiguous alignment positions using the alignment program Seaview (Galtier et al., 1996). The program Modelgenerator (Keane et al., 2004) was then used to identify the most appropriate substitution matrix and combination of gamma distribution and proportion of invariant sites (see Supplemental Figures 1 to 9 online for model details). Because many of our data sets included 100+ sequences, it was not possible to perform a full maximum likelihood (ML) phylogeny for all of the data sets. We therefore used two fastML methods with very different heuristic tree search methods: one that begins with distance-based phylogenetic tree analyses (PhyML; Guindon and Gascuel, 2003) and one that begins with parsimony-based phylogenetic tree analyses (RAxML; Stamatakis, 2006). We used both of these methods to control for the possibility that either form of the fastML search strategy might over- or underinflate the bootstrap topology support values of key topology nodes. We ran the PhyML analyses for 1000 bootstrap replicates using the model of sequence evolution identified using Modelgenerator analyses (see Supplemental Figures 1 to 9 online for model details). RAxML analysis was then used to assess bootstrap support via our easyRAx script (http://projects.exeter.ac.uk/ceem/easyRAx.html). RAxML and Modelgenerator predicted models were generally the same but, where they were not, Modelgenerator analyses were used (see Supplemental Figures 1 to 9 online). The best scoring RAxML tree was determined with the PROTMIX method, starting with 10 randomized maximum parsimony (MP) trees. Statistical support was evaluated with 100 bootstrap replicates.
MrBayes analysis (Ronquist and Huelsenbeck, 2003) was conducted for each data set with the following settings: 1,000,000 generation samples, using a substitution matrix and model of site rate variation as before, but allowing the MCMCMC to search alternative site rate variation model parameter values. The tree search included two MCMCMC searches with four chains each (three heated, heat parameter = default) with a sampling frequency of 100 generations. The likelihood values of the two MCMCMC searches were compared to check if they had converged. In all cases, a burn-in of 1000 generation samples or less was excluded and the remaining plateau sampled for the consensus Bayesian tree.
To test support for contentious branching points, we performed nonparametric branch support tests based on a Shimodaira-Hasegawa-like procedure implicated using PhyML (Anisimova and Gascuel, 2006). We used this tool to test the statistical significance of specific topological relationships over a collapsed version of the same branching relationship.
Finally, in the four cases where phylogenetic analyses supported the placement of a fungal or plant gene within a plant or fungal cluster (Figures 1 and 2), we used alternative topology statistical tests to investigate if these branching relationships were robust. We used PAUP (Swofford, 2002) to constrain the monophyly of the donor taxon group and calculated the best topology using parsimony and distance methods. We then calculated branch lengths and site likelihood values using ML in P4 (Foster, 2004) for the alternative topologies using the same models as before. We used the statistical topology comparison platform Consel (Shimodaira and Hasegawa, 2001) to investigate if we could reject the alternative tree topologies at the 5% confidence level using either the AU or SH test.
Unmasked alignment files (*.mase) incorporating details of mask selection as MASE files readable using Seaview (Galtier et al., 1996) and all masked alignments used for phylogenetic analysis as phylip files (*.phy) are presented as Supplemental Data Sets 1A to 9D online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries, accession numbers for each HGT are shown in Table 2, and all accession numbers for sequence data reported in this article can be found in Supplemental Figures 1 to 9 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Description of Phylogenetic Analysis, Including the Models Used, Sequence Database Accession Numbers, Alternative Topology Tests, and Sssessment of Hidden Paralogy for the Phylogeny Shown in Figure 1A.
Supplemental Figure 2. Description of Phylogenetic Analysis, Including the Models Used, Sequence Database Accession Numbers, Alternative Topology Tests, and Assessment of Hidden Paralogy for the Phylogeny Shown in Figure 1B.
Supplemental Figure 3. Description of Phylogenetic Analysis, Including the Models Used, Sequence Database Accession Numbers, Alternative Topology Tests, and Assessment of Hidden Paralogy for the Phylogeny Shown in Figure 1C.
Supplemental Figure 4. Description of Phylogenetic Analysis, Including the Models Used, Sequence Database Accession Numbers, Alternative Topology Tests, and Assessment of Hidden Paralogy for the Phylogeny Shown in Figure 2.
Supplemental Figure 5. Description of Phylogenetic Analysis, Including the Models Used, Sequence Database Accession Numbers, Alternative Topology Tests, and Assessment of Hidden Paralogy for the Phylogeny Shown in Figure 3A.
