Abstract
We utilized a series of analogs of D-V13K (a 26-residue amphipathic α-helical antimicrobial peptide, denoted D1) to compare and contrast the role of hydrophobicity on antifungal and antibacterial activity to the results obtained previously with Pseudomonas aeruginosa strains. Antifungal activity for Zygomycota fungi decreased with increasing hydrophobicity (D-V13K/A12L/A20L/A23L, denoted D4, the most hydrophobic analog was 6-fold less active than D1, the least hydrophobic analog). In contrast, antifungal activity for Ascomycota fungi increased with increasing hydrophobicity (D4, the most hydrophobic analog was 5-fold more active than D1). Hemolytic activity is dramatically affected by increasing hydrophobicity with peptide D4 being 286-fold more hemolytic than peptide D1.
The therapeutic index for peptide D1 is 1569-fold and 62-fold better for Zygomycota fungi and Ascomycota fungi, respectively, compared to peptide D4. To reduce the hemolytic activity of peptide D4 and improve/maintain the antifungal activity of D4, we substituted another lysine residue in the center of the nonpolar face (V16K) to generate D5 (D-V13K/V16K/A12L/A20L/A23L). This analog D5 decreased hemolytic activity by 13-fold, enhanced antifungal activity to Zygomycota fungi by 16-fold and improved the therapeutic index by 201-fold compared to D4 and represents a unique approach to control specificity while maintaining high hydrophobicity in the two hydrophobic segments on the nonpolar face of D5.
Keywords: antimicrobial peptide, antifungal activity, antibacterial activity, hemolytic activity, hydrophobicity, amphipathicity, helicity, peptide self-association
INTRODUCTION
Fungal infections can affect various parts of the body, ranging from superficial and cutaneous to deeply invasive and disseminated (1). Human mycoses include aspergillosis, blastomycosis, candidiasis, coccidioidomycosis, cryptococcosis, histoplasmosis, paracoccidiomycosis, sporotrichosis and zygomycosis. Fungal infections occur more frequently in people whose immune system is suppressed, who have been treated with broad-spectrum antibacterial agents, or who have been subject to invasive procedures (1). Fungal infections are the major cause of morbidity and mortality in patients with organ transplantation, cancer chemotherapy and the human immunodeficiency virus (HIV) (2–4). Species of Candida and Aspergillus account for more than 80% of fungal infections in patients with solid-organ transplantation (2). The systemic mycoses (cryptococcosis, histoplasmosis, and sporotrichosis) as well as the superficial and mucocutaneous mycoses (candidiasis and dermatophytosis) are the most common fungal infections in patients infected with HIV (4). Species of Candida, Aspergillus, Rhizopus and Cryptococcus neoformans are a common cause of infection in cancer patients (3).
Treatment of fungal infections has lagged behind bacterial chemotherapy and there are substantially fewer antifungal than antibacterial drugs (1). The predominant reason is that fungi are eukaryotes and thus agents that inhibit fungal protein, RNA, or DNA biosynthesis are likely to do the same in the patients, producing toxic side effects. On the other hand, more and more resistant pathogenic fungal strains have been reported (2). Therefore, the development of a new class of antibiotics is critical. Cationic antimicrobial peptides (AMPs) have unusually broad spectra of “antimicrobial” activity, especially an ability to kill or neutralize fungi (including yeasts), which make them important candidates as potential antifungal therapeutic agents.
Although the exact mode of action of antimicrobial peptides has not been established, it is generally accepted that the cytoplasmic membrane is the main target of many antimicrobial peptides. Peptide accumulation in the membrane causes increased permeability and a loss of barrier function, resulting in the leakage of cytoplasmic components and cell death. This same mechanism is employed by polyene antibiotics to kill fungi. Cationic AMPs of the α-helical class have two unique features: a net positive charge of at least +2 and an amphipathic character, with a non-polar face and a polar/charged face (5), (6). Factors believed to be important for antimicrobial activity include peptide hydrophobicity, the presence of positively charged residues, an amphipathic nature that segregates basic and hydrophobic residues, and secondary structure. Peptides with mainly antifungal activity, such as many of those isolated from plants, are generally rich in polar and neutral amino acids, which suggests a unique structure-activity relationship (7). No obvious conserved structural domains that give rise to antifungal activity are present and the mechanism of action of some antifungal peptides is still not clear (8).
In our previous research, to investigate what features of α-helical antimicrobial peptides could be changed to control specificity between prokaryotic and eukaryotic cells, we chose a peptide, V681 (Cecropin A (1–8) + Melittin B (1–18) derivative), which had excellent antimicrobial activity but also exhibited high toxicity to human red blood cells as measured by hemolytic activity (9). We showed that a single valine to lysine substitution in the center of the non-polar face dramatically reduced toxicity and increased the therapeutic index (9). This was the first report that a single substitution of a positively charged residue in the center of the non-polar face could act as a specificity determinant in an α-helical antimicrobial peptide, controlling activity between eukaryotic and prokaryotic cells. We then showed that the sole target of this peptide was the membrane by comparing the biological activities and biophysical properties of L- and D- enantiomers of peptide V13K (10). These peptides had equal activities suggesting that the antimicrobial mechanism did not involve a stereoselective interaction with a chiral enzyme, lipid or protein receptor (10). In addition, the all-D peptide was resistant to proteolytic enzyme degradation, which enhances the potential of D-V13K as a clinical therapeutic. We subsequently investigated the role of hydrophobicity of the non-polar face and showed that there was an optimum hydrophobicity of the non-polar face required to obtain the best therapeutic index (11). Increases in hydrophobicity beyond this optimum resulted in a dramatic reduction in antimicrobial activity, which correlated with an increase of peptide self-association (11). We then investigated the effects of net charge and the number of positively charged residues on the polar face to show that both are important for antimicrobial activity and hemolytic activity (12).
Our studies have now increased the list of factors important for antimicrobial activity to include: 1) the importance of lack of secondary structure in benign (non-denaturing) medium, but inducible structure in the presence of the hydrophobic environment of the membrane; 2) the presence of a positively-charged residue in the center of the non-polar face of amphipathic cyclic β-sheet and α-helical peptides as a determinant of locating the peptides to the interfacial region of prokaryotic membranes and decreasing transmembrane penetration into eukaryotic membranes; and 3) importance of peptide self-association in an aqueous environment to the biological activities of these peptides (9–11,13,14).
In the current study, we used the D-form of V13K analogs to investigate the effect of hydrophobicity of the antimicrobial peptides on their antifungal activity toward different pathogenic fungi, which include Aspergillus nidulans, Absidia corymbifera, Rhizomucor spp., Rhizopus microsporus, Rhizopus oryzae, Scedosporium prolificans and Candida albicans. These results were compared to their antibacterial activity toward different gram-negative and gram-positive bacterial strains as well as their hemolytic activity. Surprisingly, we showed that hydrophobicity has significant and different effects on antifungal activity depending on the class of fungi. In Zygomycota fungi, increasing hydrophobicity decreased antifungal activity, whereas increasing hydrophobicity increased antifungal activity for Ascomycota fungi.
MATERIALS AND METHODS
Peptide Synthesis and Purification
Synthesis of the peptides was carried out by solid-phase peptide synthesis using t-butyloxycarbonyl chemistry and 4-methylbenzhydrylamine resin (0.97 mmol/g) followed by cleavage of the peptide from the resin as described previously (9–11). Peptide purification was performed by reversed-phase high-performance liquid chromatography (RP-HPLC) on a Zorbax 300 SB-C8 column (250×9.4 mm I.D.; 6.5 µm particle size, 300 Å pore size; Agilent Technologies, Little Falls, DE, USA) with a linear AB gradient (0.1% acetonitrile/min) at a flow rate of 2 mL/min, where eluent A was 0.2% aqueous trifluoroacetic acid (TFA), pH 2, and eluent B was 0.2% TFA in acetonitrile, where the shallow 0.1% acetonitrile/min gradient started 12% below the acetonitrile concentration required to elute the peptide on injection of an analytical sample and employing a gradient of 1% acetonitrile/min (15). The purity of the peptides was verified by analytical RP-HPLC as described below and was further characterized by mass spectrometry and amino acid analysis.
Analytical RP-HPLC and Temperature Profiling of Peptides
Crude and purified peptides were analyzed on an Agilent 1100 series liquid chromatograph (Little Falls, DE, USA). Runs were performed on a Zorbax 300 SB-C8 column (150×2.1 mm I.D.; 5 µm particle size, 300 Å pore size) from Agilent Technologies using a linear AB gradient (1% acetonitrile/min) and a flow rate of 0.25 mL/min, where eluent A was 0.2% aqueous TFA, pH 2, and eluent B was 0.2% TFA in acetonitrile. Temperature profiling analyses were performed on the same column in 3°C increments, from 5°C to 80°C using a linear AB gradient of 0.5% acetonitrile/min, as described previously (9–11,13).
