Summary
Recent studies suggest that thousands of genes may contribute to breast cancer pathophysiologies when deregulated by genomic or epigenomic events. Here, we describe a model “system” to appraise the functional contributions of these genes to breast cancer subsets. In general, the recurrent genomic and transcriptional characteristics of 51 breast cancer cell lines mirror those of 145 primary breast tumors, although some significant differences are documented. The cell lines that comprise the system also exhibit the substantial genomic, transcriptional, and biological heterogeneity found in primary tumors. We show, using Trastuzumab (Herceptin) monotherapy as an example, that the system can be used to identify molecular features that predict or indicate response to targeted therapies or other physiological perturbations.
SIGNIFICANCE
The description of the in vitro breast cancer cell line system described here allows assessment of similarities and differences between the cell lines and primary human breast tumors. In general, the system seems well suited to assess the functional contributions of genome copy number abnormalities to breast cancer pathophysiologies, since most of the recurrent genomic deregulation of transcription present in primary tumors is retained in the cell lines. The genomically and biologically heterogeneous cell line system also may be used to identify molecular features that predict or indicate response (or lack thereof) to pathway-targeted therapeutic agents. These features may be assessed as candidate response predictors/indicators to guide early-phase clinical trials.
Introduction
The evolution of a normal, finite-life-span somatic epithelial cell into an immortalized, metastatic cell requires deregulation of multiple cellular processes including genome stability, proliferation, apoptosis, motility, and angiogenesis (Albertson et al., 2003; Hanahan and Weinberg, 2000). Changes in genome copy number and/or structure are particularly important as deregulating events in cancer progression (Hyman et al., 2002; Jeffrey et al., 2005; Kallioniemi et al., 1994; Loo et al., 2004; Pollack et al., 2002; Roylance et al., 1999; Tirkkonen et al., 1998), and elucidation of recurrent aberrations has revealed many important oncogenes and tumor suppressors (Neve et al., 2004). In fact, over a thousand genes have now been reported to be deregulated by recurrent genome aberrations in breast cancer alone (Fridlyand et al., 2006; Hyman et al., 2002; Pollack et al., 2002). Functional assessment of several of these genes in cell lines and xenografts has rovided invaluable insights into the roles they play in cellular physiology (Alimandi et al., 1995; Cheng et al., 2004). However, interpreting these results in the context of breast cancer pathophysiology requires an understanding of the extent to which the cell lines mirror aberrations that are present in primary tumors. To this end, we describe here a comprehensive comparison of the molecular and biological features of a collection of 51 breast cancer cell lines with those measured for primary breast tumors.
The central result of the study is a comparison of genome copy number and transcriptional profiles for the cell lines with those measured for primary breast tumors (Fridlyand et al., 2006). We also evaluated protein and phosphoprotein levels for selected genes in signaling pathways that are frequently deregulated in cancer. These analyses show that the cell lines display the same heterogeneity in copy number and expression abnormalities as the primary tumors, and they carry almost all of the recurrent genomic abnormalities associated with clinical outcome in primary tumors. In addition, the breast cancer cell lines cluster into basal-like and luminal expression subsets, as do primary tumors, although the partitioning of genome aberrations between these subsets is somewhat different than that in basal-like and luminal primary tumors. Importantly, the cell line collection exhibits heterogeneous responses to targeted therapeutics paralleling clinical observations. From these studies, we conclude that the cell line collection mirrors most of the important genomic and resulting transcriptional abnormalities found in primary breast tumors and that analysis of the functions of these genes in the ensemble of cell lines will accurately reflect how they contribute to breast cancer pathophysiologies. We also illustrate the possibility that correlative analyses of the heterogeneous responses to treatment with therapeutic agents that attack these genes may allow identification of molecular features that predict response in individual patients.
Results
Genomic features
We performed array CGH using arrays at 1 Mb resolution (see Experimental Procedures). Our analyses of genome copy number abnormalities in 51 cell lines (listed in Table 1) are provided in Table S1 in the Supplemental Data available with this article online (see also http://cancer.lbl.gov/breastcancer/data.php). As with primary tumors, cell lines exhibit pronounced genomic heterogeneity, even between lines with similar transcriptional profiles (e.g., luminal or basal-like) and biological characteristics, although the number of genome abnormalities per cell line is, on average, higher than that in primary tumors. Figure S1 shows genome copy number abnormality profiles for cell lines that exhibit different levels of genome aberration complexity, in agreement with published array CGH for these cell lines (Larramendy et al., 2000; Shadeo and Lam, 2006; Snijders et al., 2001). Several, like SUM159PT, show relatively few abnormalities. Others, like T47D, show many low-level abnormalities, and many, like BT474 and MCF7, show many abnormalities with high-level amplification. A few, like HCC1500, show extraordinary levels of abnormality not typically found in primary tumors.
Table 1.
Source, clinical, and pathological features of tumors used to derive breast cancer cell lines used in this study
| Cell line | Gene cluster | ER | PR | HER2 | TP53 | Source | Tumor type | Age (years) | Ethnicity | Culture media | Culture conditions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 600MPE | Lu | + | [−] | − | IDC | DMEM, 10% FBS | 37°C, 5% CO2 | ||||
| 2 | AU565a | Lu | − | [−] | + | +WT | PE | AC | 43 | W | RPMI, 10% FBS | 37°C, 5% CO2 |
| 3 | BT20 | BaA | − | [−] | ++WT | P.Br | IDC | 74 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 4 | BT474 | Lu | + | [+] | + | + | P.Br | IDC | 60 | W | RPMI, 10% FBS | 37°C, 5% CO2 |