Supplemental Figure 6. Description of Phylogenetic Analysis, Including the Models Used, Sequence Database Accession Numbers, Alternative Topology Tests, and Assessment of Hidden Paralogy for the Phylogeny Shown in Figure 3B.
Supplemental Figure 7. Description of Phylogenetic Analysis, Including the Models Used and Database Sequence Accession Numbers for the Phylogenetic Tree Shown in Figure 4A.
Supplemental Figure 8. Description of Phylogenetic Analysis, Including the Models Used and Database Sequence Accession Numbers for the Phylogenetic Tree Shown in Figure 4B
Supplemental Figure 9. Description of Phylogenetic Analysis, Including the Models Used and Database Sequence Accession Numbers for the Phylogenetic Tree Shown in Figure 4C.
Supplemental Table 1. Genomes Used for HGT Identification Pipeline.
Supplemental Table 2. Results of Phylogenetic Analysis of Genes Linked to the Nine Plant-Fungi HGTs on the Genome Contigs of the HGT Recipient Taxa.
Supplemental Data Set 1A. Figure 1A Unmasked (mase) Sequence Alignment.
Supplemental Data Set 1B. Figure 1A Masked (phylip) Sequence Alignment.
Supplemental Data Set 1C. Figure 1A Masked (phylip) Sequence Alignment (Continued).
Supplemental Data Set 2A. Figure 1B Unmasked (mase) Sequence Alignment.
Supplemental Data Set 2B. Figure 1B Masked (phylip) Sequence Alignment.
Supplemental Data Set 3A. Figure 1C Unmasked (mase) Sequence Alignment.
Supplemental Data Set 3B. Figure 1C Masked (phylip) Sequence Alignment.
Supplemental Data Set 4A. Figure 2 Unmasked (mase) Sequence Alignment.
Supplemental Data Set 4B. Figure 2 Unmasked (mase) Sequence Alignment (Continued).
Supplemental Data Set 4C. Figure 2 Masked (phylip) Sequence Alignment.
Supplemental Data Set 4D. Figure 2 Masked (phylip) Sequence Alignment (Continued).
Supplemental Data Set 5A. Figure 3A Unmasked (mase) Sequence Alignment.
Supplemental Data Set 5B. Figure 3A Unmasked (mase) Sequence Alignment (Continued).
Supplemental Data Set 5C. Figure 3A Masked (phylip) Sequence Alignment.
Supplemental Data Set 5D. Figure 3A Masked (phylip) Sequence Alignment (Continued).
Supplemental Data Set 6A. Figure 3B Unmasked (mase) Sequence Alignment.
Supplemental Data Set 6B. Figure 3B Unmasked (phylip) Sequence Alignment.
Supplemental Data Set 6C. Figure 3B Masked (phylip) Sequence Alignment (Continued).
Supplemental Data Set 7A. Figure 4A Unmasked (mase) Sequence Alignment.
Supplemental Data Set 7B. Figure 4A Masked (phylip) Sequence Alignment.
Supplemental Data Set 8A. Figure 4B Unmasked (mase) Sequence Alignment.
Supplemental Data Set 8B. Figure 4B Masked (phylip) Sequence Alignment.
Supplemental Data Set 9A. Figure 4C Unmasked (mase) Sequence Alignment.
Supplemental Data Set 9B. Figure 4C Masked (phylip) Sequence Alignment.
Supplementary Material
Acknowledgments
T.A.R. is supported by a Leverhulme Early Career Fellowship. D.M.S. is supported by the Biotechnology and Biological Sciences Research Council, UK. G.L. is supported by a University of Exeter Studentship. This project was funded by the Department for Environment, Food, and Rural Affairs, UK (Research Contract CPEC21 to N.J.T., C.R.T., and T.A.R.). We thank the Department of Energy Joint Genome Institute, the Broad Institute, TBestDB, and Genoscope for making genome sequence data publicly available. The authors declare no competing financial interests.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Thomas A. Richards (t.a.richards@exeter.ac.uk).
Online version contains Web-only data.
