Abstract
Previous work has shown that the essential R210 of subunit a in the E. coli ATP synthase can be switched with a conserved glutamine Q252 with retention of a moderate level of function, that a third mutation P204T enhances this function, and that the arginine Q252R can be replaced by lysine without total loss of activity. In this study, the roles of P204T and R210Q were examined. It was concluded that the threonine in P204T is not directly involved in function since its replacement by alanine did not significantly affect growth properties. Similarly, it was concluded that the glutamine in R210Q is not directly involved with function since replacement by glycine results in significantly enhanced function. Not only did the rate of ATP-driven proton translocation increase, but also the sensitivity of ATP hydrolysis to inhibition by N,N′-dicyclohexylcarbdiimide (DCCD) rose to more than 50%. Finally, mutations at position E219, a residue near the proton pathway, were used to test whether the arginine-switched mutant uses the normal proton pathway. In a wild type background, the E219K mutant was confirmed to have greater function than the E219Q mutant, as has been shown previously. This same unusual result was observed in the triple mutant background, P204T/R210Q/Q252R, suggesting that the arginine-switched mutants are using the normal proton pathway from the periplasm.
Keywords: ATP synthase, F1Fo, subunit a, proton translocation, rotary motor, ATPase
1. Introduction
During oxidative or photo-phosphorylation ATP is synthesized by the rotary enzyme ATP synthase. This enzyme is found in the membranes of mitochondria, chloroplasts, and eubacteria (For reviews see [1–3]). It is composed of two parts: an F1 sector that houses nucleotide-binding catalytic sites, and an Fo sector that conducts protons across the membrane. Each functions as a rotary motor, which allows an electrochemical ion gradient across the membrane to drive ATP synthesis, and in some situations, ATP hydrolysis by F1 is used to generate the gradient. The latter situation can arise, not only in vitro, but also if the electrochemical gradient is too low. The sectors are connected by two stalks. The central stalk consists of rotary subunits, and the peripheral stalk consists of stator subunits. In Escherichia coli, the enzyme consists of eight distinct types of subunits: F1 is formed by α3β3γδε and Fo is formed by ab2c10 [4, 5].
During ATP synthesis in E. coli, protons travel from the periplasm, through subunit a to Asp 61 of one c subunit. The c subunits are arranged in a 10-fold symmetrical ring with the conserved acidic residue situated near the center of the transmembrane region. This protonation leads to rotation of the c-oligomer, which is tightly attached to the γ and ε subunits of F1. The rotor is formed from these three types of subunits. The γ subunit contains two long α-helices that form a shaft. This shaft is surrounded by the pseudo-symmetrical hexamer of alternating α and β subunits. There are three catalytic sites, which are composed of primarily by β subunits. Rotation of the asymmetric γ subunit contributes to a series of conformational changes at the catalytic sites, which are necessary for the binding of ADP and Pi, formation of ATP, and release of product. This sequential process is described by the binding-change mechanism [6, 7]. The catalytic subunits are connected to subunit a through the peripheral stalk, which contains δ and two b subunits. The δ subunit is connected to one or more α subunits at the membrane-distal end, and the two b subunits are connected to subunit a in and near the membrane. These subunits form the stator, against which the rotary subunits, γ-ε-c10, rotate.
Details of the nucleotide binding sites, and the conformational changes that they undergo, are largely known from a series of crystallographic studies [8–13], and from intrinsic fluorescent probes [14–16]. Further insights into the relationship of substrate binding, product release, and subunit rotation have been provided by a series of single-molecule studies [17–22]. Much less information is available about the Fo motor, because no high resolution structure of an intact Fo has been solved.
Proton translocation is thought to occur exclusively through a and c subunits. In E. coli subunit a is encoded as a single polypeptide of 271 amino acids. It folds in the membrane with five transmembrane spans, with the amino terminus in the periplasm and the carboxy terminus in the cytoplasm [23–25]. In E. coli subunit c is encoded as a polypeptide of 79 amino acids, and it folds in the membrane as two anti-parallel alpha-helices with both termini in the periplasm. It forms a ring from ten monomers and interacts with subunit a at its outer surface [26, 27]. Subunit a was predicted to have two off-set half-channels that guide protons to and from the c subunits at the a-c interface [28, 29]. Recent work suggests that the periplasmic half-channel is within subunit a, but that the cytoplasmic half-channel is at the interface of subunit a and the c oligomer [30–33].
The work in this study addresses the role of the essential Arg 210 residue in subunit a. Arg 210 is found in the fourth transmembrane span (TM4), near conserved polar residues N214 (TM4) and Q252 (TM5). It has an essential interaction with Asp 61 of subunit c, and it might also interact with other conserved residues in subunit a when the c-oligomer undergoes rotation [34]. It was discovered [35] that the conserved arginine could be switched with the conserved glutamine to make a functional double mutant: R210Q/Q252R. A spontaneous mutation P204T improved the function of the ATP synthase. ATP synthesis by these mutants was demonstrated in a previous publication by this lab [36], and it was shown that lysine could substitute for the arginine in the switched construct. Since the level of function was not high, it was not clear if it was important for the essential arginine to be a part of TM4, and if it functioned through the normal proton pathway. These questions were addressed in the current study.
