Abstract
Nearly 100 genes and functional polymorphisms underlying natural variation in plant development and physiology have been identified. In crop plants, these include genes involved in domestication traits, such as those related to plant architecture, fruit and seed structure and morphology, as well as yield and quality traits improved by subsequent crop breeding. In wild plants, comparable traits have been dissected mainly in Arabidopsis thaliana. In this review, we discuss the major contributions of the analysis of natural variation to our understanding of plant development and physiology, focusing in particular on the timing of germination and flowering, plant growth and morphology, primary metabolism, and mineral accumulation. Overall, functional polymorphisms appear in all types of genes and gene regions, and they may have multiple mutational causes. However, understanding this diversity in relation to adaptation and environmental variation is a challenge for which tools are now available.
INTRODUCTION
Intraspecific natural variation (hereinafter referred to as natural variation) may be broadly defined as the within-species phenotypic variation caused by spontaneously arising mutations that have been maintained in nature by any evolutionary process including, among others, artificial and natural selection. Thus, natural variation embraces the enormous diversity present within wild plant species as well as most of the genetic variants that are found in domesticated plants. Some of the phenotypic differences existing in wild or cultivated plants are due to single-gene (monogenic) allelic variants. However, most of the natural variation is quantitative and determined by molecular polymorphisms at multiple loci and genes (multigenic), which are referred to as quantitative trait loci (QTL) and quantitative trait genes (QTGs).
The natural variation present in crop plants has been exploited since their domestication thousands of years ago by the genetic manipulation of developmental traits and physiological features related to adaptation to agriculture. Methods for genetic analysis and mapping of natural quantitative variation were developed a few decades ago for crop species, in which many more studies have been performed than in wild plants. Currently, genomic resources have been developed for several herbaceous crop plants, such as rice (Oryza sativa), barley (Hordeum vulgare), and tomato (Solanum lycopersicum), enabling the identification of the genes and nucleotide polymorphisms underlying QTL involved in domestication, yield, biotic and abiotic stress, and quality traits (Doebley et al., 2006; Sang, 2009). In addition, the recent availability of the first genome sequences of cultivated perennial woody plants, such as poplar (Populus spp) and grapevine (Vitis vinifera; Tuskan et al., 2006; Jaillon et al., 2007), is increasing the scope of traits and processes whose natural variation can be dissected molecularly.
In addition, the analysis of natural variation in wild species has begun to elucidate the molecular bases of phenotypic differences related to plant adaptation to distinct natural environments and to determine the ecological and evolutionary processes that maintain this variation (Mitchell-Olds et al., 2007). The molecular approach to these questions has been applied mainly to the wild annual crucifer Arabidopsis thaliana, which has become a model plant for the study of natural variation (Mitchell-Olds and Schmitt, 2006). Its wide geographical and environmental distribution, combined with its small genome and the availability of unprecedented genetic and genome resources, have strongly facilitated its molecular analysis in the last decade. As a result, A. thaliana has provided the largest number of genes and nucleotide polymorphisms underlying natural variation of any plant species (Alonso-Blanco et al., 2005). However, the specific ecological niche and life history of A. thaliana limits the plant traits and processes that can be approached in a single species. Therefore, new plant models phylogenetically related to A. thaliana (e.g., Arabidopsis lyrata; Clauss and Koch, 2006) as well as unrelated species (e.g., of the genera Aquilegia [Kramer, 2009], Mimulus [Wu et al., 2007], Ipomoea [Clegg and Durbin, 2003], and Helianthus [Rieseberg et al., 2003]) are beginning to be used in studies of natural variation and speciation.
Our understanding of plant biology has benefited tremendously from work done with artificially induced mutants in model species, as illustrated by A. thaliana mutants currently available for most genes, and mutant screenings continue to be useful in the identification of gene function (Page and Grossniklaus, 2002). Gene functions involved in plant survival and adaptation can be identified partially by induced mutant analyses of different wild genotypes, where mutants with reduced fitness are easily selected. However, current mutant collections have been obtained using a limited number of laboratory strains, which harbor only a small portion of A. thaliana natural variation. Interestingly, Clark et al. (2007) showed that ;9.4% of A. thaliana protein-coding genes are naturally absent or knocked out in wild accessions, limiting the mutant spectra that can be obtained from each accession. Therefore, natural variation provides a relevant complementary resource to discover novel gene functions as well as those allelic variants that specifically interact with the genetic background and/or the environment or alleles showing small effects on phenotype, particularly for traits related to plant adaptation (Benfey and Mitchell-Olds, 2008).
Genetic analyses of natural variation in plants currently are mainly performed by QTL mapping, often called linkage mapping, in which phenotypic variation is associated with allelic variation at molecular markers segregating in experimental mapping populations derived from directed crosses (Doerge, 2002). Thus, genomic regions accounting for trait variation are located in large physical intervals containing the causal QTLs. Further analyses of these regions with a combination of functional strategies allows the final identification of QTGs and nucleotide polymorphisms altering the function of those genes (reviewed in Koornneef et al., 2004; Alonso-Blanco et al., 2005; Weigel and Nordborg, 2005; González-Martínez et al., 2006). Association mapping, which involves looking for phenotype-genotype associations in a general population of individuals whose degree of relatedness or pedigree is unknown, is also becoming more popular and useful in plant systems, owing to improvements in statistical and analytical tools and in gene sequencing technology (see Myles et al., 2009).
The information gleaned from the various types of QTL analysis is extending the current concept of gene function based on a single wild-type allele and provides a more realistic and dynamic view of gene function by estimating the effects of different natural alleles depending on genetic background, environment, and evolutionary temporal scale. In addition, recent genetic analyses of large data sets generated by high-throughput profiling of mapping populations with “-omics” procedures (genomics, transcriptomics, metabolomics, proteomics, ionomics, or epigenomics) is allowing the genetic integration of several levels of molecular regulation of phenotypic trait variation. Thus, QTL and gene networks controlling natural variation are being inferred (Jansen et al., 2009).
In this review, we discuss the major contributions of the analysis of natural variation to our understanding of plant development and physiology. Given the large number of studies performed in the past, we do not aim to completely cover all traits analyzed so far, but to focus on several relevant phenotypes recently or extensively dissected in A. thaliana and major crop species. These traits include the timing of germination and flowering, plant growth and morphology, primary metabolism, and mineral accumulation. Detailed discussion of natural variation for secondary metabolites is covered by Kliebenstein (2009a), and various reviews have specifically described the natural variation involved in crop domestication and diversification (Doebley et al., 2006; Pourkheirandish and Komatsuda, 2007; Purugganan and Fuller, 2009; Sang, 2009), plant development (Alonso-Blanco et al., 2005), or resistance to pathogens (Holub, 2007). Here, we summarize the major genes and allelic variants currently known to participate in plant adaptation to different natural and agricultural environments through some modifications of plant development and physiology. The kind of mutations, molecular mechanisms, and gene networks involved in microevolution and adaptation of different plant species to changing environments are beginning to be elucidated. Consequently, new perspectives in our understanding of phenotype and pan-genome variation and evolution are emerging (Morgante et al., 2007).
DEVELOPMENTAL PROCESSES
Domestication of crop plants mainly has resulted in changes in traits related to floral and seed morphology, such as shattering, free-threshing, and seed coat color in cereals (Salamini et al., 2002, Dubcovsky and Dvorak, 2007), plant architecture-like branching patterns in maize (Zea mays) and rice (Izawa et al., 2009), fruit morphology-like size and shape in Solaneaceae crops (Tanksley, 2004), and seed dormancy, especially in domesticated cereals (Salamini et al., 2002). In addition, the analysis of natural variation has identified novel molecular mechanisms involved in the regulation of life history traits, such as seed dormancy and flowering time, by different environmental and endogenous clues. This section highlights some of the most important advances resulting from these studies.
Seed Dormancy and Germination
Most wild annual plant species, including A. thaliana, show substantial intraspecific natural variation for seed dormancy and germination properties, which is presumably involved in adaptation to different environments. Domestication has often led to a reduction of seed dormancy in crop plants, although considerable variation remains among cultivated varieties. The genetic bases of this variation have been approached mainly in A. thaliana and cereal plants, mostly in relation to environmental factors, such as temperature and light, and to endogenous hormonal signals, such as abscisic acid (ABA) and gibberellin (GA).
In A. thaliana, QTL mapping analyses of germination-related traits have been performed in relation to after-ripening (Alonso-Blanco et al., 2003; Clerkx et al., 2004), light regimes (van der Schaar et al., 1997; Laserna et al., 2008; Meng et al., 2008), low temperature (Meng et al., 2008), or GA inhibitors (van der Schaar et al., 1997). Many of the loci are only detected in specific assays or conditions, indicating strong QTL by environment interactions. However, several QTLs colocate in different crosses, suggesting the presence of moderate frequency natural alleles with pleiotropic effects on dormancy and related traits, such as cold-tolerant germination and seed longevity (Clerkx et al., 2004; Meng et al., 2008). Particularly, the locus DOG1 has been detected in three experimental populations (Alonso-Blanco et al., 2003; Clerkx et al., 2004; Laserna et al., 2008) and has been isolated, identifying a member of a small gene family of unknown molecular function (Bentsink et al., 2006; Table 1). DOG1 induces seed dormancy and is specifically expressed during seed development. In addition, it has been shown that DOG1 functional nucleotide polymorphism(s) affect its expression, dormant accessions having higher RNA levels than nondormant accessions (Bentsink et al., 2006). Furthermore, DOG1 is involved in the ABA-mediated sugar signaling pathway in seedlings since glucose induces the expression of the DOG1/GSQ5 Cvi allele, whereas Landsberg erecta and Columbia alleles do not respond (Teng et al., 2008). Thus, DOG1 has provided the first gene involved in natural variation for seed dormancy and illustrates the value of this variation to identify previously unknown genes for this trait.
Table 1.