Characterization of Helical Structure
The mean residue molar ellipticities of peptides were determined by circular dichroism (CD) spectroscopy, using a Jasco J-810 spectropolarimeter (Easton, MD, USA) at 5°C under benign (non-denaturing) conditions (50 mM NaH2PO4 / Na2HPO4 / 100 mM KCl, pH 7.0), hereafter referred to as benign buffer, as well as in the presence of an α-helix inducing solvent, 2,2,2-trifluoroethanol, TFE, (50 mM NaH2PO4 / Na2HPO4 / 100 mM KC1, pH 7.0 buffer/50% TFE). A 10-fold dilution of an approximately 500 µM stock solution of the peptide analogs was loaded into a 0.1 cm quartz cell and its ellipticity scanned from 195 to 250 nm.
Determination of Peptide Amphipathicity
Amphipathicity of peptide analogs was determined by the calculation of hydrophobic moment(16) using the software package Jemboss version 1.2.1(17), modified to include a hydrophobicity scale determined in our laboratory (18). The hydrophobicity scale used in this study is as follows: Trp, 33.0; Phe, 30.1; Leu, 24.6; Ile, 22.8; Met, 17.3; Tyr, 16.0; Val, 15.0; Pro, 10.4; Cys, 9.1; His, 4.7; Ala, 4.1; Arg, 4.1; Thr, 4.1; Gln, 1.6; Ser, 1.2; Asn, 1.0; Gly, 0.0; Glu, −0.4; Asp, −0.8; and Lys, −2.0 (18). These hydrophobicity coefficients were determined from RP-HPLC at pH 7 (10 mM Na2HPO4 buffer containing 50 mM NaCl) of a model random coil 10-residue peptide sequence, Ac-X-G-A-K-G-A-G-V-G-L-amide, where position X was substituted by all 20 naturally occurring amino acids. This HPLC-derived scale reflects the relative differences in hydrophilicity / hydrophobicity of the 20 amino acid side-chains more accurately than previously determined scales because the substitution site is unaffected by nearest-neighbor or conformational effects (18).
Fungal Strains used in this Study
The filamentous fungal and yeast strains used in this study were either purchased from ATCC or generous gifts from various institutions: Aspergillus nidulans (AZN 2867), Absida corymbifera (clinical isolate), Rhizomucor spp. (clinical isolate), Rhizopus microsporus (clinical isolate), Rhizopus oryzae (AZN 8892), Scedosporium prolificans (clinical isolate), Candida albicans (ATCC 24433).
Measurement of Antifungal Activity (MIC50 and MIC90)
Fungal spores (final concentration 104 spores/ml) were suspended in 1/2 Potato Dextrose Broth (Difco), and the yeast strains were suspended at a starting A600=0.001 in the yeast complete medium YPG (1% yeast extract, 1% peptone, 2% glucose). The medium was supplemented with tetracyclin (10 µg/ml) and cefotaxim (100 µg/ml), dispensed by aliquots of 80 µl into wells of a microplate containing 20 µl of either water or the sample to be analyzed. Growth of fungi and yeasts was evaluated after 24 h at 30°C by light microscopy and after 48 h by measuring the culture absorbance at 595 nm using a microplate reader. Under conditions where the antifungal assay was performed in the presence of salt, the 1/2 Potato Dextrose Broth medium was prepared in phosphate-buffered saline, 137 mM NaCl.
The procedure used for the determination of the minimal inhibitory concentration (MIC) was identical to that for the antifungal assay and was based on 2 sets of determinations. The MIC values are expressed as the lowest peptide concentration that causes 90% or 50% growth inhibition. The fungicidal effects of the synthetic peptides in the MIC assay were verified by reinoculation of the yeasts in potato dextrose broth at the end of the incubation time.
Measurement of Antibacterial Activity (MIC)
MICs were determined by a standard microtiter dilution method in Mueller Hinton Broth (MHB) medium and were based on 4 sets of determinations. Serial dilutions (two-fold decrease that ranged from 64 µg/ml to 0.5 µg/ml) of the 10× compound were added to the microtiter plates in a volume of 10 µL followed by 90 µL of bacteria to give a final inoculum of 5×105 colony-forming units (CFU)/mL. The plates were incubated at 37°C for 24 h, and the MICs were determined as the lowest peptide concentration that inhibited growth.
Measurement of Hemolytic Activity (HC50)
Peptide samples were added to 1% human erythrocytes in phosphate-buffered saline (100 mM NaCl, 80 mM Na2HPO4, 20 mM NaH2PO4, pH 7.4) and the reaction mixtures were incubated at 37°C for 18 h in microtiter plates. Two-fold serial dilutions (ranged from 1000 µg/ml to 2 µg/ml) with 3 sets of determinations of the peptide samples were carried out in order to determine the concentration that produced no hemolysis. This determination was made by withdrawing aliquots from the hemolysis assays and removing unlysed erythrocytes by centrifugation (800×g). Hemoglobin release was determined spectrophotometrically at 570 nm. The hemolytic activity was determined as the peptide concentration that caused 50% hemolysis of erythrocytes after 18 h (HC50). The control for no release of hemoglobin was a sample of 1% erythrocytes without any peptide added. Since erythrocytes were in an isotonic medium, no detectable release (<1% of that released upon complete hemolysis) of hemoglobin was observed from this control during the course of the assay. HC50 was determined by a plot of peptide concentration versus percent lysis.
Calculation of Therapeutic Index (HC50/MIC50 Ratio)
The therapeutic index is a widely accepted parameter to represent the specificity of antimicrobial compounds between prokaryotic and eukaryotic cells. It is calculated by the ratio of HC50 (hemolytic activity) and MIC50 (antimicrobial activity); thus, larger values of therapeutic index indicate greater antimicrobial specificity. It should be noted that both the HC and MIC values are carried out by serial two-fold dilutions; thus, for individual bacteria and individual peptides, the therapeutic index could vary as much as four-fold if the peptide is very active in both hemolytic and antimicrobial activities; of course, if a peptide has poor or no hemolytic activity, the major variation in the therapeutic index comes from the variation in the MIC value (as much as two-fold).
Stimulation of Peripheral Blood Mononuclear Cells for Cytokine Production
Isolation of peripheral blood mononuclear cells (PBMCs) from 5 healthy individuals was performed as described elsewhere (19). Briefly, venous blood was drawn into 10 ml tubes containing 0.2 mg of EDTA (Monoject's-Hertogenbosch, Netherlands). The PBMC fraction was obtained by density centrifugation of blood using Ficoll-Paque (Pharmacia Biotech AB, Sweden). The PBMCs were washed twice in saline and resuspended in culture medium (RPMI 1640 Dutch modification, ICN Biomedicals, Costa Mesa, CA), supplemented with gentamicin 1%, L-glutamine 1% and pyruvate 1%. The PBMCs were incubated in 96-well tissue culture plates (Greiner, Alphen, Netherlands) at a concentration of 5×105 cells per well in a total volume of 200 µl, in the presence or absence of a set of stimuli in different experiments. These stimuli consisted of a three concentration dose-response range of the various peptides (0.01, 1.0 and 100 µg/ml). After 24 h of incubation, the supernatants were collected and stored at −80°C until analysis. The study with the cells collected from human volunteers was approved by the ethical committee of the Radboud University, Nijmegen, The Netherlands.
Cytokine Measurements
Interleukin-6 (IL-6) and tumor necrosis factor (TNF) were measured by ELISA according to the manufacturer's protocol (Pelikine, CLB, Amsterdam, the Netherlands). Two experiments were done (Exp 1 and Exp 2 in Table 5), with cells from two different volunteers.
Table 5.