| 5 | BT483 | Lu | + | [+] | − | P.Br | IDC, pap | 23 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 6 | BT549 | BaB | − | [−] | ++M | P.Br | IDC, pap | 72 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 7 | CAMA1 | Lu | + | [−] | + | PE | AC | 51 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 8 | HBL100 | BaB | − | [−] | ++ | P.Br | N | 27 | DMEM, 10% FBS | 37°C, 5% CO2 | ||
| 9 | HCC1007d | Lu | + | [−] | [+/−] | P.Br | Duc.Ca | 67 | B | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 10 | HCC1143d | BaA | − | [−] | ++M | P.Br | Duc.Ca | 52 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 11 | HCC1187d | BaA | − | [−] | ++M | P.Br | Duc.Ca | 41 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 12 | HCC1428d | Lu | + | [+] | [+] | PE | AC | 49 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 13 | HCC1500d | BaB | − | [−] | − | P.Br | Duc.Ca | 32 | B | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 14 | HCC1569d | BaA | − | [−] | + | −M | P.Br | MC | 70 | B | RPMI, 10% FBS | 37°C, 5% CO2 |
| 15 | HCC1937d | BaA | − | [−] | [−] | P.Br | Duc.Ca | 24 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 16 | HCC1954d | BaA | − | [−] | + | [+/−] | P.Br | Duc.Ca | 61 | EI | RPMI, 10% FBS | 37°C, 5% CO2 |
| 17 | HCC202d | Lu | − | [−] | + | [−] | P.Br | Duc.Ca | 82 | W | RPMI, 10% FBS | 37°C, 5% CO2 |
| 18 | HCC2157d | BaA | − | [−] | [+] | P.Br | Duc.Ca | 48 | B | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 19 | HCC2185d | Lu | − | [−] | [+] | PE | MLCa | 49 | WH | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 20 | HCC3153d | BaA | − | [−] | [−] | RPMI, 10% FBS | 37°C, 5% CO2 | |||||
| 21 | HCC38d | BaB | − | [−] | ++M | P.Br | Duc.Ca | 50 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 22 | HCC70d | BaA | − | [−] | ++M | P.Br | Duc.Ca | 49 | B | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 23 | HS578T | BaB | − | [−] | +M | P.Br | IDC | 74 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 24 | LY2 | Lu | + | [−] | +/− | PE | IDC | 69 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 25 | MCF10Ab | BaB | − | [−] | +/−WT | P.Br | F | 36 | W | DMEM/F12* | 37°C, 5% CO2 | |
| 26 | MCF12Ab | BaB | − | [−] | + | P.Br | F | 60 | W | DMEM/F12* | 37°C, 5% CO2 | |
| 27 | MCF7 | Lu | + | [+] | +/−WT | PE | IDC | 69 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 28 | MDAMB134VI | Lu | + | [−] | +/−WT | PE | IDC | 47 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 29 | MDAMB157 | BaB | − | [−] | − | PE | MC | 44 | B | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 30 | MDAMB175VII | Lu | + | [−] | +/−WT | PE | IDC | 56 | B | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 31 | MDAMB231 | BaB | − | [−] | ++M | PE | AC | 51 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 32 | MDAMB361 | Lu | + | [−] | + | −WT | P.Br | AC | 40 | W | DMEM, 10% FBS | 37°C, 5% CO2 |
| 33 | MDAMB415 | Lu | + | [−] | + | PE | AC | 38 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 34 | MDAMB435 | BaB | − | [−] | +M | PE | IDC | 31 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 35 | MDAMB436 | BaB | [−] | [−] | [−] | PE | IDC | 43 | W | L15, 10% FBS | 37°C, no CO2 | |
| 36 | MDAMB453 | Lu | − | [−] | −WT | PF | AC | 48 | W | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 37 | MDAMB468 | BaA | [−] | [−] | [+] | PE | AC | 51 | B | L15, 10% FBS | 37°C, no CO2 | |
| 38 | SKBR3a | Lu | − | [−] | + | + | PE | AC | 43 | W | McCoys 5A, 10% FBS | 37°C, 5% CO2 |
| 39 | SUM1315MO2c | BaB | − | [−] | [+] | Sk | IDC | Ham’s F12, 5%-IE | 37°C, 5% CO2 | |||
| 40 | SUM149PTc | BaB | [−] | [−] | [+] | P.Br | Inf Duc.Ca | Ham’s F12, 5%-IH | 37°C, 5% CO2 | |||
| 41 | SUM159PTc | BaB | [−] | [−] | [−] | P.Br | AnCar | Ham’s F12, 5%-IH | 37°C, 5% CO2 | |||
| 42 | SUM185PEc | Lu | [−] | [−] | [−] | PE | Duc.Ca | Ham’s F12, 5%-IH | 37°C, 5% CO2 | |||
| 43 | SUM190PTc | BaA | − | [−] | + | [+/−] | P.Br | Inf | Ham’s F12, SF-IH** | 37°C, 5% CO2 | ||
| 44 | SUM225CWNc | BaA | − | [−] | + | ++ | CWN | IDC | Ham’s F12, 5%-IH | 37°C, 5% CO2 | ||
| 45 | SUM44PEc | Lu | [+] | [−] | [−] | PE | Ca | Ham’s F12, SF-IH** | 37°C, 5% CO2 | |||
| 46 | SUM52PEc | Lu | [+] | [−] | [−] | PE | Ca | Ham’s F12, 5%-IH*** | 37°C, 5% CO2 | |||
| 47 | T47D | Lu | + | [+] | ++M | PE | IDC | 54 | RPMI, 10% FBS | 37°C, 5% CO2 | ||
| 48 | UACC812 | Lu | + | [−] | + | −WT | P.Br | IDC | 43 | DMEM, 10% FBS | 37°C, 5% CO2 | |
| 49 | ZR751 | Lu | + | [−] | − | AF | IDC | 63 | W | RPMI, 10% FBS | 37°C, 5% CO2 | |
| 50 | ZR7530 | Lu | + | [−] | + | −WT | AF | IDC | 47 | B | RPMI, 10% FBS | 37°C, 5% CO2 |
| 51 | ZR75B | Lu | + | [−] | +/− | RPMI, 10% FBS | 37°C, 5% CO2 |
AC, adenocarcinoma; AF, ascites fluid; AnCa, anaplastic carcinoma; ASC, acantholytic squamous carcinoma; BaA, Basal A; BaB, Basal B; Ca, carcinoma; CWN, chest wall nodule; Duc.Ca, ductal carcinoma; F, fibrocystic disease; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; Inf, inflammatory; LN, lymph node; Lu, luminal; MC, metaplastic carcinoma; MLCa, metastatic lobular carcinoma; N, normal; Pap, papillary; ND, not done; P.Br, primary breast; PE, pleural effusion; Sk, skin; W, White; B, Black; H, Hispanic; EI; East Indian.
ER/PR/HER2/TP53 status: ER/PR positivity, HER2 overexpression, and TP53 protein levels and mutational status (obtained from the Sanger web site; M, mutant protein; WT, wild-type protein) are indicated. Expression data are derived from mRNA and protein levels presented in this article (Table S2 and Figure S4). Square brackets indicate that levels are inferred from mRNA levels alone where protein data is not available.
Media conditions: FBS, fetal bovine serum; I, Insulin (0.01 mg/ml); H, hydrocortisone (500 ng/ml); E, EGF (20 ng/ml); DMEM, Dulbecco’s modified Eagle’s medium, GIBCO #11965-092; RPMI, RPMI medium 1640, GIBCO #27016-021; Ham’s F12, F-12 nutrient mixture (Ham), GIBCO #11039-021; DMEM/F12, Dulbecco’s modified Eagle’s medium: Nutrient mix F-12 (D-MEM/F-12), GIBCO #11039-021; L15, Leibovitz’s L-15 medium, GIBCO #11415-064.
For MCF10A and MCF12A, supplement DMEM/F12 media with 5% horse serum, 20 ng/ml EGF, 100 ng/ml cholera toxin, 0.01 mg/ml insulin, and 500 ng/ml hydrocortisone.
For serum-free (SF) medium, supplement Ham’s F12 with 0.5 μg/ml bovine serum albumin, 5 mM ethanolamine, 10 mM HEPES, 5 μg/ml transferrin, 10 μM T3, 50 μM Se, 5 μg/ml insulin, 1 μg/ml hydrocortisone.
For serum-containing F12 media, supplement media with 5% FBS, 5 μg/ml insulin, and 1 μg/ml hydrocortisone or 10 ng/ml EGF.
AU565 and SKBR3 were derived from the same patient.
Derived from a reduction mammoplasty.
Provided by Steve Ethier (http://www.cancer.med.umich.edu/breast_cell/Production/index.html).
Provided by Adi Gazdar, now available through ATCC (Gazdar et al., 1998).