Genes and Funtional Polymorphisms Involved in Natural Variation for Different Traits and Species
| Functional Nucleotide Polymorphism
|
Allelic Dysfunction | ||||||
|---|---|---|---|---|---|---|---|
| Plant Species | Locus/Gene | Phenotypic Effects | Molecular Function | Gene Position | Mutation | References | |
| A. thaliana | DOG1 | Seed dormancy, sugar sensing | Unknown | Promoter | Unknown | ME | Bentsink et al. (2006) |
| Thale cress | FRI | Flowering | Unknown | Promoter, coding | Ins, Del, nonsense Subs | TP, ME | Johanson et al. (2000); Shindo et al. (2005) |
| FRL1 | Flowering | FRI-like | Coding | Nonsense Subs | TP | Schläppi (2006) | |
| FRL2 | Flowering | FRI-like | Coding | Missense Subs | AP | Schläppi (2006) | |
| FLC | Flowering | MADS TF | Intron, coding | TE insertion, nonsense Subs | ME, TP | Michaels et al. (2003); Werner et al. (2005a) | |
| FLW1/FLM | Flowering | MADS TF | Promoter + coding | Gene Del | Null expression | Werner et al. (2005b) | |
| ART1/HUA2 | Flowering, shoot morphology | RNA processing factor | Coding | Nonsense and missense Subs | TP, AP | Doyle et al. (2005); Wang et al. (2007) | |
| EDI/CRY2 | Flowering, seedling growth, fruit size | Photoreceptor | Coding | Missense Subs | AP | El-Assal et al. (2001, 2004) | |
| PHYD | Flowering, seedling growth | Photoreceptor | Coding | Del | TP | Aukerman et al. (1997) | |
| PHYC | Flowering, seedling growth | Photoreceptor | Coding | Nonsense Subs | TP | Balasubramanian et al. (2006) | |
| PHYA | Seedling growth | Photoreceptor | Coding | Missense Subs | AP | Maloof et al. (2001) | |
| PHYB | Seedling growth | Photoreceptor | Coding | Missense Subs | AP | Filiault et al. (2008) | |
| TZP/Light5 | Seedling growth | Zn knuckle protein | Coding | Ins | TP | Loudet et al. (2008) | |
| CAL | Flower morphology | MADS TF | Coding | Missense Subs | AP | Kempin et al. (1995) | |
| IIL1 | Leaf morphology | Leu biosynthesis enzyme | Intron | Ins, Del | ME | Sureshkumar et al. (2009) | |
| GL1 | Trichome formation | Myb TF | Coding | Del | ME | Hauser et al. (2001) | |
| PUB8 | Self-incompatibility | U box protein | Promoter | Unknown | ME | Liu et al. (2007) | |
| MUM2 | Seed mucilage | Galactosidase | Coding | Del | ME | Macquet et al. (2007) | |
| BRX | Root growth | Novel TF | Coding | Nonsense Subs | TP | Mouchel et al. (2004) | |
| LPR1 | Root growth response to P | Multicopper oxidase | Promoter | Del | ME | Svistoonoff et al. (2007) | |
| INV | Sugar composition, root growth | Invertase | Unknown | Unknown | Unknown | Sergeeva et al. (2006) | |
| Small effect QTL | Biomass accumulation | Protein kinase | Unknown | Unknown | Unknown | Kroyman and Mitchell-Olds (2005) | |
| DM1 | Hybrid incompatibility, disease resistance | NB-LRR proteins | Promoter + coding | Nonsense Subs, Del | AP | Bomblies et al. (2007) | |
| QTL3 | Hybrid incompatibility, disease resistance | NB-LRR proteins | Unknown | Unknown | Unknown | Alcázar et al. (2009) | |
| LD1. 1 | Genetic incompatiblity, embryogenesis | His biosynthesis enzyme | Promoter, coding | Nonsense Subs | ME, TP | Bikard et al. (2009) | |
| LD1.5 | Genetic incompatiblity, embryogenesis | His biosynthesis enzyme | Promoter + coding | Gene Del | Null expression | Bikard et al. (2009) | |
| APR2 | Sulfate accumulation | Sulfate reductase | Coding | Nonsense Subs | AP | Loudet et al. (2007) | |
| MOT1 | Mo accumulation | Sulfate transporter | Promoter | Del | ME | Baxter et al. (2008) | |
| HMA5 | Cu toxicity | Cu ATPase transporter | Coding | Missense Subs | AP | Kobayashi et al. (2008) | |
| HKT1 | Na accumulation | Na+ transporter | Promoter | Del | ME | Rus et al. (2006) | |
| A. lyrata | FRI | Flowering | FRI-like | Coding | Indel | AP | Kuittinen et al. (2008) |
| B. rapa | BrFLC1 | Flowering | MADS TF | Unknown | Unknown | Unknown | Kole et al. (2001) |
| Turnip | BrFLC2 | Flowering | MADS TF | Unknown | Unknown | Unknown | Schranz et al. (2002) |
| S. lycopersicum | SUN | Fruit shape | IQ67 domain protein | Coding + promotor | Gene duplication | ME | Xiao et al. (2008) |
| Tomato | OVATE | Fruit shape | Nuclear protein | Coding | Nonsense Subs | TP | Cong et al. (2002) |
| FAS | Fruit shape | Yabby TF | Intron | Ins | ME | Cong et al. (2008) | |
| Fw2.2 | Fruit size | Cell cycle control | Promoter | Subs, indel | ME | Frary et al. (2000) | |
| Sw4. 1 | Seed size | Transporter | Unknown | Unknown | Unknown | Orsi and Tanksley (2009) | |
| Brix9-2-5/LIN5 | Fruit sugar content | Invertase | Coding | Missense Subs | AP | Fridman et al. (2004) | |
| Cnr | Fruit ripening | SBP TF | Promoter | DNA methylation | ME | Manning et al. (2006) | |
| Pisum sativum | LF/PsTFL1c | Flowering | TFL1-like protein | Promoter, coding | Gene Del | Null expression | Foucher et al. (2003) |
| Pea | R | Seed composition and morphology | Starch branching enzyme | Coding | TE insertion | TP, ME | Bhattacharyya et al. (1990) |
| Lynaria vulgaris | Lcyc | Flower simetry | TCP TF | Promoter + coding | DNA methylation | ME | Cubas et al. (1999) |
| Toadflax | |||||||
| T. aestivum | VRN1 | Flowering | AP1-like MADS TF | Promoter, intron | Del, Ins | ME | Yan et al. (2003) |
| Bread wheat | VRN3 | Flowering | FT-like protein | Promoter | TE insertion | ME | Yan et al. (2006) |
| Q | Seed free-thressing | AP2-like TF | Coding | Missense Subs | AP | Simons et al. (2006) | |
| RhtB1, RhtD1 | Plant and leaf size | GAI-like protein | Coding | Nonsense Subs | TP | Peng et al. (1999) | |
| Gpc-B1/NAM-B1 | Senescence; Zn, Fe, and protein content | NAC TF | Coding | Del, gen Del | TP | Uauy et al. (2006) | |
| Triticum monococum | VRN1 | Flowering | AP1-like MADS TF | Promoter | Del | ME | Yan et al. (2003) |
| Einkorn wheat | VRN2 | Flowering | CCT domain protein | Promoter, coding | Missense Subs | AP | Yan et al. (2004a) |
| H. vulgare | VRN2 | Flowering | CCT domain protein | Promoter + coding | Gene Del | Null expression | Yan et al. (2004a) |
| Barley | VRN3 | Flowering | FT-like protein | Intron | Subs | ME | Yan et al. (2006) |
| Ppd1 | Flowering | Response regulator | Coding | Missense Subs | AP | Turner et al. (2005) | |
| Nud | Seed free-threshing | ERF TF | Promoter + coding | Gene Del | Null expression | Taketa et al. (2008) | |
| Vrs1 | Seed number, ear morphology | HD TF | Coding | Ins | TP | Komatsuda et al. (2007) | |
| Bot1 | Boron toxicity tolerance | Boron efflux transporter | Promoter + coding | Gene duplication | ME | Sutton et al. (2007) | |
| O. sativa | qLTG3 | Germination | Unknown | Coding | Del | ME | Fujino et al. (2008) |
| Rice | Rc | Germination, seed color | bHLH TF | Coding | Del | TP | Sweeney et al. (2006) |
| Hd1 | Flowering | CO-like TF | Coding | Del, TE insertion | TP, ME | Yano et al. (2000); Doi et al. (2004) | |
| Hd3a | Flowering | FT-like protein | Unknown | Unknown | ME | Kojima et al. (2002) | |
| Hd6 | Flowering | Protein kinase CK2 | Coding | Nonsense Subs | TP | Takahashi et al. (2001) | |
| Ehd1 | Flowering | Response regulator | Coding | Missense Subs | AP | Doi et al. (2004) | |
| Ghd7 | Flowering, plant height, seed yield | CCT domain protein | Coding | Gene Del, nonsense Subs | Null expression | Xue et al. (2008) | |
| sh4 | Shattering | Myb TF | Coding | Missense Subs | AP | Li et al. (2006) | |
| qSH1 | Shattering | HD TF | Promoter | Missense Subs | ME | Konishi et al. (2006) | |
| qSW5 | Seed size | Unknown | Coding, promoter | Del | ME | Shomura et al. (2008) | |
| GW2 | Seed size | RING protein | Coding | Del | TP | Song et al. (2007) | |
| GS3 | Seed size | Unknown | Coding | Missense Subs | TP | Fan et al. (2006) | |
| Gn1a | Seed number | CK oxidase/dehydrogenase | Promoter | Del | ME | Ashikari et al. (2005) | |
| Wx | Seed starch composition | Starch biosynthesis enzyme | Intron | Splice site Subs | TP | Isshiki et al. (1998) | |
| SKC1 | Salt tolerance | HKT transporter | Coding | Missense Subs | AP | Ren et al. (2005) | |
| Z. mays | Vgt1 | Flowering | AP2-like TF | Promoter enhancer | TE insertion, indel | ME | Salvi et al. (2007) |
| Maize | tb 1 | Inflorescence architecture | TCP TF | Promoter enhancer | Unknown | ME | Doebley et al. (1997) |
| tga1 | Seed morphology | SBP TF | Coding | Missense Subs | AP | Wang et al. (2005) | |
| Wx | Starch composition | Starch biosynthesis enzyme | Promoter, intron, coding | TE insertion | ME | Varagona et al. (1992) | |
TF: Transcription factor; TE: Transposable element; Subs: substitution; Ins: insertion; Del: deletion; TP: truncated protein; AP: altered protein; ME: Misexpression
In cereals, several analyses have identified seed dormancy QTLs in crosses between cultivated varieties with low dormancy and between cultivated and dormant weedy accessions or wild relative species (Lin et al., 1998; Gu et al., 2005; Hori et al., 2007; Imtiaz et al., 2008). In particular, the large-effect locus Sdr1 of rice might be orthologous to barley SD4 and wheat (Triticum aestivum) 4AL loci (Lin et al., 1998). Detailed QTL characterizations have shown that some loci may control dormancy through seed maternal tissues, while others function in offspring tissue(s) (Gu et al., 2008). As in wheat, maternally inherited seed dormancy has been associated with the seed pigmentation color variation determined by the rice Rc locus (Gu et al., 2005), which encodes a bHLH transcription factor (Sweeney et al., 2006). In addition, rice QTLs have been found for germination at low temperature, and the first of such loci, qLTG3-1, has been recently isolated (Fujino et al., 2008). qLTG3-1 encodes a protein of unknown function that is expressed in the embryo during seed germination. It has been suggested that qLTG3-1 is involved in tissue weakening during seed germination, which is reduced in the nondormant rice accession bearing a loss-of-function deletion allele.
Genetic analyses of dormancy and germination also have been performed in other crop species, such as lettuce (Lactuca sativa; Argyris et al., 2005). Interestingly, an interspecific cross between L. sativa and L. serriola identified Htg6.1 as a major effect locus involved in germination at high temperature (thermoinhibition), which colocates with germination QTLs for ABA sensitivity and GA response as well as with the ABA biosynthesis gene Ls-CED4. In addition, Ls-CED4 is induced by high temperature only in the thermoinhibited cultivated species, which strongly supports Ls-CED4 as candidate for Htg6.1 (Argyris et al., 2008).
Flowering Time
Molecular studies of the natural variation in the annual species A. thaliana, pea (Pisum sativum), rice, barley, wheat, and maize have identified ;25 different causal genes and their nucleotide polymorphisms of major effect (Table 1). In addition, genetic analyses of flowering time or related traits have been performed in other annual plants, such as tomato (González-Martinez et al., 2006), biennial species like Brassica sp (Schranz et al. 2002), short-lived perennials like Fragaria vesca (Albani et al., 2004), and several long-lived woody trees (Frewen et al., 2000; Casasoli et al., 2006). These studies are identifying multiple candidate genes and polymorphisms, which allow comparative analyses among species with rather different environmental and ecological distributions as well as physiological and developmental behaviors.
A. thaliana is a facultative long-day species widely distributed in temperate regions of the Northern hemisphere. Two extreme flowering behaviors have been described among wild genotypes grown in laboratory conditions, which are considered winter (late) and spring (early) life habits because they show strong and weak vernalization responses, respectively. Nine genes have been isolated that contribute to A. thaliana natural variation for flowering time (Table 1). Among these, FRI and FLC are involved in the regulation of flowering by vernalization and were first identified because of this natural variation. FLC encodes a transcripition factor of the MADS family that represses flowering (Michaels and Amasino, 1999). Its expression is downregulated by the low temperature of winter through epigenetic mechanisms leading to histone methylation and chromatin modifications (reviewed in Sung and Amasino, 2005). FRI encodes a protein of unknown function that also represses flowering initiation by activating the expression of FLC (Figure 1; Johanson et al., 2000). Most winter annual genotypes carry active alleles of both genes, which interact epistatically to delay flowering, while many summer annual accessions bear partial or total loss-of-function alleles in one or both genes (Werner et al., 2005a). More than 25 FRI independent loss-of-function alleles have been described, which mainly encode truncated proteins, indicating that FRI loss-of-function has evolved multiple times from a late flowering ancestor (Johanson et al., 2000; Le Corre et al., 2002; Shindo et al., 2005). By contrast, several natural loss-of-function alleles of FLC with reduced gene expression are caused by independent insertions of transposon elements within the first regulatory intron (Gazzani et al., 2003; Michaels et al., 2003). Other rare alleles are produced by nonsense or splicing mutations predicted to generate truncated proteins (Werner et al., 2005a). Interestingly, it has been suggested that additional allelic variation in FLC cis-regulatory sequences that alters the efficiency of FLC chromatin silencing might contribute to the quantitative variation for vernalization response present among late flowering genotypes (Shindo et al., 2006).
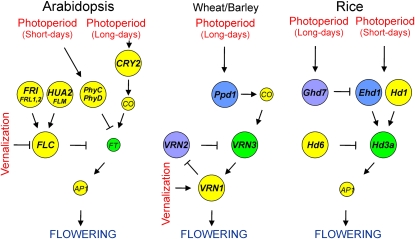
Figure 1.
Gene Networks Involved in Natural Variation for Flowering Responses to Vernalization and Photoperiod signals in A. thaliana, Wheat, and Rice.
Diagrams do not represent full molecular models of flowering regulation, but they show gene network components and branches known to contribute to the natural intraspecific variation existing in these species. Yellow color represents genes that contribute to the variation in a single species, which do not present known homologous genes contributing to natural variation in other species. Green color depicts homologous genes that contribute to the variation in several species. Blue and purple indicate genes accounting for the variation in one species, which show homologous protein domains with genes contributing to the variation in another species. Given their large functional homology, CO, FT, and AP1 homologs of the three species are included in the three diagrams, with small circles depicting genes that do not contribute to natural variation. See text for further details.
QTL mapping studies have also shown that other loci, including homologs of FRI and FLC, affect the vernalization response (Figure 1). Natural loss-of-function alleles in two FRI-like genes (FRL1 and FRL2; Schläppi, 2006) and one FLC-like gene named FLM/FLW1 (Werner et al., 2005b) have been shown to produce early flowering. Additional flowering time QTLs affecting this response have been colocated with other FLC homologs in several crosses (Alonso-Blanco et al., 1998; O'Neill et al., 2008). Furthermore, loss- and gain-of-function alleles of another FLC regulator called HUA2 likely contribute to this variation (Doyle et al., 2005; Wang et al., 2007).