Cytokine assay of D-(V13K) analogs
| A | |||||
|---|---|---|---|---|---|
| Peptide Name | Peptide Concentration (µg/ml) |
TNF (ng/ml) | IL-6 (pg/ml) | ||
| Exp 1 | Exp 2 | Exp 1 | Exp 2 | ||
| RPMI media (background) | ND | ND | ND | 18 | |
| D1 | 100 | ND | ND | ND | 570 |
| 1 | ND | ND | 4 | 8 | |
| 0.01 | ND | ND | ND | ND | |
| D2 | 100 | ND | ND | ND | ND |
| 1 | ND | ND | 121 | 5 | |
| 0.01 | ND | ND | ND | 3 | |
| D3 | 100 | ND | 0.05 | ND | 333 |
| 1 | ND | ND | ND | 18 | |
| 0.01 | ND | 0.05 | ND | ND | |
| D4 | 100 | 0.04 | ND | ND | 216 |
| 1 | 0.025 | ND | 38 | 36 | |
| 0.01 | 0.05 | ND | 5 | ND | |
| D5 | 100 | 0.07 | 0.045 | ND | ND |
| 1 | ND | ND | ND | 7 | |
| 0.01 | ND | ND | ND | ND | |
| B | ||
|---|---|---|
| Positive control | IL-6 (pg/ml) |
|
| Exp 1 | Exp 2 | |
| E.Coli LPS 10 ng/ml | 14000 | 7300 |
| RPMI media (background) | 12 | 9 |
ND: not detectable
RESULTS
Peptide Design
Antimicrobial peptides consisting of all-L-amino acids can be susceptible to proteolytic degradation by enzymes produced by the organism one is trying to kill. All-D-peptides are resistant to proteolytic enzyme degradation which enhances their potential as clinical therapeutics. Of course, all-D-peptides can only be used if it has been established that the antimicrobial mechanism of action does not involve a stereoselective interaction with a chiral enzyme or lipid or protein receptor. In our case, we have shown that for antimicrobial peptide V13K, its all-L-form (L-V13K) or all-D-form (D-V13K) were equally active, suggesting that the sole target for these peptides was the membrane (10). The parent peptide used in this study was D-V13K (D1), a 26-residue amphipathic peptide consisting of all D-amino acid residues, which adopts an α-helical conformation in a hydrophobic environment and contains a hydrophilic, positively-charged lysine residue in the center of the non-polar face (position 13) (Table 1, Figure 1) (9–11). In the present study, we used peptide D-V13K as a framework to alter peptide hydrophobicity systematically on the nonpolar face of the helix by replacing one (peptide D2), two (D3) or three (D4) alanine residues with more hydrophobic leucine residues to increase hydrophobicity. The peptide sequences are shown in Table 1, with helical wheel and helical net representations shown in Figure 1. The number of i→i+3 and i→i+4 hydrophobic interactions on the nonpolar face (a peptide sequence in an α-helical conformation allows a side-chain in position i to interact with a side-chain in position i+3 or i+4 along the sequence) increases with the addition of leucine residues (6 for D1, 9 for D2, 11 for D3 and 12 for D4) (Figure 1). In our previous studies, we showed that 1) placement of a positively charged residue in the center of the non-polar face of amphipathic α-helical (9–11) and cyclic β-sheet (20) antimicrobial peptides is a determinant of specificity between eukaryotic and prokaryotic cells; 2) increasing hydrophobicity over an optimum value decreased antibacterial activity because of strong peptide self-association, which we proposed prevents the peptide from passing through the cell wall to reach the membrane in prokaryotic cells, while increasing hydrophobicity increases hemolytic activity; and 3) increased peptide self-association had no effect on peptide access to eukaryotic membranes. Based on these observations, we hypothesize that the optimum therapeutic index could be achieved by increasing hydrophobicity to increase antimicrobial activity and maintaining poor hemolytic activity by the addition of an extra positive charge in the center of the nonpolar face. Thus, we designed peptide D5 (D-(V13K, A12L, A20L, A23L, V16K)) by replacing the hydrophobic valine residue at position 16 with a positively-charged lysine residue to give two lysine residues in the center of the nonpolar face (positions 13 and 16) (Figure 1). This additional positive charge would further disrupt the consistency of the hydrophobic surface, and prevent the high-level of self-association observed with peptide D4. This V16K substitution was designed to allow the increased hydrophobicity (A12L, A20L, A23L) to enhance antimicrobial activity without increasing hemolytic activity. In this case, the number of i→i+3 and i→i+4 potential hydrophobic interactions decreased from 12 for D4 to 10 for D5, with the continuous hydrophobic face of D4 now disrupted into two separate hydrophobic segments in D5 (Figure 1).
Table 1.
Peptides used in this study
| Peptide Name |
Substitutiona | Sequenceb |
|---|---|---|
| 1 13 26 | ||
| D1 | D-(V13K) | Ac-K-W-K-S-F-L-K-T-F-K-S-A-K-K-T-V-L-H-T-A-L-K-A-I-S-S-amide |
| D2 | D-(V13K, A20L) | Ac-K-W-K-S-F-L-K-T-F-K-S-A-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| D3 | D-(V13K, A12L, A20L) | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| D4 | D-(V13K, A12L, A20L, A23L) | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-L-I-S-S-amide |
| D5 | D-(V13K, A12L, A20L, A23L, V16K) | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-K-L-H-T-L-L-K-L-I-S-S-amide |
The D- denotes that all amino acid residues in each peptide are in the D conformation.
Peptide sequences are shown using the one-letter code for amino acid residues; Ac- denotes Nα-acetyl and -amide denotes Cα- amide. The important substitutions on the nonpolar face are bolded.
Figure 1. Helical wheel (upper panel) /helical net (lower panel) representation of the sequences of lead compound D1 and analogs shown in Table 1 .

The peptides are denoted D1, D2, D3, D4 and D5. The alanine to leucine substitutions (position 12, 20 and 23) are colored yellow. The lysine residue at position 13 and valine to lysine substitution at position 16 are denoted by blue triangles. In the helical wheel, the nonpolar face is indicated as an open arc and the polar face is shown as a solid arc. In the helical net, the amino acid residues on the polar face are circled. The i→i+3 and i→i+4 potential hydrophobic interactions along the helix are shown as black bars. The numbers of hydrophobic interactions on the nonpolar face are indicated at the bottom of each helical net. The one-letter code is used for amino acid residues. The sequences are shown in Table 1.
Peptide Hydrophobicity
RP-HPLC of peptides is a particularly good method to characterize apparent peptide hydrophobicity, and the retention times of peptides are highly sensitive to the conformational status of peptides upon interaction with the hydrophobic environment of the column matrix (9), (21). The nonpolar face of an amphipathic α-helical peptide represents a preferred binding domain for interaction with the hydrophobic matrix of a reversed-phase column (22). In this study, the observed hydrophobicity of the peptides (as expressed by RP-HPLC retention time) is in the order D1 < D5 < D2 < D3 < D4 (tR range from 76.8 min to 101.6 min, Table 2). Triple-Leu-substituted peptide D4 showed the highest hydrophobicity among the peptide analogs (tR=101.6 min; Table 2). By replacing one extra valine with lysine at position 16, the hydrophobicity decreased from 101.6 min for D4 to 80.4 min for D5, i.e., the effect of a triple Ala→Leu substitution on hydrophobicity (D1 → D4; hydrophobicity values of 76.8 min and 101.6 min, respectively, for an increase of 24.8 min) was essentially overridden by only a single Val→Lys substitution (D5 → D4; a decrease in hydrophobicity of 21.2 min). It should be noted, however, that although the overall hydrphobicity of D5 is dramatically decreased compared to D4 due to the presence of the extra Lys residue, the Leu residues are still increasing the hydrophobicity of the two individual hydrophobic segments.
Table 2.
Biophysical data of D-(V13K) analogs
| Peptide Name |
Amphipathicitya | Hydrophobicity |
Benign |
50% TFE |
PAe | ||
|---|---|---|---|---|---|---|---|
| tRb (min) | [θ]222c | %Helixd | [θ]222c | %Helixd | |||
| D1 | 4.92 | 76.8 | 1,150 | 3 | 34,100 | 81 | 2.78 |
| D2 | 5.71 | 86.7 | 2,300 | 5 | 37,550 | 89 | 4.62 |
| D3 | 5.86 | 94.8 | 4,850 | 12 | 38,450 | 91 | 7.67 |
| D4 | 6.34 | 101.6 | 10,550 | 25 | 42,050 | 100 | 9.63 |
| D5 | 5.78 | 80.4 | 3,700 | 9 | 35,500 | 84 | 4.35 |
Amphipathicity of peptide analogs was determined by calculation of hydrophobic moment (16) using the software package Jemboss version 1.2.1 (17), modified to include a hydrophobicity scale determined in our laboratory at pH 7 (18).
tR denotes retention time in RP-HPLC at pH 2 and room temperature, and is a measure of overall peptide hydrophobicity.
The mean residue molar ellipticities [θ]222 (deg cm2/dmol) at wavelength 222 nm were measured at 5 °C in benign conditions (100 mM KCl, 50 mM NaH2PO4/Na2HPO4, pH 7.0) or in benign buffer containing 50% trifluoroethanol (TFE) by circular dichroism spectroscopy.
The helical content (as a percentage) of a peptide relative to the molar ellipticity value of peptide D4 in the presence of 50% TFE.
PA denotes dimerization parameter of each peptide during RP-HPLC temperature profiling, which is the maximal retention time difference of (tRt-tR5 for peptide analogs)-(tRt-tR5 for control peptide C) within the temperature range; tRt-tR5 is the retention time difference of a peptide at a specific temperature (tRt) compared with that at 5°C (tR5). The sequence of control peptide C is Ac-ELEKGGLEGEKGGKELEK-amide.
Peptide Amphipathicity
The sequence of our lead compound, D1, even with a lysine residue in the center of the nonpolar face is still very amphipathic with a value of 4.92 (Table 2). There is an increase in amphipathicity as we increase the hydrophobicity systematically. The amiphipathicity of our analogs ranged from 4.92 to 6.34 (Table 2). By replacing valine with lysine (V16K), the amphipathicity only decreased from 6.34 for D4 to 5.78 for D5.
Enantiomeric Forms of Peptides
As mentioned above, in our previous study (10), we showed that L- and D- enantiomers of peptide V13K had equal activities, and that the all-D peptides were resistant to proteolytic enzyme degradation. The all-D peptides proved to be more active against fungi than their L- enantiomers, due to their resistance to proteolytic enzymes in the cell envelope of different fungal strains (data not shown).