References
- Almeida, F.C., Leszczyniecka, M., Fisher, P.B., and DeSalle, R. (2008). Examining ancient inter-domain horizontal gene transfer. Evol. Bioinform. Online 4 109–119. [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J.O. (2005). Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62 1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J.O., Hirt, R.P., Foster, P.G., and Roger, A.J. (2006). Evolution of four gene families with patchy phylogenetic distributions: Influx of genes into protist genomes. BMC Evol. Biol. 6 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J.O., Sjogren, A.M., Davis, L.A.M., Embley, T.M., and Roger, A.J. (2003). Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13 94–104. [DOI] [PubMed] [Google Scholar]
- Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55 539–552. [DOI] [PubMed] [Google Scholar]
- Aoki, S., and Syno, K. (1999). Horizontal gene transfer and mutation: Ngrol genes in the genome of Nicotiana glauca. Proc. Natl. Acad. Sci. USA 96 13229–13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald, J.M., Rogers, M.B., Toop, M., Ishida, K., and Keeling, P.J. (2003). Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. USA 100 7678–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko, R.G., Harlow, T.J., and Ragan, M.A. (2005). Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 102 14332–14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belbahri, L., Calmin, G., Mauch, F., and Andersson, J.O. (2008). Evolution of the cutinase gene family: Evidence for lateral gene transfer of a candidate Phytophthora virulence factor. Gene 408 1–8. [DOI] [PubMed] [Google Scholar]
- Bergthorsson, U., Adams, K.L., Thomason, B., and Palmer, J.D. (2003). Widespread horizontal gene transfer of mitochondrial genes in flowering plants. Nature 242 197–201. [DOI] [PubMed] [Google Scholar]
- Bergthorsson, U., Richardson, A.O., Young, G.J., Goertzen, L.R., and Palmer, J.D. (2004). Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. USA 101 17747–17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki, F., Shalchian-Tabrizi, K., and Pawlowski, J. (2008). Phylogenomics reveals a new 'megagroup' including most photosynthetic eukaryotes. Biol. Lett. 4 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552. [DOI] [PubMed] [Google Scholar]
- Cheshire, M.V. (1979). Nature and Origin of Carbohydrates in Soils. (London: Academic Press).
- Cho, Y., and Palmer, J.D. (1999). Multiple acquisitions via horizontal transfer of a group I intron in the mitochondrial cox1 gene during evolution of the Araceae family. Mol. Biol. Evol. 16 1155–1165. [DOI] [PubMed] [Google Scholar]
- Cusimano, N., Zhang, L.B., and Renner, S.S. (2008). Reevaluation of the cox1 group I intron in Araceae and angiosperms indicates a history dominated by loss rather than horizontal transfer. Mol. Biol. Evol. 25 265–276. [DOI] [PubMed] [Google Scholar]
- de Lorenzo, V., and Neilands, J.B. (1986). Characterization of iucA and iucC genes of the aerobactin system of plasmid Co1V–K30 in Escherichia coli. J. Bacteriol. 167 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao, X., Freeling, M., and Lisch, D. (2006). Horizontal transfer of a plant transposon. PLoS Biol. 4 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle, W.F. (1998). You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14 307–311. [DOI] [PubMed] [Google Scholar]
- Doolittle, W.F., Boucher, Y., Nesbo, C.L., Douady, C.J., Andersson, J.O., and Roger, A.J. (2003). How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 39–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, L., et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, D.A., Logue, M.E., Stajich, J.E., and Butler, G. (2006). A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, P.G. (2004). Modeling compositional heterogeneity. Syst. Biol. 53 485–495. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L., Stukenbrock, E.H., Liu, Z., Meinhardt, S., Ling, H., Faris, J.D., Rasmussen, J.B., Solomon, P.S., McDonald, B.A., and Oliver, R.P. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38 953–956. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Gouy, M., and Gautier, C. (1996). SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12 543–548. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallve, S., Romeu, A., and Palau, J. (2000). Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol. Biol. Evol. 17 352–361. [DOI] [PubMed] [Google Scholar]
- Goremykin, V.V., Salamini, F., Velasco, R., and Viola, R. (2008). MtDNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 26 99–110. [DOI] [PubMed] [Google Scholar]
- Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Gunn, F.J., Tate, C.G., and Henderson, P.J. (1994). Identification of a novel sugar-H+ symport protein, FucP, for transport of L-fucose into Escherichia coli. Mol. Microbiol. 12 799–809. [DOI] [PubMed] [Google Scholar]
- Haas, H., Eisendle, M., and Turgeon, B.G. (2008). Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46 149–187. [DOI] [PubMed] [Google Scholar]