In addition, natural variation for the flowering photoperiod response is caused by three photoreceptor genes, CRY2, PHYC, and PHYD, in A. thaliana (Table 1, Figure 1). Rare loss-of-function alleles of PHYD and PHYC as well as a gain-of-function of CRY2 produce early flowering and reduced photoperiod response (Aukerman et al., 1997; El-Assal et al., 2001; Balasubramanian et al., 2006). Association mapping analyses suggest that haplotypes of CRY2 and PHYC differing in eight amino acid substitutions might further contribute to this variation (Olsen et al., 2004; Balasubramanian et al., 2006).
QTL linkage mapping and association analyses of candidate genes also have been performed in other Brassciaceae species, showing that FLC-like genes probably contribute to the differentiation of annual and biennial habits in Brassica rapa (Schranz et al., 2002; Yuan et al., 2009) and that functional variants of a FRI-like gene of A. lyrata affects its vernalization requirement (Kuittinen et al., 2008). In addition, molecular analyses have identified the LF gene in another dicotyledonous crop species, pea, which is a long-day temperate legume that also responds to vernalization. Pea LF behaves as a flowering repressor that is homologous to A. thaliana TFL1, and a natural loss-of-function LF allele caused by a large deletion shows reduced photoperiod response (Foucher et al., 2003).
In the last few years, comparable progress has been achieved in understanding flowering time variation in several monocotyledonous grass species, including temperate cereals with similar photoperiod and vernalization responses to A. thaliana (Table 1). Two genes largely determine the life habit and vernalization response of barley and wheat. VRN1 encodes a MADS box transcription factor homologous to the meristem identity protein AP1 of A. thaliana (Yan et al., 2003), and VRN2 encodes a grass-specific putative transcription factor with a CCT domain (Yan et al., 2004b). VRN1 promotes flowering and is speculated to be directly downregulated by vernalization, whereas VRN2 is a repressor that reduces the expression of VRN1 mediated by VRN3, a feedback regulatory loop among these genes accounting for their epistatic interactions (Figure 1; Distelfeld et al., 2009a). The early flowering spring habit of wheat and barley has been associated with more than 15 independent insertion and deletions in the first regulatory intron of VRN1 (Yan et al., 2004a; Fu et al., 2005; Szucs et al., 2007) and with several loss-of-function alleles of VRN2 (Distelfeld et al., 2009b). In addition, natural variation for the photoperiod response and its interaction with vernalization is determined by VRN3 and Ppd1. VRN3, which is a homolog of A. thaliana FT encoding a long-distance flowering signal, is upregulated by long-day photoperiod and vernalization and activates VRN1 expression (Yan et al., 2006). Ppd1 encodes a pseudoresponse regulator that mediates the photoperiod upregulation of VRN3 (Turner et al., 2005). Dominant mutations in the promoter and first intron of wheat and barley VRN3 show increased expression and response to vernalization (Yan et al., 2006). By contrast, missense recessive mutations in the CCT domain of barley Ppd-H1 likely cause late flowering under long-day photoperiod (Turner et al., 2005). Moreover, another barley FT homolog has been proposed as a possible candidate for the Ppd-H2 photoperiod QTL (Faure et al., 2007; Kikuchi et al., 2009).
Extensive analyses are also identifying the molecular bases of the natural variation for flowering time of rice and maize. In contrast with A. thaliana and temperate cereals, these are tropical short-day photoperiod species with no significant response to vernalization. To date, five genes have been isolated as determining the large variation for photoperiod response in rice (Table 1). Hd1 and Hd3a are flowering promoters, homologs of A. thaliana CO and FT genes (Yano et al., 2000; Kojima et al., 2002). Several Hd1 loss-of-function alleles reduce the expression of Hd3a and produce late flowering under short days, which indicates a conservation of this photoperiod regulatory module in phylogenetically distant short- and long-day flowering species (Figure 1). In addition, EhdD1 encodes a response regulator that independently upregulates Hd3a under short days (Doi et al., 2004). By contrast, Ghd7 and Hd6 encode a CCT domain protein and a CK2 kinase, respectively, which probably repress Hd3a under long-day photoperiod (Takahashi et al., 2001; Xue et al., 2008). Loss-of-function alleles of both genes produce early flowering, but additional alleles differing in several Ghd7 missense substitutions have been suggested to further contribute to rice flowering natural variation. In maize, the Vgt1 locus has been identified as accounting for the natural variation for flowering photoperiod response (Salvi et al., 2007). Vgt1 is an ;2-kb noncoding cis-element that regulates the expression of an AP2 homolog located 70 kb downstream and behaves as a flowering repressor. Several polymorphisms in the Vgt1 region have been strongly associated with flowering time variation (Salvi et al., 2007; Ducrocq et al., 2008).
The identification of large-effect QTGs has shown that photoreceptor and regulatory genes of different classes, such as those encoding transcription factors or signal transduction components, account for a large portion of the natural variation for flowering time in different species. Comparisons among species with similar physiological and environmental requirements for flowering induction show that some homologous genes contribute to the intraspecific variation in related species of the same plant family (e.g., FLC and VRN3 in Brassicaceae and grasses, respectively). Thus, parallel evolution of flowering time by mutations in homologous conserved genes is apparent in different groups of related species. However, comparisons among distant species from the same or different phylogenetic families show that natural variation for flowering responses to vernalization and photoperiod signals is mainly generated by allelic variation at different genes in each species, which are often highly divergent among species (Figure 1). Current results suggest that such convergent evolution of flowering responses is determined by species-specific genes showing analogous network positions in distant species (e.g., FLC and VRN2) and by homologous genes with partly differentiated functions (e.g., AP1 and VRN1). However, photoperiod responses seem to involve more conserved genes among species (photoreceptors, CO, and FT) than those involved in vernalization (FLC, VRN1, and VRN2), suggesting stronger constrains for the evolution of photoperiod than vernalization responses. Given the well-documented climate changes across evolutionary time, it can be further speculated that these differential constraints have been influenced by a wider temporal prevalence of photoperiod than vernalization signals.
The availability of causal genes and polymorphisms enables the study of the evolutionary processes that generate and maintain flowering time variation within and among species (reviewed in Roux et al., 2006; Mitchell-Olds et al., 2007). In crop species, fitness effects and adaptive values of genetic variants are often documented because segregating populations are grown in their natural agricultural environments (Izawa, 2007). Detailed analyses indicate that some loci affect not only heading date but several other fitness components, as shown with the effects of Ghd7 on plant height, panicle size, and seed production (Xue et al., 2008). Geographical distributions of alleles of Hd1, Ehd1, and Dgh7 from rice (Izawa, 2007; Xue et al., 2008) or Vgt1 and Dwarf8 from maize (Camus-Kulandaivelu et al., 2006; Ducrocq et al., 2008) show latitudinal and/or altitudinal clines, which suggests that they are involved in adaptation to different world regions. Current multilocus analyses of cultivated varieties are elucidating the role of flowering time allelic variation in regional adaptation and crop expansion during the few thousand years after domestication.
Similar studies have been initiated in A. thaliana, where in contrast with crop plants, several of the natural alleles identified in CRY2, FLM, PhyC, PhyD, or HUA2 (Table 1) are found in a single accession; therefore, it is possible that they are rare deleterious alleles. However, analyses of nucleotide diversity of CRY2, PhyC, FRI, and FLC have detected highly differentiated haplotypic groups, suggesting that their allelic variation might be maintained by various types of selection (Olsen et al., 2004; Caicedo et al., 2004; Stinchcombe et al., 2004; Balasubramanian et al., 2006). Most of the large number of FRI loss-of-function alleles appear at very low frequency, suggesting that they have been generated recently, for example, after the last glaciation (Le Corre et al., 2002; Shindo et al., 2005). However, two of these alleles are widely distributed in Central Europe, supporting the notion that their frequency increased due to strong natural selection during the postglacial colonization of this region (Johanson et al., 2000; Toomajian et al., 2006). In addition, a Eurasian latitudinal cline of flowering time and vernalization sensitivity has been shown to be partly determined by interactions among FRI, FLC, and PhyC allelic variation, suggesting that these interactions contribute to latitudinal and local adaptation (Caicedo et al., 2004; Lempe et al., 2005; Balasubramanian et al., 2006). Supporting the adaptive value of this allelic variation, analysis of FRI and FLC alleles in field experiments performed under environments closer to their natural habitat than laboratory conditions show that the FRI/FLC epistatic interaction affects different fitness components depending on the season of germination (Korves et al., 2007). In addition, antagonistic pleiotropic effects of FRI allelic variation on different fitness components, such as flowering time and plant architecture, suggest that complex phenotypic pleiotropy accounts for the adaptive value of FRI alleles (Scarcelli et al., 2007). Similarly, the large pleiotropic effects described for photoreceptor genes and HUA2 (Table 1) probably also contribute to the global adaptive value of their genetic variants. Thus, several demographical and selective processes seem to determine the natural variation for flowering time in crop and wild species, but their relative significance and molecular targets are just beginning to emerge.
Plant Architecture and Morphology
More than 20 loci of major effect have now been cloned for several domestication traits related to plant architecture, revealing that loss of functions of transcription factor genes (sometimes previously unknown) are often involved (Table 1). Examples are the rice shattering locus sh4 encoding a Myb factor (Li et al., 2006), the rice shattering locus qSH1, and the six/two row locus Vrs1 of barley encoding homeodomain transcription factors (Konishi et al., 2006; Komatsuda et al. 2007) and a SBP gene underlying the teosite glume architecture1 (tga1) locus in maize (Wang et al., 2005). Gain-of-function alleles of genes encoding other transcription factors also have been identified. These are illustrated with the classical maize tb1 and wheat Rht1 genes involved in plant architecture, whose selected alleles show mutations in a cis-regulatory region or in the coding region, respectively (Doebley et al., 2006; Peng et al., 1999; Clark et al., 2006). In tomato, fruit size and shape strongly differ from the small and round fruits of ancestral wild species. Detailed QTL analyses (reviewed in Tanksley, 2004) have been followed up with the cloning of four major effect loci (Table 1). Thus, FW2.2 and OVATE identified novel classes of genes (Frary et al., 2000; Liu et al., 2002), the FAS locus has recently found to be a Yabby-type cell fate gene (Cong et al., 2008), and SUN has been shown to encode an IQ67 domain protein (Xiao et al., 2008). An ovate nonsense allele causes fruits with a pear shape (Liu et al., 2002), while different regulatory mutations producing misexpression of FW2.2, FAS, and SUN lead to large fruit phenotypes. Furthermore, seed size loci have been isolated in several species, such as the rice GW2 and GS3 loci encoding an ubiquitin-E3-ligase and a transmembrane protein, respectively (Fan et al., 2006; Song et al., 2007), or the tomato Sw4.1 locus encoding an ABC transporter (Orsi and Tanksley, 2009). On the other hand, a large number of QTL analyses have been described in many crops for other developmental and morphological traits related to yield and plant performance (e.g., http://www.gramene.org/ and http://www.sgn.cornell.edu/index.pl). The first genes underlying those loci have been identified mainly in rice (Sakamoto and Matsuoka, 2008; Weng et al., 2008) as illustrated with the Gn1a locus encoding a cytokinin dehydrogenase that affects grain number (Ashikari et al., 2005) or the pleiotropic Ghd7 locus with effects on various yield components (Xue et al., 2008).
Developmental traits related to plant architecture and organ morphology also have been studied in A. thaliana, where natural variation for seed size (Alonso-Blanco et al.,1999), inflorescence architecture (Juenger et al., 2000), leaf morphology (Pérez-Pérez et al., 2002), and flower size (Juenger et al., 2005) have been described. Although none of the underlying genes have been cloned, microsynteny comparisons between tomato and A. thaliana, combined with functional analyses, suggest that a Sw4.1 ortholog gene might underlie an A. thaliana seed size QTL (Orsi and Tanksley, 2009). Several QTLs have been detected for root growth and architecture (Mouchel et al., 2004; Loudet et al., 2005; Fitz Gerald et al., 2006), and the major effect locus BRX has identified a novel gene with unknown molecular function (Mouchel et al., 2004). In addition, developmental features present as monogenic traits in some A. thaliana accessions have identified non-sense mutations in the Myb transcription factor gene GL1 causing glabrous phenotype (Hauser et al., 2001) and in the β-galactosidase gene corresponding to the MUM2 locus, which reduces seed mucilage liberation after imbibition (Macquet et al., 2007).
Different strategies are revealing cryptic developmental genetic variation of A. thaliana that is only apparent under specific conditions (Table 1). It has been shown that differential expansion of an intronic trinucleotide repeat affects the expression of a Leu biosynthesis enzyme gene, hence causing aberrant leaf morphology defects that are mainly visible under short-day photoperiod and high temperature conditions (Sureshkumar et al., 2009). In addition, A. thaliana transgenic lines carrying the SRK and SCR genes that determine self-incompatibility in A. lyrata have been used to reveal a gene underlying pseudo-self-compatibility. Thus, it has been shown that polymorphisms in the promoter region of the previously uncharacterized gene PLANT U-BOX8 regulating SRK transcript levels reduce its expression and cause such compatibility (Liu et al., 2007). These examples show how the analysis of morphological and developmental traits in crops and wild plants are identifying not only genes with allelic variants increasing fitness under particular conditions, but also cryptic genetic variation with unknown effects on plant adaptation.
