Abstract
Previous studies from this laboratory provided evidence that the intracellular bacterial pathogen Chlamydophila (Chlamydia) pneumoniae is present in the late-onset Alzheimer’s disease (AD) brain. Here we report culture of the organism from two AD brain samples, each of which originated from a different geographic region of North America. Culturable organisms were detectable after one and two passages in HEp-2 cells, respectively, for the two samples. Both isolates, designated Tor-1 and Phi-1, were demonstrated to be authentic C. pneumoniae using PCR assays targeting the C. pneumoniae-specific genes Cpn0695, Cpn1046, and tyrP. Assessment of inclusion morphology and quantitation of infectious yields in epithelial (HEp-2), astrocytic (U-87 MG), and microglial (CHME-5) cell lines demonstrated an active, rather than a persistent, growth phenotype for both isolates in all host cell types. Sequencing of the omp1 gene from each isolate, and directly from DNA prepared from several additional AD brain tissue samples PCR-positive for C. pneumoniae, revealed genetically diverse chlamydial populations. Both brain isolates carry several copies of the tyrP gene, a triple copy in Tor-1, and predominantly a triple copy in Phi-1 with a minor population component having a double copy. This observation indicated that the brain isolates are more closely related to respiratory than to vascular/atheroma strains of C. pneumoniae.
Keywords: C. pneumoniae, Alzheimer’s disease, Pathogenesis, Inflammation, Clinical isolate, Bacterial pathogenesis
Introduction
Chlamydophila (Chlamydia) pneumoniae is a respiratory pathogen known to be a causative agent for community-acquired pneumonia (e.g. Grayston, 1992; Campbell and Kuo, 2002). Several studies also have implicated infection with this obligate intracellular eubacterium in more severe respiratory pathologies, including chronic obstructive pulmonary disease (Clementsen et al., 2002). Importantly, infection with C. pneumoniae has been associated with several non-pulmonary diseases, including atherosclerosis, giant cell (temporal) arteritis, inflammatory arthritis, and others (Schumacher et al., 1999; Wagner et al., 2000; Belland et al., 2004; Watson and Alp, 2008; see also below). Epidemiologic study has demonstrated that the prevalence of infection with this organism is high in all adult populations examined, and that the prevalence increases with increasing age; multiple re-infections appear to be common (e.g. Leinonen, 1993). As with all chlamydial species, C. pneumoniae undergoes a biphasic developmental cycle. In the first phase, the elementary body (EB), the infectious, metabolically-inactive extracellular form of the organism, attaches to a target eukaryotic host cell; these are most often epithelial cells, but many other cell types can be infected (e.g. Hatch, 1999). The organism ends up in a cytoplasmic inclusion within which it reorganizes into the metabolically active growth form of the organism, the reticulate body (RB). RBs undergo several rounds of cell division, after which 70–80% reorganize back to the EB form. Newly-formed EB exit the host cell via lysis or exocytosis or ‘extrusion’ to continue propagation of the infection (Hatch, 1999; Hybiske and Stephens, 2007). C. pneumoniae requires about 72 h to complete passage through the cycle. Many studies have shown that under certain conditions and/or within specific host cell types, C. pneumoniae can alter its biologic state to generate persistent, long-term infections. Chlamydiae undergoing such infections are morphologically aberrant and display an unusual transcriptional profile (e.g. Byrne et al., 2001; Gérard et al., 2001, 2002, 2004; Ouellette et al., 2006; Mäurer et al., 2007; see also below). The transcriptional responses of various host cell types actively or persistently infected with chlamydiae also have been investigated (e.g. Schrader et al., 2007; Rodriguez et al., 2007; Alvesalo et al., 2008). Importantly, reports indicate that the mechanisms of pathogenesis differ between actively- and persistently-infecting chlamydiae and that it is in the persistent state that these organisms elicit chronic disease (Hogan et al., 2004; Whittum-Hudson et al., 2004, 2007). Alzheimer’s disease (AD) is a neurodegenerative disorder associated with atrophy/death of neurons, leading ultimately to severe cognitive dysfunction. The disease occurs in two forms: an early-onset form that is genetically determined, and a more common late-onset (sporadic) form that is not (Keefover, 1996). The incidence of sporadic AD increases with increasing age and is now considered to be the most prevalent cause of senile dementia (Keefover, 1996). The means by which the neuropathology characteristic of late-onset AD is elicited is poorly understood, but much research indicates that neuritic senile plaques (NSP), one aspect of that pathology, are critical in neuronal degeneration (Citron, 2004). Indeed, the dominant hypothesis currently guiding AD research focuses on the role of NSP in disease genesis (Sommer, 2002). Neurofibrillary tangles (NFT) also have been demonstrated to be important in the neuropathogenesis of AD (e.g. Iqbal et al., 2003).
In previous work, we identified C. pneumoniae in a high proportion of brain tissue samples examined from patients with sporadic AD, and we showed that the organism was present in those samples almost exclusively in brain regions displaying the NSP and NFT characteristic of the disease; the organism was extremely rare in congruent brain samples from age- and sex-matched non-AD (control) individuals (Balin et al., 1998; Arking et al., 1999). We further showed that astrocytes and microglia were host cells for the organism in the AD brain, and in recent studies we and others have identified neurons as host cells for the organism as well (Balin et al., 1998; Gérard et al., 2006; Appelt et al., 2008).
In our earlier reports, we demonstrated culture of C. pneumoniae from infected AD brain tissues, but we did not provide a detailed analysis of that original isolate (Balin et al., 1998; Gérard et al., 2006). However, recent studies from a collaborating group showed that respiratory infection of mice with that brain isolate elicits some aspects of CNS neuropathology that resemble those characteristics of late-onset AD (Little et al., 2004). Isolates of C. pneumoniae and well-studied serovars of C. trachomatis have been shown to include a significant level of DNA sequence variation at several loci (e.g. Daugaard et al., 2001; Viratyosin et al., 2002; Gieffers et al., 2003; Dreses-Werringloer et al., 2003). In some cases, DNA sequence variants at specific loci have been shown to correlate with tissue tropism and/or pathogenicity for these organisms (Gieffers et al., 2003; Fehlner-Gardiner et al., 2002; Caldwell et al., 2003; Cochrane et al., 2005).
For reasons relating to the possible elicitation of pathogenesis, we are interested in determining whether C. pneumoniae in the late-onset AD brain displays a normal active or persistent growth phenotype in epithelial cells and in its host cells in the brain. Further, it is important to define whether DNA sequences in AD brain isolates show variation similar to that shown for other, well-studied strains/isolates of C. pneumoniae. We therefore cultured the organism from brain tissues of two AD patients, each from a different geographic region of North America. In this article, we provide an initial analysis of basic cell biologic aspects shown by the brain-derived isolates of C. pneumoniae during infection of 3 human cell lines, and we initiate the analysis of genetic variation in those isolates and among brain tissue samples from individuals with late-onset AD.
Materials and methods
Cell culture, control C. pneumoniae
HEp-2 cells were maintained in Iscove’s Modified Dulbecco’s Medium (Invitrogen, Carlsbad, CA, USA), supplemented with 10% FBS, 2 mM L-glutamine and 10 µg/ml gentamycin (Invitrogen). The human astrocytoma cell line U-87 MG (Lytle et al., 2005; gift of Dr. T.R. Reddy) and the human microglioma cell line CHME-5 (de Gannes et al., 2000; gift of Dr. D. Feinstein) were grown in Dulbecco’s Modified Earle’s Medium (Invitrogen) with supplements as above. C. pneumoniae strain AR-39 was grown in HEp-2 cells as described (Byrne et al., 2001).
Brain tissue samples
Post-mortem brain tissue samples from patients with confirmed late-onset AD were obtained under an approved protocol from the Canadian Brain Tissue Bank (Toronto, Ontario, Canada), the Michigan Alzheimer’s Disease Research Center (Ann Arbor, MI, USA), and from the MCP-Hahnemann School of Medicine, Dept. of Pathology (Philadelphia, PA, USA; through Dr. B.J. Balin, now at the Philadelphia College of Osteopathic Medicine). All samples were confirmed as AD through histopathologic examination by a certified neuropathologist. Table 1 summarizes information for patients from whom samples were obtained.
Table 1.
