Summary
DNA topoisomerases are enzymes that disentangle the topological problems that arise from double stranded DNA. Many of these can be solved by the generation of either single or double strand breaks. However, where there is a clear requirement to alter DNA topology by introducing transient double strand breaks, only DNA topoisomerase II (Top2) can carry out this reaction. Extensive biochemical and structural studies have provided detailed models of how Top2 alters DNA structure, and recent molecular studies have greatly expanded the biological contexts where Top2 functions, such as DNA replication, transcription and chromosome segregation, processes that are essential for preventing tumorigenesis.
Introduction
The double stranded nature of DNA creates a special set of problems for processes that require strand unwinding such as transcription and replication. The unwinding that occurs during these processes creates a topological problem because the unwinding must be compensated by overwinding elsewhere in the DNA molecule. DNA topoisomerases are enzymes that solve these difficulties by introducing transient breaks in DNA. The transient breaks allow changes in DNA topology that eliminate the overwinding. There are two classes of topoisomerases: type I enzymes introduce single strand breaks in DNA and type II topoisomerases introduce double strand breaks 1,2. Since a single unrepaired double strand break has potentially lethal consequences, type II topoisomerases might be viewed as a particularly dangerous way of dealing with the topological problems of DNA. The work of Sundin and Varshavsky showed that there were topological problems arising at the completion of replication that absolutely required a type II topoisomerase to separate replicated molecules3,4. Studies in a wide range of eukaryotes have confirmed these initial notions and have shown that type II topoisomerases are required to segregate replicated chromosomes. Moreover, type II topoisomerases participate in many of the nuclear processes that generate topological problems.
The last few years has led to an explosion in findings concerning the biochemistry and biology of type II topoisomerases. The biochemical steps in the Top2 reaction have been demonstrated, and structural studies, have provided an underpinning for understanding the enzyme reaction cycle. At the same time, the number of processes that use type II topoisomerases, especially in gene expression, have multiplied. This was driven by efforts to understand why mammalian cells express two Top2 isozymes (Top2α and Top2β, Box 1), while most other eukaryotes manage with a single Top2 enzyme. There has been continued interest in how cells regulate when and where type II enzymes act, as well as how cells insulate themselves when type II enzymes fail to function properly. This review highlights recent work on the role of Top2 in replication, transcription and chromosome structure that may be relevant to the phenotypes of cancer cells.
Box 1. The complement of type II topoisomerases in eukaryotic cells.
There are two broad classes of type II topoisomerases, type IIA topoisomerases, which include prokaryotic DNA gyrase, prokaryotic topoisomerase IV, and eukaryotic Top2, and type IIB topoisomerases, including TopoVI from plants82, and Spo11 homologs that are required to introduce double strand cleavage that initiates meiotic recombination83. In lower eukaryotes, including single cell organisms such as yeast, insects, vertebrates such as Xenopus, there is a single Top2 isoform. Mammals have two Top2 isoforms termed “α” and “β”84. Expression of Top2α is cell cycle regulated, and this enzyme is essential for the viability of all dividing cells. Many non-dividing cells lack detectable Top2α. The β isozyme is required for viability in mouse, and plays a key role in neuronal development. Embryos lacking Top2β fail to innervate the diaphragm, and die at or before birth. Since the embryos develop almost to term, it possible to isolate viable cells completely lacking Top2β. The roles of Top2α and Top2β appear to be dictated mainly by their C-terminal domains. In a conditional knockout cell system, expression of Top2β fails to complement a deficiency of Top2α, although the catalytic domains of Top2β fused to the C-terminal domain of Top2α provides complementation. Conversely, the C-terminal domain of Top2β fused to Top2α catalytic domains does not complement the conditional deficiency of Top2α85. The second class of type II topoisomerases, type IIB enzymes, are homologous to archaebacterial type II topoisomerases. Mammals as well as lower eukaryotes have a type IIB homolog Spo1186,87. This enzyme is required to initiate meiotic recombination by the generation of an enzyme-mediated double strand break. Type IIB topoisomerases play diverse physiological functions in plants82,88.
How the Top2 machine works
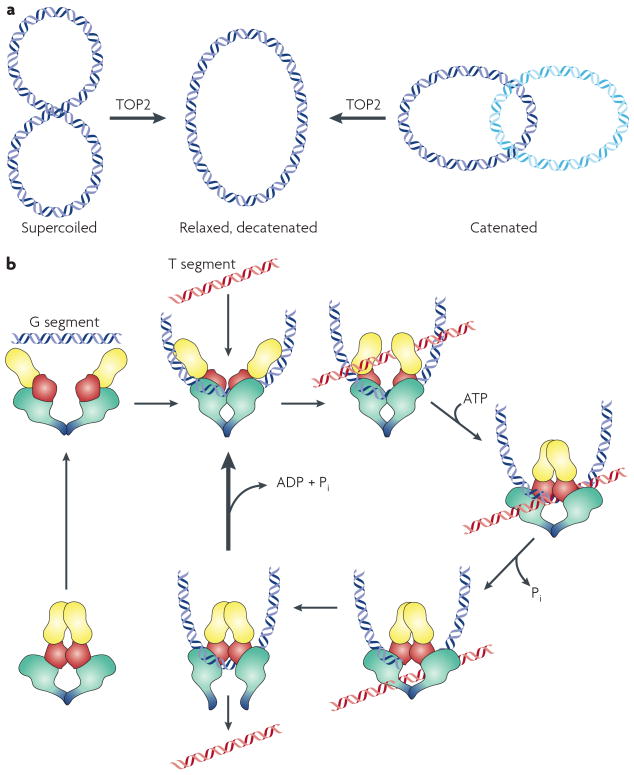
Topological changes in DNA require the introduction of DNA strand breaks, and topoisomerases provide a safe mechanism for introducing these changes. Because the strand breaks are protected (covalently bound to protein) they neither generate ends that are subject to rearrangement or recombination, nor generate DNA damage responses. A simple topological change, illustrated in Fig.1 is the decatenation of a singly linked catenane. Catenanes are the simplest topological change that can be visualized. Another important change is in the regulation of DNA supercoiling. Topoisomerases can convert DNA that is underwound to the energetically more stable state of no superhelical turns. Although it is most rigorous to discuss topological changes in the context of a circular DNA molecule, topological considerations also apply to long linear molecules, since the “break” that is the DNA end is too far away to allow the changes in winding to occur with reasonable kinetics. This is a very brief description of topological changes in DNA, and more complete and careful descriptions are available 5.
Figure 1. Mechanism of strand passage by type II topoisomerases.
A. Reactions catalyzed by eukaryotic Top2 include decatenation of linked intact double stranded DNA and relaxation of supercoiled DNA. The reaction formally requires introduction of a double strand break, strand passage, and break resealing. B. Topoisomerase II interacts with two DNA strands to effect strand passage. The enzyme introduces a double strand break in one DNA strand, termed the G or “gate segment”, and will pass a second strand termed the T segment through the break. In the presence of Mg2+, the enzyme can cleave the DNA, forming a phosphotyrosine linkage between each single strand and a tyrosine in each subunit. ATP binding causes the enzyme to form a closed clamp. The closed clamp may also capture another strand (the T strand) that will pass through the break made in the G strand. After passing through the break in the G strand, the T strand exits the enzyme through the carboxy terminus (the bottom of the enzyme as drawn). ATP hydrolysis occurs at two steps in the reaction cycle89. The first ATP hydrolyzed may assist in strand passage. The second hydrolysis step (along with release of ADP and Pi) allows the clamp to re-open, and allows release of the G segment (for a distributive reaction). Alternately, the enzyme may initiate another catalytic cycle without dissociating from the G strand.