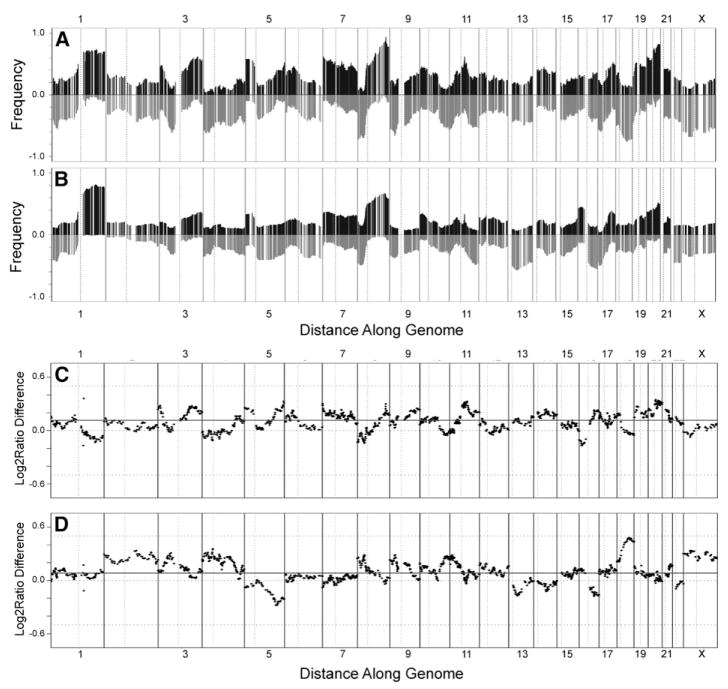
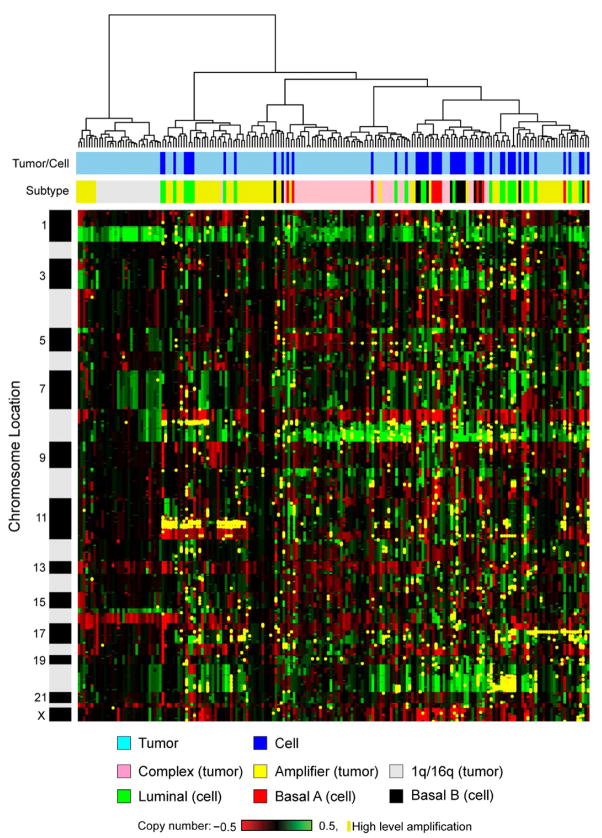
Figures 1A and 1B show that the recurrent abnormalities in the cell lines are similar to those in primary tumors, indicating that cell lines have retained most of the genomic abnormalities of the original tumors including regions of high-level amplification, and they have not selected abundant new abnormalities. Recurrent gene copy number changes in the 51 breast cancer cell lines that match recurrent aberrations in primary tumors are summarized in Figure S2. However, the agreement is not perfect, as illustrated in the comparisons of the relative frequencies of gains and losses between tumors and cell lines shown in Figures 1C and 1D, respectively. The major differences involve losses of chromosome 5q (more frequent in tumors) and chromosome 18 (more frequent in cell lines). The direct comparison of the cell line genome copy number aberration profiles with those in 145 primary tumors in Figure 2 shows that the cell line collection is overrepresented in lines with high-level amplification (i.e., in the previously reported “amplifier” genotype [Fridlyand et al., 2006]).
Figure 1. Comparison of array CGH analyses of human breast cancer cell lines and primary tumors.
A and B: Frequencies of significant increases or decreases in genome copy number are plotted as a function of genome location for 51 cell lines (A) and 145 primary tumors (Fridlyand et al., 2006) (B). Positive values indicate frequencies of samples showing copy number increases [Log2(copy number) > 0.3], and negative values indicate frequencies of samples showing copy number decreases [Log2(copy number) < −0.3].
C and D: Differences (y axis) between frequencies of gains and losses across the genome for the cell lines versus the tumors are represented in C and D, respectively. Genome copy number aberration frequencies are plotted as a function of location position in the genome beginning at 1pter to the left and ending at Xqter to the right (chromosome locations are indicated by numbers above and below graphs). Vertical lines indicate chromosome boundaries. Vertical dotted lines indicate centromere locations.
Figure 2. Unsupervised hierarchical clustering of genome aberrations in 51 breast cancer cell lines and 145 primary breast tumors.
Clusters show results at 1952 BAC probes common between the tumor and cell line CGH arrays. Each row represents a BAC probe, and each column represents a cell line or tumor sample. Green indicates increased genome copy number, and red indicates decreased genome copy number. Yellow indicates high-level amplification. The bar to the left shows chromosome locations with chromosome 1pter to the top and 22qter to the bottom. The locations of the odd-numbered chromosomes (shaded black) are indicated. The upper color bar shows columns representing tumors or cell lines. The lower color bar indicates the genomic characteristics of the cell lines and tumors from this study and for the tumors as reported (Fridlyand et al., 2006). Color codes are indicated at the bottom of the figure.
Transcription profiles
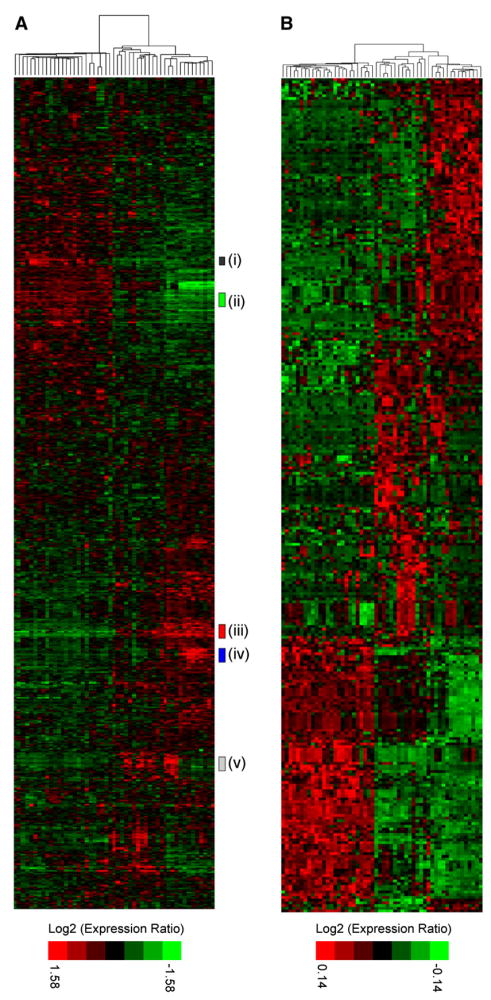
Hierarchical clustering of our analyses of transcriptional profiles (Table S2; see also http://cancer.lbl.gov/breastcancer/data.php) of the 51 breast cell lines using transcripts showing substantial variation across the samples revealed two major branches (Figure 3A). We identified one cluster as luminal [ERBB3- and ESR1-positive, (ii) and (i) in Figure 3A] and the other as basal-like [ESR1-negative, CAV1-positive, (iii) in Figure 3A; Jones et al., 2004] using published gene markers of in vivo histology (Abd El-Rehim et al., 2004; Cattoretti et al., 1988; Jones et al., 2004; Korsching et al., 2002; Nielsen et al., 2004; Simpson et al., 2004). The luminal cluster was generally uniform across all samples, whereas the basal-like cluster contained at least two major subdivisions we termed Basal “A” [KRT5-, KRT14-positive, (v) in Figure 3A] and Basal “B” [VIM-positive, (iv) in Figure 3A]. The Basal A cluster matches closely to the Perou basal-like signature (Chung et al., 2002; Perou et al., 1999, 2000; Sorlie et al., 2001), whereas the more distinct Basal B subgroup exhibits a stem-cell like expression profile and may reflect the clinical “triple-negative” tumor type. These clusters and histological associations are similar to those previously reported for tumors and cell lines (Chung et al., 2002; Perou et al., 1999, 2000; Sorlie et al., 2001), and clustering of the cell lines using gene expression of published markers of histology (Figure S3A) produced a cluster similar to those in Figure 3.