2. Materials and Methods
2.1 Materials
Synthetic oligonucleotides were purchased from Operon Technologies, Inc (Huntsville AL USA). Restriction enzymes, T4 DNA ligase, and buffers were purchased from New England Biolabs, Inc (Beverly MA, USA). DNA sequencing of plasmid DNA was performed by Lone Star Labs, Inc. (Houston TX, USA). Plasmid DNA was purified from bacteria through use of kits from Qiagen USA (Valencia CA, USA) or MO BIO Laboratories (Carlsbad CA, USA), Inc. Mouse anti-HA antibodies were supplied by Roche (Indianapolis IN, USA) for Western blots. BCIP (5-bromo-4-chloro-3-indoylphosphate-toluidine salt), NBT (p-nitro blue tetrazolium chloride), SDS-polyacrylamide gels, and low range and broad range protein molecular weight standards were obtained from Bio-Rad (Hercules CA, USA). All other chemicals were purchased from Sigma-Aldrich or Fisher.
2.2 Plasmids, mutagenesis, growth and expression
Mutations were constructed in the pTW1-HisHA plasmid, which codes for subunit a with both an HA and a His6 tag at the carboxyl-terminus. The hemaglutinin epitope, YPYDVPDYA (derived from hemaglutinin protein of the human influenza virus), was used for immuno-detection of subunit a by anti-HA antibodies. This plasmid complements strain RH305, which has a mutation in the gene for subunit a thereby fails to express it. Mutagenesis of subunit a in pTW1-HisHA was performed by the Quikchange mutagenesis kit (La Jolla CA, USA). Mutated plasmids were transferred into XL1-Blue host competent cells and grown in LB media/plates with chloramphenicol selection. Transformation included heat shocking the XL1 cells by means of a chemical transformation in a 42°C water bath. LB media consisted of 10g/L Tryptone, 5g/L yeast extract, and 5g/L NaCl.
For succinate analysis the mutated pTW1-HisHA plasmids were transformed into RH305 competent cells and growth on succinate minimal medium (0.2% succinate) plates was assayed [37]. Some mutations were transferred from pTW1-HisHA to the larger pFV2-HA, a 9.2 kb plasmid that contains all genes necessary for the ATP synthase enzyme, including an HA tag at the carboxy-terminus of subunit a. The HA tag was introduced by Quikchange mutagenesis using the following oligonucleotide, and its complement (BspE I site underlined):
5′-GTCTGAAGAACATTACCCTTACGACGTTCCGGATTACGCTTAATTTACCAACAC. It generates the seven-residue HA tag (in italics) following the terminal residue His 271: 268-SEEHYPYDVPDYA. A 707 bp fragment from pTW1-HisHA was purified following digestion with PflM I and BspE I (at the HA site). This fragment was ligated to a 8.5 kb fragment from pFV2-HA using the same enzymes. Mutations in pFV2-HA were transformed into the DK8 host competent cells, which lack all genes for uncB-C and thus lack the ATP synthase [38]. For succinate growth analysis DK8 cells were supplemented with 200mM of each isoleucine, leucine, and valine. Growth yield was measured in minimal A media supplemented with 0.2% glucose [37]. For the growth yield analysis 50 mL of glucose minimal media was inoculated with 1 mL of overnight culture, the cultures were incubated in a shaker at 37 °C, and optical density measurements were taken at A600 every half hour until the growth level reached a plateau.
Membrane vesicles were prepared from E. coli cells using a French pressure cell. The cells were grown in LB medium at 37°C until A600 = 1.0. The cells were harvested and centrifuged at 8000 rpm for 15 m using a Beckman JA20 rotor at 4°C. The cell pellet was then resuspended in French Press buffer, (50 mM Tris-Cl, 10 mM MgSO4, pH 7.5) by homogenizing in a 10 mL glass homogenizer. Once resuspended, the cells were passed through the French press under a pressure of 14,000 psi (96.5 MPa). The samples were centrifuged again at 8000 rpm for 15 min. The supernatant was collected and centrifuged at 50,000 rpm for 1 h at 4°C in a Beckman Ti 70.1 rotor. The resulting pellet was resuspended in the French Press buffer, and immediately passed through a 5 ml G-50 Sephadex column. The eluate collected from the column was then spun down in a Beckman Coulter Optima TLX ultracentrifuge using the rotor TLA 100.3, at 100,000 rpm for 1 h at 4°C. The pellet was collected and resuspended in the French Press buffer. This resulting suspension of inverted membrane vesicles was used for performing assays.
2.3 Immunoblotting
Membrane proteins obtained by the method described above were used for immunoblotting. 60 µg of protein was used per sample, following a Bio-Rad DCA protein assay using bovine albumin serum as a protein standard. These samples were incubated with 2% weight/volume sodium dodecylsulfate for 20 minutes at 37°C and then pipetted into wells of 12% acrylamide gels. The samples were electrophoresed at 150 V for 1 h, and then transferred to a nitrocellulose PVDF membrane sheet using a Trans-Blot apparatus (Bio-Rad) at 100 V for 1hr. The membranes were washed and developed as previously described, using mouse anti-HA antibodies and an alkaline phosphatase assay.