Secondary Structure of Peptides
Figure 2 shows the CD spectra of the peptide analogs in different environments, i.e., under benign conditions (non-denaturing) (Figure 2A) and in buffer with 50% TFE to mimic the hydrophobic environment of the membrane (Figure 2B). It should be noted that all-D helical peptides will exhibit a positive spectrum while all-L helical peptides will exhibit a negative spectrum (10). All peptides except D4 exhibited negligible secondary structure in benign buffer (Figure 2A and Table 2). D4, the triple-Leu-substituted peptide, exhibited an α-helix spectrum under benign conditions (25% α-helix, Table 2) compared to the spectra of the other analogs. Regardless of the different secondary structures of the peptides in benign buffer, a highly helical structure was induced by the nonpolar environment of 50% TFE, a mimic of hydrophobicity and the α-helix-inducing ability of the membrane (Figure 2B and Table 2). All the peptide analogs showed a typical α-helix spectrum with double maxima at 208 nm and 222 nm. The helicities of the peptides in benign buffer and in 50% TFE relative to that of peptide D4 in 50% TFE were determined (Table 2). From Figure 2C, it is clear that increasing peptide hydrophobicity linearly correlates with increasing α-helical structure of the peptides in hydrophobic (50% TFE) environments (R2=0.956).
Figure 2. Circular dichroism (CD) spectra of peptides D1, D2, D3, D4 and D5.
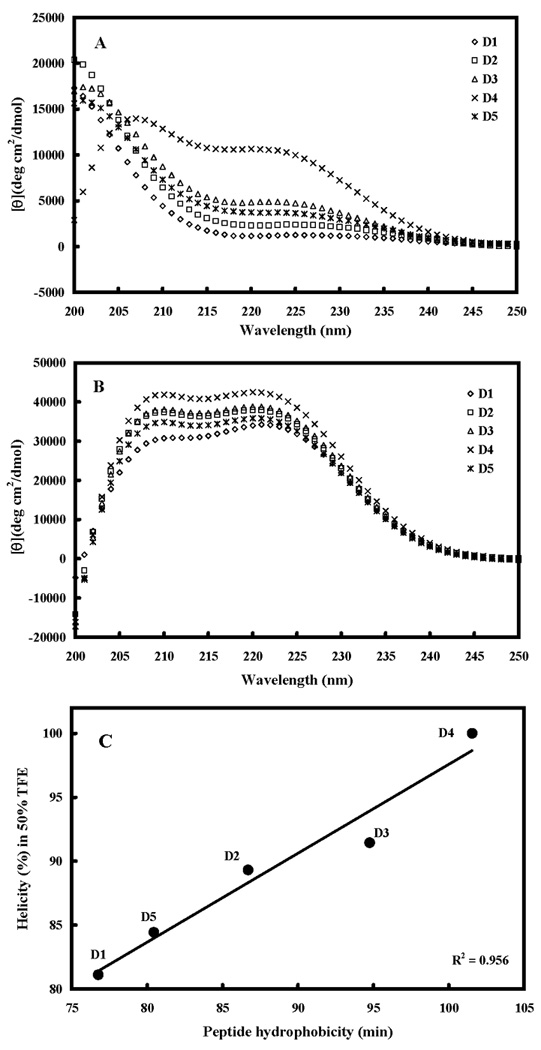
Panel A shows the CD spectra of peptide analogs in benign buffer (100 mM KCl, 50 mM NaH2PO4/Na2HPO4 at pH 7.0, 5 °C and panel B shows the spectra in the presence of buffer-trifluoroethanol (TFE) (1:1, v/v). The relationships of peptide hydrophobicity and helicity is shown in panel C. Hydrophobicity is expressed as the retention times of peptides in RP-HPLC at room temperature (Table 2).
Peptide Self-association
Peptide self-association (i.e., the ability to oligomerize / dimerize) in aqueous solution is a very important parameter for antimicrobial activity (9–11). We postulated that monomeric random-coil antimicrobial peptides are best suited to pass through the capsule and cell wall of microorganisms prior to penetration into the cytoplasmic membrane, induction of α-helical structure and disruption of membrane structure to kill target cells (11). Thus, if the self-association ability of a peptide in aqueous media is too strong (e.g., forming stable folded dimers through interaction of their non-polar faces) this could decrease the ability of the peptide to dissociate to monomer where the dimer cannot effectively pass through the capsule and cell wall to reach the membrane. The ability of the peptides in the present study to self-associate was determined by the technique of RP-HPLC temperature profiling at pH 2 (13,23,24). The reason pH 2 is used to determine self-association of cationic AMPs is that highly positively charged peptides are frequently not eluted from reversed-phase columns at pH 7 due to non-specific binding to negatively charged silanols on the column matrix. This is not a problem at pH 2 since the silanols are protonated (i.e., neutral) and non-specific electrostatic interactions are eliminated. At pH 2, the interactions between the peptide and the reversed-phase matrix involve ideal retention behavior, i.e., only hydrophobic interactions between the preferred binding domain (nonpolar face) of the amphipathic molecule and the hydrophobic surface of the column matrix are present (22). Figure 3A shows the retention behavior of the peptides after normalization to their retention times at 5°C. Control peptide C shows a linear decrease in retention time with increasing temperature and is representative of peptides which have no ability to self-associate during RP-HPLC. Control peptide C is a monomeric random coil peptide in both aqueous and hydrophobic media; thus, its linear decrease in peptide retention behavior with increasing temperature within the range of 5°C to 80°C represents only the general effects of temperature due to greater solute diffusivity and enhanced mass transfer between the stationary and mobile phase at higher temperatures (25). To allow for these general temperature effects, the data for the control peptide was subtracted from each temperature profile as shown in Figure 3B. Thus, the peptide self-association parameter, PA, represents the maximum change in peptide retention time relative to the random coil peptide C. Note that the higher the PA value, the greater the self-association.
Figure 3. Peptide self-association ability as monitored by RP-HPLC temperature profiling.
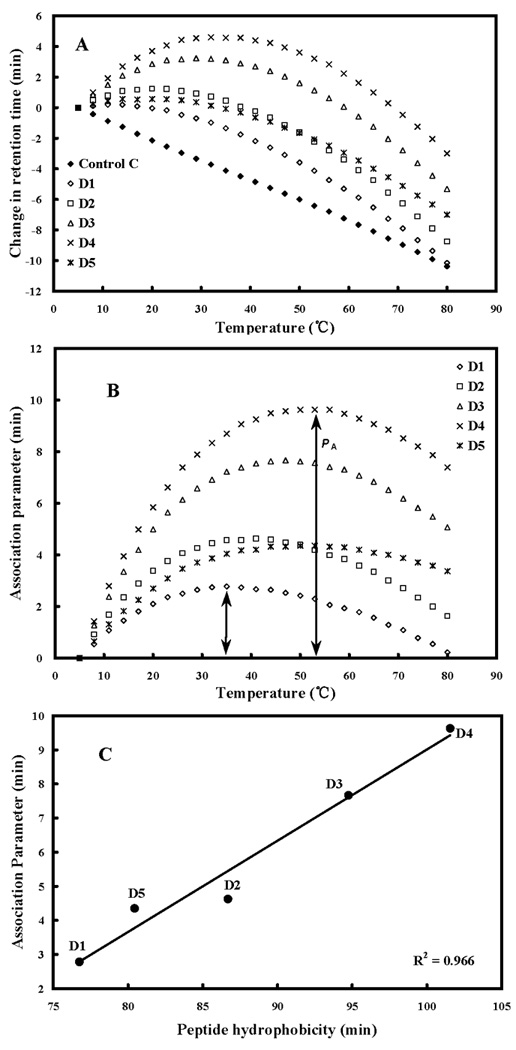
In panel A, the retention time of peptides are normalized to 5°C through the expression (tRt–tR5), where tRt is the retention time at a specific temperature of an antimicrobial peptide or control peptide C, and tR5 is the retention time at 5°C. In panel B, the retention behavior of the peptides was normalized to that of control peptide C through the expression (tRt–tR5 for peptides D1-D5)–(tRt-tR5 for control peptide C). The maximum change in retention time from the control peptide C defines the peptide association parameter, denoted PA. The relationship of peptide hydophobicity and association ability is shown in panel C. Hydrophobicity is expressed as the retention times of peptides in RP-HPLC at room temperature (Table 2).
By replacing a single valine with lysine in the center of the nonpolar face (V13K, D1 in the present study), there was a dramatic decrease in self-association (9,10). However, by systematically increasing the hydrophobicity of the nonpolar face (from peptide D1 to D4), the self-association ability also increased (indeed, Figure 3C shows a linear increase in self-association ability with increasing hydrophobicity of the non-polar face (R2=0.966). By replacing a second valine with lysine in the center of the nonpolar face (position 16) of D4 generating D5, there was a dramatic decrease in self-association ability (Figure 3B), i.e., the substantial positive effect of a triple Ala→Leu substitution (D4) on self-association was overridden by a single V16K substitution (D5). Thus, peptide D5 maintains the three Leu residues and an increase in hydrophobicity in the two hydrophobic patches (Figure 1) while maintaining low self-association compared to peptide D4 (Table 2, Figure 3C).