Summary of AD patient characteristics.
| Patient | Sample Designated |
Sample source | Age at death/ sex |
Tissue | Postmortem interval (h) |
|---|---|---|---|---|---|
| AD1 | TOR-a | Toronto | 81/M | hippocampus | 2 |
| AD2 | TOR-b | Toronto | 90/F | hippocampus | 14 |
| AD3 | TOR-c | Toronto | 80/M | hippocampus | 4 |
| AD4 | PHI-a | Philadelphia | 87/F | temporal cortex | 8 |
| AD5 | PHI-b | Philadelphia | 70/F | hippocampus | 24 |
| AD6 | AA-a | Ann Arbor | 61/F | hippocampus | 8.75 |
| AD7 | AA-b | Ann Arbor | 67/M | hippocampus | 13 |
| AD8 | AA-c | Ann Arbor | 72/M | hippocampus | 13 |
| AD9 | AA-d | Ann Arbor | 78/F | hippocampus | 12 |
Culture of C. pneumoniae from AD brain tissues
Culture from brain tissues was performed in HEp-2 cells maintained as above. 300–400 mg of brain tissue were homogenized with 3–4 ml SPG buffer in a Dounce homogenizer with loose pestle. Various volumes of 1:10-diluted homogenates were added to confluent HEp-2 cell monolayers in six-well plates, centrifuged for 1 h at 750 × g at 30°C, then incubated for 3 days or more at 35°C in medium with 1 µg/ml cycloheximide. To monitor development of chlamydial inclusions, samples of putatively infected cultures were stained after each growth cycle with a FITC-labeled genus-specific antibody (Pathfinder™; BioRad, Hercules, CA, USA). Putatively infected cultures were harvested after 3–4 days incubation, lysed by sonication with subsequent passage of the cell lysates to fresh monolayers. If no obvious infection was identified, incubation times were extended for at least another growth cycle. For large-scale preparation of EB from AD brain-derived isolates, 12–16 six-well plates were infected with lysates from highly infected cells; harvested cells were sonicated to release chlamydiae. To remove cell debris, lysates were centrifuged for 20 min at 830 × g at 4°C. EB were concentrated from supernatants by centrifugation for 1 h at 67,000 × g, washed in SPG/reharvested by centrifugation and the resulting pellets resuspended in SPG for storage at −80°C. Processing of the two brain samples, and culture of chlamydiae from those tissues, were done completely independently to avoid cross contamination. A negative control was run in parallel during the entire process for each brain sample. Infectivity of the preparations was determined as described below. The C. pneumoniae isolate from the Toronto brain tissue sample (AD1) was designated Tor-1; that from the Philadelphia sample (AD4) was designated Phi-1 (Table 1).
Infection of human cells with brain isolates
Cells were infected for immunofluorescence assays on glass cover slips in 24-well plates. Briefly, HEp-2 and CHME-5 cells were seeded at 2×105 cells/well, U-87 MG at 4×105 cells/well, and grown for 24 h. Cells were inoculated with strain AR-39, Phi-1, and Tor-1 EB at a multiplicity of infection (MOI) of 0.5 for HEp-2, 1 for CHME-5 and U-87 MG cells, via centrifugation for 50 min at 750 × g at 30°C. After 2 h at 37°C, cells were washed 3× with HBSS, then incubated at 35°C in growth medium. Medium was supplemented with 1 µg/ml cycloheximide for HEp-2 cells. Cells were stained for chlamydial inclusions at 24, 48, and 72 h post-infection (p.i.) with the Pathfinder™mAb and analyzed using a Nikon E600 microscope with epifluorescence.
Infectious yield of C. pneumoniae in HEp-2, U-87 MG, and CHME-5 cultures
Six-well plates were seeded at 8×105 cells/well for HEp-2 and CHME-5 cells, and 1.2×106 cells/well for U-87 MG cells, then grown overnight. Cells were inoculated at a MOI of 0.5 (HEp-2) and 1 (CHME-5, U-87 MG) with C. pneumoniae EB as above, harvested by trypsination at 24, 48, and 72 h p.i., and frozen at −80°C. Chlamydial infectivity was determined by titration of cell lysates on HEp-2 monolayers. Cell suspensions were thawed on ice, sonicated, and vortexed with glass beads to disrupt cells. At least 3 different dilutions per cell lysate were prepared and centrifuged as duplicates onto HEp-2 monolayers in 96-well plates with incubation at 35°C for 3 days. Inclusions were visualized by staining with the Pathfinder™ mAb, counted, and expressed as IFU/ml.
Nucleic acid analyses
Nucleic acid preparations were made from Tor-1 and Phi-1 EB and from brain tissue samples as described (e.g. Gérard et al., 2002). C. trachomatis-, C. pneumoniae-, C. caviae-, and C. psittaci-specific PCRs were performed with primers listed in Table 2. In other experiments, DNA from culture isolates and brain tissues was used as template to amplify the omp1 gene for sequence analyses. Amplifications were done using Platinum High Fidelity Taq polymerase (Invitrogen); a nested PCR reaction sometimes was necessary to obtain a usable product from brain tissues for sequencing. The primers were: (outer) 5’-aaggccgtttttcaatgataagagcttcct-3’ and 5’-gtcttgatgtgatgtgaagtagg-3’; (inner) 5’-tgcctgcaggatatcttgtc-3’ and 5’-tgtagactctgataacaaggtgaggag-3’. Purified PCR products were cloned into the pGEM-T Easy vector (Promega Biotech, Madison, WI, USA). The DNA sequences of clone inserts from transformants in E. coli were determined at the core DNA sequencing facility of Wayne State University. Real-time PCR analyses were done as described (Gérard et al., 2005).
Table 2.
Chlamydial genes targeted and primers used for PCR and/or RT-PCR assays.
| C. pneumoniaea | Primers Employed | Product size |
|---|---|---|
| ompA (Cpn0695) | 5'-tattatccgccgcatttg-3' | 535 bp |
| 5'-gaggtgtctgtgtaaagttc-3' | ||
| Cpn1046 | 5'-gaaggttgctccactgatatg-3' | 729 bp |
| 5'-agtgagttctaccagttcatc-3' | ||
| C. trachomatis | ||
| plasmid ORF4 (pCt02) | 5’-ttccccttgtaattcgttgc-3’ | 201 bp |
| 5’-tagtaactgccacttcatca-3 | ||
| C. caviaeb | ||
| incA | 5'-gctgctgctgtaggggctaaaacggc-3' | 849 bp |
| 5'-ctctgcaccatcctgaaccataacacc-3' | ||
| C. psittacic | ||
| Cps0300 | 5'-catcgagtcgctatctct-3' | 871 bp |
| 5'-ccagttcgtccacctc-3' |
Gene sequence for each C. pneumoniae and C. trachomatis gene or transcript examined available at www.stdgen.lanl.gov; see also Stephens et al. (1998) and Read et al. (2000).
Full genome sequence, including the sequence for the incA gene, available at GenBank accession number AE015925.
Gene sequence generously supplied to A.P.H. by G.S.A. Myers, pers. comm.
tyrP analyses
Approximately 400 ng of DNA from EB of the Tor-1 and Phi-1 isolates and from strain AR-39 was digested with Nru1 (NE Biolabs, Beverly, MA, USA), separated on an agarose gel, and transferred to nylon filters (Roche, Indianapolis, IN, USA). A digoxigenin-labeled DNA probe whose sequence was localized within tyrP was used to determine the copy number of that gene in each DNA preparation. The probe was prepared by amplifying a 1-kb fragment of tyrP as described (Gieffers et al., 2003) with subsequent digoxigenin-labeling using the DIG-High Prime labeling and starter Kit I (Roche). Hybridization and detection were done using manufacturer’s instructions.
Results
Culture of C. pneumoniae from AD brain tissue samples
For the present study, tissue samples from the hippocampus (patient AD1, Table 1, Tor-1 isolate) and temporal cortex (patient AD4, Table 1, Phi-1 isolate) from individuals with documented AD were subjected to culture in HEp-2 cells. For the hippocampal specimen, inclusions were identified after a single growth cycle of 3 days. With that sample, an infection rate of 1–2% of host cells was achieved using 0.5 ml 1:10-diluted tissue homogenate as initial inoculum. For the temporal cortex sample, a longer culture period was required to detect inclusions. Immunofluorescence staining of host cells with the mAb was difficult to assess after one passage due to the large amount tissue debris present. Unambiguous detection of a few inclusions was achieved for this sample after 2 growth cycles (not shown). Assessment of inclusion presence for both samples was done by staining with the Pathfinder™ mAb (see below).