The catalytic reaction
Eukaryotic type II topoisomerases are large homodimeric enzymes. The overall reaction strategy is the generation of a transient double strand break, with each subunit breaking one DNA strand. The enzyme will pass an unbroken strand through the transient break, and then reseal the break. The detailed reaction mechanism of Top2 is presented in Fig. 1. DNA cleavage by Top2 uses a tyrosine that is activated to attack the phosphodiester backbone of DNA and form a phosphotyrosine linkage. For Top2, cleavage requires a collaboration between the active site tyrosine and other residues including the TOPRIM domain6. The TOPRIM domain includes an acidic triad of residues that is involved in complexing a divalent cation, which is absolutely required for the cleavage reaction. Because the energy of the phosphodiester bond is conserved in the phosphotyrosine bond, the cleavage reaction can be reversed without a high-energy co-factor, leading to restoration of the phosphodiester bond and the free enzyme. This mechanism of DNA cleavage provides several distinct advantages including the protection of DNA ends and the ability to quickly and efficiently religate the DNA strand break. It is this reaction that is exploited by many drugs that target Top2. Agents such as etoposide and mAMSA inhibit the religation step and trap Top2 as a covalent complex with the enzyme covalently bound to DNA with broken DNA strands. An important property of the covalent complex is that in most instances it remains freely reversible. Removal of the drug allows the enzyme to rapidly and efficiently reseals the DNA break.
Early studies of Top2 targeting drugs relied on enzyme denaturation to trap the drug-stabilized complex. Since the breaks and covalently bound protein were only efficiently detected in the presence of a protein denaturant, it was formally possible that the denaturant somehow induced the strand breaks. Therefore the intermediate was termed a “cleavable complex”. Since many studies have demonstrated the presence of cleaved DNA in the absence of denaturants, terms such as “cleaved complex” are more precise, although cleavable complex continues to be used for historical reasons.
Structural analyses of Top2
Although the mechanics of DNA cleavage and strand passage were originally studied biochemically, a series of elegant structural studies has provided support and elaboration of the enzyme mechanisms described above. The model for most structural studies has been the type II topoisomerase from yeast. However, eukaryotic type II topoisomerases are highly conserved, and the structural insights from the yeast enzyme are also likely to apply to the human enzyme. The N-terminal domain of the protein carries the ATP binding domain. A central portion of the protein includes the TOPRIM domain, followed by the breakage reunion domain, which carries the active site tyrosine. The C-terminal domain of the protein is not conserved between the type II topoisomerase from different species, nor is it conserved between Top2α and Top2β. The C-terminal domain is likely required for nuclear localization, regulation of enzyme activity by post-translational modification, and regulation of enzyme function by protein:protein interactions. The size and flexibility of Top2 has prevented the determination of structures of the intact enzyme. Therefore, much of the structural information has been obtained from structures of portions of the protein. The structures that have been determined for yeast Top2 include the amino terminal domain of the protein7 (the human α amino terminal domain has also been reported8), and three separate structures of the breakage-reunion domain, including a recent structure that includes this domain bound to DNA9. These protein structures have provided a rich source of insights into Top2 function and have been reviewed comprehensively10,11. Therefore, only key highlights of the structures are described (Fig. 2).
Figure 2. Structure of eukaryotic Top2.
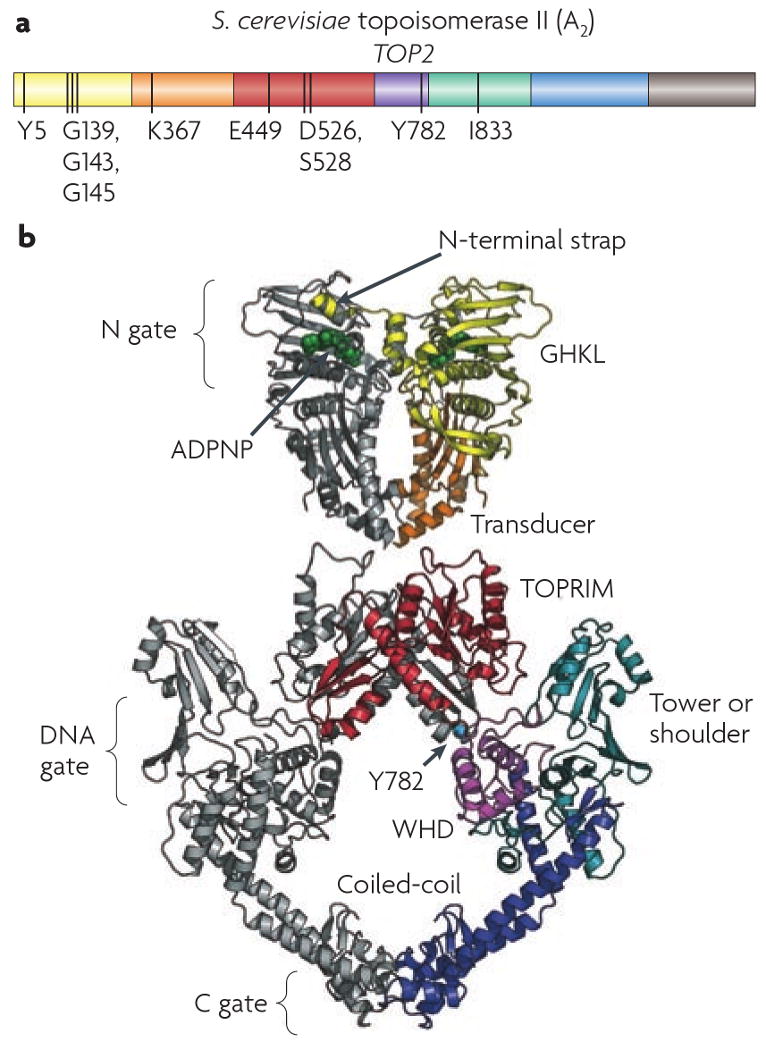
A. Domain structure of a eukaryotic Top2. Domains are indicated in color, and key residues are indicated. The residues marked include G139, G143 and G145 in the ATP binding domain, Lys367, a transducer domain residue that contributes to the ATPase; Glu449, Asp526, and Asp 528 the acidic triad involved in binding a divalent cation; Tyr782, the residue that makes a covalent attachment with DNA, and Ile833, a tower domain residue that is involved in DNA interaction. B. Structure of yeast Top2 based on structures for the ATPase domain and the breakage reunion domain7,90. The GHKL and transducer domain is shown in yellow and orange, TOPRIM, winged helix, tower, and coiled coil are shown in red, purple, teal and blue, and Tyr782 is shown as a cyan sphere. The figure is from James Berger 11.