- Intrieri, M.C., and Buiatti, M. (2001). The horizontal transfer of Agrobacterium rhizogenes genes and the evolution of the genus Nicotiana. Mol. Phylogenet. Evol. 20 100–110. [DOI] [PubMed] [Google Scholar]
- Jain, R., Rivera, M.C., and Lake, J.A. (1999). Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. USA 96 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R., Rivera, M.C., Moore, J.E., and Lake, J.A. (2003). Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20 1598–1602. [DOI] [PubMed] [Google Scholar]
- James, T.Y., et al. (2006). Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443 818–822. [DOI] [PubMed] [Google Scholar]
- Jurka, J., Kapitonov, V.V., Pavlicek, A., Klonowski, P., Kohany, O., and Walichiewicz, J. (2005). Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110 462–467. [DOI] [PubMed] [Google Scholar]
- Keane, T.M., Creevey, C.J., Naughton, T.J., Pentony, M.M., Naughton, T.J., and Mcinerney, J.O. (2004). Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, P.J., and Inagaki, Y. (2004). A class of eukaryotic GTPase with a punctate distribution suggesting multiple functional replacements of translation elongation factor 1alpha. Proc. Natl. Acad. Sci. USA 101 15380–15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, P.J., and Palmer, J.D. (2008). Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9 605–618. [DOI] [PubMed] [Google Scholar]
- Kim, K.K., Song, H.K., Shin, D.H., Hwang, K.Y., Choe, S., Yoo, O.J., and Suh, S.W. (1997). Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity. Structure 5 1571–1584. [DOI] [PubMed] [Google Scholar]
- Kim, M., Cui, M.L., Cubas, P., Gillies, A., Lee, K., Chapman, M.A., Abbott, R.J., and Coen, E. (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science 322 1116–1119. [DOI] [PubMed] [Google Scholar]
- Koski, L.B., and Golding, G.B. (2001). The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 52 540–542. [DOI] [PubMed] [Google Scholar]
- Li, L., Stoeckert, C.J., Jr., and Roos, D.S. (2003). OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, B., et al. (2005). The genome of the protist parasite Entamoeba histolytica. Nature 433 865–868. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2005). CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 33 D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., and Penny, D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa, A., Reyes-Prieto, A., and Bhattacharya, D. (2008). Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS One 3 e2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, E.A., Koski, L.B., Zhang, Y., Yang, L., Wang, E., Gray, M.W., Burger, G., and Lang, B.F. (2007). TBestDB: A taxonomically broad database of expressed sequence tags (ESTs). Nucleic Acids Res. 35 D445–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirozynski, K.A., and Malloch, D.W. (1975). The origin of land plants: A matter of mycotrophism. Biosystems 5 153–164. [DOI] [PubMed] [Google Scholar]
- Ragan, M.A. (2001. a). On surrogate methods for detecting lateral gene transfer. FEMS Microbiol. Lett. 201 187–191. [DOI] [PubMed] [Google Scholar]
- Ragan, M.A. (2001. b). Detection of lateral gene transfer among microbial genomes. Curr. Opin. Genet. Dev. 11 620–626. [DOI] [PubMed] [Google Scholar]
- Richards, T.A., Dacks, J.B., Campbell, S.A., Blanchard, J.L., Foster, P.G., McLeod, R., and Roberts, C.W. (2006. b). Evolutionary origins of the eukaryotic shikimate pathway: Gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5 1517–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, T.A., Dacks, J.B., Jenkinson, J.M., Thornton, C.R., and Talbot, N.J. (2006. a). Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr. Biol. 16 1857–1864. [DOI] [PubMed] [Google Scholar]
- Richards, T.A., Hirt, R.P., Williams, B.A., and Embley, T.M. (2003). Horizontal gene transfer and the evolution of parasitic protozoa. Protist 154 17–32. [DOI] [PubMed] [Google Scholar]
- Richardson, A.O., and Palmer, J.D. (2007). Horizontal gene transfer in plants. J. Exp. Bot. 58 1–9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta, N., Brinkmann, H., Burey, S.C., Roure, B., Burger, G., Loffelhardt, W., Bohnert, H.J., Philippe, H., and Lang, B.F. (2005). Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15 1325–1330. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sano, T., Kuraya, Y., Amino, S., and Nagata, T. (1999). Phosphate as a limiting factor for the cell division of tobacco BY-2 cells. Plant Cell Physiol. 40 1–8. [DOI] [PubMed] [Google Scholar]
- Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51 492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira, H., and Hasegawa, M. (2001). CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics 17 1246–1247. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 2688–2690. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates).
- Wang, B., and Qiu, Y.L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16 299–363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.