Figure 3. Gene expression profiles of 51 human breast cancer cell lines.
A: Hierarchical cluster analysis of breast cancer cell lines with subclusters [(i) through (v)] indicated by colored bars at side. Genes were restricted to those showing significant variance across all samples, resulting in clustering of 1438 probe sets (see Experimental Procedures).
B: Cell lines clustered by genes selected by PAM analysis representing the luminal, Basal A, and Basal B clusters. Clustering was performed as described in the Experimental Procedures. Each row represents a gene, and each column represents a cell line sample. As shown in the color bar, black represents no change, red represents upregulation, and green represents downregulation of gene expression.
To identify genes that classify the luminal, Basal A, and Basal B subtypes, PAM analysis was performed (see Experimental Procedures) (Tibshirani et al., 2002). Table S3 lists 305 classifier genes, and Figure 3B shows the breast cancer cell lines clustered by those genes. These genes are likely to be intimately involved in the differentiation status of the cell types and/or tumor biology.
In addition to published histological markers, luminal A cell lines also preferentially expressed genes, such as GATA3, TOB1, ERBB3, and SPDEF, that have been associated with a more differentiated, noninvasive phenotype (Beck et al., 2001; Charafe-Jauffret et al., 2006; Feldman et al., 2003; Lim et al., 2000). Basal B cell lines were more clearly distinct from luminal cells than those in the Basal A cluster and preferentially expressed genes such as CD44, MSN, TGFBR2, CAV1/2, VIM, SPARC, and AXL, while CD24 was weakly expressed. Interestingly, MCF10A and MCF12A (two immortal, nontransformed cell lines) share transcriptional characteristics with all other identified subtypes and had many features of basal progenitor cells (Dontu et al., 2003a; Stingl et al., 1998), suggesting that these cells may represent a multipotent lineage. In contrast to Perou et al. (2000), we did not find a distinct HER2 cluster. Rather, HER2-amplified cells were scattered across the luminal cluster and the Basal A cluster.
Although the gene expression patterns generally reflected the major transcriptional classes found in primary tumors, the differences in frequency of genome copy number abnormalities between basal-like and luminal cell lines were different than those between luminal and basal-like primary tumors (Fridlyand et al., 2006). For example, luminal tumors (Figure 4A) showed fewer genome aberrations than basal tumors (Figure 4B) overall, and basal-like tumors carried higher frequencies of copy number gains involving chromosomes 10p and 22q and losses of 5q, 12q, and 15p compared to luminal tumors (Figure 4C). However, luminal cell lines (Figure 4D) showed about the same frequency of genome copy number abnormalities as basal-like cell lines (Figure 4E). In addition, luminal cell lines showed more copy number gains involving chromosomes 12q and fewer copy number gains involving 19p relative to basal-like cell lines (Figure 4F).
Figure 4. Comparative analyses of aberration frequencies in basal and luminal primary tumors and cell lines.
A–C: A and B show frequencies of genome copy number gains and losses in luminal and basal breast tumors, respectively (Fridlyand et al., 2006). C shows univariate statistical assessments of differences between the two tumor types.
D–F: D and E show frequencies of genome copy number gains and losses in luminal and Basal B breast cancer cell lines, respectively. F shows univariate statistical assessments of differences between the two cell line types. p values of 0.05 and 0.01 are indicated as in C. All data are plotted beginning at chromosome 1pter to the left and ending at Xqter to the right. Vertical solid lines indicate chromosome boundaries. Vertical dashed lines indicate centromere locations. p values of 0.05 and 0.01 are indicated by dashed horizontal lines in C and F.
Genomic deregulation of gene expression
Our analyses of gene expression and copy number in the 51 cell lines revealed 1778 gene transcripts whose levels were correlated with genome copy number (Pearson’s correlation ≥ 0.5, Holm-adjusted p value ≤ 0.05), suggesting that expression levels were deregulated by genomic aberrations. Table S4 summarizes the statistically significant genome copy number versus gene expression correlations discovered in this study. A similar analysis in primary breast tumors (Fridlyand et al., 2006) identified 1182 significantly correlated genes (Pearson’s correlation ≥0.5, Holm-adjusted p value ≤0.05). We assessed the agreement between the tumor and cell line correlation data sets and found that 72% of the genes scored as significantly deregulated in primary tumors also were significantly deregulated in the cell line set (odds ratio for agreement of 16 for correlation > 0.7). This indicates that the cell lines retain most of the genome-aberration-mediated gene deregulation present in primary tumors (Table S4).
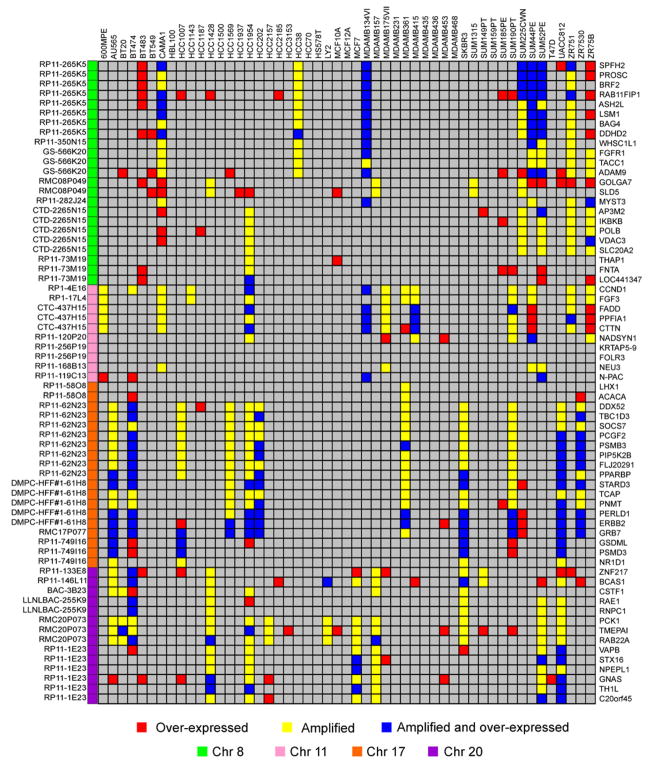
Sixty-six of the deregulated genes in the tumors were in regions of high-level amplification associated with reduced survival duration and so are both markers for tumors that are resistant to current therapies and candidate therapeutic targets. We identified cell lines in which the 66 candidate therapeutic target genes were amplified and overexpressed by clustering the cell lines using probe sets matched to these genes. Figure 5 indicates the cell lines in which these genes are amplified and overexpressed. Combined, these data show that 88% (55/66) of these genes are amplified and overexpressed in at least one cell line. These cell lines should be useful models for assessment of the roles that gene amplification and overexpression play in breast cancer pathophysiology.
Figure 5. A functional model to investigate lead candidate therapeutic targets.
Gene expression and copy number for the 66 candidate therapeutic genes in the 51 breast cancer cell lines. Genes were selected for their overexpression and association with outcome in human breast tumors (Chin et al., 2006). High gene expression (≥2-fold over mean gene expression for all the samples) is shown in red, and high-level amplification (≥0.9 Log2 ratio) is shown in yellow for each cell line. Genes that have high expression and gene copy number are shown in blue. Gene HUGO names are shown to the right, and the corresponding BAC clone ID and chromosome are shown to the left.