Antibacterial Activity
From our previous studies, the all-L forms of our peptide analogs (L1, L2, L3 and L4, with the same sequences as D1, D2, D3 and D4, respectively) demonstrated that an optimum hydrophobicity on the non-polar face was required to obtain the best antimicrobial activity (indicated by the arrow) against six clinical-isolate strains of P. aeruginosa (11) (Figure 4). Increasing hydrophobicity beyond the optimum value dramatically decreased antimicrobial activity (peptide L4, Fig. 4). Similarly, decreasing the hydrophobicity beyond peptide L1 dramatically decreased antimicrobial activity. Thus, there is a window of hydrophobicity (indicated by the shaded area in Fig. 4) for maintaining good antimicrobial activity. This window of hydrophobicity allows one to select the peptide hydrophobicity that provides the best therapeutic index (see hemolytic activities described below).
Figure 4. Correlation of peptide hydrophobicity and antibacterial activity (MIC) for six clinical isolates of Pseudomonas aeruginosa.
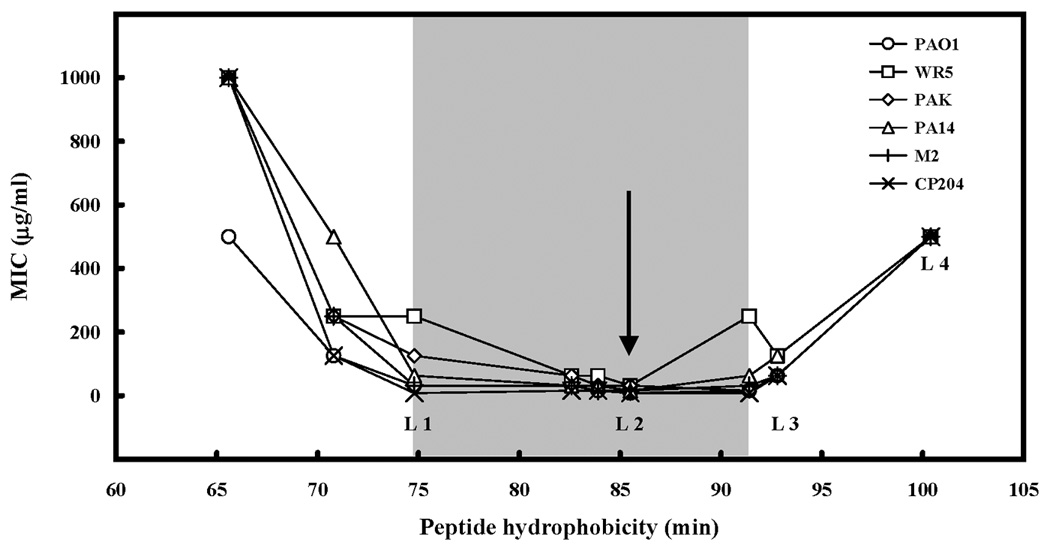
Hydrophobicity is expressed as the retention times of peptides in RP-HPLC at room temperature (11). The shaded area shows the optimal hydrophobicity zone for antimicrobial activity. The arrow denotes the optimal antimicrobial activity. The peptides denoted by L1, L2, L3 and L4 are identical in sequence to D1, D2, D3 and D4, respectively (Table 1), where L and D denote the all L form and all D form of the peptides, respectively.
The antibacterial activities against six gram-negative bacteria/strains and six gram-positive bacteria/strains are compared in Table 3. Geometric mean of MIC was calculated to provide an overall view of antimicrobial activity of different analogs. It is clear that our peptides were effective in killing the microorganisms tested. The tested gram-negative bacteria showed a similar correlation between MIC values and peptide hydrophobicity (Figure 5A) as seen previously for P. aeruginosa (Figure 4): increasing the peptide hydrophobicity from 76.8 min for D1 to 101.6 min for D4 resulted in a reduction in antibacterial activity, albeit the magnitude of the effect differed for each bacterium/strain; for instance, little change was seen for E. coli C857 over the entire range of peptide hydrophobicity for D1 to D4. For the gram-positive bacteria (Figure 5B), the results were more complex, with antibacterial activity for three of the bacteria first increasing with increasing peptide hydrophobicity and then decreasing with a further increase in hydrophobicity. However, one of the bacteria (B. subtilus C971) showed relatively little change over the hydrophobicity range. By replacing another valine with lysine in the center of the nonpolar face (position 16), D5 exhibited an increase in antibacterial activity 2-fold greater compared to D4 for gram-negative and gram-positive bacteria (Table 3). It should be noted that the effects of hydrophobicity for peptide L4 (Figure 4) was an order of magnitude greater than the effects of increasing hydrophobicity on the gram-negative and gram-positive bacteria shown in Figure 5.
Table 3.
Biological activity of D-(V13K) analogs against different Gram-negative (A), and Gram-positive (B) bacteriaa
| A | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide Name |
Hemolytic activity |
Antimicrobial activity against Gram-negative bacteria |
Therapeutic Index |
|||||||||
| HC50b (µg/ml) |
Foldc | MICd (µg/ml) |
GMe | Foldf | HC50/MICg | Foldh | ||||||
| P.aeruginosa | E.coli | E.coli | E.coli | S.typhimurium | S.typhimurium | |||||||
| C498 | C857 | C587 | ||||||||||
| PAO1 | UB1005 | UB1005 | DH5a | 14208S | 14208S | |||||||
| D1 | 1000 | 286 | 8 | 4 | 2 | 2 | 32 | 4 | 5.0 | 4.5 | 198.4 | 1281 |
| D2 | 83 | 24 | 8 | 8 | 4 | 2 | 64 | 8 | 8.0 | 2.8 | 10.4 | 67 |
| D3 | 14 | 4 | 16 | 16 | 8 | 2 | 64 | 16 | 12.7 | 1.8 | 1.1 | 7 |
| D4 | 3.5 | 1 | 16 | 32 | 16 | 4 | >64 | 32 | 22.6 | 1.0 | 0.2 | 1 |
| D5 | 44 | 13 | 8 | 8 | 8 | 2 | >64 | 8 | 10.1 | 2.2 | 4.4 | 28 |
| B | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide Name |
Hemolytic activity |
Antimicrobial activity against Gram-positive bacteria |
Therapeutic Index |
|||||||||
| HC50b (µg/ml) |
Foldc | MICd (µg/ml) |
GMe | Foldf | HC50/MICg | Foldh | ||||||
| S.aureus | S.aureus | S.epidermidis | B.subtilus | B.subtilus | E.faecalis | |||||||
| K147 | C622 | C621 | C626 | C971 | C625 | |||||||
| ATCC3923 | Clinical Isolate | Enviromental Isolate |
ATCC6633 | ATCC29212 | ||||||||
| D1 | 1000 | 286 | 64 | 64 | 4 | 32 | 2 | 32 | 18.0 | 0.9 | 55.7 | 255 |
| D2 | 83 | 24 | 16 | 8 | 8 | 16 | 4 | 16 | 10.1 | 1.6 | 8.2 | 38 |
| D3 | 14 | 4 | 16 | 8 | 8 | 8 | 8 | 32 | 11.3 | 1.4 | 1.2 | 6 |
| D4 | 3.5 | 1 | 32 | 16 | 16 | 16 | 4 | 32 | 16.0 | 1.0 | 0.2 | 1 |
| D5 | 44 | 13 | 8 | 4 | 8 | 8 | 16 | 16 | 9.0 | 1.8 | 4.9 | 22 |
Antibacterial activity is given as the most frequently observed value of 4 sets of determinations. For example, observed values of 4,4,4,8, the value of 4 is recorded. We also included different cultures of the same strains.
HC50 is the maximal peptide concentration that produces 50% hemolysis of human red blood cells after 18 h in the standard microtiter dilution method.
The fold improvement in HC50 compared to that of D4.
MIC is minimal inhibitory concentration that inhibited growth of different strains in Mueller-Hinton (MH) medium at 37°C after 24h. MIC is given based on four sets of determinations.
GM, geometric mean of the MIC values. When no detectable antimicrobial activity was observed at 64 µg/mL, a value of 128 µg/mL was used for calculation of the GM value.
The fold improvement in antimicrobial activity (geometric mean data) compared to that of D4.
Therapeutic index is the ratio of the HC50 value (µg/mL) over the geometric mean MIC value (µg/mL). Large values indicate greater antimicrobial specificity.
The fold improvement in therapeutic index compared to that of D4.
Figure 5. Correlation of peptide hydrophobicity and antibacterial activity (MIC) for gram-negative bacteria (A) and gram-positive bacteria (B).