Confirmation that culture isolates are authentic C. pneumoniae
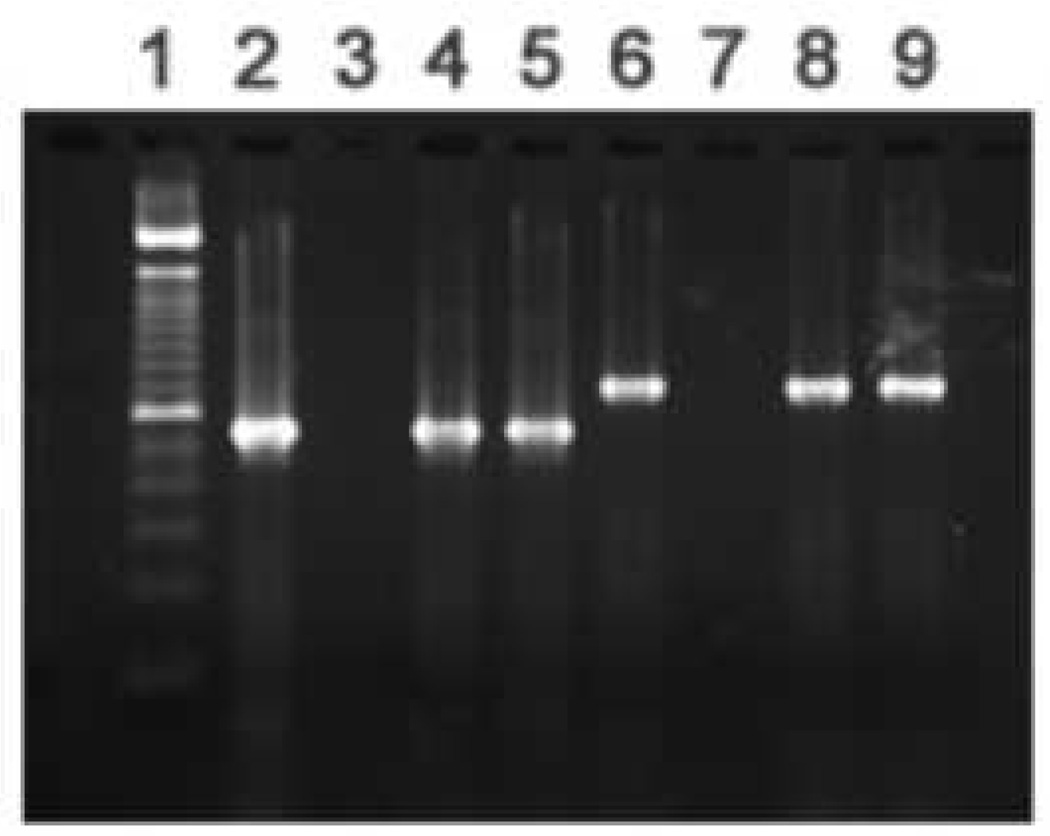
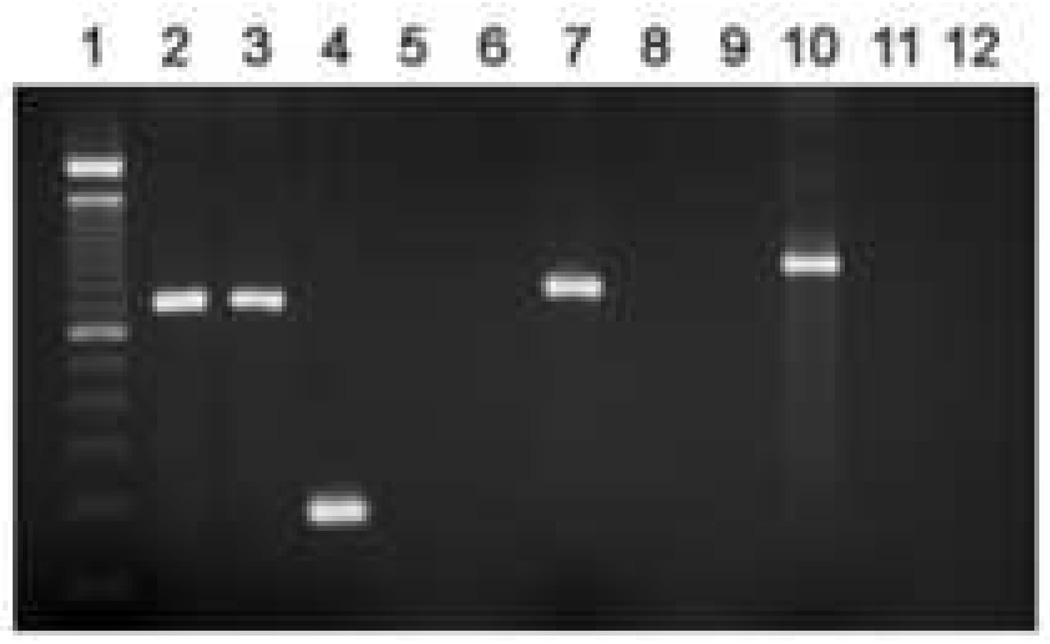
The Pathfinder™ mAb targets genus-specific LPS and thus cannot define the chlamydial species present in a given sample. Species definition for the brain isolates was done initially using PCR assays targeting genes specific to various chlamydial species. The data provided in Fig. 1A shows that PCR assays targeting the omp1 gene of C. pneumoniae (Cpn0695, encoding the major outer membrane protein, MOMP) and those targeting the Cpn1046 gene (encoding a probable aromatic amino acid hydroxylase not present in C. trachomatis; Read et al., 2000) were both positive for the Tor-1 and Phi-1 isolates; the C. pneumoniae-specific tyrP gene also was identified in both isolates (see below). In contrast, the data given in Fig. 1B demonstrate that PCR assays targeting ORF4 (pCTO2; specific to the C. trachomatis plasmid), incA of C. caviae, and Cps0300 of C. psittaci (strain 6BC) all yielded no amplification products. Sequence data from omp1 of the Tor-1 and Phi-1 isolates is given below and clearly identified the MOMP of C. pneumoniae. Thus, the organisms cultured from AD brain tissues were authentic C. pneumoniae and not another chlamydial or other species.
Fig. 1.
Representative PCR screening assays targeting C. pneumoniae genes (Panel A), and genes from other chlamydial species (Panel B), using DNA prepared from the Tor-1 and Phi-1 isolates. EBs were prepared from each isolate as described in ‘Materials and methods’. DNA preparation from each isolate and PCR assay systems used in these analyses, also are described in ‘Materials and methods’ and Table 2. (A) Lanes in the gel in Panel A are: 1, 100 bp size standards; amplifications targeting 2, Cpn0695 (ompA) using C. pneumoniae strain AR-39 DNA as template (positive control); 3, Cpn0695 using water as template (negative control); 4, Cpn0695 using Tor-1 DNA, and 5, Phi-1 DNA as template; 6, Cpn1046 using C. pneumoniae strain AR-39 DNA as template (positive control); 7, Cpn1046 using water as template (negative control); 8, Cpn1046 using Tor-1 DNA, and 9, Phi-1 DNA as template. (B) Lanes in the gel in Panel B are: 1, 100 bp size standards; amplifications targeting 2, Cpn1046 using Tor-1 DNA as template; 3, Cpn1046 using Phi-1 DNA as template; 4, pCt02 (plasmid ORF4 C. trachomatis) using C. trachomatis serovar K, and 5, Tor-1, and 6, Phi-1 DNA as template; 7, incA (C. caviae) using C. caviae (GPIC), and 8, Tor-1, and 9, Phi-1 DNA as template; 10, Cps0300 (C. psittaci) using C. psittaci strain 6BC, and 11, Tor-1, and 12, Phi-1 DNA as template. Amplification product sizes are given in Table 2.
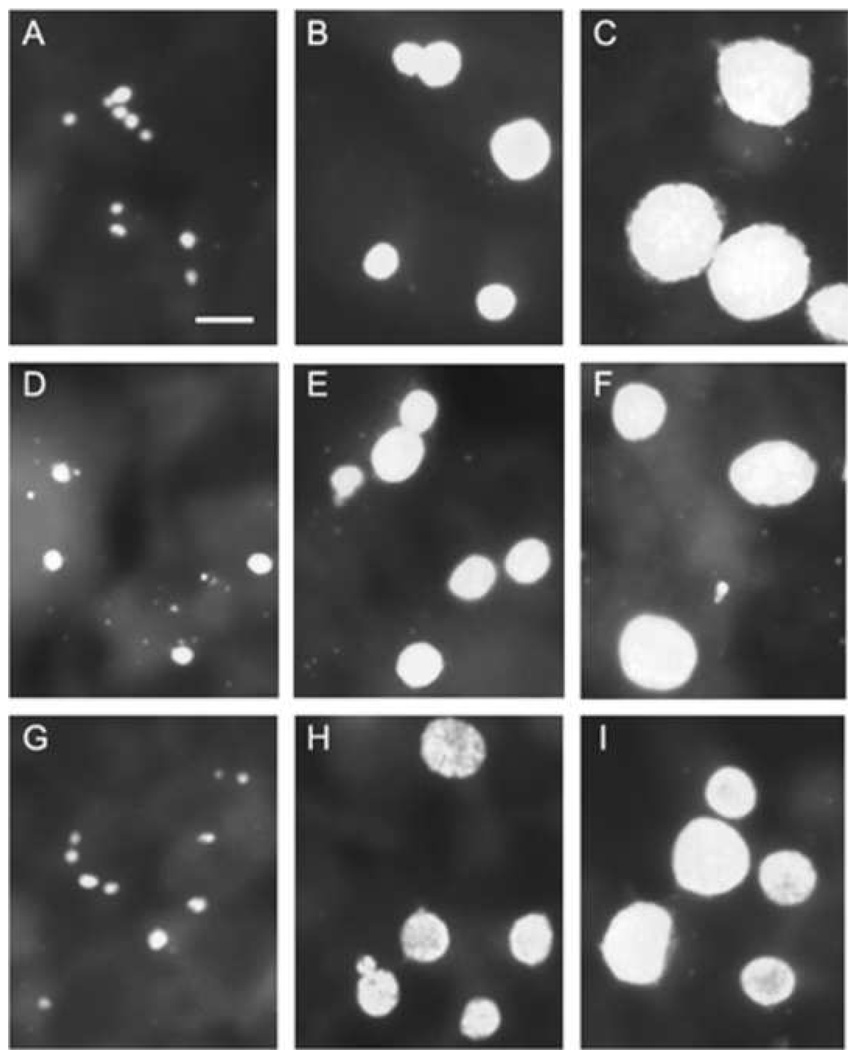
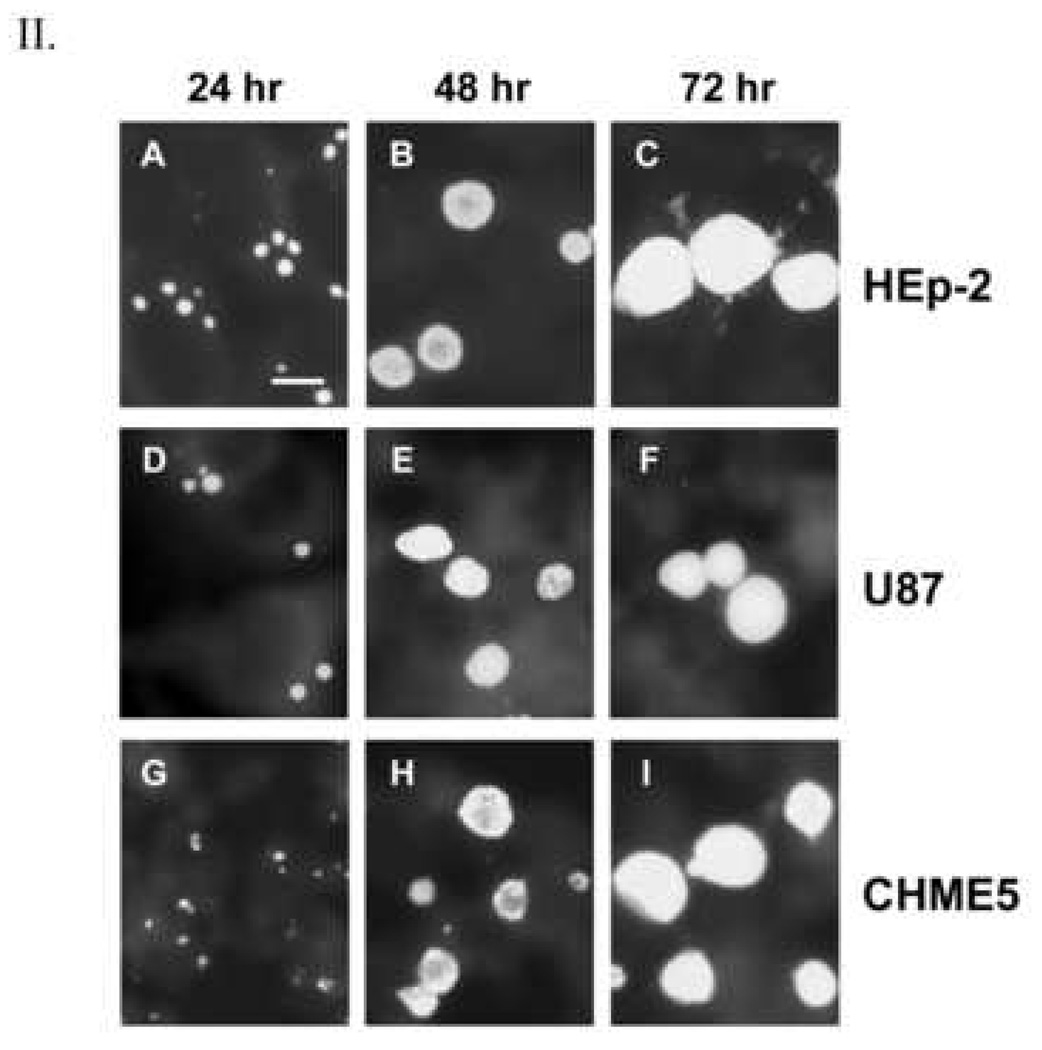
Analysis of Tor-1- and Phi-1-infected HEp-2, U-87 MG, and CHME-5 cells
Studies from this laboratory showed that astrocytes and microglia serve as host cells for C. pneumoniae in the AD brain (Balin et al., 1998; Gérard et al., 2006); however, that work provided no insight into the cell biologic or molecular characteristics of such infections for either cell type. To provide initial characterization of C. pneumoniae infection of those cell types, human astrocytoma (U-87 MG) and microglial (CHME-5) cell lines were inoculated with Tor-1 and Phi-1 EB; the resulting inclusion size and morphology were then determined and compared to