VEGETATIVE GROWTH AND PHYSIOLOGY
Natural variation has been described extensively for general growth traits (e.g., biomass accumulation) as well as primary metabolism and mineral uptake and accumulation, and the complex molecular bases underlying these traits are beginning to be elucidated. Such studies are aiding our understanding of hybrid vigor (positive heterosis) and hybrid incompatibility (negative heterosis), which are of primary concern in agronomy, as well as specific metabolic pathways and physiological processes that affect crop growth and/or crop quality.
Growth and Biomass Accumulation
The complexity underlying the regulation of biomass accumulation is illustrated with the precise dissection of a small-effect QTL for this trait in A. thaliana, which includes two closely linked QTLs that have opposing effects on biomass accumulation within a small genomic region of 210 kb (Kroymann and Mitchell-Olds, 2005). In addition, the effect of both loci depends on the genetic background. Thus, a highly polygenic architecture involving genetic interactions has been speculated for biomass accumulation (Kroymann and Mitchell-Olds, 2005). Recently, significant progress has been made in understanding the genetic and molecular bases of growth traits that show positive or negative heterosis, as well as in finding genes contributing to the variation for growth interactions with abiotic environmental factors.
A large number of QTL studies are currently dissecting the genetic bases of positive heterosis or hybrid vigor, which is a growth trait observed in many hybrids of most species and a major plant breeding objective. Numerous QTLs with different levels of dominant, overdominant, and epistatic effects have been mapped for heterosis in maize (Frascaroli et al., 2007), rice (Li et al., 2001), tomato (Semel et al., 2006), rapeseed (Brassica napus) (Radoev et al., 2008), and A. thaliana (Kusterer et al., 2007; Melchinger et al., 2007). Comparisons of genomic sequences among maize inbred lines have shown extensive within-species lost of gene colinearity, with a large portion of genes being absent in some individuals (Fu and Dooner, 2002; Song and Messing, 2003). This lack of colinearity has led to a molecular hypothesis for the dominance model of heterosis and its counterpart inbreeding depression, where the presence of distinct genes in different lines might cause hybrid vigor (Fu and Dooner, 2002). In addition, overdominance frequently has been detected at the level of gene expression in hybrids of maize and A. thaliana (Song and Messing, 2003; Vuylsteke et al., 2005).
On the other hand, analyses of A. thaliana hybrids are identifying the molecular bases of a related fundamental evolutionary phenomenon, namely, negative heterosis or hybrid incompatibility (Table 1). Nearly 2% of A. thaliana intraspecific crosses yield hybrid plants expressing severe growth defects and leaf necrosis, which are due to epistatic interactions between two or three genes fitting the Dobshansky-Muller model of hybrid postzygotic isolation (Bomblies et al., 2007; Alcázar et al., 2009). Alleles of two different tandem repeats of NB-LRR disease resistance genes differing in sequence and gene copy number have been shown to cause these hybrid incompatibilities (Bomblies et al., 2007; Alcázar et al., 2009). Incompatible combinations are characterized by autoimmune-like responses and enhanced disease resistance, which depend on growth temperature and activation of the salicylic acid stress signaling pathway (Alcázar et al., 2009). In addition, embryo lethality segregating in crosses among wild A. thaliana genotypes is determined by an epistatic interaction between two paralogous genes encoding a key enzyme involved in His biosynthesis (Bikard et al., 2009). Thus, divergent evolution of particular duplicated genes appears as the main mechanism causing genetic incompatibilities at different postzygotic levels.
Several QTLs have been isolated that contribute to natural variation for growth responses to light or soil nutrient availability in A. thaliana. Analyses of light responses have shown that amino acid substitutions in photoreceptors, including phytochrome A (Maloof et al., 2001), phytocrome B (Filiault et al., 2008), cryptochome 2 (Botto et al., 2003), and probably phytochrome C (Balasubramanian et al., 2006), affect hypocotyl elongation in different light conditions. In addition, a tandem zinc knuckle/PLU3 (TZP/LIGHT5) domain-encoding gene has been shown to promote growth, associated with an insertion mutation appearing as a rare natural loss-of-function allele (Loudet et al., 2008). Furthermore, it has been shown that natural polymorphisms affecting the expression of LPR1, which encodes a multicopper oxidase, strongly influences the primary root growth response to phosphate starvation (Svistoonoff et al., 2007). Hence, it appears that mainly signal perception and transduction components have been involved in natural variation for growth environmental responses.
Primary Metabolism
Primary metabolism has been defined as “those essential reactions involving compounds that are formed as part of the normal anabolic and catabolic processes, which result in assimilation, respiration, transport, and differentiation processes that take place in most, if not all, cells of an organism” (Fernie and Schauer, 2009). Besides a wide range of intermediate compounds (e.g., compounds from glycolysis or Krebs cycle), primary metabolites include end products of metabolic pathways that accumulate in (harvestable) sink organs (e.g., seeds, fruits, and tubers) determining relevant crop quality traits related to nutritional content and composition. Recent molecular analyses of natural variation have identified a large number of genes encoding enzymes and have associated primary metabolism with phenotypic variation for morphology and growth-related traits (Lisec et al., 2008; Schauer et al., 2008).
Two decades ago, analyses of starch content in seeds of pea and cereals identified the genes underlying classical loci, such as Mendel's pea rugosus (R) and maize Waxy. These pioneering works illustrate the pleiotropic effects of primary metabolism on plant development. The R locus encodes a starch branching enzyme, whose recessive allele carrying a transposon insertion in the coding sequence has been widely spread in European cultivars due to the associated sweetness of the wrinkled pea phenotype (Bhattacharyya et al., 1990). Maize Waxy loci encode granule-bound starch biosynthesis enzymes, where multiple loss-of-function alleles caused by transposon insertions lead to a waxy appearance of kernels that reflect the lack of amylose in endosperm (Varagona et al., 1992). Moreover, a single splice donor site mutation in a homologous Waxy gene is responsible for the absence of amylose in glutinous rice varieties, which has been widely spread in East Asia associated with human cultural behaviors (Olsen and Purugganan, 2002).
Recent quantitative analyses of natural variation for the content of primary metabolites in fruits of tomato interspecific crosses (Schauer et al., 2006) and in potato tubers (Solanum tuberosum; Menéndez et al., 2002) have found a large number of QTLs underlying the content of other carbohydrate metabolites. Isolation of a large-effect QTL for tomato sugar content identified the invertase encoding gene LIN5, where a single amino acid substitution alters its activity (Fridman et al., 2004). Based on colocation and genetic association, two tandemly repeated invertase genes, invGE and invGF, have been also suggested to underlie a cold-sweetening QTL in potato (Li et al., 2005).
Several studies have dissected the variation for primary metabolism of A. thaliana through the analysis of enzymatic activities and metabolite contents in different vegetative organs. QTLs have been found for the activity of multiple carbohydrate metabolism enzymes, some of them colocating with the corresponding structural genes, for example, phosphogluco-mutase (PGM) and hexokinase, which indicates cis-regulatory variation (Mitchell-Olds and Pedersen, 1998; Keurentjes et al., 2008b). Histochemical analysis further shows that certain PGM QTLs are tissue specific (Sergeeva et al., 2004). However, several enzymatic activity QTLs are located in different genomic regions, identifying trans-acting sequences. These loci might simultaneously coregulate several enzymes, hence revealing novel modes of carbohydrate metabolic regulation (Mitchell-Olds and Pedersen, 1998; Sergeeva et al., 2004; Keurentjes et al., 2008b). In addition, a vacuolar invertase has been found that contributes specifically to the natural variation for root elongation but not for hypocotyl length (Sergeeva et al., 2006). Similarly, QTLs have been identified for nitrogen- and sulfur-containing compounds, such as nitrate, sulfate, and free amino acids (Loudet et al., 2003, 2007). A large-effect QTG has been isolated for sulfate content, which corresponds to the APR2 sulfate reductase gene (Loudet et al., 2007). APR2 activity is altered by a single amino acid substitution, and the effects of this polymorphism interact with nitrogen availability, hence illustrating the interaction between sulfate assimilation and nitrogen metabolism. Furthermore, as described in the previous section, genes involved in Leu or His biosynthesis account for natural variation for severe zygotic (embryo) and postzygotic morphological defects (Bikard et al., 2009; Sureshkumar et al., 2009).
A. thaliana natural variation for untargeted metabololites from vegetative tissues has been dissected by quantifying high-throughput metabolic profiles in different mapping populations (Keurentjes et al., 2006; Meyer et al., 2007; Rowe et al., 2008). A large number of metabolite content QTLs (mQTLs) have been mapped, demonstrating that two- and three-way epistatic interactions determine a substantial proportion of this variation (Lisec et al., 2008; Rowe et al., 2008). Candidate genes have been proposed for a large proportion of detected mQTL based on the enzymatic activities involved in different biochemical pathways (Lisec et al., 2008). In addition, comparative analyses have shown colocation of QTLs for biomas accumulation and mQTL, supporting a link between growth and metabolic profiles (Lisec et al., 2008).
Mineral Accumulation
Most nutrients that plants need for growth and development are supplied as minerals to the roots, and they are classified as macronutrients (Ca, K, Mg, N, P, and S) or micronutrients (B, Cl, Fe, Mn, Co, Cu, Mo, Ni, and Zn) depending on the necessary quantities. The composition of mineral nutrients and trace elements (i.e., the inorganic component of an organism) is now referred to as the ionome (Salt et al., 2008). There is substantial natural variation for mineral use efficiency, root uptake, translocation from roots to shoots, and accumulation in the seed as storage and supply for the germinating seedling. This variation has been reported in many species, leading to breeding programs such as those aiming to improve zinc and iron status of cereal grains or tuber crops (www.harvestplus.org). Recent QTL analyses have identified six genes involved in the complex mechanisms controlling the variation for plant nutrition and mineral homeostasis (Table 1; Ghandilyan et al., 2006)
QTL studies in crop species have focused on seed or leaf mineral concentrations, mostly in rice, wheat, barley, Brassica species, and soybean (Glycine max). In cereals, mostly rice, QTLs have been mapped for grain minerals (Joppa et al., 1997; Garcia-Oliveira et al., 2009), flag leaf nitrogen (N) (Ishimaru et al., 2001) or phosphorous (P) uptake (Wissuwa et al., 2002), salt tolerance related to Na/K homeostasis (Lin et al., 2004), and grain accumulation of toxic Cd (Ueno et al., 2009). In Brassica crops, loci have been reported for phosphate concentrations in leaves and seeds of B. rapa (Zhao et al., 2008), for Ca and Mg concentrations in leaves of B. oleracea (Broadley et al., 2008), and for leaf mineral concentrations in B. rapa (Wu et al., 2008). In soybean, QTLs have been found for Ca and S concentrations in seeds (Panthee et al., 2006; Zhang et al., 2009). Three large-effect QTLs have been isolated in crops, which have identified several molecular mechanisms linking mineral content and other physiological traits. The rice SKC gene encodes a Na+ selective transporter, whose more active allele determined by missense mutations increases salt tolerance (Ren et al., 2005). A boron tolerance locus of barley corresponds to the boron efflux transporter gene Bot1 that appears duplicated and overexpressed in a B tolerant accession (Sutton et al., 2007). In addition, the Gpc-B1/NAM-B1 locus of wheat encodes a NAC transcription factor that regulates nutrient remobilization from leaves to developing grains (Uauy et al., 2006). It has been shown that nonfunctional alleles caused by different deletions are present in cultivated varieties, leading to delayed senescence and reduced protein, Zn, and Fe grain content.
Detailed analyses of the ionome in A. thaliana have shown considerable variation for leaf mineral concentrations under various mineral/metal supply conditions (Salt et al., 2008). QTLs have been identified for accumulation of different elements (Ca, Cu, Fe, K, Mg, Mn, Na, P, S, and Zn) in seeds, siliques, leaves, and roots under different growth conditions (Vreugdenhil et al., 2004; Waters and Grusak, 2008; Ghandilyan et al., 2009). Overall, these analyses show that mineral concentrations are controlled by a large number of loci with mostly moderate effect, which often interact strongly with growth environment. Interestingly, a strong P QTL has been colocated with a previously identified locus for phytate (myo-inositol-1,2,3,4,5,6-hexakisphosphate or IP6), the storage compound for P in seeds (Bentsink et al., 2003). However, only the ERECTA gene, encoding a receptor-like kinase for which a mutant allele segregates in mapping populations, has been demonstrated to affect mineral concentrations in these studies (Waters and Grusak, 2008; Ghandilyan et al., 2009).
In addition, A. thaliana QTL analyses have been focused on accumulation of specific minerals, including N (as nitrate; Loudet et al., 2003; Harada et al., 2004), K (Harada and Leigh, 2006), Cu (Kobayashi et al., 2008), Mo (Baxter et al., 2008), and Na (Rus et al., 2006). These studies have led to the isolation of three genes underlying large-effect QTLs, which also encode different mineral transport components. A root copper tolerance locus corresponds to the HMA5 gene encoding a Cu-transporting ATPase. Several natural alleles differing in missense mutations in conserved motifs show lower activity and Cu translocation to the shoot (Kobayashi et al., 2008). A mitochondrial molybdenum transporter encoded by the nuclear MOT1 gene underlies a shoot Mo concentration QTL. A deletion in MOT1 promoter region has been associated with low gene expression and low shoot Mo concentration, suggesting that this regulatory mutation is the causal nucleotide polymorphism (Baxter et al., 2008). Finally, HKT1 encodes a Na+ transporter for which two loss-of-function alleles associated with promoter deletions produce lower root expression and enhanced shoot Na+ levels in two coastal accessions (Rus et al., 2006).