2.4 Functional assays
Proton translocation by inverted membranes was measured by the fluorescence quenching of ACMA (9-amino 3-chloro 2-methoxy acridine) after addition of NADH or ATP. The amount of protein used was approximately 500 µg in 2 mL of solution (50 mM MOPS, pH 7.3, 10 mM MgCl2, 1 µM ACMA, and 0.5 mM ATP or 0.5 mM NADH). The excitation wavelength was 410 nm and the emission wavelength was 490 nm. Once the level of fluorescence reached a plateau, 0.1 mM FCCP was added, resulting in a sharp increase in fluorescence back to almost the starting point. To demonstrate that the membranes were not leaky to protons, NADH was added in place of the ATP to measure the rate of quenching. For all mutants, the level of quenching was nearly identical to the wild type (data not shown). Sensitivity of the quenching to treatment of membranes with DCCD (N, N′-dicyclohexyl carbodiimide) was also measured. 20 µM DCCD was added to each sample cuvette and incubated for 30 min at room temperature prior to the assay. Rates of ATP hydrolysis were measured using a coupled enzyme assay. For each assay 80 µg of inverted membranes were used in a volume of 1 mL. The reaction medium was 50 mM Tris-Cl ,12 mM MgSO4 , pH 7.7, 20 U each of lactate dehydrogenase and pyruvate kinase, and 10 mM KCN. To initiate the reaction 10 mM ATP and 0.28 mM NADH were added.
3. Results
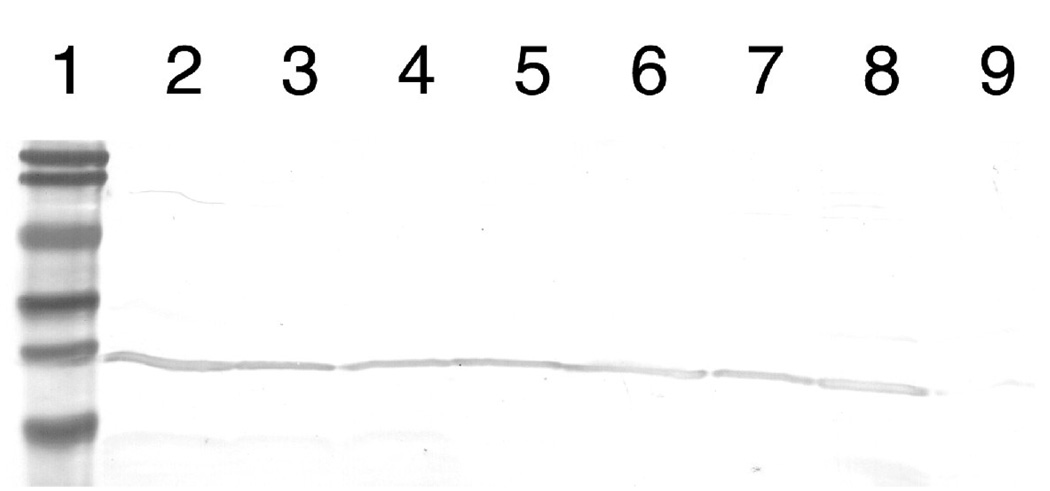
Mutations in subunit a were constructed at three different positions, in either a wild type background, or in an “Arginine-switched” background that includes the essential arginine residue relocated from position 210 to position 252. All mutations were constructed initially in pTW1-HisHA, a plasmid that encodes only subunit a, or in a variant of pTW1-HisHA that contained a mutant allele of subunit a. Initial functional analysis, which was assessment of growth on succinate minimal medium, was done after transforming RH305, a strain that lacks subunit a expression. All mutations were transferred to pFV2-HA, a plasmid that encodes all eight subunits of the ATP synthase, with an HA-epitope tag at the C-terminus of subunit a, and a His-tag at the N-terminus of the β subunit. These plasmids were used to transform strain DK8, which lacks genes for all eight subunits of the ATP synthase [38]. Immunoblots from membrane preparations of all of the mutants described in this study, using an anti-HA antibody, are shown in Fig. 1. All showed a level of expression of subunit a similar to that of the wild type.
Figure 1.
Immunoblots of membrane proteins from mutants. Subunit a was detected by a mouse anti-HA antibody. 60 µg of protein was loaded per lane. All mutations are found in the pFV2-HA plasmid. Lane 1, molecular weight standards: 104 kD, 83 kD, 49 kD, 36 kD, 28 kD, and 19 kD. Lane 2 - pFV2 wild type subunit a, lane 3 -P204T/R210Q/Q252R, lane 4 - P204T/R210G/Q252R, lane 5 - E219K in background of P204T/R210Q/Q252R, lane 6 - E219Q in background of P204T/R210Q/Q252R, lane 7 -E219K in wild type background, lane 8 - E219Q in wild type background, and lane 9 – strain DK8 (no plasmid).
As originally described by Hatch et al. [35], the P204T mutation was discovered as a spontaneous partial suppressor mutation in the background of the “Arginine-switched” construct R210Q/Q252R, because it enhanced growth properties on succinate minimal medium. This finding was confirmed recently by Ishmukhametov et al. [36]. The basis of the enhanced function of the P204T mutation in the context of the “Arginine-switched” background had not been addressed previously. To probe the role of the P204T substitution in the context of R210Q/Q252R, two new mutations were constructed: P204A/R210Q/Q252R and P204S/R210Q/Q252R. The results of growth analysis on succinate minimal medium, and growth yield measurements on limiting glucose medium are presented in Table 1. Visual inspection of growth on succinate minimal plates indicated that all three triple mutants grew at a similar rate, and all were superior to the double mutant R210Q/Q2525R. In the growth yield analysis on limiting glucose, all three triple mutants had a similar growth yield, relative to the wild type. The results suggest that the threonine residue is not essential for the observed enhancement of growth, but that it might be somewhat superior to other small amino acids.
TABLE 1.