The N-terminal- domain of the ATPase region consists of a GHKL fold that is found in a variety of ATPases10. An important characteristic of the ATP binding site is that both subunits contribute to its overall architecture7. The collaboration between the subunits couples ATP binding to dimer formation. Similarly, after ATP hydrolysis and release of ADP and Pi, dimerization at the amino terminus is destabilized. The C-terminal part of the ATPase has been termed the transducer domain. The transducer domain plays the role of signaling ATP binding to the breakage reunion domain. It appears to do so by undergoing a shift in position upon ATP binding that may trigger other conformational changes in the breakage reunion domain. Importantly, the transducer domain contributes amino acids that participate in ATP binding and hydrolysis. This may allow ATP hydrolysis to influence progression of the enzyme through the catalytic cycle (in addition to its role in amino terminal dimerization).
The breakage-reunion domain consists of a large heart shaped structure with a large central cavity. The amino terminal portion of the protein consists of the TOPRIM domain. The active site tyrosine is part of a winged helix domain that is similar to the catabolite activator protein (CAP-like domain). Adjacent to the CAP-like domain is a “tower” that leads into a long coiled coil that terminates in another dimer interface12. This initial structure, confirmed by a subsequent structure exhibited several features consistent with the two-gate model described above, especially in the C-terminal dimer interface that was likely to represent the exit point for the T-segment. The structure raised several questions, including the large separation between the tyrosine residues of the two subunits that was too great for interaction with B-DNA. A reasonable interpretation was that this structure represents an open state, where the enzyme has introduced a break in the DNA and separated the two strands to allow for passage of the T-segment. This point of view was supported by a second structure with a reduced separation between the active site tyrosines13. This second structure also showed a substantial conformational shift suggesting that this structure represented an intermediate between the open structure and the structure prior to cleavage. A second aspect of the two structures was that the TOPRIM domains were located too far from the active site tyrosines to participate in DNA cleavage, as suggested by biochemical data. Some of the questions raised by were answered in a third structure that included a nicked DNA molecule bound to the breakage reunion domain. In this structure, the TOPRIM domain is brought near to the active site tyrosine, allowing collaboration for DNA cleavage.
There are two other noteworthy aspects of the Top2:DNA binary complex9. First, the path of the DNA bound to TOP2 is sharply bent by 150°. The DNA between the active site tyrosines is in an A form helix. The detection of a bend in the DNA provided strong support for a model explaining a peculiar property of type II topoisomerases. DNA topoisomerase I, when carrying out relaxation generates a series of topoisomers centered around the lowest free energy state. The distribution of topoisomers follows a Boltzman distribution, consistent with the free energy associated with DNA supercoiling. By contrast, the distribution of topoisomers formed by Top2 relaxation is much sharper than expected. The biological significance of this reaction is that Top2 needs to perform complete separation of catenated molecules prior to mitosis and the presence of a single link would be sufficient to prevent proper segregation. Therefore this property of Top2, termed topology simplification helps to insure a complete decatenation reaction14. Cozzarelli and colleagues proposed a model for topology simplification that required that Top2 introduce a strong bend in the G-segment15. The reported structure fulfills this expectation. Finally, the Top2 structure shows that the carboxy terminal dimerization found in the other two structures to be disrupted. In other words, this structure shows the C-terminal gate open, in support of the prediction that the T-segment exits the enzyme through this gate.
Biological functions of Top
A key question in the biology of Top2 proteins is how the protein is localized to where it needs to perform its important functions. Since genetic analysis by loss-of-function mutants is difficult for proteins that are essential for all cells, proteomic approaches have also been used to dissect processes that use Top2 isozymes. An additional difficulty is that ectopic expression of Top2α has been difficult to achieve, and overexpression of the enzyme induces apoptosis16. This problem has been ameliorated by expression of N-terminal EGFP Top2 fusions17, however, it remains possible that the N-terminal tag significantly affects the function of the protein. Table 1 presents a compilation of proteins that have been described in the literature that physically interact with Top2 isozymes. Not all of the interactions listed in Table 1 have been demonstrated to have physiological relevance.
Table 1.
Proteins and protein complexes interacting with Top2 in mammalian cells
| Gene | Postulated function with Top2 | Isozyme | Reference |
|---|---|---|---|
| 14-3-3 ε | Modulates Top2 cleavage activity by an unknown mechanism, 14-3-3 proteins play roles in cell signaling | α | 91 |
| APC | adenomatous polyposis coli, important regulator of mitotic proteins | α | 92 |
| Aurora B | Protein kinase, regulator of mitotic events | α | 93 |
| BRCA1 | Tumor suppressor, activates Top2 decatenation activity perhaps by mono-ubiquitination | α | 94 |
| Casein kinase II (CK2) | Protein kinase with diverse functions, activates Top2 (phosphorylation by the enzyme not required) | α/β | 95-100 |
| p34CDC2 | Protein kinase regulator of cell cycle progression | α | 101 |
| CHRAC | Chromatin remodeling complex | Found in Drosophila but not human cells | 102,103 |
| CRM1 | Nuclear export protein, may stimulate elimination of Top2 from the nucleus under some conditions | α | 104,105 |
| HDAC1 | Histone deacetylase, gene repression | α/β | 106,107 |
| HDAC2 | Histone deacetylase, gene repression | α/β | 106,107 |
| Jab1/CSN5 | Regulation of Top2 stability | α | 108 |
| Ku70/Ku80 | Non-homologous end-joining, telomere metabolism, transcription | α/β | 56,57,109 |
| p53 | Tumor suppressor with diverse functions | α/β | 110 |
| PARP | poly(ADP-ribose) polymerase-1, multiple ce3llular functions including DNA repair | β | 56,57 |
| PCNA | DNA clamp required for DNA replication and repair | α | 111 |
| PIN1 | Interacts with both CK2 and Top2α, involved in replication termination and chromosome condensation | α | 41,112,113 |
| PLCR1 | PLCR1 encodes phospholipid scramblase 1, its possible roles in Top2 function are unknown | α/β | 114 |
| RanBP2 | SUMO E3 ligase, modifies Top2α | α | 39,115 |
| RARα | Retinoic acid receptor α, gene regulation | β | 59,99 |
| RHA1 | RNA helicase I | α | 116 |
| SUMO 1/2/3 | Small ubiquitin-like modifier, diverse cellular functions | α/β | 35,117-119 |
| TCF4 | beta-catenin/T-cell factor-4 (TCF-4) nuclear complex, transcription | α | 120 |
| TOPBP1 | DNA damage checkpoint protein, homolog of the yeast replication protein Dpb11 | β | 80,121 |
| Toposome | Multi-protein complex that includes RHA1, a protein kinase SRPK1, and other proteins. Function not yet determined. | α | 122 |
Role of Top2 in replication
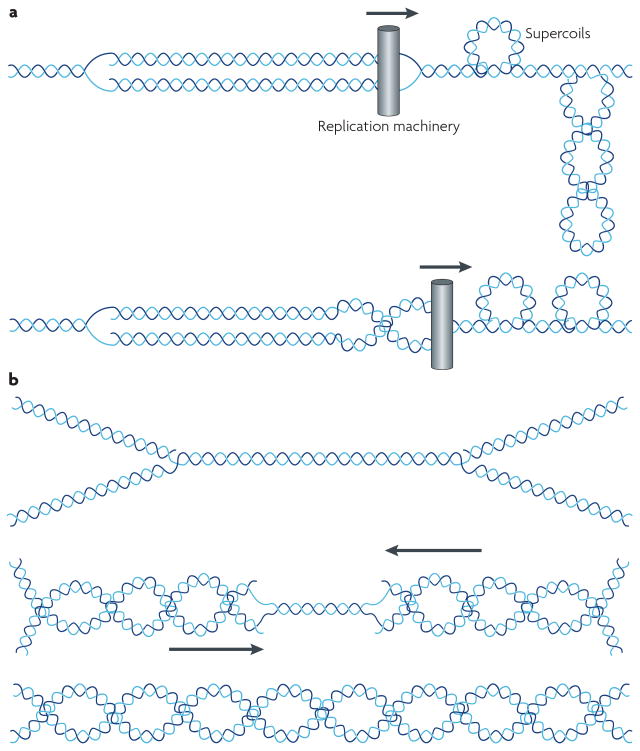
One of the central roles of DNA topoisomerases is to solve the topological problems associated with replication. Semi-conservative replication involves the unwinding of duplex DNA and copying of each strand. In the absence of a topoisomerase activity the unwinding of the parental duplex leads to the accumulation of positively supercoiled DNA in front of the replication fork, which can be relaxed by either Top1 or Top2. In addition to the generation of positive supercoiling in front of the fork, the positively supercoiled DNA at the replication fork can isomerize by migration of the positive supercoiling into wrapping of the two replicated strands (Fig. 3). This structure called a precatenane, is a substrate for Top2 mediated DNA catenation, and may represent a plausible mechanism for Top2 action during replication elongation18. Studies in bacterial replication have provided clear evidence for precatenane formation 19 and it is likely that this mechanism is also important in eukaryotic cells18,20. In the latter stages of replication, when two replication forks impinge on each other, there is no longer room for a type I topoisomerase to relax positive supercoils, and completion of replication leads to two interlinked catenanes. This catenated dimer requires Top2 for resolution (Fig. 3).