Studies of A. thaliana mineral accumulation have inspired research on natural variation for exceptional zinc and/or cadmium accumulation in leaves of the related metal hyperaccumulator species Arabidopsis halleri and Thlaspi caerulescens. Several loci have been mapped in populations derived from crosses between A. halleri and A. lyrata (Courbot et al., 2007), one of them colocalizing with HMA4, a locus containing three copies of the Ah-HMA4 gene that is essential for Zn hyperaccumulation and Cd tolerance in A. halleri (Hanikenne et al., 2008). In addition, multiple QTLs have been located for root or shoot Zn or Cd accumulation in F3 populations of T. caerulescens (Assunção et al., 2006; Deniau et al., 2006). Thus, comparative analyses with A. thaliana are not only assisting the isolation of genes underlying those loci but will also enable identification of the molecular bases of interspecific variation present in wild crucifers.
REVEALING THE VARIOUS LEVELS OF PHENOTYPIC REGULATION: NATURAL VARIATION FOR MOLECULAR TRAITS
The path from genotype to phenotype includes several molecular intermediate steps in the translation of genetic information, with regulation occurring at each of these levels (Keurentjes et al., 2008a). Extensive natural variation has been described for the amount of molecular components, such as gene transcripts (Vuylsteke et al., 2005; Kliebenstein et al., 2006a), protein abundance and activity (Chevalier et al., 2004; Cross et al., 2006), and metabolites (Kliebenstein et al., 2001; Cross et al., 2006). Quantitative genetic analyses of “-omics” data collected from mapping populations by so-called genetical genomics approaches (Jansen and Nap, 2001) have shown that much of this natural variation is genetically determined by specific expression QTL (eQTL), mQTL, or protein QTL (pQTL) (Keurentjes et al., 2008a). In addition, a portion of this variation is determined by epigenetic mechanisms of regulation, such as methylation and chromatin remodeling (He et al., 2004; Vaughn et al., 2007), which in turn can also result from DNA sequence polymorphisms (Johannes et al., 2008).
The genetic bases of natural variation for genome-wide gene expression have been approached systematically in A. thaliana, barley, and maize (Schadt et al., 2003; Vuylsteke et al., 2006; Keurentjes et al., 2007; West et al., 2007; Potokina et al., 2008b). Expression variation of ;30 to 40% of the segregating genes is regulated by an eQTL colocating with the gene encoding the mRNA, most of those local eQTLs being the result of cis-regulated genes (i.e., allele-specific expression). However, most variably expressed genes are regulated in trans by eQTLs located in distant genomic regions (trans-eQTLs) that often appear concentrated in a small number of regulatory hot spots (Keurentjes et al. 2007; West et al., 2007; Fu et al., 2009). cis-eQTLs show larger average effects than trans-eQTLs probably due to the stronger impact of direct cis-regulation on the target gene. In addition, it has been shown that QTLs for gene expression variation might depend on spatial and temporal regulation (Potokina et al., 2008a). The molecular identification of regulatory eQTLs is currently facilitated by statistical methods that combine the functional annotation of biological relevant gene sets, the genomic position of genes and eQTLs, and the coexpression of candidate regulators and target genes. In this way, complex genetic networks are constructed, which reveal many novel putative regulatory steps together with strong interconnectivity and epistasis (Kliebenstein et al., 2006b; Keurentjes et al., 2007; Druka et al., 2008). As described above, large-scale untargeted analyses of metabolic profiles have also revealed extensive genetic variation for metabolite content in A. thaliana and tomato (Keurentjes et al., 2006; Schauer et al., 2006; Lisec et al., 2008). Large numbers of mQTLs have been mapped, although they show a lower average heritability than eQTL (Rowe et al., 2008).
The genetic dissection of natural variation for developmental and physiological traits is beginning to link phenotypic QTLs for complex traits with eQTL, mQTL, and pQTL (Keurentjes, 2009). Several studies, mostly in A.thaliana, have integrated information from different levels of molecular regulation to improve the elucidation of genetic control of quantitative traits (Hirai et al., 2004; Tohge et al., 2005; Meyer et al., 2007; Wentzell et al., 2007; Druka et al., 2008; Lisec et al., 2008). Thus, it has been shown that plant growth and development are interwoven with primary metabolism, regulation occurring at different levels, including gene expression, enzyme activity, and metabolite abundance (Keurentjes et al., 2008b). Although these studies often reveal direct relationships between the various levels of biological information, such a relationship is not always linear or present. This was illustrated recently with a system-wide analysis, wherein much of the natural genetic variation affecting gene expression or functional protein differences did not to appear to lead to phenotypic variation, a phenomenon known as phenotypic buffering (Fu et al., 2009). Interestingly, several loci have been identified as contributing to natural variation for the ability to buffer developmental processes through environmental canalization (Hall et al., 2007; Sangster et al., 2008). Hence, integrative analyses of multiple phenotypic and molecular regulatory levels are elucidating the mechanisms controlling developmental buffering and canalization, which are presumed to contribute to biological robustness of plant species and facilitate evolvability in changing environments (Queitsch et al., 2002; Kitano, 2004).
MOLECULAR TARGETS OF SELECTION FOR PLANT ADAPTATION: CURRENT AND FUTURE PROSPECTS
Nearly 100 genes underlying natural variation for plant development and growth have been identified in crop plants and A. thaliana. Most of them correspond to QTLs with large effects on trait variation and provide a first inventory of mechanisms, genes, and functional nucleotide polymorphisms that might be involved in plant adaptation to different agricultural or natural environments. They belong to all types of ontological classes, including transcription factors, signal transduction components, enzymes of primary and secondary metabolism, hormone metabolism and signaling, metal transporters, as well as genes with unknown functions. Interestingly, some of these genes had not been found previously in mutant screenings because common laboratory strains carry natural loss-of-function alleles, such as FLC, FRI, or DOG1 in A. thaliana.
Nucleotide polymorphisms affecting the function of these genes are identifying the mutational mechanisms that generate natural variation. A large proportion of natural alleles carry null loss-of-function mutations, which are often produced by structural nonsense or insertion and deletion (indel) mutations (Alonso-Blanco et al., 2005). A particular class of these alleles corresponds to complete gene deletions or duplications caused by retrotransposition, recombination, and other molecular mechanisms, which may play a major role in heterosis of crop and wild species (Fu and Dooner, 2002; Song and Messing, 2003). A second type of common allele is a change-of-function allele produced by a missense mutation altering protein structure and function, such as described for photoreceptor genes (Table 1). In addition, multiple change-of-function alleles carrying regulatory mutations in three types of genomic regions affecting gene expression have been described for many traits and species. Most of these alleles show indels in their nearby promoter regions, in agreement with the large number of cis-eQTLs identified (Keurentjes et al., 2007; West et al., 2007). Second, gene expression changes are caused by unknown mutations located in distant 5′-regions of the genes as illustrated for maize Tb1 (Clark et al., 2006) and Vgt1 (Salvi et al., 2007). Third, expression alterations also appear to be produced by intronic mutations, including insertions of tranposons or differential expansion of microsatellite repeated sequences (Sureshkumar et al., 2009). Finally, although most natural alleles that have been described are caused by variation at the level of nucleotide sequence, natural epigenetic variation also contributes to heritable developmental and physiological variation, as indicated by stably inherited epialleles in the LCyc gene involved in Linaria flower morphology (Cubas et al., 1999) and in an SBP-box gene affecting tomato fruit ripening (Manning et al., 2006). This is supported by the considerable amount of natural variation found for DNA methylation (Riddle and Richards, 2005; Zhang et al., 2006b; Vaughn et al., 2007), which is partly mediated by small interfering RNAs also differing among accessions (Zhai et al., 2008). It is hypothesized that methylation and heterochromatinization mediated by different kinds of small interfering RNAs might cis- and trans-regulate gene expression and function in a more quantitative and reversible manner than sequence variation (Zhai et al., 2008).
All of these studies provide information on the particular classes of genes and alleles that might have been naturally or artificially selected for different traits (reviewed in Alonso-Blanco et al., 2005). For instance, as described by Purugganan and Fuller (2009), it seems that domestication is mainly associated with transcriptional activators and changes in transcriptional networks. By contrast, crop diversification after domestication involves a large proportion of genes encoding enzymes, with loss-of-function alleles differently contributing to crop domestication and diversification. Thus, we are beginning to understand how natural variation is generated and maintained depending on plant species, traits, environments, and selective forces. However, conclusive generalizations on evolutionary processes await systematic extensions of the following aspects of these analyses: (1) the number of species analyzed, particularly of wild plants; (2) the number of genes identified in each species; (3) the spectrum of allelic effects per gene; and (4) the estimates of genetic and environmental interactions.
Understanding the ecological and evolutionary significance of natural variation requires precise evaluation of their role in adaptation to particular environments, which is just beginning to be approached. Based on their presence in a single wild genotype and their relative large effects, some of the A. thaliana natural alleles are probably deleterious variants segregating in natural populations. This has been suggested for loss-of-function alleles of HUA2 and LIGHT5, found only in a subset of lines derived from the same parental accession (Doyle et al., 2005; Loudet et al., 2008). Similarly, the BRX loss-of-function allele has not been found in genotypes collected in the same geographical site where this variant was originally sampled many years before (Shindo et al., 2008). As described above for flowering time, demonstrating that natural variation in specific genes is involved in adaptation is currently addressed by combining population and evolutionary genetic analyses with detailed phenotypic studies of fitness-related traits. Analysis of the amount and pattern of nucleotide diversity may show potential signatures of selection, hence suggesting different evolutionary mechanisms to maintain such variation. This is illustrated with the identification of selective sweeps in several domestication genes of crop plants (Purugganan and Fuller, 2009) and with the highly differentiated haplotypes found in some A. thaliana photoreceptor genes (Olsen et al., 2004; Balasubramanian et al., 2006). However, demographical history also affects the patterns of DNA sequence diversity; therefore, other studies are necessary to test adaptive effects of such alleles. On one hand, detailed phenotypic characterizations under natural and agricultural conditions are detecting fitness effects of particular alleles or genotypes (Tian et al., 2003; Shindo et al., 2008; Wilczek et al., 2009). In addition, comparisons of differentiation among local populations for quantitative traits (QST) and nucleotide polymorphisms (FST) might further assist the detection of selection signatures on particular genes (Le Corre, 2005; Stenoien et al., 2005). On the other hand, association analyses of nucleotidic and phenotypic diversity with environmental variation might identify environmental factors driving phenotypic selection. It is expected that new extensive collections of A. thaliana wild genotypes recently developed from several world regions (Jorgensen and Mauricio, 2004; Stenoien et al., 2005; Schmid et al., 2006; Beck et al., 2008; Picó et al., 2008) will facilitate estimations of allele frequencies, geographical distributions, and genetic associations with environmental factors.
In the past decade, the analysis of natural variation in crop plants and A. thaliana has provided an unprecedented amount of information on the genetic and molecular mechanisms that determine intraspecific variation and adaptation. It can be anticipated that this trend will continue in the next decade, especially with the broad implementation of “-omics” technologies for the precise analysis of natural variation at different levels. First, high-throughput genotyping will enable the development of new genetic mapping strategies, such as genome-wide association (Azanzana et al. 2005; Nordborg and Weigel, 2008; Waugh et al., 2009), which is expected to provide higher mapping resolution. Second, comparisons of thousands of whole-genome sequences from genotypes of the same species will provide a dynamic perspective of a species' genome in terms of pan-genomes (Morgante et al., 2007). Third, common integration of QTLs and genes causing phenotypic variation with those affecting the various levels of molecular variation will provide complete (genome-wide and whole organisms) phenotypic regulatory networks. The latter should include not only eQTL, mQTL, and pQTL estimated at particular points but also molecular fluxes in those networks. Finally, extension of the analysis of natural variation to other crop and wild species will allow deep comparisons among species. Such comparative intraspecific and interespecific studies will elucidate the mechanisms involved in the evolution of plant development and physiology at higher levels, including those of speciation and species diversification in relation to earth changing environments.
Acknowledgments
We apologize to those authors whose work could not be discussed due to space limitations. Research in our laboratories was supported by the European Research Area-Plant Genomics program Grant 034B ARABRAS and the Max Planck Society (M.R. and M.K.), Grants GEN2006-27786-E/VEG and BIO2007-62632 from the Ministerio de Ciencia e Innovación of Spain (C.A.-B.), the Netherlands Organization for Scientific Research, VENI scheme (JK and LB), and the Centre for Biosystems Genomics (Netherlands Genomics Initiative; J.J.B.K. and D.V.).