Analysis of subunit a mutants used in this study
| Mutation | Growth on succinate minimal medium1 |
Growth yield on limiting glucose-minimal medium2 |
ATP Hydrolysis (µmoles/min/mg protein) 3 |
Sensitivity to DCCD4 |
|---|---|---|---|---|
| wild type | ++++ | 100 | 0.80 ± 0.04 | 70 ± 7 % |
| P204T/R210Q/Q252R | ++ | 63± 4 | 0.21 ± 0.01 | 11 ± 3 % |
| P204A/R210Q/Q252R | ++ | 61± 1 | ND5 | ND |
| P204S/R210Q/Q252R | ++ | 59 ± 2 | ND | ND |
| R210Q/Q252R | + | 57 ± 2 | ND | ND |
| P204T/R210G/Q252R | +++ | 79 ± 2 | 0.51 ± 0.02 | 53 ± 3 % |
| E219K | +++ | 80 ± 2 | 0.81 ± 0.19 | ND |
| E219Q | − | 52 ± 2 | 0.23 ± 0.02 | ND |
| E219K (P204T/R210Q/Q252R) | + | 55 ± 2 | 0.23 ± 0.03 | ND |
| E219Q (P204T/R210Q/Q252R) | − | 51 ± 2 | 0.23 ± 0.02 | ND |
| wild type | ++++ | 100 | 0.80 ± 0.04 | 70 ± 7 % |
| null | − | 49 ± 1 | ND | ND |
For growth on succinate minimal medium, the mutations were constructed in the pTW1-HisHA plasmid, which encodes only subunit a, and the background strain was RH305, which fails to express subunit a. RH305 without a plasmid represents the null sample, and RH305 with pTW1-HisHA represents the wild type. The size of the colonies after growth for 48 h at 37 °C is indicated by the number of + signs. The – sign indicates no growth.
Growth yield measurements were done in Minimal A medium with limiting glucose (6 mM), as measured by absorbance of liquid culture at 600 nm. For these measurements, the mutations were transferred to the pFV2-HA plasmid, which encodes all of the ATP synthase structural genes including an HA epitope-tagged subunit a. The background strain was DK8, which is deleted for the 8 structural genes of the atp operon. DK8 with plasmid pFV2-HA that has an internal deletion in the gene for subunit a from the 2 BamH I sites, represents the null sample, and DK8 with pFV2-HA represents the wild type.
Rates were determined from two independent membrane preparations. The mean and the standard error are indicated.
DCCD sensitivity reflects the remaining fraction of activity after an incubation with 20 µM DCCD for 30 minutes at 23°C.
ND, Not determined.
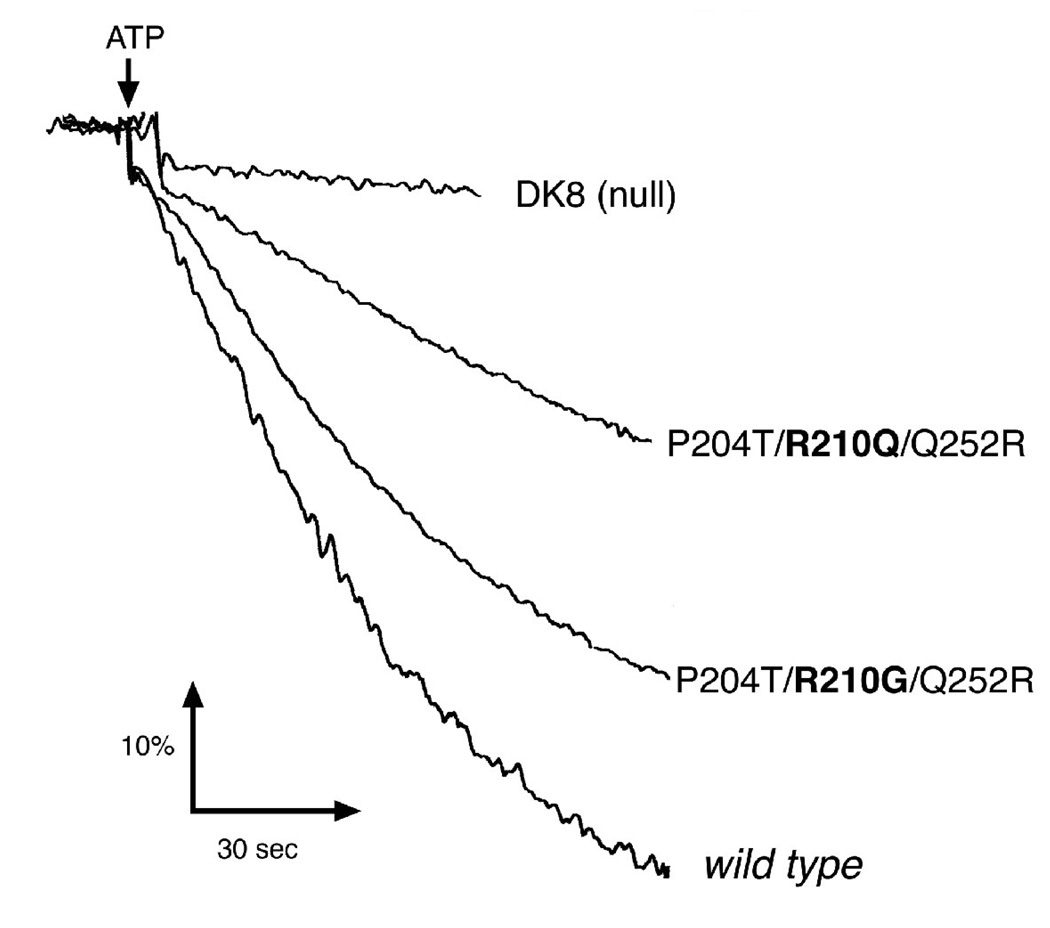
The role of R210Q in the context of the “Arginine-switched” mutant P204T/R210Q/Q252R was examined. Saturation mutagenesis was applied to position 210 while retaining P204T and Q252R. Initially, several mutations constructed in pTW1-HisHA, and identified by DNA sequencing, were used to transform strain RH305. Of this group, only glycine and isoleucine permitted growth on succinate minimal medium, indicating an active ATP synthase. Since the glycine construct (P204T/R210G/Q252R) showed greater growth on succinate than did the original mutant (P204T/R210Q/Q252R), this triple mutation was transferred to pFV2-HA for further analysis, and the results are shown in Table 1. In addition to enhanced growth on succinate, the growth yield of the R210G mutant was 80% of wild type, as compared to the original R210Q mutant, which was only 63%. These results suggested that the glutamine residue in the “Arginine-switched” construct was not important for function, since replacement by glycine enhanced growth properties. To further investigate this, membrane vesicles were prepared from cells bearing both sets of triple mutations: P204T/R210Q/Q252R and P204T/R210G/Q252R. ATP-driven proton translocation assays were performed with each, and compared to the wild type, as shown in Fig. 2. As was shown previously in this lab, membranes from the P204T/R210Q/Q252R mutant showed modest fluorescence quenching of the acridine dye, ACMA, indicating proton translocation. By comparison, membranes from the P204T/R210G/Q252R mutant were much more similar to the wild type rate. NADH-driven proton translocation rates were similar in all cases, indicating that membranes from the mutants were not particularly leaky to protons. In addition, fluorescence quenching was totally sensitive to pretreatment of the membranes with DCCD, and to addition of a classical uncoupler, FCCP, to the assay chamber (not shown).