Figure 3. Roles of Top2 in replication.
A. Partition of superhelical strain during replication fork progression in vivo. During replication helicase action on DNA creates positive superhelical stress on the DNA. This results in positive supercoils in front of the fork, as shown in A. The structure shown in A can isomerize into intertwinings of the daughter duplexes, generating precatenanes, as shown in B. (Figure is from Postow and Cozzarelli, Bioessays 21:805, 1999). B. At early steps in replication, when forks are widely separated, either Top1 or Top2 can function as a replication swivel. Top1 acts by relaxing positive supercoils while Top2 unlinks precatenanes. Note that Top2 also should be able to relax positive supercoils, and does not require the isomerization to precatenanes for unlinking replicated strands. As the replication forks converge, there is a limited ability to generate positive supercoiling, and complete unlinking absolutely requires Top2.
The products of replication of a small circular DNA in vitro in the absence of Top2 are catenated dimeric plasmids. A requirement for Top2 in this reaction with chromosomal DNA was first observed in yeast. Yeast cells having Top2 as the only active topoisomerase are viable, and undergo normal DNA replication21,22, without activation of any S phase dependent checkpoints 23. In the absence of Top2 (e.g., using temperature sensitive yeast mutants), cells complete DNA replication, and die when they enter mitosis24. Interestingly, the effects on replication differ between yeast cells completely lacking any Top2 protein, and cells carrying an enzymatically inactive protein25. Cells depleted of Top2 using a conditionally degradable Top2 protein could complete replication, but not chromosome decatenation, and in agreement with results obtained with temperature sensitive proteins, lose viability at mitosis. Expression of a catalytically dead protein generated a different phenotype, a failure to complete replication at sites where two replication forks meet. A plausible model for these results is that Top2 is normally recruited to act where replication forks meet. In the complete absence of the protein, replication is complete, with the products of the reaction being catenated sister chromatids. The presence of a catalytically inactive protein interferes with the completion of replication, leading to checkpoint induction.
Experiments in mammalian cells using conditionally expressed Top2α26 or siRNA knockdown of Top2α27 (see Box 1) support this model for Top2 action. Since many cell types can be recovered from top2β homozygous knockout mice and grown in culture, it is unlikely that top2β plays a critical role in replication. Studies using RNAi of Top2α generally fail to reveal an indispensable role during replication, although recent experiments suggest that phosphorylation of Top2α during S phase is required for normal S phase progression27. Interestingly, biochemical analysis of the human Top2α has shown that the protein is much more active in relaxing positively supercoiled substrates than negatively supercoiled substrates. This property is not found in Top2β, nor is it seen with lower eukaryotic type II topoisomerases28. Since positive supercoiling is expected to be generated in advance of a replication fork, this preferential activity has been suggested to imply an important role for Top2α at some point in replication.
The role of Top2 in chromosome segregation
Although catenation of replicated chromosomes is presented as a “problem”, the generation of catenated sister chromatids may assist in the proper segregation of duplicated chromosomes. After replication, sister chromatids must stay together until mitosis. Precocious separation leads to inaccurate chromosome transmission. While early models of sister chromatid cohesion posited a role for catenanes in cohesion maintenance, subsequent studies showed that specialized protein complexes called cohesins were essential for keeping sister chromatids together29,30. Surprisingly, while mutation of cohesins diminished cohesion, some cohesion was still evident 31,32. One possible explanation for these results is that cohesion can be maintained by multiple mechanisms, with catenanes representing one of several mechanisms.
Support for a role for catenation in chromosome cohesion came from seminal studies on regulation of yeast Top2 by the ubiquitin like modified SUMO33. Mutation of the SMT4, the isopeptidase that deconjugates SUMO leads to precocious sister chromatid separation. The defect in cohesion was specific for regions near the yeast centromere. This defect could be suppressed either by overexpression of yeast Top2, or by mutating all candidate sites on Top2 that could be modified by SUMO. One explanation for these results is that SUMO modification blocks the ability of Top2 to maintain cohesion at chromosomes. An economical explanation is that SUMO modification inhibits decatenation (or promotes catenation) by Top2. Since Top2 plays roles in chromosome structure, the SUMO modification may impart a structural alteration required for maintaining cohesion at centromeres. Other recent studies in mammalian cells support the hypothesis that Top2 has a function at centromeres34, although the full details remain to be elaborated.
Protein modification of Top2α
SUMO modification of Top2α is crucial in mammalian cells. Initial experiments in Xenopus suggested that PIASγ is a major SUMO E3 ligase35. Depletion of PIASγ from Xenopus extracts leads to metaphase arrest, and depletion of sumoylated proteins from the inner centromere. Support for this hypothesis was obtained using siRNA directed against PIASγ in human cells, finding a lack of Top2α localization to centromeres in PIASγ deficient cells 36. Surprisingly, a chromosome segregation defect was not seen in a mouse knockout of PIASγ37,38. Other recent results have called into question the role of PIASγ in sumoylation of Top2. RanBP2 is a nucleoporin with SUMO E3 ligase activity. The gene is essential in mouse, and hypomorphs show a defect in chromosome segregation, generation of anaphase bridges, induction of high levels of aneuploidy, and elevated spontaneous and chemical induced tumorigenesis39. In vitro analysis demonstrated that RanBP2 hypomorphs are defective in Sumo modification of Top2α with a failure to localize Top2α to centromeres. Ectopic expression of either RanBP2 or a SUMO-Top2 fusion restores Top2 localization to centromeres. The same authors also showed that PIASγ deficient MEFs do not show a defect in Top2α localization, nor do they show a defect in Top2α sumoylation. These results provide overwhelming support for the hypothesis that RanBP2 is the major SUMO E3 ligase for Top2α. What might be the importance of PIASγ? Although it may not participate in sumoylation of Top2, many other centromere proteins are also sumoylated, and the defects observed in Xenopus extracts may reflect roles in sumoylating other proteins. The experiments with RanBP1 highlight the potential importance of Top2 in chromosome stability. RanBP2 likely has other important targets besides Top2α that may contribute to high levels of aneuploidy and tumorigenesis. However, the results suggest the interesting possibility that prevention of aneuploidy rightly qualifies Top2α as a tumor suppressor.