similarly infected HEp-2 cells at 24, 48, and 72 h p.i.. Fig. 2 provides representative immunofluorescence images of all 3 cell types inoculated with Phi-1 (Group I panels) and with Tor-1 (Group II panels). In both brain-related cell lines, inclusion size and shape were virtually identical to those in infected HEp-2 cells at all times examined p.i. for both isolates. Thus, human microglia and astrocytes can be infected with the C. pneumoniae AD brain isolates. Inclusions produced by Tor-1 and Phi-1 infections of HEp-2, U-87 MG, and CHME-5 cells were essentially identical in size and morphology to those produced by infection of these cell lines with AR-39, a respiratory strain of C. pneumoniae (Dreses-Werringloer et al., 2006).
Fig. 2.
Immunofluorescence analysis of inclusion size and shape in HEp-2 (Panels A–C), U-87 MG (Panels D–F), and CHME-5 (Panels G–I) cells infected with C. pneumoniae isolates Phi-1 (Group I[A–I], left) and Tor-1 (Group II[A–I], right) at 24 h p.i. (Panels A, D, G), 48 h p.i. (Panels B, E, H), and 72 h p.i. (Panels C, F, I). Cells were infected on glass cover slips at MOI 0.5 for HEp-2 cells and MOI 1 for U-87 MG and CHME-5 cells. Infected cells were stained with the Pathfinder™ mAb, which targets the chlamydial LPS. Infected cells were visualized using epifluorescence on a Nikon E600 microscope. The scale bar in Panel A in each group represents 10 µm, and all photomicrographs were taken at the same magnification in both large groups.
Active infection of human glial cells by AD brain-derived isolates
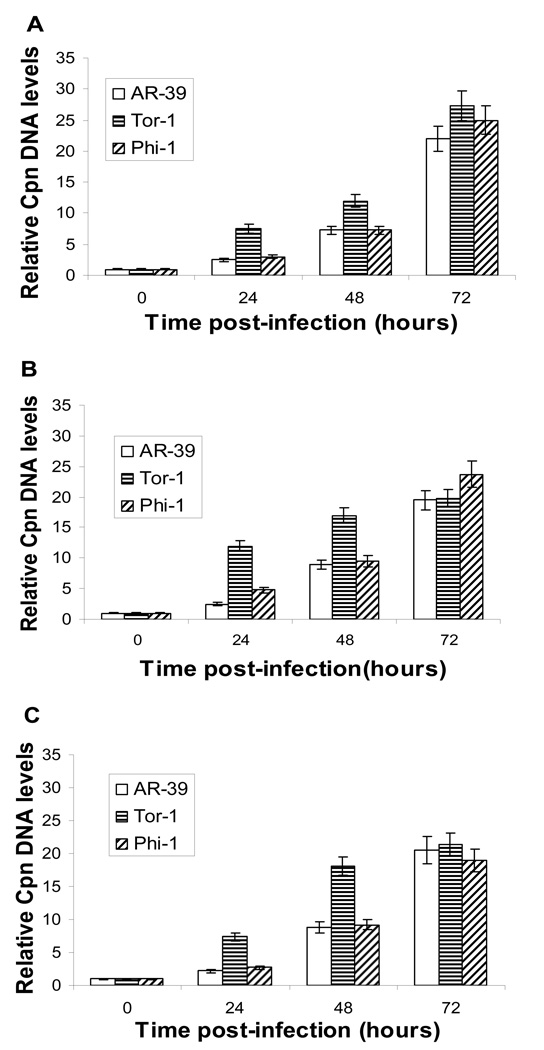
In active chlamydial infection, the developmental cycle terminates with release of newly-formed EB (Hatch, 1999); in persistent infection, replication of RB is interrupted, engendering attenuated EB production (e.g. Beatty et al., 1994; Gérard et al., 2001). The similar attributes observed in Tor-1- and Phi-1-infected HEp-2, U-87 MG, and CHME-5 cells in the analyses above, and the similarity of those attributes to those characteristic of AR-39 infection of HEp-2 cells, suggested that both brain-related cell lines undergo active, rather than persistent, infection in the cell lines employed. A study from this group showed that accumulation of bacterial chromosome is far slower in persistently- than in actively-infecting chlamydiae (Gérard et al., 2001). Real-time PCR to assess chlamydial chromosome accumulation for Tor-1 and Phi-1 infecting each cell line indicated that such accumulation by strain AR-39 and the Phi-1 isolate was similar at all times examined in each of the 3 host cell lines (Fig. 3). We note that at 24 h and 48 h p.i., the relative level of DNA in the Tor-1 isolate was several-fold higher than those of AR-39 or Phi-1, although that level was congruent with those of the other two strains at the final time point examined, 72 h p.i. (see Discussion).
Fig. 3.
Representative quantitative real-time PCR analyses of relative levels of chromosomal DNA from C. pneumoniae isolates Phi-1 and Tor-1 as a function of time post-infection in HEp-2 cells (Panel A), U-87 MG cells (Panel B), and CHME-5 cells (Panel C). Cells were infected, nucleic acids prepared and analyzed, as given in ‘Materials and methods’. Assays were run 3 times independently and in triplicate each time, from 2 independent experiments. Data were normalized to host 18S rDNA levels and indexed to the level at t=0. The bars indicate standard error.