Figure 2.
Proton Translocation Assays of R210G. Fluorescence quenching of ACMA is shown as a function of time, initiated by the addition of ATP to membrane vesicles. Wild type is DK8/pFV2-HA. DK8 is the atp deletion strain. The two triple mutants are found in the pFV2-HA plasmid. The four samples all showed similar rates of quenching in response to the addition of NADH, and the ATP-dependent quenching of all were inhibited by pretreatment with 20 µM DCCD, and the quenching returned to the initial level upon addition of FCCP (not shown). Other details are in the Materials and Methods.
Finally, the P204T/R210G/Q252R mutant was tested for rates of ATP hydrolysis in membrane vesicles. A striking feature of the original Arginine-switched mutant, P204T/R210Q/Q252R, was that ATP hydrolysis was inhibited by DCCD to a very small extent, no more than 5%, as shown previously. In Table 1 a similarly low level of DCCD sensitivity is shown for this mutant. In contrast, the sensitivity of the glycine mutant (P204T/R210G/Q252R) is 53% as compared to the typical wild type level of 70%. The low rates of ATP hydrolysis by membranes containing the original arginine-switched mutant (P204T/R210Q/Q252R) are not entirely due to lack of binding F1. In a previous paper [36] we showed that in the presence of the detergent lauryl dimethylamine N-oxide, which releases F1 from the membranes, the rate of ATP hydrolysis increased to 60–70% of the wild type level.
The last question addressed in this study of the Arginine-switched mutant was whether it used the normal proton pathway. Protons are thought to enter Fo through a channel formed by three transmembrane helices in subunit a, before protonating Asp 61 of subunit c. One residue of subunit a, Glu 219, is near this channel [30], and it is also known that mutations of this residue have distinct phenotypes. The mutant E219Q cannot grow on succinate minimal medium, and membranes show little or no ATP-driven proton translocation [39]. In contrast, E219K, exhibits nearly wild type behavior [29, 40, 41]. Therefore, these two mutations were constructed in the backgrounds of both wild type and P204T/R210Q/Q252R. The results of growth analysis of these mutants are presented in Table 1. Consistent with previous results, the E219K mutant grows well on a succinate minimal medium, and has a growth yield of 80% of the wild type. The E219Q mutant does not grow on succinate medium, and has a growth yield of 52% of the wild type, only marginally more than the null strain (49%). These mutations behave in a similar fashion in the background of P204T/R210Q/Q252R. The E219K mutant grows slowly on a succinate medium, while the E219Q mutant does not grow. The growth yields on minimal glucose are 55% and 51%, respectively, compared to 63% for the background strain. In addition, the rates of ATP hydrolysis were measured in membrane vesicles. In this case, both the wild type and the single mutant E219K had similar rates of about 0.8 µmoles ATP hydrolyzed per min per mg protein. The single mutant E219Q, and the mutants from the P204T/R210Q/Q252R background all had similar, lower levels of ATP hydrolysis, consistent with previous studies.