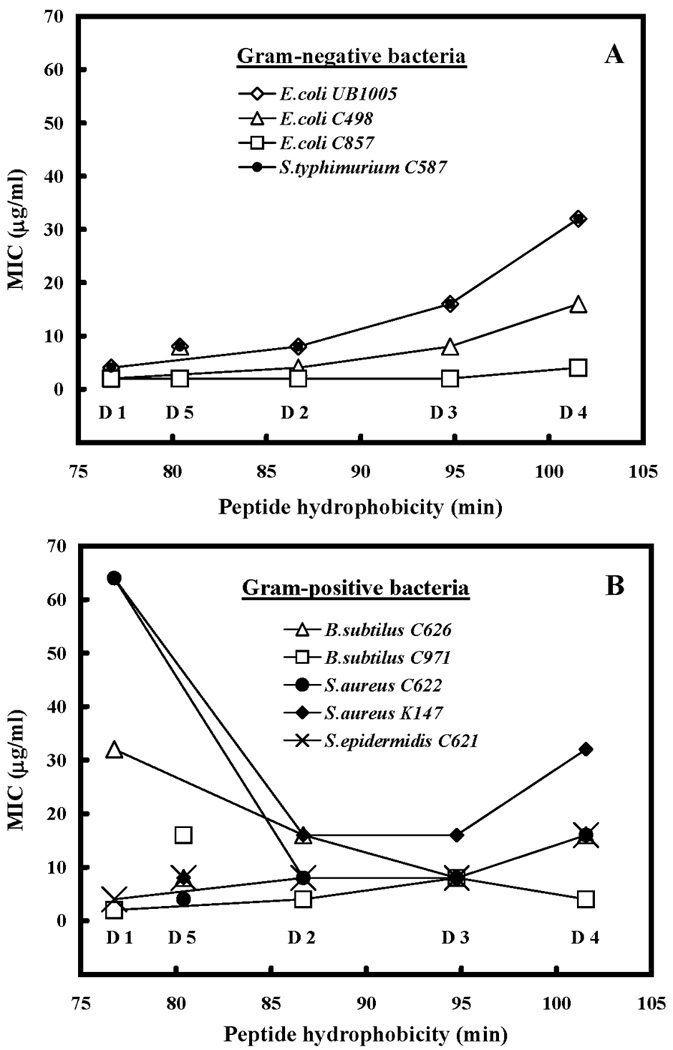
Hydrophobicity is expressed as the retention times of peptides in RP-HPLC at room temperature (Table 2). Lines are drawn through peptides D1 to D4 only, since these peptides systematically increase in hydrophobicity as shown in Figure 1 and Table 1.
Antifungal activity
MIC50 values, the minimal inhibitory concentration of peptide that inhibits 50% of fungal growth, were evaluated for seven different fungal strains (Table 4): both filamentous fungi (A. nidulans, A. corymbifera, Rhizomucor spp., R. microsporus, R. oryzae, S. prolificans) and encapsulated yeast (C. albicans). All seven are fungal pathogens: A. corymbifera, Rhizomucor spp., R. microsporus, R. oryzae belong to the phylum of zygomycota and could cause zygomycosis; A. nidulans, S. prolificans and C. albicans belong to the phylum of ascomycota and could cause aspergillosis, ascomycota and candidiasis, respectively.
Table 4.
Biological activity of D-(V13K) analogs against zygomycota fungi (A) and ascomycota fungi (B) strains a
| A | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide Name |
Hemolytic activity |
Antifungal activity against zygomycota fungi (µg/ml) |
Therapeutic index |
|||||||||||
| HC50b (µg/ml) |
Foldc | A. corymbifera | Rhizomucor spp. | R. microsporus | R. oryzae | GMe | Foldf | HC50/MIC50g | Foldh | |||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |||||||
| D1 | 1000.0 | 286 | 12.5 | 12.5 | 4.7 | 6.3 | 6.3 | 12.5 | 18.8 | 25.0 | 9.1 | 5.5 | 109.9 | 1569 |
| D2 | 83 | 24 | 9.4 | 12.5 | 3.1 | 3.1 | 6.3 | 6.3 | 12.5 | 25.0 | 6.9 | 7.2 | 12.0 | 171 |
| D3 | 14 | 4 | 50.0 | 50.0 | 4.7 | 50.0 | 6.3 | 6.3 | 50.0 | 50.0 | 16.5 | 3.0 | 0.9 | 12 |
| D4 | 3.5 | 1 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 1.0 | 0.07 | 1 |
| D5 | 44 | 13 | 3.1 | 3.1 | 1.6 | 1.6 | 3.1 | 3.1 | 6.3 | 6.3 | 3.1 | 16.0 | 14.1 | 201 |
| B | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide Name |
Hemolytic activity |
Antifungal activity against zygomycota fungi (µg/ml) |
Therapeutic index |
|||||||||
| HC50b (µg/ml) |
Foldc | A. nidulans | S. prolificans | C. albicans | GMe | Foldf | HC50/MIC50g | Foldh | ||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |||||||
| D1 | 1000.0 | 286 | 9.4 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 28.6 | 0.2 | 34.9 | 62 |
| D2 | 83 | 24 | 4.7 | 25.0 | 12.5 | 25.0 | 50.0 | 50.0 | 14.3 | 0.4 | 5.8 | 10 |
| D3 | 14 | 4 | 3.1 | 12.5 | 6.3 | 12.5 | 25.0 | 50.0 | 7.9 | 0.8 | 1.8 | 3 |
| D4 | 3.5 | 1 | 3.1 | 6.3 | 6.3 | 6.3 | 12.5 | 25.0 | 6.3 | 1.0 | 0.6 | 1 |
| D5 | 44 | 13 | 1.6 | 2.1 | 4.7 | 6.3 | 50.0 | 50.0 | 7.2 | 0.9 | 6.2 | 11 |
Antifungal activity is given as mean value of 2 sets of determinations. Any variation was within the accepted two-fold variation observed with MIC values. (In most cases, the values observed were identical.)
HC50 is the maximal peptide concentration that produces 50% hemolysis of human red blood cells after 18 h in the standard microtiter dilution method.
The fold improvement in HC50 compared to that of D4.
MIC50 (or MIC90) are the defined as the peptide concentration (µg/ml) that inhibits 50% (or 90%) of fungal growth.
GM, geometric mean of the MIC50 values.
The fold improvement in antifungal activity (geometric mean data) compared to that of D4.
Therapeutic index is the ratio of the HC50 value (µg/mL) over the geometric mean MIC50 value (µg/mL). Large values indicate greater antimicrobial specificity.
The fold improvement in therapeutic index compared to that of D4.
Figure 6A shows the relationship between MIC50 values of zygomycota fungi and peptide hydrophobicity. A systematic increase in hydrophobicity (from peptide D1 to D4) resulted in a 6-fold reduction in antifungal activity (Figure 6A, Table 4A). However, for the ascomycota fungi, the same series of peptides generated different results: increasing peptide hydophobicity generally led to a continuous increase in antifungal activity with peptide D4 having a 5-fold increase in antifungal activity over peptide D1 (Figure 6B, Table 3B).
Figure 6. Correlation of peptide hydrophobicity and antifungal activity (MIC50) for zygomycota fungi (A) and ascomycota fungi (B).
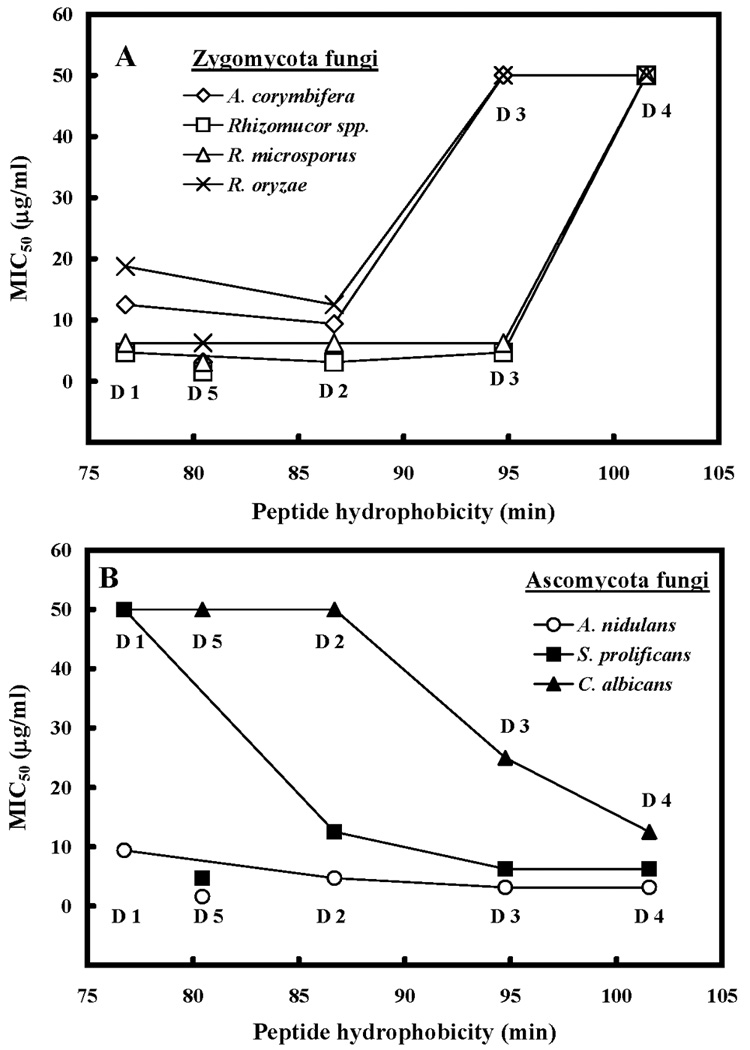
Hydrophobicity is expressed as the retention times of peptides in RP-HPLC at room temperature (Table 2). Lines are drawn through peptides D1 to D4 only, since these peptides systematically increase in hydrophobicity as shown in Figure 1 and Table 1.