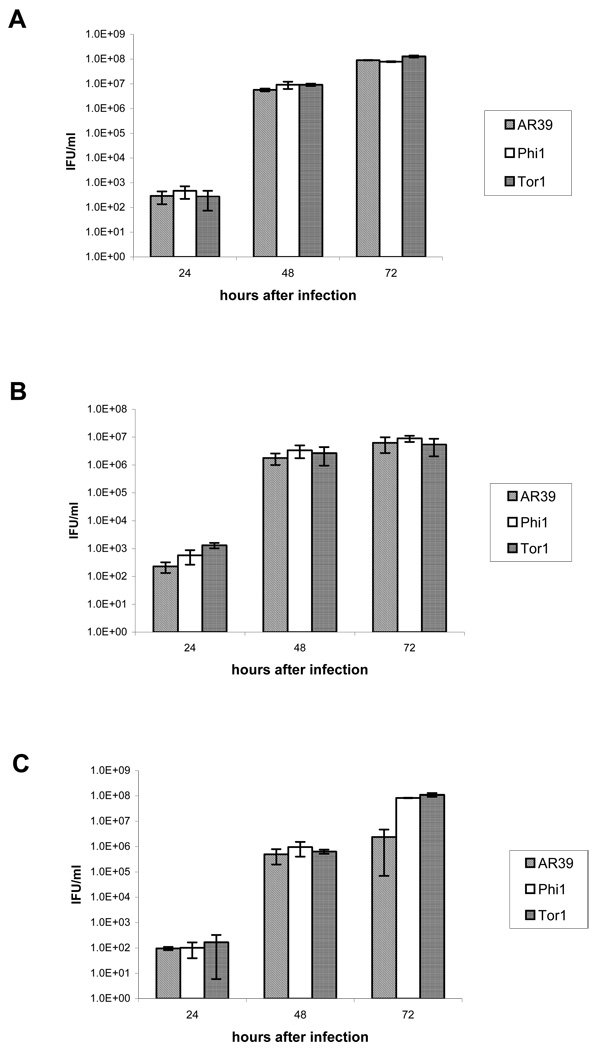
The data provided in Fig. 4 demonstrate that both glial cell lines allowed efficient production of infectious progeny from each of the 3 C. pneumoniae strains/isolates examined. HEp-2 cells were the most productive, given the lower MOI (0.5) used compared to that (1) used for the glial cell lines (compare Fig. 4A to 4B). Interestingly, the level of infectious EB produced by the Tor-1 isolate was congruent to those produced by AR-39 and the Phi-1 isolate at both the 24 h and 48 h time points p.i., despite the relatively higher level of chromosomal DNA from that isolate at 24 h and 48 h p.i. (see Discussion). The infectious yield in HEp-2 cells was, however, ~10-fold lower than that for both the Tor-1 and Phi-1 isolates at 72 h p.i. in CHME-5 cells. Regardless, these assays demonstrate that the brain isolates undergo active infection in each of the 3 cell lines/types assessed.
Fig. 4.
Recovery of infectious EB from HEp-2 cells (Panel A), U-87 MG cells (Panel B), and CHME-5 cells (Panel C) at 24, 48, and 72 h p.i. with the AR-39 strain of C. pneumoniae and the Tor-1 and Phi-1 isolates of that organism from AD brain tissue samples. Cultures were inoculated, and the infectious yields from each assessed as given in ‘Materials and methods’. Data are expressed as average IFU/ml of triplicate samples.
omp1 DNA sequence in AD brain isolates and AD brain tissues
The sequence of omp1 traditionally has been used to differentiate strains/serovars of C. trachomatis. We determined the sequence of that gene from the Tor-1 and Phi-1 isolates and from C. pneumoniae DNA amplified directly from nucleic acid preparations from infected brain tissue samples from additional AD patients. The published omp1 DNA sequence from strain AR-39 was used as reference (Read et al., 2000). Table 3 provides a summary of sequence results from the brain isolates and AR-39. These data showed that both isolates and AR-39 harbored substantial nucleotide variation in omp1, indicating that all three are non-clonal populations. Not all base changes identified lead to amino acid substitutions, and no common substitutions occurred among Tor-1, Phi-1, and AR-39. When amino acid substitutions were engendered by nucleotide change, the nature of the substituted amino acids was diverse, including conservative changes (e.g., lys→arg, thr→ala), and a more drastic change from basic lys→acidic glu. All variations, except that in codon 6 in the leader sequence, were in conserved, not variable, domains.
Table 3.
Nucleotide variation in the omp1 gene from AR-39 and the AD brain isolates Tor-1 and Phi-1 compared to the published omp1 sequence for AR-39a.
| Isolate/clone # | Position of nucleotide change (sequence change) |
Resulting amino acid change |
Location |
|---|---|---|---|
| AR-39 | |||
| AR-39/5 | 453 (GCG→GCA) | silent | CDb II |
| AR-39/9 | no variation | ||
| AR-39/21 | 1164 (AGA→AGG) | silent | CD V |
| AR-39/22 | no variation | ||
| AR-39/23 | 455 (TTC→TCC) | Phe152→Ser | CD II |
| AR-39/24 | 424 (AAT→GAT) | Asn142→Asp | CD II |
| Tor-1 | |||
| Tor-1/4 | no variation | ||
| Tor-1/6 | 653 (AAA→AGA) | Lys218→Arg | CD III |
| Tor-1/7 | 155 (TGC→TAC) | Cys52→Tyr | CD I |
| 409 (ACT→GCT) | Thr137→Ala | CD II | |
| Tor-1/8 | no variation | ||
| Tor-1/9 | no variation | ||
| Tor-1/12 | 83 (AAC→AGC) | Asn28→Ser | CD I |
| Phi-1 | |||
| Phi-1/1 | no variation | ||
| Phi-1/2 | 831 (AGA→AGG) | silent | CD I |
| Phi-1/3 | no variation | ||
| Phi-1/5 | 16 (AAG→GAG) | Lys6→Glu | leader sequence |
| 380 (AAC→AGC) | Asn127→Ser | CD II | |
| Phi-1/8 | 376 (TTA→CTA) | silent | CD II |
| Phi-1/9 | 561 (TCT→TCC) | silent | CD III |
Numbering started at the first translated codon.
conserved domain
Data from omp1 amplified directly from AD brain tissues are presented in Table 4. Nine specimens were processed, from patients from three areas of North America. As with Tor-1 and Phi-1, these DNA sequence determinations showed a high level of nucleotide variation. The majority of changes identified in the directly-amplified samples resulted in amino acid substitutions; however, in contrast to data above, the variations were more evenly distributed over the omp1 sequence; 17 changes were located in conserved and 11 in variable domains, and 8 were found in the leader sequence. We identified a couple amino acid substitutions held in common among different brain samples. That is, the leu281→phe and ser11→tyr exchanges were found in the TOR-a and PHI-a samples, and the thr→ala change in codon 163 in PHI-b and AA-a. Three cloned inserts, two of AA-a and one of AA-b, had lys161→arg. The lys6→glu and asn127→ser changes were present in 4 inserts derived from sample AA-b. Whereas AA-b/10 had only these two changes, inserts AA-b/11 and AA-b/24 carried an additional variation in codon 34, a change from leu→ser. A fourth change (asp31→gly), not seen in any other insert, was identified in clone AA-b/24. The variations found in codons 6 and 380 in three inserts of sample AA-b were present in one insert of the Phi-1 isolate (Table 3 and Table 4; see Discussion). No other nucleotide changes identified in omp1 from the brain isolates were found in inserts from direct omp1 amplifications from brain tissues.
Table 4.
Nucleotide variation in the omp1 gene amplified from DNA preparations from AD brain tissues compared to the published omp1 sequence from strain AR-39a.