Protein profiles
We measured levels of 49 gene products or posttranslationally modified gene products associated with aspects of signal transduction and cell cycle regulation and/or frequently found to be aberrant in human cancers using western analysis. These analyses revealed cell lines in which these regulatory processes may be aberrant and allowed an initial assessment of the extent to which genome aberrations affected the protein/phosphoprotein levels. These data also allowed us to assess the extent to which the RNA levels in the cell lines reflected protein levels. Western blots for the cell line collection are shown in Figure S4, and semiquantitative measures of protein levels are summarized in Table S5. Figures S3B and S3C show that the degree of concordance between semiquantitative measures of protein levels from the western blots and RNA expression levels from the Affymetrix expression array analyses varied considerably among the genes (Souchelnytskyi, 2002). We found that concordance for 54% was strong (e.g., ESR1, CDKN1B), while 46% showed low or no concordance (e.g., PTEN). This finding is not surprising considering the high degree of posttranslational processing and degradation that occurs in signaling pathways that regulate proliferation and survival.
Biological and molecular associations
One important use of the cell line collection is identification of molecular events that are associated with biological phenotype. Establishing such associations is a first step in the development of a molecular understanding of the biological phenotype. The molecular diversity between the cell lines allows this to be accomplished in a robust manner.
Morphology and invasion
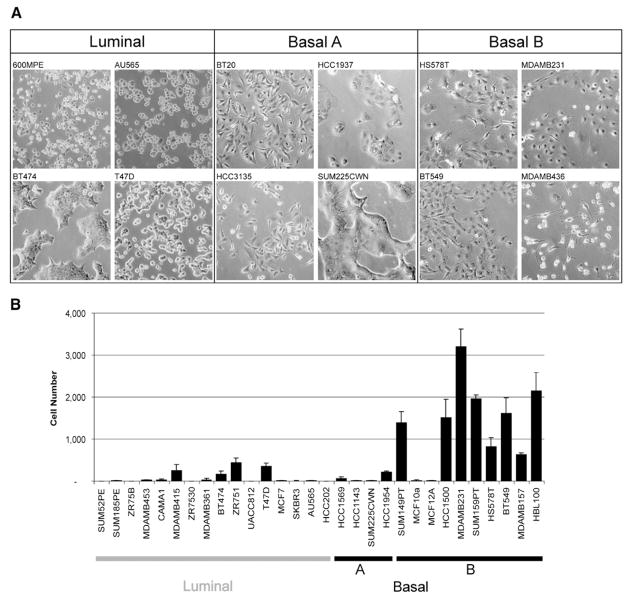
One of the clearest associations in the cell line collection is the relationship between the transcriptionally defined subgroups and distinctive biological characteristics such as morphology and invasive potential illustrated in Figure 6. Figure 6A shows that luminal cells appear more differentiated and form tight cell-cell junctions, while the Basal B cells appear less differentiated and have a more mesenchymal-like appearance. Basal A cells may have either luminal-like or basal-like morphologies. Similar stratification was noted in three-dimensional cultures (data not shown). Figure 6B shows that Basal B cells are much more frequently highly invasive in Boyden chamber assays than Basal A and luminal cells.
Figure 6. Relationship of transcriptional profiles to biological function.
A: Morphology of cell lines grown in tissue culture on plastic.
B: Invasive potential of 30 breast cancer cell lines as measured by modified Boyden chamber assays (see Experimental Procedures). Each data point represents the mean ± SD of three wells.
Predictors of therapeutic response
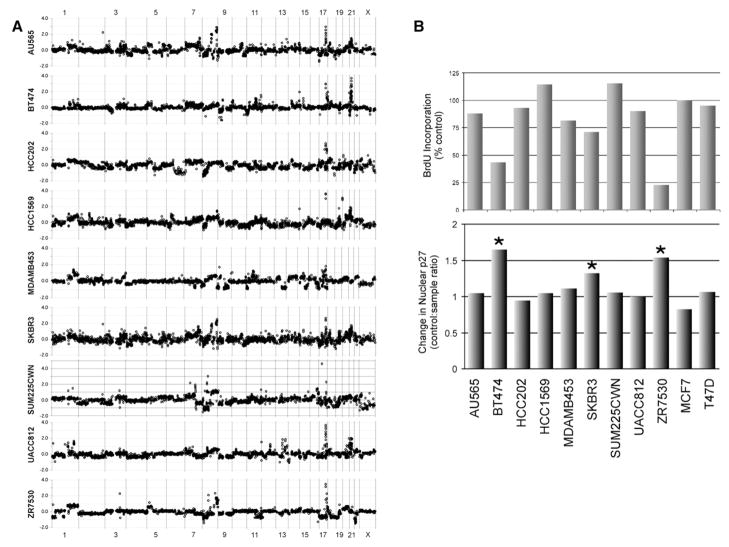
One of the promising potential applications of association analysis using the cell line system is identification of molecular signatures that predict responses to therapies that target genes that are deregulated by genome abnormalities. To illustrate this application, we assessed biological responses to Trastuzumab in nine HER2-amplified cell lines and two control cell lines. Genome copy number profiles for the HER2-amplified cell lines are shown in Figure 7A. Figure 7B shows that the non-amplified control cell lines, MCF7 and T47D, were unaffected by Trastuzumab as expected. However, this figure also shows that only three of the nine HER2-amplified lines exhibited a robust response to Trastuzumab as measured by inhibition of BrdUrd incorporation. This frequency of response is similar to that reported in clinical evaluations of Trastuzumab monotherapy (Vogel et al., 2005). Pearson’s correlations between molecular signatures and biological response to Trastuzumab in HER2-amplified cell lines revealed associations with Trastuzumab response. These are summarized in Table S6. Protein levels most strongly correlated with response included increased levels of MEK (S217/219), ESR1, TYK2, FASN, GRB7, and MAPK1/3 (Thr202/Tyr204). Protein levels associated with resistance included high levels of SFN, CAV2, GRB2, RB1, and FLNA. Genomic regions 12q13 and 19q13 were correlated with sensitivity, while 1p36, 11q14, and 17p11 were associated with resistance. From ontologic analysis of gene expression, it appears that upregulation of genes involved in insulin/MAPK signaling predicts response to Herceptin, whereas the mTOR pathway, Toll-like receptor pathway, N-Glycan biosynthesis, and inositol-phosphate signaling are associated with resistance. This analysis suggests that assessment of these molecular features in primary tumors will more precisely identify patients that will respond to Trastuzumab.
Figure 7. Indicators of therapeutic response to Trastuzumab in HER2-positive breast cancer cell lines.
A: CGH profiles of nine breast cancer cell lines overexpressing HER2.
B: Response of HER2-overexpressing cell lines to 48 hr treatment with 21 μg/ml Trastuzumab (Herceptin) as measured by BrdUrd incorporation (top panel) and relocalization of p27KIP1 (lower panel).
Indicators of therapeutic response
Association studies also identify molecular events that change in response to treatment with targeted therapies. A previous study suggested that regulation of p27KIP1 is critical in mediating response to Trastuzumab (Nahta et al., 2004). However, this study was limited to clonal Trastuzumab-resistant variants of SKBR3 cells. Our analyses of the molecular and biological responses of HER2 amplified cell lines to Trastuzumab, shown in Figures 7B and 7C (or to 4D5; data not shown), confirm that association. Specifically, we found that increases in the levels of p27KIP1 (CDKN1B) protein and translocation of p27KIP1 to the nucleus were associated with cell cycle arrest as measured by inhibition of BrdUrd incorporation—probably due to inhibition of the formation of CCNE1-CDK2 complexes (Lane et al., 2000; Nahta et al., 2004; Neve et al., 2000). Importantly, while the steady-state level of p27KIP1 tended to be lower in Trastuzumab-responsive cell lines, it was not significantly predictive of overall response. These analyses suggest that measurement of the nuclear localization p27KIP1 in clinical specimens (e.g., in fine needle aspirates or core biopsies) taken during early stages of treatment with Trastuzumab will be an early indication that patients are responding to the treatment.