With the extra valine to lysine substitution in the center of the nonpolar face (position 16) of D4 to generate D5, antifungal activity increased by 16-fold for zygomycota fungi and maintained the same level for ascomycota fungi (Table 4). Overall, D5 is the best analog in our series for most of the tested fungal strains.
Hemolytic activity
The hemolytic activities of the peptides against human erythrocytes were determined as a measure of peptide toxicity toward higher eukaryotic cells. The effect of peptide concentration on erythrocyte hemolysis is shown in Figure 7A. From these plots the peptide concentration that produced 50% hemolysis was determined (HC50). D4 showed the strongest hemolytic activity, while D1 showed the weakest. Hemolytic activity for peptides D2, D3, D4 and D5 increased in a hyperbolic fashion with increasing peptide concentration and all plateaued at 100% lysis when the peptide concentration was high enough. By comparison, hemolytic activity for peptide D1 increased in a linear fashion with increasing peptide concentration (Fig. 7A).
Figure 7. The hemolytic activity of peptides D1 and analogs.
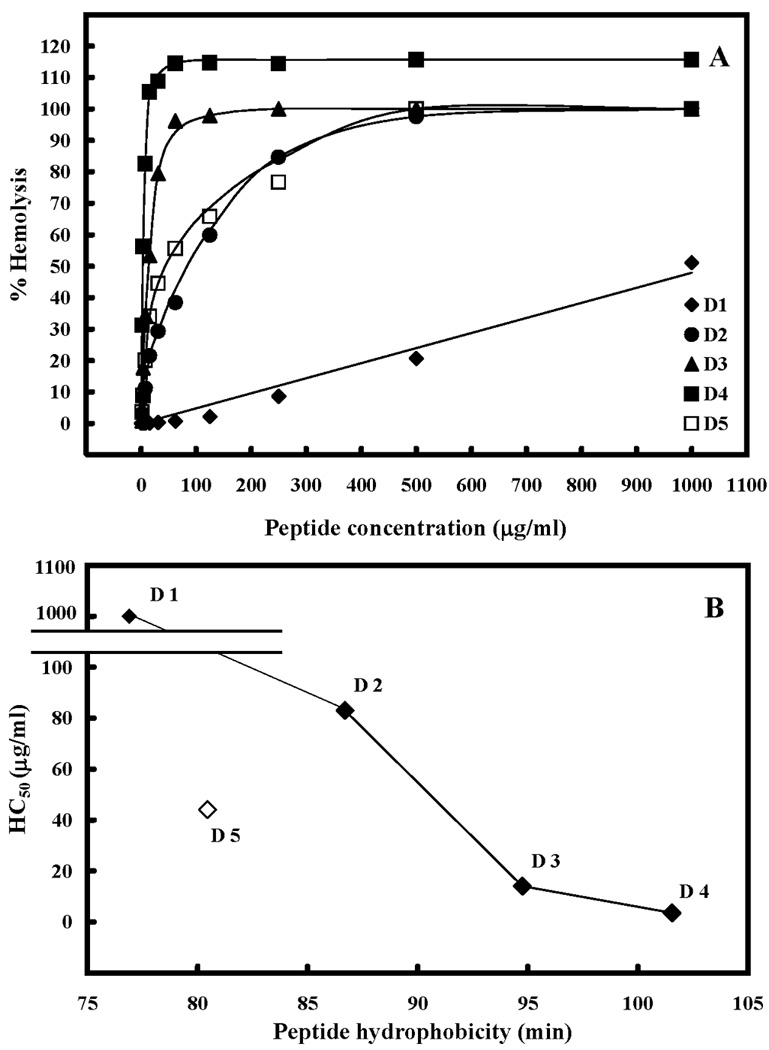
The concentration-response curves of peptides for lysis of human red blood cells (hRBC) are shown in panel A. The relationship of peptide hydrophobicity and HC50 (peptide concentration that causes 50% hemolysis) is shown in panel B. Hydrophobicity is expressed as the retention times of peptides in RP-HPLC at room temperature (Table 2). Lines are drawn through peptides D1 to D4 only, since these peptides systematically increase in hydrophobicity as shown in Figure 1 and Table 1.
Hemolytic activity represented as HC50 is shown in Table 4 and Figure 7B. Increasing peptide hydrophobicity by replacing one, two or three alanine residues with leucine residues, decreased the HC50 values from 1000 µg/ml for D1 to 83 µg/ml, 14 µg/ml and 3.5 µg/ml for D2, D3 and D4 respectively (Table 4, Figure 7B), i.e., a 286-fold increase in hemolysis compared to that of the parent peptide, D1. By replacement of a second valine with lysine at position 16, to produce D5, hemolytic activity was decreased by 13-fold relative to D4 (from 3.5 µg/ml for D4 to 44 µg/ml for D5).
Therapeutic Index
The therapeutic indices for different fungal strains are shown in Table 4. The geometric mean MIC50 value for zygomycota and ascomycota fungi was used to give an overall view of therapeutic index in fungi. Compared to that of the parent peptide, D1, triple-Leu-substituted peptide D4 showed a decrease in therapeutic index by more than 1569-fold and 62-fold for zygomycota and ascomycota fungi, respectively, relative to peptide D4 (Table 4). Replacing a second valine with lysine at position 16 (D5) increased the therapeutic index by more than 200-fold and 11-fold for zygomycota and ascomycota fungi, respectively (Table 4). Zygomycota and ascomycota fungi exhibited different responses in MIC50 to an increase in peptide hydrophobicity (Figure 6); however, with the factor of hemolytic activity, the therapeutic index of both zygomycota and ascomycota fungi express similar responses to an increase in peptide hydrophobicity (Table 4).
The therapeutic indices for different bacteria strains are shown in Table 3. Peptide D4, with the highest hydrophobicity among all analogs, exhibits the lowest therapeutic index: about 0.2 for both gram-negative bacteria and gram-positive bacteria. For peptide D5, the therapeutic index increased by 28- and 22-fold relative to peptide D4 for gram-negative and gram-positive bacteria, respectively.
Cytokine Production
Antibacterial and/or antifungal peptides may stimulate cytokine production, which could have potential serious side-effects in patients, through their strong systemic inflammatory reactions. Thus, we tested our peptide analogs for production of tumor necrosis factor (TNF) and interleukin-6 (IL-6), two of the most potent proinflammatory cytokines (Table 5). There was only a very low stimulation of IL-6 production when very high concentrations of some of the peptides were used. However, for the positive control (LPS stimulation, a standard cytokine stimulus), a 1000-fold lower concentration than the peptides would give 10- to 100-fold higher cytokine stimulation. Thus, the peptides are very ineffective at stimulating cytokine production and even if very high concentrations of some (not all) of the peptides are used, a potential patient may only experience slight fever reactions, as seen with other medications (e.g., amphotericin B, interferon-gamma).
DISCUSSION
Most antifungal agents interact with or inhibit ergosterol, the major sterol in the fungal plasma membrane (1,26). The polyene antibiotics, such as amphotericin B, the “gold standard” for the treatment of invasive fungal infections, bind to the membrane ergosterol, causing membrane leakage and cell death (1). Amphotericin B has higher affinity for ergosterol than cholesterol, the sterol counterpart in mammalian cells, thus limiting their toxicity to the patients (1). The azole derivatives affect ergosterol biosynthesis (1). Overall, since ergosterol is a key target for most antifungal drugs, their toxicity in mammalian cells would be limited considerably. However, for the membrane-permeabilizing peptides, their interaction with the cell membrane is non-specific, and ergosterol is not the only target of antimicrobial peptides. Zwitterionic phosphatidylcholine (PC) and phosphatidylethanolamine (PE) are the major phospholipid classes in fungi, with smaller amounts of negatively charged phosphatidylinositol (PI, 3–10%), phosphatidylserine (PS) and diphosphatidylglycerol (DPG, 2–5%) (27). Compared to hRBC (28), fungi have a higher amount of negatively charged PI and DPG. Such differences may result in higher susceptibility of fungal cells to antimicrobial peptides than red blood cells.
The cell wall or cell envelope is a barrier, which can hinder AMPs from reaching the cell membrane. Once close to the microbial surface, AMPs must traverse capsular polysaccharides (LPS) on the outer membrane before they can interact with the inner membrane of gram-negative bacteria; on the other hand, AMPs have to traverse capsular polysaccharides, teichoic acids and lipoteichoic acids in order to interact with the membrane of gram-positive bacteria (29). The fungal cell wall has been shown to be primarily composed of chitin, glucans, mannans and glycoproteins; and there is evidence of extensive cross-linking between these components (30). Thus, the fungal cell wall is a barrier even more difficult to cross by AMPs than the bacterial cell envelope. According to our previous results (11), if the self-association ability of a peptide in aqueous media is too strong (e.g., forming stable folded dimers), it could decrease the ability of the peptide to dissociate and pass through the capsule and cell wall of microorganisms and, hence, prevent penetration into the cytoplasmic membrane to kill target cells. In our current experiments, peptide D4, which has the highest self-association ability (Table 2, Figure 3), overall exhibits the worst antimicrobial activity.