| Sample/clone | Position of nucleotide change (sequence change) |
Resulting amino acid change |
Location |
|---|---|---|---|
| TOR-ab | |||
| TOR-a/4 | no variation | ||
| TOR-a/5 | no variation | ||
| TOR-a/8 | 390 (GAT→GAC) | silent | CDc II |
| TOR-a/10 | 843 (TTA→TTC) | Leu281→Phe | CD IV |
| TOR-a/11 | no variation | ||
| TOR-a/14 | 32 (TCC→TAC) | Ser11→Tyr | leader sequence |
| TOR-b | |||
| TOR-b/1 | 567 (GTA→GTG) | silent | CD III |
| TOR-b/3 | no variation | ||
| TOR-b/8 | no variation | ||
| TOR-b/9 | no variation | ||
| TOR-b/18 | no variation | ||
| TOR-c | |||
| TOR-c/7 | 925 (GCT→ACT) | Ala309→Thr | VD IV |
| TOR-c/17 | 1078 (ACT→GCT) | Thr360→Ala | CD V |
| TOR-c/25 | no variation | ||
| TOR-c/34 | no variation | ||
| TOR-c/36 | 590 (GCC→GTC) | Ala197→Val | CD III |
| PHI-a | |||
| PHI-a/3 | no variation | ||
| PHI-a/9 | no variation | ||
| PHI-a/14 | 32 (TCC→TAC) | Ser11→Tyr | leader sequence |
| PHI-a/18 | no variation | ||
| PHI-a/19 | 843 (TTA→TTC) | Leu281→Phe | CD IV |
| PHI-a/20 | no variation | ||
| PHI-b | |||
| PHI-b/2 | no variation | ||
| PHI-b/10 | 292 (ACT→GCT) | Thr98→Ala | VD I |
| PHI-b/6 | 298 (GCC→ACC) | Ala100→Thr | VD I |
| 487 (ACT→GCT) | Thr163→Ala | VD II | |
| PHI-b/11 | no variation | ||
| AA-a | |||
| AA-a/1 | no variation | ||
| AA-a/2 | 482 (AAA→AGA) | Lys161→Arg | VD II |
| AA-a/4 | 513 (CCA→CCG) | silent | VD II |
| AA-a/5 | no variation | ||
| AA-a/11 | 487 (ACT→GCT) | Thr163→Ala | VD II |
| 902 (ATT→ACT) | Ile301→Thr | CD IV | |
| 1124 (TTA→TCA) | Leu375→Ser | CD V | |
| AA-b | |||
| AA-b/1 | 482 (AAA→AGA) | Lys161→Arg | VD II |
| AA-b/10 | 16 (AAG→GAG) | Lys6→Glu | leader sequence |
| 380 (AAC→AGC) | Asn127→Ser | CD II | |
| AA-b/11 | 16 (AAG→GAG) | Lys6→Glu | leader sequence |
| 101 (AAC→AGC) | Leu34→Ser | CD I | |
| 380 (AAC→AGC) | Asn127→Ser | CD II | |
| AA-b/24 | 16 (AAG→GAG) | Lys6→Glu | leader sequence |
| 92 (GAG→GGT) | Asp31→Gly | CD I | |
| 101 (AAC→AGC | Leu34→Ser | CD I | |
| 380 (AAC→AGC) | Asn127→Ser | CD II | |
| AA-b/28 | 16 (AAG→GAG) | Lys6→Glu | leader sequence |
| 380 (AAC→AGC) | Asn127→Ser | CD II | |
| 750 (GCT→GCC) | silent | VD III | |
| AA-c | |||
| AA-c/3 | no variation | ||
| AA-c/4 | 87 (CCT→CCG) | silent | CD I |
| AA-c/5 | 58 (TCC→CCC) | Ser20→Pro | leader sequence |
| 935 (AAC→ACC) | Asn312→Thr | VD IV | |
| AA-c/8 | no variation | ||
| AA-c/9 | no variation | ||
| AA-d | |||
| AA-d/8 | no variation | ||
| AA-d/24 | no variation | ||
| AA-d/26 | 70 (TTG→CTG) | silent | CD I |
| 490 (ATG→GCT) | Thr164→Ala | VD II | |
| AA-d/37 | no variation | ||
| AA-d/38 | 48 (GGT→GGC) | silent | leader sequence |
Numbering started at the first translated codon.
TOR indicates samples from the Toronto source, PHI from Philadelphia, and AA from the Ann Arbor source.
CD, conserved domain; VD, variable domain
Analysis of tyrP
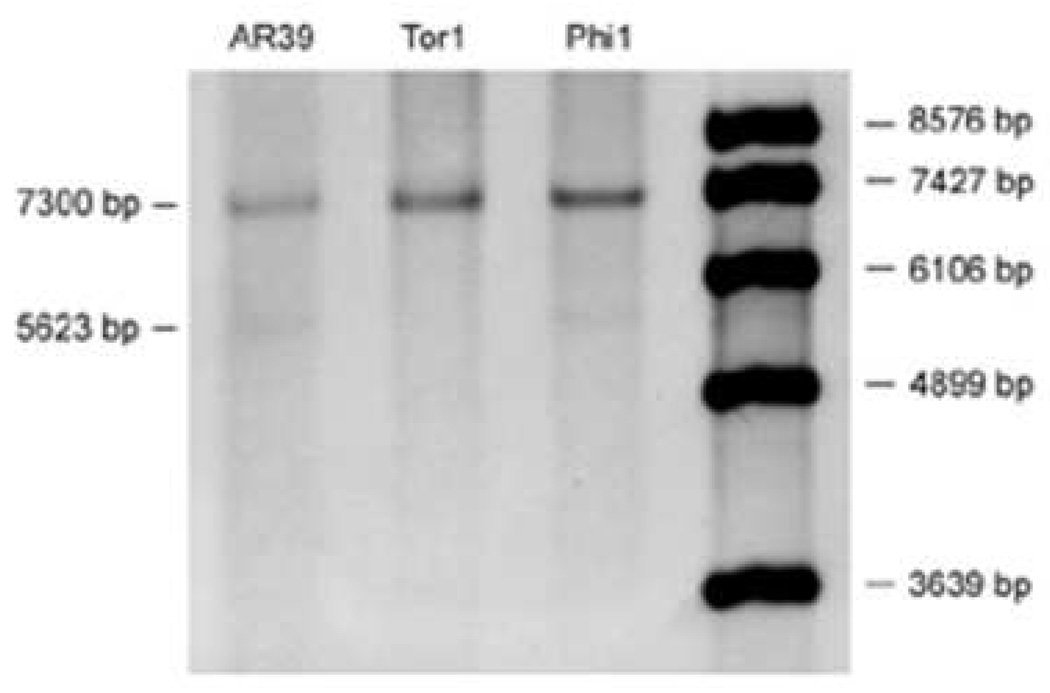
The tyrP gene of C. pneumoniae, encoding a tyr and trp permease, has been shown to differ among various strains/isolates of this organism. It is present in a single copy in strain J138, in two copies in TW-183, and in 3 copies in AR-39 and CWL029 (Gieffers et al., 2003; Shirai et al., 2000). The double and triple copies result from duplication or triplication of a 1649-bp fragment containing tyrP and two truncated genes, glmS and yccA. As shown in Fig. 5, both brain isolates and AR-39 each carried a triple copy of tyrP. A weak band of 5623 bp, representing a double copy, was also detected in AR-39 and Phi-1 but not in Tor-1. Thus, Tor-1 comprises a population harboring a triple copy at tyrP, while the Phi-1 isolate is a more mixed population at this locus, comprised of organisms with double or triple gene copies. These data suggest that both Tor-1 and Phi-1 are not only non-clonal, but also not identical to one another or to strain AR-39. The data further suggest that both the brain isolates are more closely related to respiratory isolates such as TW-183 and AR-39 than to atheroma isolates such as J138.
Fig. 5.
Southern blot analysis of the tyrP locus in AR-39 and the brain isolates Tor-1 and Phi-1. DNA was prepared from the AD brain isolates, and blotting and analyses were done as given in ‘Materials and methods’. A 5623-bp fragment indicates a double copy, the 7300-bp fragment a triple copy of the gene. AR-39 and Phi-1 were mixtures of double and triple copy population; Tor-1 is a population carrying only the triple copy.
Discussion
In this report, we describe the successful culture of C. pneumoniae from postmortem brain tissues from 2 patients with sporadic AD, and we provide initial characterization of those two isolates. The observations presented demonstrate that this pathogen can be cultured from infected brain tissue of AD patients, that infection of cultured human astrocyte and microglial cells, as well as human epithelial cells, with each of the AD brain isolates displays an active, rather than a persistent, phenotype, that the brain-derived organism is not identical to respiratory or atheroma-derived strains of C. pneumoniae, and that it is genetically heterogenous within individual patients.
Recovery of C. pneumoniae from clinical samples is often unsuccessful; e.g., isolation of this organism from atheromatous materials has been achieved by only a few groups, and in some cases detection of organism required 8 passages (Apfalter et al., 2000). In the cultures from AD brain tissues, the organism was detected after a single passage for one isolate and two passages for the other. Previous analyses of brain tissues from patient AD1, whose tissue produced detectable C. pneumoniae (Tor-1) after one passage, indicated a heavy burden of organism, partly explaining the ease of culture (Gérard et al., 2005). The load of organism in atherosclerotic plaques may be low, and if so that low burden might explain in part the variable success in isolation from that site. Another explanation for culture difficulties may be that C. pneumoniae persistently, rather than actively, infects those tissues. The successful culture of C. pneumoniae from AD brain specimens suggests active infection in those tissues in vivo, although further study of this issue is required.
In our studies, the amount of homogenized brain tissue used for initial inoculation of HEp-2 cells was critical to the outcome of the analyses. Lower tissue concentrations resulted in earlier detection of chamydial inclusions in the host cells, probably due at least in part to lower levels of obstruction of the infection process by tissue debris. We have not compared systematically the efficiency of extended culture vs. serial passage for isolation of C. pneumoniae, but our observations suggest that the latter approach improved culture sensitivity.
We recently reported that the respiratory strain AR-39 undergoes active growth in the U-87 MG and CHME-5 cell lines (Dreses-Werringloer et al., 2006). Here, we quantified the yield of infectious progeny produced by AR-39 and the brain isolates in all 3 cell lines. Even though HEp-2 cells were the most susceptible host cell type for infection by each of the three 3 strains/isolates, the glial cell lines produced high levels of EB. It is interesting to note that, while the relative level of chromosomal DNA produced by the Tor-1 isolate was higher than those from AR-39 and Phi-1 at 24 h and 48 h p.i. in each of the 3 host cell types, that DNA level was congruent with all others at the final time point examined in all those host cells. In contrast, production of infectious progeny at each of the time points examined was virtually identical for each strain/isolate in each host cell type. We suspect that this inconsistency results from a somewhat increased doubling rate for Tor-1 RB during active infection compared to those of AR-39 and the Phi-1 isolate, combined with a relatively normal rate of RB to EB ‘dedifferentiation’; i.e., a rate similar to those characteristic of AR-39, Phi-1, and other C. pneumoniae strains and isolates. We intend to examine this possible explanation using genetic means. Regardless, it seems clear that the Tor-1 developmental cycle differs in some respects from those of AR-39 and other strains.