References
- Albani, M.C., Battey, N.H., and Wilkinson, M.J. (2004). The development of ISSR-derived SCAR markers around the SEASONAL FLOWERING LOCUS (SFL) in Fragaria vesca. Theor. Appl. Genet. 109 571–579. [DOI] [PubMed] [Google Scholar]
- Alcázar, R., García, A.V., Parker, J.E., and Reymond, M. (2009). Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. USA 106 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco, C., Bentsink, L., Hanhart, C.J., Blankestijn-De Vries, H., and Koornneef, M. (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164 711–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C.J., and Koornneef, M. (1999). Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 4710–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco, C., El-Assal, S.E., Coupland, G., and Koornneef, M. (1998). Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco, C., Mendez-Vigo, B., and Koornneef, M. (2005). From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int. J. Dev. Biol. 49 717–732. [DOI] [PubMed] [Google Scholar]
- Aranzana, M.J., et al. (2005). Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1 e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris, J., Dahal, P., Hayashi, E., Still, D.W., and Bradford, K.J. (2008). Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 148 926–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris, J., Truco, M.J., Ochoa, O., Knapp, S.J., Still, D.W., Lenssen, G.M., Schut, J.W., Michelmore, R.W., and Bradford, K.J. (2005). Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor. Appl. Genet. 111 1365–1376. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E.R., Qian, Q., Kitano, H., and Matsuoka, M. (2005). Cytokinin oxidase regulates rice grain production. Science 309 741–745. [DOI] [PubMed] [Google Scholar]
- Assunção, A.G., Pieper, B., Vromans, J., Lindhout, P., Aarts, M.G., and Schat, H. (2006). Construction of a genetic linkage map of Thlaspi caerulescens and quantitative trait loci analysis of zinc accumulation. New Phytol. 170 21–32. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., Hirschfeld, M., Wester, L., Weaver, M., Clack, T., Amasino, R.M., and Sharrock, R.A. (1997). A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, S., Sureshkumar, S., Agrawal, M., Michael, T.P., Wessinger, C., Maloof, J.N., Clark, R., Warthmann, N., Chory, J., and Weigel, D. (2006). The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, I., Muthukumar, B., Park, H.C., Buchner, P., Lahner, B., Danku, J., Zhao, K., Lee, J., Hawkesford, M.J., Guerinot, M.L., and Salt, D.E. (2008). Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter MOT1. PLoS Genet. 4 e1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, J.B., Schmuths, H., and Schaal, B.A. (2008). Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects pleistocene glacial dynamics. Mol. Ecol. 17 902–915. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., and Mitchell-Olds, T. (2008). From genotype to phenotype: Systems biology meets natural variation. Science 320 495–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink, L., Jowett, J., Hanhart, C.J., and Koornneef, M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink, L., Yuan, K., Koornneef, M., and Vreugdenhil, D. (2003). The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor. Appl. Genet. 106 1234–1243. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, M.K., Smith, A.M., Ellis, T.H., Hedley, C., and Martin, C. (1990). The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60 115–122. [DOI] [PubMed] [Google Scholar]
- Bikard, D., Patel, D., Le Mette, C., Giorgi, V., Camilleri, C., Bennett, M.J., and Loudet, O. (2009). Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323 623–626. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J.L., and Weigel, D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, J.F., Alonso-Blanco, C., Garzaron, I., Sanchez, R.A., and Casal, J.J. (2003). The Cape Verde Islands allele of cryptochrome 2 enhances cotyledon unfolding in the absence of blue light in Arabidopsis. Plant Physiol. 133 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley, M.R., Hammond, J.P., King, G.J., Astley, D., Bowen, H.C., Meacham, M.C., Mead, A., Pink, D.A.C., Teakle, G.R., Hayden, R.M., Spracklen, W.P., and White, P.J. (2008). Shoot calcium and magnesium concentrations differ between subtaxa, are highly heritable, and associate with potentially pleiotropic loci in Brassica oleracea. Plant Physiol. 146 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A.L., Stinchcombe, J.R., Olsen, K.M., Schmitt, J., and Purugganan, M.D. (2004). Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus-Kulandaivelu, L., Veyrieras, J.B., Madur, D., Combes, V., Fourmann, M., Barraud, S., Dubreuil, P., Gouesnard, B., Manicacci, D., and Charcosset, A. (2006). Maize adaptation to temperate climate: Relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172 2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasoli, M., Derory, J., Morera-Dutrey, C., Brendel, O., Porth, I., Guehl, J.M., Villani, F., and Kremer, A. (2006). Comparison of quantitative trait loci for adaptive traits between oak and chestnut based on an expressed sequence tag consensus map. Genetics 172 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, F., Martin, O., Rofidal, V., Devauchelle, A.D., Barteau, S., Sommerer, N., and Rossignol, M. (2004). Proteomic investigation of natural variation between Arabidopsis ecotypes. Proteomics 4 1372–1381. [DOI] [PubMed] [Google Scholar]
- Clark, R.M., Wagler, T.N., Quijada, P. (2006). A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescence architecture. Nat. Genet. 38 594–597. [DOI] [PubMed] [Google Scholar]
- Clark, R.M., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342. [DOI] [PubMed] [Google Scholar]
- Clauss, M.J., and Koch, M.A. (2006). Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci. 11 449–459. [DOI] [PubMed] [Google Scholar]
- Clegg, M.T., and Durbin, M.L. (2003). Tracing floral adaptations from ecology to molecules. Nat. Rev. Genet. 4 206–215. [DOI] [PubMed] [Google Scholar]
- Clerkx, E.J.M., El Lithy, M.E., Vierling, E., Ruys, G.J., Blankestijin-De Vries, H., Groot, S.P.C., Vreugdenhil, D., and Koornneef, M. (2004). Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 135 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, B., Barrero, L.S., and Tanksley, S.D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40 800–804. [DOI] [PubMed] [Google Scholar]
- Courbot, M., Willems, G., Motte, P., Arvidsson, S., Roosens, N., Saumitou-Laprade, P., and Verbruggen, N. (2007). A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4, a gene encoding a heavy metal ATPase. Plant Physiol. 144 1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, J.M., von Korff, M., Altmann, T., Bartzetko, L., Sulpice, R., Gibon, Y., Palacios, N., and Stitt, M. (2006). Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 142 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P., Vincent, C., and Coen, E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401 157–161. [DOI] [PubMed] [Google Scholar]
- Deniau, A., Pieper, B., Ten Bookum, W., Lindhout, P., Aarts, M., and Schat, H. (2006). QTL analysis of cadmium and zinc accumulation in the heavy metal hyperaccumulator Thlaspi caerulescens. Theor. Appl. Genet. 113 907–920. [DOI] [PubMed] [Google Scholar]
- Distelfeld, A., Li, C., and Dubcovsky, J. (2009. a). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12 178–184. [DOI] [PubMed] [Google Scholar]
- Distelfeld, A., Tranquilli, G., Li, C., Yan, L., and Dubcovsky, J. (2009. b). Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 149 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J.F., Gaut, B.S., and Smith, B.D. (2006). The molecular genetics of crop domestication. Cell 127 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doerge, R.W. (2002). Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 3 43–52. [DOI] [PubMed] [Google Scholar]
- Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., Yano, M., and Yoshimura, A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, M.R., Bizzell, C.M., Keller, M.R., Michaels, S.D., Song, J., Noh, Y.S., and Amasino, R.M. (2005). HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 41 376–385. [DOI] [PubMed] [Google Scholar]
- Druka, A., et al. (2008). Towards systems genetic analyses in barley: Integration of phenotypic, expression and genotype data into GeneNetwork. BMC Genet. 9 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316 1862–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrocq, S., Madur, D., Veyrieras, J.B., Camus-Kulandaivelu, L., Kloiber-Maitz, M., Presterl, T., Ouzunova, M., Manicacci, D., and Charcosset, A. (2008). Key impact of Vgt1 on flowering time adaptation in maize: Evidence from association mapping and ecogeographical information. Genetics 178 2433–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal, E.-D.S., Alonso-Blanco, C., Peeters, A.J.M., Raz, V., and Koornneef, M. (2001). A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29 435–440. [DOI] [PubMed] [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X., and Zhang, Q. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112 1164–1171. [DOI] [PubMed] [Google Scholar]
- Faure, S., Higgins, J., Turner, A., and Laurie, D.A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie, A.R., and Schauer, N. (2009). Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 25 39–48. [DOI] [PubMed] [Google Scholar]
- Filiault, D.L., Wessinger, C.A., Dinneny, J.R., Lutes, J., Borevitz, J.O., Weigel, D., Chory, J., and Maloof, J.N. (2008). Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc. Natl. Acad. Sci. USA 105 3157–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz Gerald, J.N., Lehti-Shiu, M.D., Ingram, P.A., Deak, K.I., Biesiada, T., and Malamy, J.E. (2006). Identification of quantitative trait loci that regulate Arabidopsis root system size and plasticity. Genetics 172 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher, F., Morin, J., Courtiade, J., Cadioux, S., Ellis, N., Banfield, M.J., and Rameau, C. (2003). DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15 2742–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary, A., Nesbitt, T.C., Grandillo, S., Knaap, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K.B., and Tanksley, S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289 85–88. [DOI] [PubMed] [Google Scholar]
- Frascaroli, E., Cane, M.A., Landi, P., Pea, G., Gianfranceschi, L., Villa, M., Morgante, M., and Pe, M.E. (2007). Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics 176 625–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frewen, B.E., Chen, T.H., Howe, G.T., Davis, J., Rohde, A., Boerjan, W., and Bradshaw, H.D., Jr. (2000). Quantitative trait loci and candidate gene mapping of bud set and bud flush in populus. Genetics 154 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E., Carrari, F., Liu, Y.S., Fernie, A.R., and Zamir, D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305 1786–1789. [DOI] [PubMed] [Google Scholar]
- Fu, D., Szucs, P., Yan, L., Helguera, M., Skinner, J.S., von Zitzewitz, J., Hayes, P.M., and Dubcovsky, J. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273 54–65. [DOI] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., et al. (2009). System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat. Genet. 41 166–167. [DOI] [PubMed] [Google Scholar]
- Fujino, K., Sekiguchi, H., Matsuda, Y., Sugimoto, K., Ono, K., and Yano, M. (2008). Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 105 12623–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Oliveira, A.L., Tan, L., Fu, Y., and Sun, C. (2009). Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J. Integr. Plant Biol. 51 84–92. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Gendall, A.R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandilyan, A., Ilk, N., Hanhart, C., Mbengue, M., Barboza, L., Schat, H., Koornneef, M., El-Lithy, M., Vreugdenhil, D., Reymond, M., and Aarts, M.G.M. (2009). A strong effect of growth medium and organ type on the identification of QTLs for phytate and mineral concentrations in three Arabidopsis thaliana RIL populations. J. Exp. Bot. 60 1409–1425. [DOI] [PubMed] [Google Scholar]
- Ghandilyan, A., Vreugdenhil, D., and Aarts, M.G.M. (2006). Progress in the genetic understanding of plant iron and zinc nutrition. Physiol. Plant. 126 407–417. [Google Scholar]
- González-Martinez, S.C., Krutovsky, K.V., and Neale, D.B. (2006). Forest-tree population genomics and adaptive evolution. New Phytol. 170 227–238. [DOI] [PubMed] [Google Scholar]
- Gu, X.Y., Kianian, S.F., Hareland, G.A., Hoffer, B.L., and Foley, M.E. (2005). Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa). Theor. Appl. Genet. 110 1108–1118. [DOI] [PubMed] [Google Scholar]
- Gu, X.Y., Turnipseed, E.B., and Foley, M.E. (2008). The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics 179 2263–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, M.C., Dworkin, I., Ungerer, M.C., and Purugganan, M. (2007). Genetics of microenvironmental canalization in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne, M., Talke, I.N., Haydon, M.J., Lanz, C., Nolte, A., Motte, P., Kroymann, J., Weigel, D., and Krämer, U. (2008). Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453 391–395. [DOI] [PubMed] [Google Scholar]
- Harada, H., Kuromori, T., Hirayama, T., Shinozaki, K., and Leigh, R.A. (2004). Quantitative trait loci analysis of nitrate storage in Arabidopsis leading to an investigation of the contribution of the anion channel gene, AtCLC-c, to variation in nitrate levels. J. Exp. Bot. 55 2005–2014. [DOI] [PubMed] [Google Scholar]
- Harada, H., and Leigh, R.A. (2006). Genetic mapping of natural variation in potassium concentrations in shoots of Arabidopsis thaliana. J. Exp. Bot. 57 953–960. [DOI] [PubMed] [Google Scholar]
- Hauser, M.-T., Harr, B., and Schlötterer, C. (2001). Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: Molecular analysis of the candidate gene GLABROUS1. Mol. Biol. Evol. 18 1754–1763. [DOI] [PubMed] [Google Scholar]
- He, Y., Doyle, M.R., and Amasino, R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, M.Y., Yano, M., Goodenowe, D.B., Kanaya, S., Kimura, T., Awazuhara, M., Arita, M., Fujiwara, T., and Saito, K. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101 10205–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub, E.B. (2007). Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 10 415–424. [DOI] [PubMed] [Google Scholar]