Top2 and chromosome structure
Classical studies indicated that Top2 plays a key role in chromosome structure and chromosome condensation40,41. Early studies using specific extraction procedures identified a chromosome scaffold that included Top2α and an additional complex now termed condensin29,42-44. A role for Top2 in condensation was certainly plausible based on possible topological constraints as chromatin is compacted. A detailed description of current issues related to chromosome structure and condensation is beyond the scope of this review. Many current issues relate to what steps absolutely require Top2, given that chromosome condensation can occur in many contexts where Top2 is absent. The ability to examine the roles of Top2 in physiological settings by RNAi or by conditional replacement using mutant alleles of Top2 will be critical in understanding these processes, and how they connect to other cellular events including decatenation and faithful chromosome segregation.
Transcription
In yeast, loss of either topoisomerase does not block DNA replication or transcription, but both processes are strongly inhibited if both enzyme activities are absent 21,22,45-47. The effect on transcription in yeast is mainly on transcription by RNA polymerase I; overall levels of polymerase II transcription are affected to a much lesser extent. It has recently been suggested that in yeast, Top2 may be more active in relaxing supercoils in chromatin than topoisomerase I48, although this property has not yet been associated with any unique phenotypic consequences. It was initially reported that there may be a unique requirement for a type II topoisomerase for transcription in vitro on chromatin templates, based in part on the association of Top2α with a multi-subunit RNA polymerase II holoenzyme49. Subsequent work indicated that either a type I or a type II topoisomerase could support transcription on chromatin templates50. This finding suggests that the critical function provided by the topoisomerase is relaxation of DNA supercoiling. It should be noted that Top1 functions as a basal transcription factor in vitro, but this function can also be carried out by Top1 protein that is catalytically inactive due to an active site mutation51-53. Therefore, these functions of topoisomerases in transcription differ from the structural role of Top1 previously described. An important model for generation of supercoiling during transcription has been described by Liu and Wang, and posits that the tracking of RNA polymerase leads to transient positive supercoiling ahead of the transcriptional machinery and negative supercoiling behind it; the generation of supercoils could reasonably be exacerbated by the presence of chromatin54,55. The transcriptional supercoiling model provides an important basis for a requirement of a topoisomerase during transcriptional elongation. Whether there are contexts where a specific topoisomerase is preferentially utilized remains to be determined.
Recent work has also provided evidence for a specific role for mammalian Top2β in transcription initiation. Ju et al. used chromatin IP (ChIP) to demonstrate that Top2β localizes to promoters of genes whose expression is activated by nuclear hormone receptors (but not to many other promoters undergoing active transcription)56,57. They showed that Top2β associates with signal dependent promoters as part of a complex that includes several proteins important for DNA repair, and that the enzymatic activity of Top2β was required for efficient transcriptional activation. It is very important to note that although the complex that associates with the promoter includes several proteins that play key roles in DNA repair, such as PARP, DNA dependent protein kinase, and Ku70/Ku86; the presence of the repair proteins does not seem to be required to “repair” the Top2β induced break. Rather, Top2β is recruited to a subset of promoters, in a complex that includes DNA repair proteins. The enzymatic function of Top2β is required at the promoter, rather than the enzyme acting in a purely structural role as as occurs with Top1. The break induced by Top2β is the normal cleavage of the enzyme reaction cycle, as shown in figure 1. The enzymatic function of PARP also appears to be required, but it may function in ways that are distinct from the function of this enzyme in DNA repair. The importance of this finding is that it establishes a specific role for the enzymatic activity of a type II topoisomerase in transcriptional regulation. Several interesting questions are raised by this work including identifying the determinants that lead to recruitment of Top2β, and assessing whether recruitment of a topoisomerase other than Top2β can also lead to transcriptional activation.
While it is easy to appreciate that Top2 activity may be required for activation of transcription58, a recent result suggests that Top2β can participate in repression of transcription. Miller and colleagues showed that Top2β can negatively regulate RARα transcriptional activation59. They hypothesize that in this context Top2β is part of a distinct complex from the one described by Ju and colleagues.
Further support for a specific role of Top2β in transcriptional regulation has been provided by Lyu and colleagues60. Since Top2β plays key roles in neural development61,62, they reasoned that loss-of-function of Top2β might lead to alterations in gene expression in neural tissue. Mice carrying homozygous deletions of Top2β are inviable due to multiple neuronal deficits including a failure of motor neurons to innervate the diaphragm. Using microarray analysis, they determined that approximately 1-4% of expressed genes showed changes in expression in Top2β -/- mice. Importantly, they were also able to demonstrate localization of Top2β to various genes including many developmentally regulated genes. Taken together the studies described above clearly indicate important contexts where Top2β influences regulation of gene expression. It will be interesting to determine in what other contexts type II topoisomerases contribute to gene regulation, especially in pathways related to cancer development.
When Topoisomerase II fails: Checkpoints for insuring correct Top2 function
Key events in progression through the cell cycle are monitored through a series of checkpoints. Checkpoints assess the integrity of critical events during the cell cycle, such as the completion of DNA replication, and the presence of an appropriate mitotic spindle63. Since topoisomerase II carries out a reaction that is essential for chromosome separation at mitosis, a plausible hypothesis is that cells can monitor the successful completion of topoisomerase II decatenation, and arrest cell cycle progression if decatenation (or chromosome condensation) is incomplete. Early studies using S. cerevisiae and S. pombe argued against this possibility, since conditional top2 mutants showed minimal cell cycle delay, and instead accumulated broken chromosomes at the time of mitosis 24,64-66. Topoisomerase II poisons generate DNA damage, in addition to inhibition of enzyme activity, and would be expected to delay cell cycle progression due to DNA damage checkpoints.67-69 The demonstration by Andoh and colleagues that bisdioxopiperazines were specific catalytic inhibitors of eukaryotic topoisomerase II70 allowed a test in mammalian cells for the presence of a checkpoint for topoisomerase II function. Downes and colleagues found that bisdioxopiperazines such as ICRF-187 and ICRF-193 were able to elicit a caffeine sensitive delay of entry into mitosis71. Subsequent work using ICRF-187 demonstrated a mitotic delay that was dependent on ATR and BRCA172, but apparently independent of both DNA damage checkpoints and the spindle checkpoint73. As carefully noted by Downes and colleagues, the checkpoint they identified depended on the properties of bisdioxopiperazines71. Since they were able to show distinct differences between etoposide (as a standard Top2 poison) and ICRF-193, they concluded that the effects of ICRF-193 arose from a lack of Top2 activity.