Production of new C. pneumoniae EB was more efficient in the glial cell lines than has been reported in other host cell types (e.g. Gaydos et al., 1996). Boelen et al. (2007) reported active infection of a murine astrocyte cell line with a respiratory C. pneumoniae strain, but in contrast to our observations that cell line displayed only a low-level productive infection. Another study that used a mouse microglial cell line described a more productive infection, but it was less effective than our infection of CHME-5 cells (Ikejima et al., 2006). All these observations indicate that quite significant cell line-dependent variations in susceptibility to C. pneumoniae exist. That infection of human glial cells in vitro by the AD brain isolates showed an active growth phenotype confirms that this is not solely a specific attribute of AR-39 or other respiratory strains of C. pneumoniae. Active infection of astrocytes and microglia may have implications for neuropathogenesis in AD, since the developmental cycle often ends with lysis of the host cell and release of new EB. We note, however, that EB can and often are released from infected host cells by exocytosis/’extrusion’, leaving the host cell intact (e.g. Hybiske and Stephens, 2007). Some loss of associated astrocytes, which are important multifunctional housekeeping cells crucial to support of normal neuronal function, could abet neuronal loss. Death of some neurons may also result from C. pneumoniae-mediated host cell lysis in the AD brain. Infection of microglia, as the main phagocytic cells in the brain, may contribute to pathogenic processes in the CNS, such as triggering inflammatory events.
Sequence data from the omp1 gene of Tor-1, Phi-1, and AR-39, as well as DNA amplified from AD brain tissues of 7 additional patients from 3 different geographic regions in North America, revealed a relatively high level of single nucleotide polymorphisms in all samples. The variations showed no general pattern, but rather were diverse in both location within the gene and in the nature of predicted amino acids substituted at codons with a non-silent nucleotide change. These observations were surprising, considering that sequencing of 4 C. pneumoniae strains showed identical DNA sequences of omp1, with the exception of one synonymous variation described in one strain (Kalman et al., 1999; Read et al., 2000; Shirai et al., 2000). This level of published sequence conservation cannot mean that little or no sequence variation exists in nature for this gene, since we and others have identified substantial variation in that sequence. For one example, Molestina et al. (1998) showed that the coronary strain A-03 displayed differences from respiratory strains in its omp1 sequence. Also, C. pneumoniae from carotid plaques displayed variation in variable domain IV of omp1 (Cochrane et al., 2005). All variants identified in omp1 from the coronary strains were different from those identified here in the AD brain isolates. Most, but not all, predicted amino acid substitutions identified here in the omp1 genes were conservative. One substitution, identified in a single clone insert from the Tor-1 isolate, was that in which cys was replaced by tyr in codon 52. Cys residues are crucial for disulfide bond formation to establish internal cross-linking within the MOMP and cross-linking with other cysteine-rich proteins in the EB outer membrane. Loss of a cysteine may engender decreased structural rigidity in the outer membrane of the organism possessing that substitution. However, prediction of whether any of our amino acid changes will result in a meaningful phenotype is simply not possible. Also, we do not know the abundance within the overall population of organisms having any particular DNA sequence at omp1; thus, potential structural effects due to amino acid variations may be masked by organisms having more prevalent wild-type or other sequences.
We identified a range of predicted amino acid substitutions in the MOMP of the brain isolates, but some changes were held in common among different samples. For example, overlapping variations were found in specimens originating from different geographic regions, some of which were identified in the Phi-1 isolate and in 4 clones of the AA-b specimen. These observations do not, of course, prove the existence of a neurotropic C. pneumoniae strain. However, they may indicate that the populations detected in the various AD patients have something in common that differentiates them from respiratory strains.
Analysis of tyrP demonstrated that the Tor-1 isolate comprises a population harboring only a triple copy of the gene, whereas the Phi-1 isolate represents a mixed population carrying predominantly the triple copy, with a minority of the double copy, as was found for the AR-39 strain as well. A recent report showed that respiratory strains contained multiple tyrP copies, whereas vascular isolates harbored a single copy only (Gieffers et al., 2003). The data presented here suggest that Tor-1 and Phi-1 are more similar to respiratory than to vascular isolates, at least in terms of tyrP copy number. The results of additional analyses for single nucleotide polymorphisms and additional DNA sequence analyses in both isolates support this contention (U.D.-W., A.P.H., manuscript in preparation). Those studies also indicated strongly that additional analyses should be performed on homogeneous bacterial populations.
Our earlier demonstrations of the presence of this unusual pathogen in the brain did not demonstrate a role for the organism in AD neuropathogenesis (Balin et al., 1998; Gérard et al., 2006). However, the observations that C. pneumoniae can be cultured from the AD brain relatively easily and that two common host cell types for the organism in the CNS can be actively infected by AD brain-derived or respiratory isolates of the organism both support such a role. We currently are investigating mechanisms of neuropathogenesis possibly elicited by this pathogen, using both in vitro and animal model systems.
Acknowledgments
This work was supported by NIH grants AI-44055 (A.P.H.), AR-47186 (H.C.G.), HL-71735 (R.J.B.), and AI-44493 and AR-48331 (both J.A.W.-H.) from the US National Institutes of Health. The Michigan Alzheimer’s Disease Research Center is supported by NIH grant AG-08671. We are grateful to the families that donated tissues to the sources listed here for research in Alzheimer’s disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apfalter P, Loidl M, Nadrchal R, Makristathis A, Rotter M, Bergmann M, Polterauer P, Hirschl AM. Isolation and continuous growth of Chlamydia pneumoniae from arterectomy specimens. Eur. J. Clin. Microbiol. Infect. Dis. 2000;19:305–308. doi: 10.1007/s100960050481. [DOI] [PubMed] [Google Scholar]
- Appelt DM, Roupas MR, Way DS, Bell MG, Albert EV, Hammond CJ, Balin BJ. Inhibition of apoptosis in neuronal cells infected with Chlamydophila (Chlamydia) pneumoniae. BMC Neurosci. 2008;24:9–13. doi: 10.1186/1471-2202-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvesalo J, Greco D, Leinonen M, Raitila T, Vuorela P, Auvinen P. Microarray analysis of a Chlamydia pneumoniae-infected human epithelial cell line by use of gene ontology hierarchy. J. Infect. Dis. 2008;197:156–162. doi: 10.1086/524142. [DOI] [PubMed] [Google Scholar]
- Arking EJ, Appelt DM, Abrams JT, Kolbe S, Hudson AP, Balin BJ. Ultrastructural analysis of Chlamydia pneumoniae in the Alzheimer’s brain. Pathogenesis. 1999;1:201–211. [PMC free article] [PubMed] [Google Scholar]
- Balin BJ, Gerard HC, Arking EJ, Appelt DM, Branigan PJ, Abrams JT, Whittum-Hudson JA, Hudson AP. Identification and localization of Chlamydia pneumoniae in the Alzheimer’s brain. Med. Microbiol. Immunol. 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- Beatty WL, Morrison RP, Byrne GI. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Ouellette SP, Gieffers J, Byrne GI. Chlamydia pneumoniae and atherosclerosis. Cell. Microbiol. 2004;6:117–127. doi: 10.1046/j.1462-5822.2003.00352.x. [DOI] [PubMed] [Google Scholar]
- Boelen E, Steinbusch HWM, van der Ven AJ, Grauls G, Bruggeman CJ, Stassen FR. Chlamydia pneumoniae infection of brain cells: an in vitro study. Neurobiol. Aging. 2007;28:524–532. doi: 10.1016/j.neurobiolaging.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Byrne GI, Ouellette SP, Wang Z, Rao JP, Lu L, Beatty WL, Hudson AP. Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect. Immun. 2001;69:5423–5429. doi: 10.1128/IAI.69.9.5423-5429.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 2003;111:1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LA, Kuo CC. Chlamydia pneumoniae pathogenesis. J. Med. Microbiol. 2002;51:623–625. doi: 10.1099/0022-1317-51-8-623. [DOI] [PubMed] [Google Scholar]
- Citron M. Beta-secretase inhibition for the treatment of Alzheimer’s disease – promise and challenge. Trends Pharmacol. 2004;25:92–97. doi: 10.1016/j.tips.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Clementsen P, Permin H, Norn S. Chlamydia pneumoniae infection and its role in asthma and chronic obstructive pulmonary disease. J. Investigat. Allergol. Clin. Immunol. 2002;12:73–79. [PubMed] [Google Scholar]