- Hori, K., Sato, K., and Takeda, K. (2007). Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor. Appl. Genet. 115 869–876. [DOI] [PubMed] [Google Scholar]
- Imtiaz, M., Ogbonnaya, F.C., Oman, J., and van Ginkel, M. (2008). Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru, K., Kobayashi, N., Ono, K., Yano, M., and Ohsugi, R. (2001). Are contents of Rubisco, soluble protein and nitrogen in flag leaves of rice controlled by the same genetics? J. Exp. Bot. 52 1827–1833. [DOI] [PubMed] [Google Scholar]
- Isshiki, M., Morino, K., Nakajima, M., Okagaki, R.J., Wessler, S.R., Izawa, T., and Shimamoto, K. (1998). A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 15 133–138. [DOI] [PubMed] [Google Scholar]
- Izawa, T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58 3091–3097. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Konishi, S., Shomura, A., and Yano, M. (2009). DNA changes tell us about rice domestication. Curr. Opin. Plant Biol. 12 185–192. [DOI] [PubMed] [Google Scholar]
- Jaillon, O., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467. [DOI] [PubMed] [Google Scholar]
- Jansen, R.C., and Nap, J.P. (2001). Genetical genomics: The added value from segregation. Trends Genet. 17 388–391. [DOI] [PubMed] [Google Scholar]
- Jansen, R.C., Tesson, B.M., Fu, J., Yang, Y., and McIntyre, L.M. (2009). Defining gene and QTL networks. Curr. Opin. Plant Biol. 12 241–246. [DOI] [PubMed] [Google Scholar]
- Jiménez-Gomez, J.M., Alonso-Blanco, C., Borja, A., Anastasio, G., Angosto, T., Lozano, R., and Martinez-Zapater, J.M. (2007). Quantitative genetic analysis of flowering time in tomato. Genome 50 303–315. [DOI] [PubMed] [Google Scholar]
- Johannes, F., Colot, V., and Jansen, R.C. (2008). Epigenome dynamics: A quantitative genetics perspective. Nat. Rev. Genet. 9 883–890. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347. [DOI] [PubMed] [Google Scholar]
- Joppa, L.R., Du, C., Hart, G.A., and Hareland, G.A. (1997). Mapping gene(s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 37 1586–1589. [Google Scholar]
- Jorgensen, S., and Mauricio, R. (2004). Neutral genetic variation among wild North American populations of the weedy plant Arabidopsis thaliana is not geographically structured. Mol. Ecol. 13 3403–3413. [DOI] [PubMed] [Google Scholar]
- Juenger, T., Pérez-Pérez, J.M., Bernal, S., and Micol, J.L. (2005). Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: Evidence for modular genetic architecture. Evol. Dev. 7 259–271. [DOI] [PubMed] [Google Scholar]
- Juenger, T., Purugganan, M., and Mackay, T.F.C. (2000). Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics 156 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes, J.J.B. (2009). Genetical metabolomics: closing in on phenotypes. Curr. Opin. Plant Biol. 12 223–230. [DOI] [PubMed] [Google Scholar]
- Keurentjes, J.J.B., Fu, J., de Vos, C.H., Lommen, A., Hall, R.D., Bino, R.J., van der Plas, L.H.W., Jansen, R.C., Vreugdenhil, D., and Koornneef, M. (2006). The genetics of plant metabolism. Nat. Genet. 38 842–849. [DOI] [PubMed] [Google Scholar]
- Keurentjes, J.J.B., Fu, J., Terpstra, I.R., Garcia, J.M., van den Ackerveken, G., Snoek, L.B., Peeters, A.J.M., Vreugdenhil, D., Koornneef, M., and Jansen, R.C. (2007). Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 104 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes, J.J.B., Koornneef, M., and Vreugdenhil, D. (2008. a). Quantitative genetics in the age of omics. Curr. Opin. Plant Biol. 11 123–128. [DOI] [PubMed] [Google Scholar]
- Keurentjes, J.J.B., Sulpice, R., Gibon, Y., Steinhauser, M.C., Fu, J., Koornneef, M., Stitt, M., and Vreugdenhil, D. (2008. b). Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol. 9 R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, R., Kawahigashi, H., Ando, T., Tonooka, T., and Handa, H. (2009). Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, H. (2004). Biological robustness. Nat. Rev. Genet. 5 826–837. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J. (2009. a). Advancing genetic theory and application by metabolic QTL analysis. Plant Cell 21 1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J. (2009. b). Quantitative genomics: Analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu. Rev. Plant Biol. 60 93–114. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., and Mitchell-Olds, T. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., West, M.A., van Leeuwen, H., Kim, K., Doerge, R.W., Michelmore, R.W., and St Clair, D.A. (2006. a). Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., West, M.A., van Leeuwen, H., Loudet, O., Doerge, R.W., and St Clair, D.A. (2006. b). Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kuroda, K., Kimura, K., Southron-Francis, J.L., Furuzawa, A., Kimura, K., Iuchi, S., Kobayashi, M., Taylor, G.J., and Koyama, H. (2008). Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 148 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Kole, C., Quijada, P., Michael, S.D., Amasino, R., and Osborn, T.C. (2001). Evidence for homology of flowering-time genes VFR2 from Brassica rapa and FLC from Arabidopsis thaliana. Theor. Appl. Genet. 102 425–430. [Google Scholar]
- Komatsuda, T., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, S., Izawa, T., Lin, S.Y., Ebana, K., Fukuta, Y., Sasaki, T., and Yano, M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312 1392–1396. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Korves, T.M., Schmid, K.J., Caicedo, A.L., Mays, C., Stinchcombe, J.R., Purugganan, M.D., and Schmitt, J. (2007). Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 169 E141–E157. [DOI] [PubMed] [Google Scholar]
- Kramer, E.M. (2009). Aquilegia: A new model for plant development, ecology, and evolution. Annu. Rev. Plant Biol. 60 261–277. [DOI] [PubMed] [Google Scholar]
- Kroymann, J., and Mitchell-Olds, T. (2005). Epistasis and balanced polymorphism influencing complex trait variation. Nature 435 95–98. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., Niittyvuopio, A., Rinne, P., and Savolainen, O. (2008). Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol. Biol. Evol. 25 319–329. [DOI] [PubMed] [Google Scholar]
- Kusterer, B., Muminovic, J., Utz, H.F., Piepho, H.-P., Barth, S., Heckenberger, M., Meyer, R.C., Altmann, T., and Melchinger, A.E. (2007). Analysis of a triple testcross design with recombinant inbred lines reveals a significant role of epistasis in heterosis for biomass-related traits in Arabidopsis. Genetics 175 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laserna, M.P., Sanchez, R.A., and Botto, J.F. (2008). Light-related loci controlling seed germination in Ler x Cvi and Bay-0 x Sha recombinant inbred-line populations of Arabidopsis thaliana. Ann. Bot. (Lond.) 102 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre, V. (2005). Variation at two flowering time genes within and among populations of Arabidopsis thaliana: Comparison with markers and traits. Mol. Ecol. 14 4181–4192. [DOI] [PubMed] [Google Scholar]
- Le Corre, V., Roux, F., and Reboud, X. (2002). DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: Extensive nonsynonymous variation is consistent with local selection for flowering time. Mol. Biol. Evol. 19 1261–1271. [DOI] [PubMed] [Google Scholar]
- Lempe, J., Balasubramanian, S., Sureshkumar, S., Singh, A., Schmid, M., and Weigel, D. (2005). Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Zhou, A., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311 19361939. [DOI] [PubMed] [Google Scholar]
- Li, L., Strahwald, J., Hofferbert, H.-R., Lubeck, J., Tacke, E., Junghans, H., Wunder, J., and Gebhardt, C. (2005). DNA variation at the invertase locus invGE/GF is associated with tuber quality traits in populations of potato breeding clones. Genetics 170 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.K., Luo, L.J., Mei, H.W., Wang, D.L., Shu, Q.Y., Tabien, R., Zhong, D.B., Ying, C.S., Stansel, J.W., Khush, G.S., and Paterson, A.H. (2001). Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158 1737–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H.X., Zhu, M.Z., Yano, M., Gao, J.P., Liang, Z.W., Su, W.A., Hu, X.H., Ren, Z.H., and Chao, D.Y. (2004). QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 108 253–260. [DOI] [PubMed] [Google Scholar]
- Lin, S.Y., Sasaki, T., and Yano, M. (1998). Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor. Appl. Genet. 96 997–1003. [Google Scholar]
- Lisec, J., Meyer, R.C., Steinfath, M., Redestig, H., Becher, M., Witucka-Wall, H., Fiehn, O., Torjek, O., Selbig, J., Altmann, T., and Willmitzer, L. (2008). Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 53 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Van Eck, J., Cong, B., and Tanksley, S.D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 99 13302–13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Sherman-Broyles, S., Nasrallah, M.E., and Nasrallah, J.B. (2007). A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet, O., Chaillou, S., Merigout, P., Talbotec, J., and Daniel-Vedele, F. (2003). Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol. 131 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet, O., Gaudon, V., Trubuil, A., and Daniel-Vedele, F. (2005). Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor. Appl. Genet. 110 742–753. [DOI] [PubMed] [Google Scholar]
- Loudet, O., Michael, T.P., Burger, B.T., Le Metté, C., Mockler, T.C., Weigel, D., and Chory, J. (2008). A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105 17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet, O., Saliba-Colombani, V., Camilleri, C., Calenge, F., Gaudon, V., Koprivova, A., North, K.A., Kopriva, S., and Daniel-Vedele, F. (2007). Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat. Genet. 39 896–900. [DOI] [PubMed] [Google Scholar]
- Macquet, A., Ralet, M.-C., Loudet, O., Kronenberger, J., Mouille, G., Marion-Poll, A., and North, H.M. (2007). A naturally occurring mutation in an Arabidopsis accession affects a β-D-falactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19 3990–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof, J.N., Borevitz, J.O., Dabi, T., Lutes, J., Nehring, R.B., Redfern, J.L., Trainer, G.T., Wilson, J.M., Asami, T., Berry, C.C., Weigel, D., and Chory, J. (2001). Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29 441–446. [DOI] [PubMed] [Google Scholar]
- Manning, K., Tor, M., Poole, M., Hong, Y., Thompson, A.J., King, G.J., Giovannoni, J.J., and Seymour, G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38 948–952. [DOI] [PubMed] [Google Scholar]
- Melchinger, A.E., Piepho, H.-P., Utz, H.F., Muminovic, J., Wegenast, T., Torjek, O., Altmann, T., and Kusterer, B. (2007). Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics 177 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez, C.M., Ritter, E., Schäfer-Pregl, R., Walkemeier, B., Kalde, A., Salamini, F., and Gebhardt, C. (2002). Cold sweetening in diploid potato: mapping quantitative trait loci and candidate genes. Genetics 162 1423–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, P.H., Macquet, A., Loudet, O., Marion-Poll, A., and North, H. (2008). Analysis of natural allelic variation controlling Arabidopsis thaliana seed germinability in respons to cold and dral: Identification of three major quantitative trait loci. Mol. Plant 1 145–154. [DOI] [PubMed] [Google Scholar]
- Meyer, R.C., Steinfath, M., Lisec, J., Becher, M., Witucka-Wall, H., Torjek, O., Fiehn, O., Eckardt, A., Willmitzer, L., Selbig, J., and Altmann, T. (2007). The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., He, Y., Scortecci, K.C., and Amasino, R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds, T., and Pedersen, D. (1998). The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds, T., and Schmitt, J. (2006). Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441 947–952. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds, T., Willis, J.H., and Goldstein, D.B. (2007). Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 8 845–856. [DOI] [PubMed] [Google Scholar]
- Morgante, M., De Paoli, E., and Radovic, S. (2007). Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 10 149–155. [DOI] [PubMed] [Google Scholar]
- Mouchel, C.L.F., Briggs, G.C., and Hardtke, C.S. (2004). Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles, S., Peiffer, J., Brown, P., Ersoz, E., Zhang, Z., Costich, D.E., and Buckler, E.S. (2009). Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21 in press. [DOI] [PMC free article] [PubMed]
- Nordborg, M., and Weigel, D. (2008). Next-generation genetics in plants. Nature 456 720–723. [DOI] [PubMed] [Google Scholar]
- O'Neill, C., Morgan, C., Kirby, J., Tschoep, H., Deng, P., Brennan, M., Rosas, U., Fraser, F., Hall, C., Gill, S., and Bancroft, I. (2008). Six new recombinant inbred populations for the study of quantitative traits in Arabidopsis thaliana. Theor. Appl. Genet. 116 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.M., Halldorsdottir, S.S., Stinchcombe, J.R., Weinig, C., Schmitt, J., and Purugganan, M.D. (2004). Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.M., and Purugganan, M.D. (2002). Molecular evidence on the origin and evolution of glutinous rice. Genetics 162 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi, C.H., and Tanksley, S.D. (2009). Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genet. 5 e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, D.R., and Grossniklaus, U. (2002). The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 3 124–136. [DOI] [PubMed] [Google Scholar]
- Panthee, D., Pantalone, V., Sams, C., Saxton, A., West, D., Orf, J., and Killam, A. (2006). Quantitative trait loci controlling sulfur containing amino acids, methionine and cysteine, in soybean seeds. Theor. Appl. Genet. 112 546–553. [DOI] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez, J.M., Serrano-Cartagena, J., and Micol, J.L. (2002). Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162 893–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picó, F.X., Mendez-Vigo, B., Martinez-Zapater, J.M., and Alonso-Blanco, C. (2008). Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics 180 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potokina, E., Druka, A., Luo, Z., Moscou, M., Wise, R., Waugh, R., and Kearsey, M. (2008. a). Tissue-dependent limited pleiotropy affects gene expression in barley. Plant J. 56 287–296. [DOI] [PubMed] [Google Scholar]