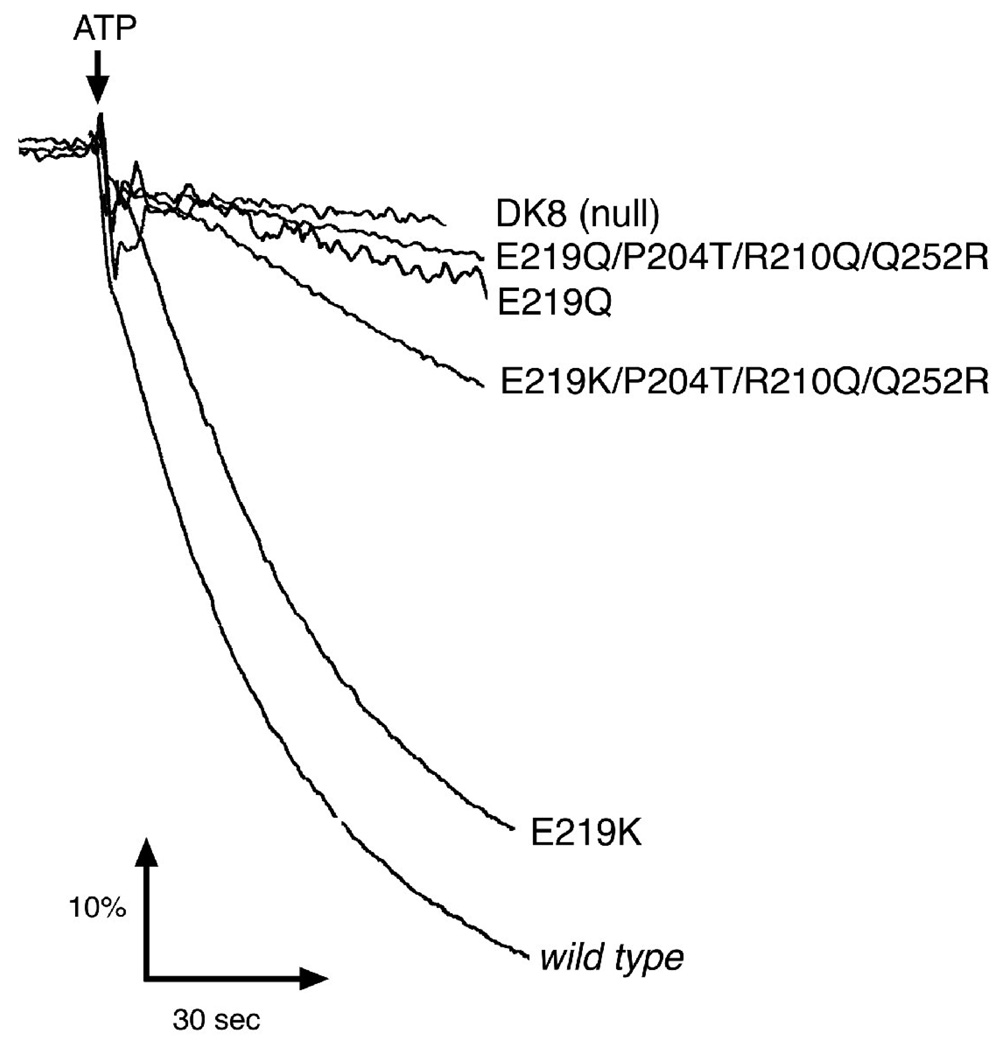
In proton translocation assays shown in Fig. 3, the E219K mutant functioned better than the E219Q mutant, in both backgrounds: wild type and P204T/R210Q/Q252R. The single mutant E219K had a rate of ATP-driven proton translocation that was nearly as great as the wild type. These results were all consistent with the growth analysis shown in Table 1.
Figure 3.
Proton Translocation Assays of E219K and E219Q. Fluorescence quenching of ACMA is shown as a function of time, initiated by the addition of ATP to membrane vesicles. All details are the same as in Figure 2.
4. Discussion
Proton translocation through Fo is thought to occur in three stages. First, protons enter from the periplasm through a channel in subunit a that extends about half-way through the membrane. This allows protonation of an Asp 61 residue in one subunit c. Second, the proton travels via rotation of the c subunit oligomer, relative to subunit a. Finally, the proton exits to the cytoplasm along the interface of subunit a and the c subunit oligomer. The key residue in subunit a is Arg 210, which is thought to engage residue Asp 61 of each of the c subunits in turn during rotation. Upon protonation of an Asp 61, its ionic interaction with Arg 210 would be significantly diminished, facilitating rotation of the c subunit oligomer, and allowing the Arg 210 to be attracted to the next, still ionized Asp 61. The crystal structure of the c subunit oligomer from Ilyobacter tartaricus [42] shows that the key carboxyl group is not entirely exposed on the external surface, but rather is found within a crevice. This would suggest that Arg 210 must move in and out during the steps of rotation. Results presented in this paper suggest such motion of Arg 210 are not coupled directly to other possible conformational changes in TM4 of subunit a, since robust function can occur when the essential arginine is part of TM5.
The identification and further analysis of mutants in which the key Arg 210 is displaced from TM4 to TM5 raised questions about the roles of the other mutated residues in the triple mutant. In this study three questions were raised with respect to the switched mutant: P204T/R10Q/Q252R. The first question was whether the threonine substitution for proline was important, or was it simply beneficial to eliminate the proline. This was accomplished by measuring the growth yields of two new substitutions, alanine and serine, along with those for proline and threonine. The issue was not so simple to resolve, since in our background the differences between the proline and threonine substitutions were not so great. Even so, the ability of alanine to substitute at position 204 with no significant reduction of growth yield indicates that the removal of proline at position 204 is probably sufficient to improve the functioning of the switched arginine mutant R210Q/Q252R.