- Cochrane M, Walker P, Gibbs H, Timms P. Multiple genotypes of Chlamydia pneumoniae identified in human carotid plaque. Microbiology. 2005;151:2285–2290. doi: 10.1099/mic.0.27781-0. [DOI] [PubMed] [Google Scholar]
- Daugaard L, Christiansen G, Birkelund S. Characterization of a hypervariable region in the genome of Chlamydia pneumoniae. FEMS Microbiol. Lett. 2001;203:241–248. doi: 10.1111/j.1574-6968.2001.tb10848.x. [DOI] [PubMed] [Google Scholar]
- de Gannes FM, Leducq N, Diolez P, Belloc F, Merle M, Canioni P, Voisin PJ. Mitochondrial impairment and recovery after heat shock treatment in a human microglial cell line. Neurochem. Int. 2000;36:233–241. doi: 10.1016/s0197-0186(99)00118-7. [DOI] [PubMed] [Google Scholar]
- Dreses-Werringloer U, Padubrin I, Köhler L, Hudson AP. Detection of nucleotide variability in rpoB in both rifampin-sensitive and rifampin-resistant isolates of Chlamydia trachomatis. Antimicrob. Agents Chemother. 2003;47:2316–2318. doi: 10.1128/AAC.47.7.2316-2318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreses-Werringloer U, Gérard HC, Whittum-Hudson JA, Hudson AP. C. pneumoniae infection of human astrocytes and microglial cells in culture displays an active, rather than a persistent, growth phenotype. Am. J. Med. Sci. 2006;332:168–174. doi: 10.1097/00000441-200610000-00003. [DOI] [PubMed] [Google Scholar]
- Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 2002;277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- Gaydos CA, Summersgill JT, Sahney NN, Ramirez JA, Quinn TC. Replication of Chlamydia pneumoniae in vitro in human macrophages, endothelial cells, and aortic artery smooth muscle cells. Infect. Immun. 1996;64:1614–1620. doi: 10.1128/iai.64.5.1614-1620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard HC, Krauße-Opatz B, Wang Z, Rudy D, Rao JP, Zeidler H, Schumacher HR, Whittum-Hudson JA, Köhler L, Hudson AP. Expression of Chlamydia trachomatis genes required for DNA synthesis and cell division in active vs. persistent infection. Mol. Microbiol. 2001;41:731–741. doi: 10.1046/j.1365-2958.2001.02550.x. [DOI] [PubMed] [Google Scholar]
- Gérard HC, Freise J, Wang Z, Roberts G, Rudy D, Krauße-Opatz B, Köhler L, Zeidler H, Schumacher HR, Whittum-Hudson JA, Hudson AP. Chlamydia trachomatis genes whose products are related to energy metabolism are expressed differentially in active vs. persistent infection. Microb. Infect. 2002;4:13–22. doi: 10.1016/s1286-4579(01)01504-0. [DOI] [PubMed] [Google Scholar]
- Gérard HC, Whittum-Hudson JA, Schumacher HR, Hudson AP. Chlamydiae and inflammatory arthritis. In: Columbus F, editor. Focus on Arthritis Research. New York: Nova Science Publishers; 2004. pp. 175–199. [Google Scholar]
- Gérard HC, Wildt KS, Whittum-Hudson JA, Lai Z, Ager J, Hudson AP. The load of Chlamydia pneumoniae in the Alzheimer’s brain varies with APOE genotype. Microb. Pathogen. 2005;39:19–26. doi: 10.1016/j.micpath.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Gérard HC, Dreses-Werringloer U, Wildt KS, Oszust C, Balin BJ, Frey WH, Bordayo EZ, Whittum-Hudson JA, Hudson AP. Chlamydia (Chlamydophila) pneumoniae in the Alzheimer’s brain. FEMS Immunol. Med. Microbiol. 2006;48:355–366. doi: 10.1111/j.1574-695X.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- Gieffers J, Durling L, Ouellette SP, Rupp J, Maass M, Byrne GI, Caldwell HD, Belland RJ. Genotypic differences in the Chlamydia neumoniae tyrP locus related to vascular tropism and pathogenicity. J. Infect. Dis. 2003;188:1085–1093. doi: 10.1086/378692. [DOI] [PubMed] [Google Scholar]
- Grayston JT. Chlamydia pneumoniae, strain TWAR pneumonia. Annu. Rev. Med. 1992;43:317–323. doi: 10.1146/annurev.me.43.020192.001533. [DOI] [PubMed] [Google Scholar]
- Hahn DL. Chlamydia pneumoniae, asthma, and COPD: what is the evidence? Ann. Allergy Asthma Immunol. 1999;83:271–288. doi: 10.1016/S1081-1206(10)62666-X. [DOI] [PubMed] [Google Scholar]
- Hatch TP. Developmental biology. In: Stephens RS, editor. Chlamydia – Intracellular Biology, Pathogenesis, and Immunity. Washington DC: ASM Press; 1999. pp. 29–67. [Google Scholar]
- Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. (USA) 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikejima H, Friedman H, Yamamoto Y. Chlamydia pneumoniae infection of microglial cells in vitro: a model of microbial infection for neurological disease. J. Med. Microbiol. 2006;55:947–952. doi: 10.1099/jmm.0.46348-0. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Alonso AC, El-Akkad E, Gong CX, Haque N, Khatoon S, Pei JJ, Tanimukai H, Tsujio I, Wang JZ, Grundke-Iqbal I. Alzheimer neurofibrillary degeneration: therapeutic targets and high-throughput assays. J. Mol. Neurosci. 2003;20:425–429. doi: 10.1385/jmn:20:3:425. [DOI] [PubMed] [Google Scholar]
- Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens RS. Comparative genomes of Chlamydia pneumoniae and Chlamydia trachomatis. Nat. Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- Keefover RW. The clinical epidemiology of Alzheimer's disease. Neurol. Clin. 1996;14:337–351. doi: 10.1016/s0733-8619(05)70260-7. [DOI] [PubMed] [Google Scholar]
- Leinonen M. Pathogenetic mechanisms and epidemiology of Chlamydia pneumoniae. Eur. Heart J. 1993;14:57–61. [PubMed] [Google Scholar]
- Little CS, Hammond CJ, MacIntyre A, Balin BJ, Appelt DM. Chlamydia pneumoniae induces Alzheimer-like amyloid plaques in brains of BALB/c mice. Neurobiol. Aging. 2004;25:419–429. doi: 10.1016/S0197-4580(03)00127-1. [DOI] [PubMed] [Google Scholar]
- Lytle RA, Jiang Z, Xheng X, Higashikubo R, Rich KM. Retinamide-induced apoptosis in gliobastomas is associated with down-regulation of Bcl-xL and Bcl-2 proteins. Neurooncol. 2005;74:225–232. doi: 10.1007/s11060-005-7305-z. [DOI] [PubMed] [Google Scholar]
- Mäurer AP, Mehlitz A, Mollenhopf HJ, Meyer TF. Gene profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathogens. 2007;3:752–769. doi: 10.1371/journal.ppat.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molestina RE, Dean D, Miller RD, Ramirez JA, Summersgill JT. Characterization of a strain of Chlamydia pneumoniae isolated from a coronary atheroma by analysis of the omp1 gene and biological activity in human endothelial cells. Infect. Immun. 1998;66:1370–1376. doi: 10.1128/iai.66.4.1370-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette SP, Hatch TP, AbdelRahman YR, Rose LA, Belland RJ, Byrne GI. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol. Microbiol. 2006;62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N, Mages J, Dietrich H, Wantia N, Wagner H, Lang R, Miethke T. MyD88-dependent changes in the pulmonary transcriptome after infection with Chlamydia pneumoniae. Physiol. Genomics. 2007;30:134–145. doi: 10.1152/physiolgenomics.00011.2007. [DOI] [PubMed] [Google Scholar]
- Schrader S, Klos A, Hess S, Zeidler H, Kuipers JG, Rihl M. Expression of inflammatory host genes in Chlamydia trachomatis-infected human monocytes. Arthritis Res. Ther. 2007;9:R54. doi: 10.1186/ar2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher HR, Gérard HC, Arayssi TK, Pando JA, Branigan PJ, Saaibi DL, Hudson AP. Lower prevalence of Chlamydia pneumoniae DNA compared with Chlamydia trachomatis DNA in synovial tissue of arthritis patients. Arthritis Rheum. 1999;42:1889–1893. doi: 10.1002/1529-0131(199909)42:9<1889::AID-ANR13>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K, Shiba T, Ishii K, Hattori H, Kuhara S, Nakazawa T. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 2000;28:2311–2314. doi: 10.1093/nar/28.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B. Alzheimer’s disease and the amyloid cascade hypothesis: ten years on. Curr. Opin. Pharmacol. 2002;2:87–92. doi: 10.1016/s1471-4892(01)00126-6. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Viratyosin W, Campbell LA, Kuo C-C, Rockey DD. Intrastrain and interstrain genetic variation within a paralogous gene family in Chlamydia pneumoniae. BMC Microbiol. 2002;2:38–49. doi: 10.1186/1471-2180-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AD, Gérard HC, Fresemann T, Schmidt WA, Gromnica-Ihle E, Hudson AP, Zeidler H. Detection of Chlamydia pneumoniae in giant cell vasculitis and correlation with the topographical arrangement of tissue-infiltrating dendritic cells. Arthritis Rheum. 2000;43:1543–1551. doi: 10.1002/1529-0131(200007)43:7<1543::AID-ANR19>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Watson C, Alp NJ. Role of Chlamydia pneumoniae in atherosclerosis. Clin. Sci. 2008;114:509–531. doi: 10.1042/CS20070298. [DOI] [PubMed] [Google Scholar]
- Whittum-Hudson JA, Schumacher HR, Hudson AP. Chlamydia pneumoniae and inflammatory arthritis. In: Yamamoto Y, Friedman H, Bendinelli M, editors. Chlamydia pneumoniae Infection and Diseases. New York: Kluwer/Academic Press; 2004. pp. 227–238. [Google Scholar]
- Whittum-Hudson JA, Gérard HC, Schumacher HR, Hudson AP. Pathogenesis of Chlamydia-associated arthritis. In: Bavoil P, Wyrick P, editors. Chlamydia – Genomics and Pathogenesis. Malden (MA): Blackwell Publishing Inc.; 2007. pp. 475–504. www.stdgen.lanl.gov. [Google Scholar]