Figures 4, 5, 6 and 7 show the relationship between peptide hydrophobicity and antimicrobial and hemolytic activity. Different microorganisms and different strains within the same organism have different responses to increasing peptide hydrophobicity. Clearly, increasing hydrophobicity has the most dramatic effect on eukaryotic cells (as measured by hemolytic activity) compared to prokaryotic cells. By increasing the peptide hydrophobicity from D1 to D4, hemolytic activity increased 286-fold (Table 4). In the case of P. aeruginosa, increasing hydrophobicity from L1 to L2 resulted in a 3-fold increase in anti-pseudomonas activity (Figure 4). However, a continuing increase in hydrophobicity from L2 to L4 resulted in a dramatic decrease (32-fold) in anti-pseudomonas activity due to increased peptide self-association (Figure 4). In fact, L4 was essentially inactive with an MIC value of 500 µg/ml. Although the same trend of decreasing activity with increasing hydrophobicity over and above D1 was observed for other gram-negative bacteria (Figure 5A), the magnitude of this effect was at least 10-fold smaller compared to the P. aeruginosa results (Figure 4). A similar trend was also observed for gram-positive bacteria (Figure 5B), with the magnitude of this effect being similar to the gram-negative organisms (Figure 5A).
In the case of Zygomycota fungi, a loss of activity with increasing peptide hydrophobicity was also observed for the most hydrophobic peptide, D4 (Figure 6A). Thus, overall, increasing hydrophobicity beyond a certain point has a negative impact on antimicrobial activity, which can best be explained by peptide self-association. The only exception that we observed was with Ascomycota fungi, where increasing peptide hydrophobicity to D4 resulted in improved activity (Figure 6B).
Overall, when taking into account gram-negative bacteria, gram-positive bacteria and fungi, D1 is the best compound in terms of therapeutic index. However, in the case of Ascomycota fungi, D1 was 5-fold less active than D4 (Table 4). This led us to the challenge of maintaining the activity of D4 for these fungi whilst increasing the therapeutic index by decreasing the hemolytic activity. Therefore, we designed a peptide with an extra Lys residue in place of Val in the center of the non-polar face to generate peptide D5. D5 was 16-fold more active than D4 for Zygomycota fungi, and similar to D4 for Ascomycota fungi but had the advantage of a 200-fold improvement in therapeutic index for Zygomycota fungi and an 11-fold improvement for Ascomycota fungi. The almost complete inability of these peptides to induce the release of proinflammatory cytokines is also beneficial, as it excludes inflammatory side effects such as seen with other defensins or even antifungal therapies (e.g. amphotericin B).
Finally, although D5 is 3–4 times more active than peptide D1 against fungi, D1 has the highest therapeutic index due to its extremely low hemolytic activity. Peptides D1 and D5 remain as potential lead compounds for broad-spectrum antimicrobial compounds.
Acknowledgements
This research was supported by a NIH grant from the National Institute of Allergy and Infectious Diseases (NIAID) R01 AI067296 and the John Stewart Chair in Peptide Chemistry to R.S.H. R.E.W.H. was supported by the Advanced Foods and Materials Network and holds a Canada Research Chair. J.H. holds a Commonwealth Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID or NIH.
References
- 1.Georgopapadakou NH, Walsh TJ. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 2.Paya CV. Fungal infections in solid-organ transplantation. Clin Infect Dis. 1993;16:677–688. doi: 10.1093/clind/16.5.677. [DOI] [PubMed] [Google Scholar]
- 3.Brown AE. Overview of fungal infections in cancer patients. Semin Oncol. 1990;17:2–5. [PubMed] [Google Scholar]
- 4.Durden FM, Elewski B. Fungal infections in HIV-infected patients. Semin Cutan Med Surg. 1997;16:200–212. doi: 10.1016/s1085-5629(97)80043-0. [DOI] [PubMed] [Google Scholar]
- 5.Hancock RE. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 6.Hoges RS, Jiang Z, Whitehurst J, MC T. Development of antimicrobial peptides as therapeutic agents. In: Leventhal M, Gad SC, editors. Development of therapeutic agents. Hoboken, NJ: John Wiley & Sons, Inc.; 2008. in press. [Google Scholar]
- 7.Lustig F, Hoebeke J, Ostergren-Lunden G, Velge-Roussel F, Bondjers G, Olsson U, Ruetschi U, Fager G. Alternative splicing determines the binding of platelet-derived growth factor (PDGF-AA) to glycosaminoglycans. Biochemistry. 1996;35:12077–12085. doi: 10.1021/bi960118l. [DOI] [PubMed] [Google Scholar]
- 8.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Vasil AI, Rehaume L, Mant CT, Burns JL, Vasil ML, Hancock RE, Hodges RS. Comparison of biophysical and biologic properties of alpha-helical enantiomeric antimicrobial peptides. Chem Biol Drug Des. 2006;67:162–173. doi: 10.1111/j.1747-0285.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother. 2007;51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Z, Vasil AI, Hale JD, Hancock RE, Vasil ML, Hodges RS. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers. 2008;90:369–383. doi: 10.1002/bip.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DL, Mant CT, Hodges RS. A novel method to measure self-association of small amphipathic molecules: temperature profiling in reversed-phase chromatography. J Biol Chem. 2003;278:22918–22927. doi: 10.1074/jbc.M301777200. [DOI] [PubMed] [Google Scholar]
- 14.Lee DL, Hodges RS. Structure-activity relationships of de novo designed cyclic antimicrobial peptides based on gramicidin S. Biopolymers. 2003;71:28–48. doi: 10.1002/bip.10374. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Mant CT, Hodges RS. Preparative reversed-phase high-performance liquid chromatography collection efficiency for an antimicrobial peptide on columns of varying diameters (1mm to 9.4mm I.D.) J Chromatogr A. 2007;1140:112–120. doi: 10.1016/j.chroma.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 17.Carver T, Bleasby A. The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics. 2003;19:1837–1843. doi: 10.1093/bioinformatics/btg251. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs JM, Mant CT, Hodges RS. Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side chains in peptides in the absence of nearest-neighbor or conformational effects. Biopolymers. 2006;84:283–297. doi: 10.1002/bip.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drenth JP, Van Uum SH, Van Deuren M, Pesman GJ, Van der Ven-Jongekrijg J, Van der Meer JW. Endurance run increases circulating IL-6 and IL-1ra but downregulates ex vivo TNF-alpha and IL-1 beta production. J Appl Physiol. 1995;79:1497–1503. doi: 10.1152/jappl.1995.79.5.1497. [DOI] [PubMed] [Google Scholar]
- 20.Kondejewski LH, Jelokhani-Niaraki M, Farmer SW, Lix B, Kay CM, Sykes BD, Hancock RE, Hodges RS. Dissociation of antimicrobial and hemolytic activities in cyclic peptide diastereomers by systematic alterations in amphipathicity. J Biol Chem. 1999;274:13181–13192. doi: 10.1074/jbc.274.19.13181. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Mant CT, Hodges RS. Determination of stereochemistry stability coefficients of amino acid side-chains in an amphipathic alpha-helix. J Pept Res. 2002;59:18–33. doi: 10.1046/j.1397-002x.2001.10994.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou NE, Mant CT, Hodges RS. Effect of preferred binding domains on peptide retention behavior in reversed-phase chromatography: amphipathic alpha-helices. Pept Res. 1990;3:8–20. [PubMed] [Google Scholar]
- 23.Mant CT, Chen Y, Hodges RS. Temperature profiling of polypeptides in reversed-phase liquid chromatography. I. Monitoring of dimerization and unfolding of amphipathic alpha-helical peptides. J Chromatogr A. 2003;1009:29–43. doi: 10.1016/s0021-9673(03)00621-6. [DOI] [PubMed] [Google Scholar]
- 24.Mant CT, Tripet B, Hodges RS. Temperature profiling of polypeptides in reversed-phase liquid chromatography. II. Monitoring of folding and stability of two-stranded alpha-helical coiled-coils. J Chromatogr A. 2003;1009:45–59. doi: 10.1016/s0021-9673(03)00919-1. [DOI] [PubMed] [Google Scholar]
- 25.Dolan JW. Temperature selectivity in reversed-phase high performance liquid chromatography. J Chromatogr A. 2002;965:195–205. doi: 10.1016/s0021-9673(01)01321-8. [DOI] [PubMed] [Google Scholar]
- 26.Kontoyiannis DP, Lewis RE. Antifungal drug resistance of pathogenic fungi. Lancet. 2002;359:1135–1144. doi: 10.1016/S0140-6736(02)08162-X. [DOI] [PubMed] [Google Scholar]
- 27.Prasad R, Ghannoum MA. Lipids of Pathogenic Fungi. CRC press; 1996. [Google Scholar]
- 28.Gabriel GJ, Som A, Madkour AE, Eren T, Tew GN. Infectious Disease: Connecting Innate Immunity to Biocidal Polymers. Mater Sci Eng R Rep. 2007;57:28–64. doi: 10.1016/j.mser.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 30.Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28:799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]