An alternate approach to assessing whether cells monitor the completion of Top2 function is to completely deplete Top2 protein before mitosis. This has been done both in yeast and mammalian cells. As described above in the section on replication, a complete depletion of Top2 does not lead to a delay in mitosis in yeast cells25, while expression of an inactive Top2 does lead to a mitotic delay. This finding is in agreement with a previous hypothesis that yeast cells carrying a temperature sensitive Top2 fails to arrest at mitosis because the presence of Top2 is needed to trigger the delay74. However, the results of Diffley and colleagues suggest that the arrest seen in yeast is due to a problem with replication rather than decatenation. In mammalian cells, whether mitotic delay is induced by Top2α depletion is a point of controversy. Removal of Top2 using a conditional Tet-off system showed that loss of Top2 protein led to mitotic delay. By contrast, no delay was seen in cells depleted for Top2α using siRNA27. In the latter experiments, loss of cell viability was clearly seen in cells depleted for Top2α. In experiments from other laboratories using siRNA directed against Top2α, no phenotype was observed presumably because the knockdown of Top2α was insufficient. Additional experiments, perhaps with primary cells will be useful in demarcating the types of cells that can carry out mitotic delay in response to insufficient Top2 activity. It is interesting to note that recent experiments indicate a lack of a mitotic delay induced by bisdioxopiperazines in embryonic stem cells and hematopoetic progenitor cells75,76.
The checkpoint induced by bisdioxopiperazines is termed a decatenation checkpoint, but it would more accurately be termed a Top2 checkpoint, since there is no direct evidence that the mitotic delay monitors chromatid decatenation. It is not clear how the cell could assess sister chromatid catenation. The presence of catenanes is a property of a chromosomal domain, and it does not generate obvious local consequences. For example, cells might assess DNA supercoiling by “counting” crossing of the DNA double helix (formally writhe), but there is no obvious way to assess writhe that is specifically associated with catenanes. Assessment of catenation state may depend more on structural alterations, perhaps at centromeres.
There has been interest in determining whether the topoisomerase II checkpoint can be exploited for cancer therapy. A small molecule inhibitor of a bisdioxopiperazines induced checkpoint has been described, although the molecular target of the small molecule is unknown77. This may be of particular use in concert with potent Top2 catalytic inhibitors. In any case, perturbing Top2 checkpoints are unlikely to be a major determinant of response to Top2 poisons, which depend more on DNA damage checkpoints for their efficacy.
Top2 is required in many biological contexts
The original impetus for studying Top2 came in part from the mysterious and complicated reactions the enzyme carries out. Therefore, early studies concentrated especially on the biochemical and structural aspects of the enzyme. These studies have now concluded with a detailed understanding of many critical issues of Top2 biochemistry. Top2 was expected to be important in chromosome replication and segregation, but the recent work suggests that decatenation of replicated chromosomes requires a precise choreography that includes regulating Top2 action both spatially and temporally. Importantly, cells may have the means of ensuring that these processes have occurred correctly, although how cells assess proper Top2 function remains unclear.
A recent surprise has been the unique roles played by Top2β. It was surprising that Top2β is specifically required in certain neuronal cells, and the finding that this enzyme is required for transcription of some genes will lead to further unappreciated biological roles for both Top2 isoforms. Although not discussed in detail here, Top2 has also been proposed to play roles in DNA repair78-80, especially in the ability of DNA lesions such as abasic sites to generate enzyme mediated DNA damage81. Other possible functions of Top2 will depend on a better understanding of the protein complexes that include Top2. As with other proteomic studies, identification of the relevant protein complexes is only the first step in understanding the relevant biological processes.
As described in the accompanying review, Top2 is especially relevant in cancer because it is the target of many active anti-cancer agents. At present, most drugs targeting Top2 kill cells by generating enzyme mediated DNA damage, rather than by inhibiting enzyme activity. The importance of Top2 in proliferating cells, as well as its roles in transcription, suggest that catalytic inhibition may also be a useful anti-cancer strategy. If this strategy proves useful, a more complete understanding of Top2 biological functions will be critical.
At a glance.
Type II topoisomerases change DNA topology by generating transient DNA double strand breaks. Type II topoisomerases are essential for all eukaryotic cells.
Mammalian cells carry two Top2 isoforms, Top2α and Top2β. Top2α is essential for all cells, and is essential for separating replicated chromosomes. Top2β is required for normal development, but is dispensable in some cell types. Type II topoisomerases are required for other processes such as transcription, and the precise roles of the two isoforms in these processes are a subject of current studies.
Type II topoisomerases use a “two gate” mechanism for carrying out topological changes in DNA. The enzyme requires ATP hydrolysis for its reaction. ATP hydrolysis is used for for conformational changes of the enzyme, and is not directly involved in DNA breakage or resealing.
Structures of several domains of yeast Top2 have provided additional information about how the enzyme carries out its reactions. A recent structure of the breakage-reunion domain of yeast Top2 bound to DNA has shown that the enzyme induces a large bend in the DNA that is cleaved by the enzyme.
Biological functions of Top2 isoforms are modulated be a variety of protein:protein interactions. Some of these interactions may affect enzyme activity, stability, and localization.
Top2 activity is also modulated by post-translational modification. In addition to phosphorylation, a critical post-translational modification of Top2 is sumoylation. Failure to sumoylate Top2α or to remove the SUMO mopdification disrupts the ability of Top2α to separate replicated chromosomes.
Top2β plays a key role in the survival of some neural cells. Top2β is important in transcriptional regulation, and it is likely that Top2β enzyme activity is specifically required.
Some aspects of Top2 function during the cell cycle are monitored by checkpoints. It has been hypothesized that a major role of checkpoints are to monitor the completion of decatenation. If so, then Top2 dependent checkpoints may be critical for normal chromosome segregation and genome stability.
Acknowledgments
Dr. James Berger, University of California Berkeley kindly provided figures that were of great use, and also provide useful discussion. I also thank Dr. Yves Pommier for encouragement. Work in the author's laboratory is supported by grants from the National Cancer Institute (CA82313 and CA21765) and the American Lebanese Syrian Associated Charities (ALSAC).
Glossary terms
- Bisdioxopiperazines
A class of small molecules, which includes ICRF-159, ICRF-187, and MST-16, that inhibit the catalytic activity of Top2 and do not stabilize the Top2 cleaved complex. Bisdioxopiperazines are the most commonly used catalytic inhibitors of type II topoisomerases
- Catenanes
Circles linked as in a chain, and clearly the two links cannot be separated without breaking one of the two molecules. A closely related structure termed a pre-catenane refers to the interwinding of DNA strands behind a replication fork. Pre-catenanes interconvert with positive supercoils that arise in front of a replication fork
- TOPRIM domain
A conserved domain found in topoisomerases, primases, and other DNA metabolic enzymes. The Toprim domain adopts a Rossman fold, and is involved in divalent cation binding
References cited
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q Rev Biophys. 1998;31:107–44. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 3.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–69. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 4.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21:103–14. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 5.Bates AD, Maxwell A. DNA topology. Oxford University Press; Oxford: 2005. [Google Scholar]
- 6.Aravind L, Leipe DD, Koonin EV. Toprim - a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Research. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc Natl Acad Sci U S A. 2003;100:10629–34. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. Journal of Biological Chemistry. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 9.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–5. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 10.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 11.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 12.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–32. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]; published erratum appears in Nature. 1996 Mar 14;380(6570):179. doi: 10.1038/380092b0.