Secondly, the importance of glutamine was examined in the switched construct R210Q/Q252R. Initially a saturation mutagenesis was planned to discover which, if any, other amino acids could substitute for the glutamine with retention of function. One of the first substitutions isolated was that of glycine, which improved the growth yield. This indicated that no side chain was necessary at position 210 for the arginine switched mutant to function, and so the original plan was abandoned, and this mutant was examined more carefully. Measurement of ATP-driven proton translocation showed that the P204T/R210G/Q252R mutant functioned nearly as well as the wild type. The sensitivity of ATP hydrolysis to inhibition by DCCD also increased to a level near that of wild type. It seems likely that the glutamine provided steric hindrance to the moved arginine, either with respect to its interactions with c subunits, or to its motion during rotation. It is interesting to note that after moving the essential arginine from position 210 in TM4 to TM5, function was improved by removing a proline and introducing a glycine in TM4. Both changes could make a more flexible polypeptide chain.
Third, the question of whether the switched arginine mutant used the normal proton pathway was addressed. It seemed possible at the outset of these experiments that the rather low rates of ATP synthesis seen with the P204T/R210Q/Q252R mutant might be accomplished through an altered path of protons, such as along the a–c interface from the periplasm, rather than through subunit a. Since the E219Q mutant was known to be nonfunctional [39], and this residue is likely near the normal proton pathway [30], this mutation was constructed in the background of the Arginine switched mutant. If the protons were able to take an alternative path the combined mutant might have shown some level of function. Not only was the E219Q mutant nonfunctional, but the E219K mutation retained function in the background of the arginine switched mutant, as it did in the wild type [29, 40, 41]. These results suggest that the normal proton pathway from the periplasm to the c subunit is used by the Arginine switched mutant. The finding that the R210G mutation further enhances the activity of the Arginine switched mutant makes it even less likely that an alternative pathway for protons is followed.
The results of his paper further support the original finding that a functional proton channel, or proton translocation mechanism, does not require that the essential positively-charged residue, normally Arg 210, be directly attached to the fourth TM helix. The robustness of the triple mutant described here, P204T/R210G/Q252R, and its ability to function along with a fourth mutation E219K, suggests that the proton pathway from the periplasm to residue Asp 61 of subunit c is not directly linked to the conformation of the key arginine. Furthermore, the results remind that while residues have been identified in subunit a that line the proton pathway from the periplasm to Asp 61, have been identified [30–32, 43], the proton pathway remains poorly understood. The indication [44] that a hydronium ion might be bound at Asp 61 provides another way to think about how the side chains might be packed, such that lysine or glycine can substitute for glutamic acid at position 219. These issues await further study.
Acknowledgments
This work was supported by grant GM40508 from the National Institutes of Health, and grant N-1378 from the Welch Foundation. We thank Nantha Surkunalingam for construction of pFV2-HA.
Abbreviations
- ACMA
9-amino-6-chloro-2-methoxyacridine
- DCCD
N,N’-dicyclohexylcarbodiimide
- FCCP
carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone
- HA
Hemaglutinin epitope
- TM4
the fourth transmembrane span of subunit a, for example
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK. The rotary mechanism of the ATP synthase. Arch. Biochem. Biophys. 2008;476:43–50. doi: 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vik SB. ATP synthesis by oxidative phosphorylation. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, editors. EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. Washington D.C.: ASM Press; 2007. [Google Scholar]
- 3.von Ballmoos C, Cook GM, Dimroth P. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- 4.Bragg PD, Hou C. Subunit composition, function, and spatial arrangement in the Ca2+-and Mg2+-activated adenosine triphosphatases of Escherichia coli and Salmonella typhimurium. Arch. Biochem. Biophys. 1975;167:311–321. doi: 10.1016/0003-9861(75)90467-1. [DOI] [PubMed] [Google Scholar]
- 5.Foster DL, Fillingame RH. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J. Biol. Chem. 1982;257:2009–2015. [PubMed] [Google Scholar]
- 6.Boyer PD. A model for conformational coupling of membrane potential and proton translocation to ATP synthesis and to active transport. FEBS Lett. 1975;58:1–6. doi: 10.1016/0014-5793(75)80212-2. [DOI] [PubMed] [Google Scholar]
- 7.Boyer PD. The binding change mechanism for ATP synthase-some probabilities and possibilities. Biochim. Biophys. Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 8.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 9.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 11.Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Saxena AK, Simcoke WN, Garboczi DN, Pedersen PL, Ko YH. Mitochondrial ATP synthase. Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism. J. Biol. Chem. 2006;281:13777–13783. doi: 10.1074/jbc.M513369200. [DOI] [PubMed] [Google Scholar]
- 13.Bowler MW, Montgomery MG, Leslie AG, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 2007;282:14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 14.Weber J, Wilke-Mounts S, Lee RS, Grell E, Senior AE. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 1993;268:20126–20133. [PubMed] [Google Scholar]