Discussion
Breast cancer is a remarkably heterogeneous disease, but subsets of tumors show recurrent patterns of transcriptional, genomic, and biological abnormality. Understanding how genes in these “patterns” collectively function in an otherwise heterogeneous biological setting to enable progression and modulate response to therapy is critical to improving management of the disease. Association studies in primary tumors provide clues about molecular events that may be important in cancer pathophysiology, but more formal proof requires model systems that mirror both the heterogeneity and recurrent molecular aberrations found in primary tumors and that can be manipulated to test associations. The comparisons between cell lines and primary tumors in this study show that the cell line collection, as a system, mirrors many but not all of the biological and genomic properties of primary tumors.
In general, the cell lines mirror both the genomic heterogeneity (Figure S1) and the recurrent genome copy number abnormalities found in primary tumors with high fidelity (Figures 1 and 2). This is remarkable, considering the fact that many of the cell lines have been carried in culture for many years or decades. This indicates that they have not accumulated substantial new recurrent aberrations during extended culture and is supported by our own analysis showing stable genomic and expression patterns in the cell lines over multiple passages. In addition, important genome aberration “landmarks” like the high-level amplifications associated with poor outcome in primary tumors are well represented. That said, the cell lines carry more aberrations, on average, than primary tumors, and high-level amplification is more frequent in the cell lines, while cell lines with simple “1q/16” genotypes (Fridlyand et al., 2006) are missing in the cell line collection (Figure 2). This might be explained by the fact that the cell lines have been derived predominantly from late-stage tumors or pleural effusions, while the tumors against which they were compared were predominantly early stage (Fridlyand et al., 2006). Alternately, high-level amplification may provide a selective advantage for growth in vitro, so cell lines with high-level amplification were isolated preferentially. The associations between gene expression and copy number in primary tumors are mostly preserved in the cell lines, although the number of genes showing significant associations with copy number is greater in the cell lines than in primary tumors. The increased number of significant associations in the cell lines may be because the cell cultures are not contaminated by normal epithelial or nonepithelial cells that may introduce confounding expression patterns, thereby decreasing some associations to the point where they are no longer significant. Overall, however, these data argue that the roles in breast cancer pathophysiology of genome aberrations captured in the cell line collection can be elucidated by manipulating the expression levels of deregulated genes in the cell lines.
However, some aspects of primary tumor cancer genomics will be difficult to study using the current collection. For example, additional cell lines derived from early-stage breast cancers will be needed to study aberration patterns such as the “1q/16q” breast tumor subtype. This may require development of cell culture conditions that are permissive to the growth of these cells. It is noteworthy in this context that the HCC cell lines (Gazdar et al., 1998; Larramendy et al., 2000) preferentially populate the basal-like lineage, while other cell lines (e.g., the SUM cells [Ethier et al., 1993]) are represented across all the lineage subtypes. This suggests the possibility that culture conditions may bias selection of breast tumor subtypes. Alternatively, the laboratory-specific lineage dependence of the derived cell lines may be explained by the tissue origin. For example, the HCC cell lines were typically derived from primary breast tumors, while many of the other cell lines were derived from pleural effusions (Table 1).
Other aspects of cancer biology also are more or less accurately represented by the cell line system. For example, the cell lines can be classified into luminal and basal-like subtypes as found in primary tumors (Figure 3). However, the two luminal subsets evident in tumors are not apparent in the cell lines, and the basal-like cell lines resolve into two distinctive clusters (Basal A and Basal B) that are not apparent in analyses of primary tumors. Similar discrepancies have been noted in earlier studies (Perou et al., 1999). Again, this might be due to the fact that the cell line expression profiles are not “contaminated” with normal epithelial or stromal cells so that the clusters resolve more clearly in the cell lines, or that the differences are due to the absence of stromal or physiological interactions and/or signaling in cell culture (Kuperwasser et al., 2004; Radisky and Bissell, 2004). Arguing against this, however, is our observation that the differences between the genome aberration patterns for the basal-like and luminal clusters in the cell line system don’t match differences in these subtypes in primary tumors (Figure 4). This suggests that the cell lines may be derived from subpopulations of tumor cells that are selected because they grow well. Intriguingly, in this regard, the highly invasive Basal B cells carry the CD44+/CD24−/low phenotype associated with the subpopulation of tumorigenic stem cells recently identified in breast cancer (Al-Hajj et al., 2003; Dontu et al., 2003b).
The high fidelity to which genome-aberration-induced transcriptional changes are preserved in the cell lines and the existence of substantial genomic, transcriptional, translational, and biological heterogeneity in the overall system support the idea that assessment of responses to inhibitors of the resulting dominant or dominant-negative genes will reveal molecular events that predict response/resistance. This concept is supported by studies of responses to Iressa of lung cancer cell lines (Tracy et al., 2004; Zhao et al., 2004) and leukemia cells (Carter et al., 2005; Mahon et al., 2000). Our analyses of the subset of HER2-amplified breast cancer cell lines show variable response to treatment with Trastuzumab as observed in the clinical trials (Vogel et al., 2001) and identify molecular features that may allow more precise identification of HER2-positive patients that will respond to therapeutic protocols containing Trastuzumab. Specifically, increased protein levels of ESR1, TYK2, FASN, GRB7, MEK (S217/219), and MAPK1/3 (Thr202/Tyr204) predict Trastuzumab sensitivity, whereas increased SFN, CAV2, GRB2, RB1, and FLNA expression is associated with Trastuzumab resistance. Many of these genes are known signaling targets or signal integrators of HER2 (Hynes and Lane, 2005), and its downstream pathways, PI3K and MAPK; therefore, mutations in these pathways may be responsible for loss of HER2 oncogene “addiction” and may modulate therapeutic response. In support of this, gene expression profiles indicate that increased expression of several insulin/MAPK pathway genes predicts response, whereas increased mTOR, Toll-like receptor, N-Glycan biosynthesis, and inositol-phosphate signaling predicts resistance. These and subsequent studies set the stage for detailed study of mechanisms of resistance, development of markers that predict or indicate response, and potential new therapeutic targets.
In sum, we have cataloged the genomic and molecular properties of a panel of cell lines and demonstrated a fidelity to those found in primary breast tumors. Recurrent genome aberrations and the resulting transcriptional changes are well preserved in the cell line collection. Thus, the cell lines seem well suited to assessment of the functional consequences of genome-aberration-mediated gene deregulation and to identification of molecular features that predict resistance/sensitivity to agents that target these aberrations. Continuing characterization of these cell lines, development of more cell lines and more realistic cell culture environments, and assessment of multiple aberration-targeted agents should provide an increasingly useful resource for the assessment of how genome aberrations contribute to breast cancer pathophysiology. This will facilitate our understanding of the mechanisms of tumorigenesis and stimulate development of new therapies targeted to selectively interfere with one or more of these processes.
Experimental procedures
Cell culture
Breast cancer cell lines were obtained from the ATCC or from collections developed in the laboratories of Drs. Steve Ethier and Adi Gazdar. Cell lines were obtained from these sources to avoid errors that occur when obtaining lines through “secondhand” sources. Since we acknowledge the existence of multiple clonal variants of some cell lines throughout the scientific community, all results presented here are reflective of the cell lines we have in our collection. To maintain the collections’ integrity, cell lines have been carefully maintained in culture, and stocks of the earliest-passage cells have been stored. Quality control is maintained by careful analysis and reanalysis of morphology, growth rates, gene expression, and protein levels. Cell lines can be accurately identified by CGH analysis. All extracts were made from subconfluent cells in the exponential phase of growth in full media. Information about the biological characteristics of the cell lines and the culture conditions are summarized in Table 1 and are available at http://cancer.lbl.gov/breastcancer/data.php.