- 13.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat Struct Biol. 1999;6:322–6. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 14.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 15.Vologodskii AV, et al. Mechanism of topology simplification by type II DNA topoisomerases. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPherson JP, Goldenberg GJ. Induction of apoptosis by deregulated expression of DNA topoisomerase IIalpha. Cancer Research. 1998;58:4519–24. [PubMed] [Google Scholar]
- 17.Mo YY, Ameiss KA, Beck WT. Overexpression of human DNA topoisomerase II alpha by fusion to enhanced green fluorescent protein. Biotechniques. 1998;25:1052–7. doi: 10.2144/98256cr04. [DOI] [PubMed] [Google Scholar]
- 18.Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. Embo Journal. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–27. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 20.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: Conformations at the fork. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–6. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]; published erratum appears in Nature. 1987 Apr 23-29;326(6115):812.
- 22.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol. 1989;208:257–67. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 23.Bermejo R, et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–36. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–63. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 25.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol Biol Cell. 2004;15:5700–11. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIalpha in cell cycle progression. J Biol Chem. 2008;283:6209–21. doi: 10.1074/jbc.M709007200. [DOI] [PubMed] [Google Scholar]
- 28.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase II alpha rapidly relaxes positively supercoiled DNA - Implications for enzyme action ahead of replication forks. Journal of Biological Chemistry. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 29.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes & Development. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 30.Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Chromosome cohesion - rings, knots, orcs and fellowship. J Cell Sci. 2008;121:2107–14. doi: 10.1242/jcs.029132. [DOI] [PubMed] [Google Scholar]
- 31.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 32.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philosophical Transactions of the Royal Society B-Biological Sciences. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–82. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 34.Porter AC, Farr CJ. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 2004;12:569–83. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- 35.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. Embo J. 2005;24:2172–82. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Martinez LA, et al. PIASgamma is required for faithful chromosome segregation in human cells. PLoS ONE. 2006;1:e53. doi: 10.1371/journal.pone.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth W, et al. PIASy-deficient mice display modest defects in IFN and Wnt signaling. Journal of Immunology. 2004;173:6189–6199. doi: 10.4049/jimmunol.173.10.6189. [DOI] [PubMed] [Google Scholar]
- 38.Wong KA, et al. Protein inhibitor of activated STAT y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Molecular and Cellular Biology. 2004;24:5577–5586. doi: 10.1128/MCB.24.12.5577-5586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawlaty MM, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–15. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belmont AS. Mitotic chromosome structure and condensation. Current Opinion in Cell Biology. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Xu YX, Manley JL. The prolyl isomerase Pin1 functions in mitotic chromosome condensation. Mol Cell. 2007;26:287–300. doi: 10.1016/j.molcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 42.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell. 2003;4:467–80. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 43.Adachi Y, Kas E, Laemmli UK. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. Embo J. 1989;8:3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188:613–29. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 45.Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–41. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 46.Goto T, Wang JC. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc Natl Acad Sci U S A. 1985;82:7178–82. doi: 10.1073/pnas.82.21.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci U S A. 1992;89:11461–5. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. Embo J. 2006;25:2575–83. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mondal N, Parvin JD. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–8. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- 50.Mondal N, et al. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 2003;31:5016–24. doi: 10.1093/nar/gkg705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci U S A. 1993;90:11508–12. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–32. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 53.Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 54.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 56.Ju BG, Rosenfeld MG. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle. 2006;5:2557–60. doi: 10.4161/cc.5.22.3497. [DOI] [PubMed] [Google Scholar]
- 57.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 58.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nature Reviews Molecular Cell Biology. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 59.McNamara S, Wang H, Hanna N, Miller WH., Jr Topoisomerase IIbeta negatively modulates retinoic acid receptor alpha function: a novel mechanism of retinoic acid resistance. Mol Cell Biol. 2008;28:2066–77. doi: 10.1128/MCB.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyu YL, et al. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26:7929–41. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc Natl Acad Sci U S A. 2003;100:7123–8. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–4. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 63.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 64.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–68. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. Embo J. 1984;3:1737–44. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uemura T, et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–25. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 67.Downes CS, Mullinger AM, Johnson RT. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc Natl Acad Sci U S A. 1991;88:8895–9. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clarke DJ, Johnson RT, Downes CS. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J Cell Sci. 1993;105(Pt 2):563–9. doi: 10.1242/jcs.105.2.563. [DOI] [PubMed] [Google Scholar]
- 69.Clifford B, Beljin M, Stark GR, Taylor WR. G2 arrest in response to topoisomerase II inhibitors: the role of p53. Cancer Res. 2003;63:4074–81. [PubMed] [Google Scholar]
- 70.Ishida R, et al. Inhibition of intracellular topoisomerase II by antitumor bis(2,6- dioxopiperazine) derivatives: mode of cell growth inhibition distinct from that of cleavable complex-forming type inhibitors. Cancer Res. 1991;51:4909–16. [PubMed] [Google Scholar]
- 71.Downes CS, et al. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–70. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 72.Deming PB, et al. The human decatenation checkpoint. Proc Natl Acad Sci U S A. 2001;98:12044–9. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol Cell. 2004;15:977–90. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 74.Andrews CA, et al. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 2006;20:1162–74. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Damelin M, Bestor TH. Decatenation checkpoint deficiency destabilizes the stem cell genome. Cell Cycle. 2006;5:345–346. doi: 10.4161/cc.5.4.2480. [DOI] [PubMed] [Google Scholar]
- 76.Damelin M, Sun YE, Sodja VB, Bestor TH. Decatenation checkpoint deficiency in stem and progenitor cells. Cancer Cell. 2005;8:479–484. doi: 10.1016/j.ccr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Haggarty SJ, et al. Small molecule modulation of the human chromatid decatenation checkpoint. Chem Biol. 2003;10:1267–79. doi: 10.1016/j.chembiol.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 78.Nitiss JL. DNA topoisomerases in DNA repair and DNA damage tolerance. In: Nickoloff JA, Hoekstra MF, editors. DNA damage and Repair, Vol. 2; DNA repair in Higher Eukaryotes. Humana Press; Totawa, NJ: 1998. pp. 517–537. [Google Scholar]