- 15.Weber J, Wilke-Mounts S, Hammond ST, Senior AE. Tryptophan substitutions surrounding the nucleotide in catalytic sites of F1-ATPase. Biochemistry. 1998;37:12042–12050. doi: 10.1021/bi981089c. [DOI] [PubMed] [Google Scholar]
- 16.Weber J, Senior AE. Bi-site catalysis in F1-ATPase: does it exist? J. Biol. Chem. 2001;276:35422–35428. doi: 10.1074/jbc.M104946200. [DOI] [PubMed] [Google Scholar]
- 17.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 18.Kato-Yamada Y, Noji H, Yasuda R, Kinosita K, Jr, Yoshida M. Direct observation of the rotation of ε subunit in F1-ATPase. J. Biol. Chem. 1998;273:19375–19377. doi: 10.1074/jbc.273.31.19375. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 20.Adachi K, Yasuda R, Noji H, Itoh H, Harada Y, Yoshida M, Kinosita K., Jr Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7243–7247. doi: 10.1073/pnas.120174297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K., Jr Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Masaike T, Koyama-Horibe F, Oiwa K, Yoshida M, Nishizaka T. Cooperative three-step motions in catalytic subunits of F1-ATPase correlate with 80° and 40° substep rotations. Nat. Struct. Mol. Biol. 2008;15:1326–1333. doi: 10.1038/nsmb.1510. [DOI] [PubMed] [Google Scholar]
- 23.Valiyaveetil FI, Fillingame RH. Transmembrane topography of subunit a in the Escherichia coli F1Fo ATP synthase. J. Biol. Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 24.Wada T, Long JC, Zhang D, Vik SB. A novel labeling approach supports the five-transmembrane model of subunit a of the Escherichia coli ATP synthase. J. Biol. Chem. 1999;274:17353–17357. doi: 10.1074/jbc.274.24.17353. [DOI] [PubMed] [Google Scholar]
- 25.Long JC, Wang S, Vik SB. Membrane topology of subunit a of the F1Fo ATP synthase as determined by labeling of unique cysteine residues. J. Biol. Chem. 1998;273:16235–16240. doi: 10.1074/jbc.273.26.16235. [DOI] [PubMed] [Google Scholar]
- 26.Jiang W, Fillingame RH. Interacting helical faces of subunits a and c in the F1Fo ATP synthase of Escherichia coli defined by disulfide cross-linking. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6607–6612. doi: 10.1073/pnas.95.12.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vorburger T, Ebneter JZ, Wiedenmann A, Morger D, Weber G, Diederichs K, Dimroth P, von Ballmoos C. Arginine-induced conformational change in the c-ring/a-subunit interface of ATP synthase. FEBS J. 2008;275:2137–2150. doi: 10.1111/j.1742-4658.2008.06368.x. [DOI] [PubMed] [Google Scholar]
- 28.Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 29.Vik SB, Antonio BJ. A mechanism of proton translocation by F1Fo ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 30.Angevine CM, Fillingame RH. Aqueous access channels in subunit a of rotary ATP synthase. J. Biol. Chem. 2003;278:6066–6074. doi: 10.1074/jbc.M210199200. [DOI] [PubMed] [Google Scholar]
- 31.Angevine CM, Herold KA, Fillingame RH. Aqueous access pathways in subunit a of rotary ATP synthase extend to both sides of the membrane. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13179–13183. doi: 10.1073/pnas.2234364100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angevine CM, Herold KAG, Vincent OD, Fillingame RH. Aqueous access pathways in ATP synthase subunit a: Reactivity of cysteine substituted into transmembrane helices 1, 3, and 5. J. Biol. Chem. 2007;282:9001–9007. doi: 10.1074/jbc.M610848200. [DOI] [PubMed] [Google Scholar]
- 33.Moore KJ, Fillingame RH. Structural interactions between transmembrane helices 4 and 5 of subunit a and the subunit c ring of Escherichia coli ATP synthase. J. Biol. Chem. 2008;283:31726–31735. doi: 10.1074/jbc.M803848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jonge MR, Koymans LHM, Guillemont JEG, Koul A, Andries K. A computational model of the inhibition of Mycobacterium tuberculosis ATPase by a new drug candidate R207910. Proteins. 2007;67:971–980. doi: 10.1002/prot.21376. [DOI] [PubMed] [Google Scholar]
- 35.Hatch LP, Cox GB, Howitt SM. The essential arginine residue at position 210 in the a subunit of the Escherichia coli ATP synthase can be transferred to position 252 with partial retention of activity. J. Biol. Chem. 1995;270:29407–29412. doi: 10.1074/jbc.270.49.29407. [DOI] [PubMed] [Google Scholar]
- 36.Ishmukhametov RR, Pond JB, Al-Huqail A, Galkin MA, Vik SB. ATP synthesis without R210 of subunit a in the Escherichia coli ATP synthase. Biochim. Biophys. Acta. 2008;1777:32–38. doi: 10.1016/j.bbabio.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 38.Klionsky DJ, Brusilow WS, Simoni RD. In vivo evidence for the role of the ε subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J. Bacteriol. 1984;160:1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cain BD, Simoni RD. Interaction between Glu-219 and His-245 within the a subunit of F1Fo-ATPase in Escherichia coli. J. Biol. Chem. 1988;263:6606–6612. [PubMed] [Google Scholar]
- 40.Valiyaveetil FI, Fillingame RH. On the role of Arg-210 and Glu-219 of subunit a in proton translocation by the Escherichia coli FoF1-ATP synthase. J. Biol. Chem. 1997;272:32635–32641. doi: 10.1074/jbc.272.51.32635. [DOI] [PubMed] [Google Scholar]
- 41.Hatch LP, Cox GB, Howitt SM. Glutamate residues at positions 219 and 252 in the a-subunit of the Escherichia coli ATP synthase are not functionally equivalent. Biochim. Biophys. Acta. 1998;1363:217–223. doi: 10.1016/s0005-2728(97)00101-1. [DOI] [PubMed] [Google Scholar]
- 42.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 43.Moore KJ, Angevine CM, Vincent OD, Schwem BE, Fillingame RH. The cytoplasmic loops of subunit a of Escherichia coli ATP synthase may participate in the proton translocating mechanism. J. Biol. Chem. 2008;283:13044–13052. doi: 10.1074/jbc.M800900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Ballmoos C, Dimroth P. Two distinct proton binding sites in the ATP synthase family. Biochemistry. 2007;46:11800–11809. doi: 10.1021/bi701083v. [DOI] [PubMed] [Google Scholar]