Nucleic acid isolation
DNA isolation
Cells growing exponentially in culture were washed in phosphate buffered saline (PBS), pelleted by centrifugation, resuspended in PBS, and pelleted again. Pellets were either frozen for long-term storage or used to extract genomic DNA directly. Genomic DNA was extracted using the Wizard DNA Purification Kit (Promega), further purified with a phenol/chloroform extraction, and quantified using a fluorimeter. Phenol/chloroform extraction of the resulting DNA increased measurement precision significantly in some experiments, presumably by removing proteins that interfered with DNA labeling and hybridization.
RNA isolation
Total RNA was extracted from cell lines using Trizol, according to standard protocols (Invitrogen). RNA integrity was assessed by denaturing formaldehyde agarose gel electrophoresis or by microanalysis (Agilent Bioanalyzer, Palo Alto, CA).
Cell lysates
Protein lysates were prepared from cells at 50%–75% confluency. The cells were washed in ice-cold PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and then with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 25 mM β-glycerophosphate, 25 mM NaF, 5 mM EGTA, 1 mM EDTA, 15 mM pyrophosphate, 2 mM sodium orthovanadate, 10 mM sodium molybdate, leupeptin (10 μg/ml), aprotinin (10μg/ml), and 1 mM PMSF. Cells were extracted in the same buffer containing 1% Nonidet-P40. Lysates were then clarified by centrifugation and frozen at −80°C. Protein concentrations were determined using the Bio-Rad protein assay kit.
Immunochemical techniques and immunoblot quantification
Immunoblot analyses were performed using 20 μg cleared cell lysates. This material was electrophoretically resolved on denaturing sodium doedecyl sulfate (SDS)-polyacrylamide gels (4%–12%), transferred to polyvinylidene difluoride membranes (PVDF; Millipore), and probed with specific antisera using standard techniques. Bound antibodies on immunoblots were detected by either chemiluminescent (ECL, Pierce) or infrared (LiCor, Odyssey) imaging. Images were recorded as TIFF files for quantitation (see below). Immunoblots analysis of each protein was performed at least twice in all cases to ensure reproducibility. Antibodies used in these western analysis are described in Table S7.
Protein quantification
Protein levels were measured by quantifying emitted chemiluminescence or infrared radiation recorded from labeled antibodies using Scion Image (http://www.scioncorp.com/) or Odyssey software (http://www.licor.com/). For each protein, the blots were made for 4 sets of 11 cell lines, each set including the same pair (SKBR3 and MCF12A) to permit intensity normalization across sets. A basic multiplicative normalization was carried out by fitting a linear mixed-effects model to log intensity values and adjusting within each set to equalize the log intensities of the pair of reference cell lines across the sets.
Boyden chamber invasion assays
Assays were performed in modified Boyden chambers with 8 μm pore filter inserts for 24-well plates (BD Bioscience). Filters were coated with 12.5 μl of ice-cold 20% basement membrane extract (Matrigel, BD Bioscience). Epithelial cells were added to the upper chamber in 300 μl of serum-free medium. For the invasion assay, 7.5 × 104 cells were seeded on the 20% Matrigel-coated filters and incubated for 24 hr. The lower chamber was filled with 300 μl of full medium. After incubation, epithelial cells on the underside of the filter were fixed with 2.5% glutaraldehyde in PBS and stained with 0.5% toluidine blue in 2% Na2CO3. Cells that remained in the gel or attached to the upper side of the filter were removed with cotton tips. Cells on the underside of the filter were counted using light microscopy. Assays were performed in triplicate or quadruplicate. The results were expressed as an average ± one standard deviation.
Comparative genomic hybridization
Each sample was analyzed using Scanning and OncoBAC arrays. Scanning arrays were comprised of 2464 BACs selected at approximately megabase intervals along the genome as described previously (Hodgson et al., 2001; Snijders et al., 2001). OncoBAC arrays were comprised of 1860 P1, PAC, or BAC clones. About three-quarters of the clones on the OncoBAC arrays contained genes and STSs implicated in cancer development or progression. All clones were printed in quadruplicate. Data presented are the union of these two data sets. Arrays were prepared as described (Fridlyand et al., 2006; Snijders et al., 2001). Briefly, we random-prime labeled 500~1000 ng of test (cell line) and reference (normal female, Promega) genomic DNA with CY3-dUTP and CY5-dUTP (Amersham), respectively, using Bioprime kit (In-vitrogen). Labeled DNA samples were coprecipitated with 50 μg of human Cot-1 DNA (Invitrogen), denatured, hybridized to BAC arrays for 48–72 hr, washed, and counterstained with DAPI. Most of the data presented are based on the results of a single hybridization. Repeated measurements of genome aberrations in other experiments show that the results are highly reproducible.
Data processing
Array CGH data image analyses were performed as described previously (Jain et al., 2002). In this process, an array probe was assigned a missing value for an array if there were fewer than two valid replicates or the standard deviation of the replicates exceeded 0.3. Array probes missing in more than 50% of samples in the OncoBAC or scanning array data sets were excluded in the corresponding set. Array probes representing the same DNA sequence were averaged within each data set and then between the two data sets. Finally, the two data sets were combined, and the array probes missing in more than 25% of the samples, unmapped array probes, and probes mapped to chromosome Y were eliminated. The final data set contained 2696 unique probes representing a resolution of 1 Mb.
Affymetrix microarray analysis
Total RNA was prepared from samples using Trizol reagent (GIBCO BRL Life Technologies), and quality was assessed on the Agilent Bioanalyser 2100. Preparation of in vitro transcription (IVT) products, oligonucleotide array hybridization, and scanning were performed according to Affymetrix (Santa Clara, California) protocols. In brief, 5 μg of total RNA from each breast cancer cell line and T7-linked oligo-dT primers were used for first-strand cDNA synthesis. IVT reactions were performed to generate biotinylated cRNA targets, which were chemically fragmented at 95°C for 35 min. Fragmented biotinylated cRNA (10 μg) was hybridized at 45°C for 16 hr to Affymetrix high-density oligonucleotide array human HG-U133A chip. The arrays were washed and stained with streptavidin-phycoerythrin (SAPE; final concentration 10 μg/ml). Signal amplification was performed using a biotinylated anti-streptavidin antibody. The array was scanned according to the manufacturer’s instructions (2001 Affymetrix Genechip Technical Manual). Scanned images were inspected for the presence of obvious defects (artifacts or scratches) on the array. Defective chips were excluded, and the sample was reanalyzed.
Data processing
Probe set based gene expression measurements were generated from quantified Affymetrix image files (“.CEL” files) using the RMA algorithm (Irizarry et al., 2003) from the BioConductor (http://www.bioconductor.org/) tools suite. All 51 CEL files were analyzed simultaneously, creating a data matrix of probe sets by cell lines in which each value is the calculated log abundance of each probe set gene for each cell line. Probe sets were annotated with Unigene annotations from the July 2003 mapping of the human genome (http://genome.ucsc.edu/), resulting in 19,764 annotated probe sets representing 13,406 unique unigenes. Gene expression values were centered by subtracting the mean value of each probe set across the cell line set from each measured value. The gene expression data were organized using hierarchical clustering to facilitate visualization of commonalities and differences in gene expression across the set of cell lines. These analyses were restricted to the set of genes that showed substantial variation across the data set by selecting all probe sets that had at least four measurements that varied by more than Log2 1.89. This resulted in 1438 probe sets corresponding to 1213 unigenes. This variation restriction was arbitrary but did not affect the outcome of the eventual analysis. Probe sets corresponding to the same gene were down-weighted inversely proportional to their frequency prior to clustering (Wouters et al., 2003). Agglomerative clustering (Eisen et al., 1998) was applied to probe sets and cell lines using the uncentered Pearson’s correlations. Resulting clusters were visualized using Java TreeView (Saldanha, 2004). All expression data, array CGH data, and cluster files are available at http://cancer.lbl.gov/breastcancer/data.php.