- Potokina, E., Druka, A., Luo, Z., Wise, R., Waugh, R., and Kearsey, M. (2008. b). Gene expression quantitative trait locus analysis of 16,000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. Plant J. 53 90–101. [DOI] [PubMed] [Google Scholar]
- Pourkheirandish, M., and Komatsuda, T. (2007). The importance of barley genetics and domestication in a global perspective. Ann. Bot. (Lond.) 100 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M.D., and Fuller, D.Q. (2009). The nature of selection during plant domestication. Nature 457 843–848. [DOI] [PubMed] [Google Scholar]
- Queitsch, C., Sangster, T.A., and Lindquist, S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417 618–624. [DOI] [PubMed] [Google Scholar]
- Radoev, M., Becker, H.C., and Ecke, W. (2008). Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Z.-H., Gao, J.-P., Li, L.-G., Cai, X.-L., Huang, W., Chao, D.-Y., Zhu, M.-Z., Wang, Z.-Y., Luan, S., and Lin, H.-X. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 1141–1146. [DOI] [PubMed] [Google Scholar]
- Riddle, N.C. and Richards, E.J. (2005). Genetic variation in epigenetic inheritance of ribosomal RNA gene methylation in Arabidopsis. Plant J. 41 524–532. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H., Raymond, O., Rosenthal, D.M., Lai, Z., Livingstone, K., Nakazato, T., Durphy, J.L., Schwarzbach, A.E., Donovan, L.A., and Lexer, C. (2003). Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301 1211–1216. [DOI] [PubMed] [Google Scholar]
- Roux, F., Touzet, P., Cuguen, J., and Le Corre, V. (2006). How to be early flowering: An evolutionary perspective. Trends Plant Sci. 11 375–381. [DOI] [PubMed] [Google Scholar]
- Rowe, H.C., Hansen, B.G., Halkier, B.A., and Kliebenstein, D.J. (2008). Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus, A., Baxter, I., Muthukumar, B., Gustin, J., Lahner, B., Yakubova, E., and Salt, D.E. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T. and Matsuoka, M. (2008). Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11 209–214. [DOI] [PubMed] [Google Scholar]
- Salamini, F., Ozkan, H., Brandolini, A., Schafer-Pregl, R., and Martin, W. (2002). Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 3 429–441. [DOI] [PubMed] [Google Scholar]
- Salt, D.E., Baxter, I., and Lahner, B. (2008). Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59 709–733. [DOI] [PubMed] [Google Scholar]
- Salvi, S., et al. (2007). Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, T. (2009). Genes and mutations underlying domestication transitions in grasses. Plant Physiol. 149 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster, T.A., Salathia, N., Undurraga, S., Milo, R., Schellenberg, K., Lindquist, S., and Queitsch, C. (2008). HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc. Natl. Acad. Sci. USA 105 2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli, N., Cheverud, J.M., Schaal, B.A., and Kover, P.X. (2007). Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus. Proc. Natl. Acad. Sci. USA 104 16986–16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E.E., et al. (2003). Genetics of gene expression surveyed in maize, mouse and man. Nature 422 297–302. [DOI] [PubMed] [Google Scholar]
- Schauer, N., Semel, Y., Balbo, I., Steinfath, M., Repsilber, D., Selbig, J., Pleban, T., Zamir, D., and Fernie, A.R. (2008). Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer, N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24 447–454. [DOI] [PubMed] [Google Scholar]
- Schläppi, M.R. (2006). FRIGIDA LIKE 2 is a functional allele in Landsberg erecta and compensates for a nonsense allele of FRIGIDA LIKE 1. Plant Physiol. 142 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K.J., Torjek, O., Meyer, R., Schmuths, H., Hoffmann, M.H., and Altmann, T. (2006). Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 112 1104–1114. [DOI] [PubMed] [Google Scholar]
- Schranz, M.E., Quijada, P., Sung, S.B., Lukens, L., Amasino, R., and Osborn, T.C. (2002). Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel, Y., Nissenbaum, J., Menda, N., Zinder, M., Krieger, U., Issman, N., Pleban, T., Lippman, Z., Gur, A., and Zamir, D. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103 12981–12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva, L.I., Keurentjes, J.J., Bentsink, L., Vonk, J., van der Plas, L.H., Koornneef, M., and Vreugdenhil, D. (2006). Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc. Natl. Acad. Sci. USA 103 2994–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva, L.I., Vonk, J., Keurentjes, J.J.B., van der Plas, L.H.W., Koornneef, M., and Vreugdenhil, D. (2004). Histochemical analysis reveals organ-specific quantitative trait loci for enzyme activities in Arabidopsis. Plant Physiol. 134 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C., Aranzana, M.J., Lister, C., Baxter, C., Nicholls, C., Nordborg, M., and Dean, C. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C., Bernasconi, G., and Hardtke, C.S. (2008). Intraspecific competition reveals conditional fitness effects of single gene polymorphism at the Arabidopsis root growth regulator BRX. New Phytol. 180 71–80. [DOI] [PubMed] [Google Scholar]
- Shindo, C., Lister, C., Crevillen, P., Nordborg, M., and Dean, C. (2006). Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20 3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K.J., Fellers, J.P., Trick, H.N., Zhang, Z., Tai, Y.S., and Faris, J.D. (2006). Molecular characterization of the major wheat domestication gene Q. Genetics 172 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and Messing, J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z., and Lin, H.X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 623–630. [DOI] [PubMed] [Google Scholar]
- Stenoien, H.K., Fenster, C.B., Tonteri, A., and Savolainen, O. (2005). Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Mol. Ecol. 14 137–148. [DOI] [PubMed] [Google Scholar]
- Stinchcombe, J.R., Weinig, C., Ungerer, M., Olsen, K.M., Mays, C., Halldorsdottir, S.S., Purugganan, M.D., and Schmitt, J. (2004). A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. USA 101 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2005). Remembering winter: Toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56 491–508. [DOI] [PubMed] [Google Scholar]
- Sureshkumar, S., Todesco, M., Schneeberger, K., Harilal, R., Balasubramanian, S., and Weigel, D. (2009). A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323 1060–1063. [DOI] [PubMed] [Google Scholar]
- Sutton, T., Baumann, U., Hayes, J., Collins, N.C., Shi, B.-J., Schnurbush, T., Hay, A., Mayo, G., Pallotta, M., Tester, M., and Langridge, P. (2007). Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318 1446–1449. [DOI] [PubMed] [Google Scholar]
- Svistoonoff, S., Creff, A., Reymond, M., Sigoillot-Claude, C., Ricaud, L., Blanchet, A., Nussaume, L., and Desnos, T. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39 792–796. [DOI] [PubMed] [Google Scholar]
- Sweeney, M.T., Thomson, M.J., Pfeil, B.E., and McCouch, S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szucs, P., Skinner, J.S., Karsai, I., Cuesta-Marcos, A., Haggard, K.G., Corey, A.E., Chen, T.H., and Hayes, P.M. (2007). Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol. Genet. Genomics 277 249–261. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Shomura, A., Sasaki, T., and Yano, M. (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 98 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa, S., et al. (2008). Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. USA 105 4062–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16 S181–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S., Rognoni, S., Bentsink, L., and Smeekens, S. (2008). The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J. 55 372–381. [DOI] [PubMed] [Google Scholar]
- Tian, D., Traw, M.B., Chen, J.Q., Kreitman, M., and Bergelson, J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423 74–77. [DOI] [PubMed] [Google Scholar]
- Tohge, T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Toomajian, C., Hu, T.T., Aranzana, M.J., Lister, C., Tang, C., Zheng, H., Zhao, K., Calabrese, P., Dean, C., and Nordborg, M. (2006). A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4 e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A., Beales, J., Faure, S., Dunford, R.P., and Laurie, D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034. [DOI] [PubMed] [Google Scholar]
- Tuskan, G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, D., Kono, I., Yokosho, K., Ando, T., Yano, M., and Ma, J.F. (2009). A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol. 182 644–653. [DOI] [PubMed] [Google Scholar]
- van der Schaar, W., Alonso-Blanco, C., Leon-Kloosterziel, K.M., Jansen, R.C., an Ooijen, J.W., and Koornneef, M. (1997). QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 79 190–200. [DOI] [PubMed] [Google Scholar]
- Varagona, M.J., Purugganan, M., and Wessler, S.R. (1992). Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, M.W., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5 e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreugdenhil, D., Aarts, M.G.M., Koornneef, M., Nelissen, H., and Ernst, W.H.O. (2004). Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ. 27 828–839. [Google Scholar]
- Vuylsteke, M., Daele, H., Vercauteren, A., Zabeau, M., and Kuiper, M. (2006). Genetic dissection of transcriptional regulation by cDNA-AFLP. Plant J. 45 439–446. [DOI] [PubMed] [Google Scholar]
- Vuylsteke, M., van Eeuwijk, F., Van Hummelen, P., Kuiper, M., and Zabeau, M. (2005). Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 171 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Nussbaum-Wagler, T., Li, B., Zhao, Q., Vigouroux, Y., Faller, M., Bomblies, K., Lukens, L., and Doebley, J.F. (2005). The origin of the naked grains of maize. Nature 436 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Sajja, U., Rosloski, S., Humphrey, T., Kim, M.C., Bomblies, K., Weigel, D., and Grbic, V. (2007). HUA2 caused natural variation in shoot morphology of A. thaliana. Curr. Biol. 17 1513–1519. [DOI] [PubMed] [Google Scholar]
- Waters, B.M., and Grusak, M.A. (2008). Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia, Landsberg erecta, Cape Verde Islands, and the mutant line ysl1ysl3. New Phytol. 177 389–405. [DOI] [PubMed] [Google Scholar]
- Waugh, R., Jannink, J.-L., Muehlbauer, G.J., and Ramsay, L. (2009). The emergence of whole genome association scans in barley. Curr. Opin. Plant Biol. 12 218–222. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nordborg, M. (2005). Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol. 138 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, J., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18 1199–1209. [DOI] [PubMed] [Google Scholar]
- Wentzell, A.M., Rowe, H.C., Hansen, B.G., Ticconi, C., Halkier, B.A., and Kliebenstein, D.J. (2007). Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 3 e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.D., Borevitz, J.O., Uhlenhaut, N.H., Ecker, J.R., Chory, J., and Weigel, D. (2005. a). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.D., Borevitz, J.O., Warthmann, N., Trainer, G.T., Ecker, J.R., Chory, J., and Weigel, D. (2005. b). Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 102 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M.A., Kim, K., Kliebenstein, D.J., van Leeuwen, H., Michelmore, R.W., Doerge, R.W., and St Clair, D.A. (2007). Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek, A.M., et al. (2009). Effects of genetic perturbation on seasonal life history plasticity. Science 323 930–934. [DOI] [PubMed] [Google Scholar]
- Wissuwa, M., Wegner, J., Ae, N., and Yano, M. (2002). Substitution mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet. 105 890–897. [DOI] [PubMed] [Google Scholar]
- Wu, C.A., Lowry, D.B., Cooley, A.M., Wright, K.M., Lee, Y.W., and Willis, J.H. (2007). Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100 220–230. [DOI] [PubMed] [Google Scholar]
- Wu, J., Yuan, Y.-X., Zhang, X.-W., Zhao, J., Song, X., Li, Y., Li, X., Sun, R., Koornneef, M., Aarts, M., and Wang, X.-W. (2008). Mapping QTLs for mineral accumulation and shoot dry biomass under different Zn nutritional conditions in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Soil 310 25–40. [Google Scholar]
- Xiao, H., Jiang, N., Schaffner, E., Stockinger, E.J., and van der Knaap, E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319 1527–1530. [DOI] [PubMed] [Google Scholar]
- Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., Zhou, H., Yu, S., Xu, C., Li, X., and Zhang, Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40 761–767. [DOI] [PubMed] [Google Scholar]
- Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., Sanchez, A., Valarik, M., Yasuda, S., and Dubcovsky, J. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Helguera, M., Kato, K., Fukuyama, S., Sherman, J., and Dubcovsky, J. (2004. a). Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 109 1677–1686. [DOI] [PubMed] [Google Scholar]
- Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., Bennetzen, J.L., Echenique, V., and Dubcovsky, J. (2004. b). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y.X., Wu, J., Sun, R.F., Zhang, X.W., Xu, D.H., Bonnema, G., and Wang, X.W. (2009). A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J., Liu, J., Liu, B., Li, P., Meyers, B.C., Chen, X., and Cao, X. (2008). Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 4 e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Chen, P., Shi, A., Hou, A., Ishibashi, T., and Wang, D. (2009). Putative quantitative trait loci associated with calcium content in soybean seed. J. Hered. 100 263–269. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006. b). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Jamar, D.C.L., Lou, P., Wang, Y., Wu, J., Wang, X., Bonnema, G., Koornneef, M., and Vreugdenhil, D. (2008). Quantitative trait loci analysis of phytate and phosphate concentrations in seeds and leaves of Brassica rapa. Plant Cell Environ. 31 887–900. [DOI] [PubMed] [Google Scholar]