- 79.Kingma PS, Osheroff N. The response of eukaryotic topoisomerases to DNA damage. Biochimica Et Biophysica Acta. 1998;1400:223–232. doi: 10.1016/s0167-4781(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 80.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–66. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilstermann AM, Osheroff N. Base excision repair intermediates as topoisomerase II poisons. J Biol Chem. 2001;276:46290–6. doi: 10.1074/jbc.M105733200. [DOI] [PubMed] [Google Scholar]
- 82.Corbett KD, Berger JM. Emerging roles for plant topoisomerase VI. Chem Biol. 2003;10:107–11. doi: 10.1016/s1074-5521(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 83.Lichten M. Meiotic recombination: breaking the genome to save it. Curr Biol. 2001;11:R253–6. doi: 10.1016/s0960-9822(01)00131-2. [DOI] [PubMed] [Google Scholar]
- 84.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase II beta. Bioessays. 1998;20:215–26. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 85.Linka RM, et al. C-terminal regions of topoisomerase II alpha and II beta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Research. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keeney S, et al. A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics. 1999;61:170–82. doi: 10.1006/geno.1999.5956. [DOI] [PubMed] [Google Scholar]
- 87.Romanienko PJ, Camerini-Otero RD. Cloning, characterization, and localization of mouse and human SPO11. Genomics. 1999;61:156–69. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 88.Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol. 2002;12:1782–6. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- 89.Harkins TT, Lindsley JE. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis. Biochemistry. 1998;37:7292–8. doi: 10.1021/bi9729099. [DOI] [PubMed] [Google Scholar]
- 90.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–32. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 91.Kurz EU, Leader KB, Kroll DJ, Clark M, Gieseler F. Modulation of human DNA topoisomerase IIalpha function by interaction with 14-3-3epsilon. J Biol Chem. 2000;275:13948–54. doi: 10.1074/jbc.275.18.13948. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, Azuma Y, Moore D, Osheroff N, Neufeld KL. Interaction between Tumor Suppressor Adenomatous Polyposis Coli and Topoisomerase II alpha: Implication for the G2/M Transition. Molecular Biology of the Cell. 2008;19:4076–4085. doi: 10.1091/mbc.E07-12-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morrison C, et al. Proteomic analysis of human metaphase chromosomes reveals topoisomerase II alpha as an Aurora B substrate. Nucleic Acids Res. 2002;30:5318–27. doi: 10.1093/nar/gkf665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat Struct Mol Biol. 2005;12:589–93. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- 95.Ackerman P, Glover CV, Osheroff N. Phosphorylation of DNA topoisomerase II by casein kinase II: modulation of eukaryotic topoisomerase II activity in vitro. Proc Natl Acad Sci U S A. 1985;82:3164–8. doi: 10.1073/pnas.82.10.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DeVore RF, Corbett AH, Osheroff N. Phosphorylation of topoisomerase II by casein kinase II and protein kinase C: effects on enzyme-mediated DNA cleavage/religation and sensitivity to the antineoplastic drugs etoposide and 4′-(9- acridinylamino)methane-sulfon-m-anisidide. Cancer Res. 1992;52:2156–61. [PubMed] [Google Scholar]
- 97.Redwood C, Davies SL, Wells NJ, Fry AM, Hickson ID. Casein kinase II stabilizes the activity of human topoisomerase IIalpha in a phosphorylation-independent manner. J Biol Chem. 1998;273:3635–42. doi: 10.1074/jbc.273.6.3635. [DOI] [PubMed] [Google Scholar]
- 98.Cardenas ME, Gasser SM. Regulation of topoisomerase II by phosphorylation: a role for casein kinase II. J Cell Sci. 1993;104(Pt 2):219–25. doi: 10.1242/jcs.104.2.219. [DOI] [PubMed] [Google Scholar]
- 99.Isaacs RJ, et al. Physiological regulation of eukaryotic topoisomerase II. Biochim Biophys Acta. 1998;1400:121–37. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 100.Ahn BH, Kim TH, Bae YS. Mapping of the interaction domain of the protein kinase CKII beta subunit with target proteins. Mol Cells. 2001;12:158–63. [PubMed] [Google Scholar]
- 101.Wells NJ, Hickson ID. Human topoisomerase II alpha is phosphorylated in a cell-cycle phase- dependent manner by a proline-directed kinase. Eur J Biochem. 1995;231:491–7. doi: 10.1111/j.1432-1033.1995.tb20723.x. [DOI] [PubMed] [Google Scholar]
- 102.Poot RA, et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. Embo J. 2000;19:3377–87. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Varga-Weisz PD, et al. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 104.Turner JG, Engel R, Derderian JA, Jove R, Sullivan DM. Human topoisomerase IIalpha nuclear export is mediated by two CRM-1-dependent nuclear export signals. J Cell Sci. 2004;117:3061–71. doi: 10.1242/jcs.01147. [DOI] [PubMed] [Google Scholar]
- 105.Mirski SE, et al. Topoisomerase II binds importin alpha isoforms and exportin/CRM1 but does not shuttle between the nucleus and cytoplasm in proliferating cells. Exp Cell Res. 2007;313:627–37. doi: 10.1016/j.yexcr.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 106.Tsai SC, et al. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet. 2000;26:349–53. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 107.Johnson CA, Padget K, Austin CA, Turner BM. Deacetylase activity associates with topoisomerase II and is necessary for etoposide-induced apoptosis. J Biol Chem. 2001;276:4539–42. doi: 10.1074/jbc.C000824200. [DOI] [PubMed] [Google Scholar]
- 108.Yun J, Tomida A, Andoh T, Tsuruo T. Interaction between glucose-regulated destruction domain of DNA topoisomerase IIalpha and MPN domain of Jab1/CSN5. J Biol Chem. 2004;279:31296–303. doi: 10.1074/jbc.M401411200. [DOI] [PubMed] [Google Scholar]
- 109.Matheos D, Ruiz MT, Price GB, Zannis-Hadjopoulos M. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochim Biophys Acta. 2002;1578:59–72. doi: 10.1016/s0167-4781(02)00497-9. [DOI] [PubMed] [Google Scholar]
- 110.Cowell IG, et al. Human topoisomerase IIalpha and IIbeta interact with the C-terminal region of p53. Exp Cell Res. 2000;255:86–94. doi: 10.1006/excr.1999.4772. [DOI] [PubMed] [Google Scholar]
- 111.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma. 2001;110:102–14. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 112.Messenger MM, et al. Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. J Biol Chem. 2002;277:23054–64. doi: 10.1074/jbc.M200111200. [DOI] [PubMed] [Google Scholar]
- 113.Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev. 2008;22:860–5. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wyles JP, Wu Z, Mirski SE, Cole SP. Nuclear interactions of topoisomerase II alpha and beta with phospholipid scramblase 1. Nucleic Acids Res. 2007;35:4076–85. doi: 10.1093/nar/gkm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Navarro MS, Bachant J. RanBP2: a tumor suppressor with a new twist on TopoII, SUMO, and centromeres. Cancer Cell. 2008;13:293–5. doi: 10.1016/j.ccr.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 116.Zhou K, et al. RNA helicase A interacts with dsDNA and topoisomerase IIalpha. Nucleic Acids Res. 2003;31:2253–60. doi: 10.1093/nar/gkg328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275:26066–73. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 118.Agostinho M, et al. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 2008;68:2409–18. doi: 10.1158/0008-5472.CAN-07-2092. [DOI] [PubMed] [Google Scholar]
- 119.Takahashi Y, Strunnikov A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2008;117:189–98. doi: 10.1007/s00412-007-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang L, et al. Functional interaction of DNA topoisomerase II alpha with the beta-catenin and T-Cell factor-4 complex. Gastroenterology. 2007;133:1569–1578. doi: 10.1053/j.gastro.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 121.Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem. 1997;250:794–9. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- 122.Lee CG, Hague LK, Li H, Donnelly R. Identification of toposome, a novel multisubunit complex containing topoisomerase IIalpha. Cell Cycle. 2004;3:638–47. [PubMed] [Google Scholar]