PAM analysis
Analysis was performed in R (http://www-stat.stanford.edu/%7Etibs/PAM/Rdist/index.html) following the instructions therein (http://www-stat.stanford.edu/%7Etibs/PAM/Rdist/doc/readme.html) (Tibshirani et al., 2002). Three classifiers were defined (luminal, Basal A, and Basal B, as determined from the hierarchical clustering of the cell line expression data). Classifier training, crossvalidation, and calculation of false discovery rates were performed, resulting in 396 genes identified at a threshold of 4.0. Subsequently, a better threshold scaling was calculated, and a threshold of 2.8 chosen based on the false discovery rate resulted in the 305 gene classifier.
Association of copy number with expression
The presence of an overall dosage effect was assessed by subdividing each chromosomal arm into nonoverlapping 20 Mb bins and computing the average of cross-Pearson’s-correlations for all gene-clone pairs that mapped to that bin. The average cross-correlations between clones and genes mapping to the same bin were significantly higher than those between clones and genes mapping to unlinked bins (p value < 10−16, Wilcoxon rank sum test). Pearson’s correlations and corresponding p values between expression level and copy number also were calculated for each gene. Each gene was assigned an observed copy number of the nearest mapped BAC array probe. Eighty percent of genes had a nearest clone within 1 Mbp, and 50% had a clone within 400 kb. Correlation between expression and copy number was only computed for the mapped genes whose absolute assigned copy number exceeded 0.2 in at least five samples. This was done to avoid spurious correlations in the absence of real copy number changes. The Holm p value adjustment was applied to correct for multiple testing. Genes with an adjusted p value < 0.05 were considered to have expression levels that were significantly affected by gene dosage. This corresponded to a minimum Pearson’s correlation of 0.44.
Microarray data
The raw data for expression profiling are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-TABM-157.
All expression data, array CGH data, and cluster files are also available at the CaBIG repository (http://caarraydb.nci.nih.gov/caarray/publicExperimentDetailAction.do?expId=1015897590151581), at http://cancer.lbl.gov/breastcancer/data.php, and in the Supplemental Data.
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (contract DE-AC03-76SF00098); the National Institutes of Health under grants CA 58207, CA112970, and CA090788; and the Avon Foundation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. For full disclaimer, see http://www-library.lbl.gov/public/tmRco/howto/RcoBerkeleyLabDisclaimer.htm.
Footnotes
Supplemental data
The Supplemental Data include four supplemental figures and seven supplemental tables and can be found with this article online at http://www.cancercell.org/cgi/content/full/10/6/515/DC1/.
Accession numbers
The raw data for expression profiling are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-TABM-157.
References
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203:661–671. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP, Kraus MH. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene. 1995;10:1813–1821. [PubMed] [Google Scholar]
- Beck GR, Jr, Zerler B, Moran E. Gene array analysis of osteoblast differentiation. Cell Growth Differ. 2001;12:61–83. [PubMed] [Google Scholar]
- Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, Milanov ZV, Atteridge CE, Biggs WH, III, Edeen PT, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Andreola S, Clemente C, D’Amato L, Rilke F. Vimentin and p53 expression on epidermal growth factor receptor-positive, oestrogen receptor-negative breast carcinomas. Br J Cancer. 1988;57:353–357. doi: 10.1038/bjc.1988.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, Bertucci F. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman P, Roydasgupta R, Kuo W-L, Lapuk A, Neve R, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell . 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. this issue. [DOI] [PubMed] [Google Scholar]
- Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nat Genet. 2002;32(Suppl):533–540. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003a;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003b;36(Suppl 1):59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993;53:627–635. [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003;63:4626–4631. [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–332. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey SS, Lonning PE, Hillner BE. Genomics-based prognosis and therapeutic prediction in breast cancer. J Natl Compr Canc Netw. 2005;3:291–300. doi: 10.6004/jnccn.2005.0016. [DOI] [PubMed] [Google Scholar]
- Jones C, Mackay A, Grigoriadis A, Cossu A, Reis-Filho JS, Fulford L, Dexter T, Davies S, Bulmer K, Ford E, et al. Expression profiling of purified normal human luminal and myoepithelial breast cells: identification of novel prognostic markers for breast cancer. Cancer Res. 2004;64:3037–3045. doi: 10.1158/0008-5472.can-03-2028. [DOI] [PubMed] [Google Scholar]
- Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola J, van Diest PJ, Brandt B, Boecker W, Buerger H. Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest. 2002;82:1525–1533. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Beuvink I, Motoyama AB, Daly JM, Neve RM, Hynes NE. ErbB2 potentiates breast tumor proliferation through modulation of p27(Kip1)-Cdk2 complex formation: receptor overexpression does not determine growth dependency. Mol Cell Biol. 2000;20:3210–3223. doi: 10.1128/mcb.20.9.3210-3223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larramendy ML, Lushnikova T, Bjorkqvist AM, Wistuba II, Virmani AK, Shivapurkar N, Gazdar AF, Knuutila S. Comparative genomic hybridization reveals complex genetic changes in primary breast cancer tumors and their cell lines. Cancer Genet Cytogenet. 2000;119:132–138. doi: 10.1016/s0165-4608(99)00226-5. [DOI] [PubMed] [Google Scholar]
- Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- Mahon FX, Deininger MW, Schultheis B, Chabrol J, Reiffers J, Goldman JM, Melo JV. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: Diverse mechanisms of resistance. Blood. 2000;96:1070–1079. [PubMed] [Google Scholar]
- Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res. 2004;64:3981–3986. doi: 10.1158/0008-5472.CAN-03-3900. [DOI] [PubMed] [Google Scholar]
- Neve RM, Sutterluty H, Pullen N, Lane HA, Daly JM, Krek W, Hynes NE. Effects of oncogenic ErbB2 on G1 cell cycle regulators in breast tumour cells. Oncogene. 2000;19:1647–1656. doi: 10.1038/sj.onc.1203470. [DOI] [PubMed] [Google Scholar]
- Neve RM, Schaefer C, Buetow K, Gray JW. Genomic events in breast cancer progression. In: Harris JR, Lippman ME, Morrow M, Osborne CK, editors. Diseases of the Breast. 3. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 383–395. [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Bissell MJ. Cancer. Respect thy neighbor! Science. 2004;303:775–777. doi: 10.1126/science.1094412. [DOI] [PubMed] [Google Scholar]
- Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res. 1999;59:1433–1436. [PubMed] [Google Scholar]
- Saldanha AJ. Java treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Shadeo A, Lam WL. Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res. 2006;8:R9. doi: 10.1186/bcr1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Steele D, Cossu A, Budroni M, Palmieri G, Lakhani SR. Distribution and significance of 14-3-3sigma, a novel myoepithelial marker, in normal, benign, and malignant breast tissue. J Pathol. 2004;202:274–285. doi: 10.1002/path.1530. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchelnytskyi S. Proteomics in studies of signal transduction in epithelial cells. J Mammary Gland Biol Neoplasia. 2002;7:359–371. doi: 10.1023/a:1024029930563. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Janne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, et al. First-line Herceptin monotherapy in metastatic breast cancer. Oncology. 2001;61(Suppl 2):37–42. doi: 10.1159/000055400. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Reddy JC, Reyno LM. Efficacy of trastuzumab. Cancer Res. 2005;65:2044. doi: 10.1158/0008-5472.CAN-04-3943. [DOI] [PubMed] [Google Scholar]
- Wouters L, Gohlmann HW, Bijnens L, Kass SU, Molenberghs G, Lewi PJ. Graphical exploration of gene expression data: a comparative study of three multivariate methods. Biometrics. 2003;59:1131–1139. doi: 10.1111/j.0006-341x.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]