Abstract
Highly-activated transcription is associated with eukaryotic genome instability, resulting in elevated rates of mitotic recombination and mutagenesis. The association between high transcription and genome stability is likely due to a variety of factors including an enhanced accumulation of DNA damage, transcription-associated supercoiling, collision between replication forks and the transcription machinery, and the persistence of RNA-DNA hybrids 1. In the case of transcription-associated mutagenesis (TAM), we previously showed that there is a direct proportionality between the level of transcription and the mutation rate in the yeast Saccharomyces cerevisiae2, and that the molecular nature of mutations is affected by highly-activated transcription 2 3. In the work presented here, we find that the accumulation of apurinic/apyrimidinic (AP) sites is greatly enhanced in highly-transcribed yeast DNA. We further demonstrate that most AP sites in highly-transcribed DNA are derived from the removal of uracil, the presence of which is linked to direct incorporation of dUTP in place of dTTP. These results reveal an unexpected relationship between transcription and the fidelity of DNA synthesis, and raise intriguing cell biological issues with regard to nucleotide pool compartmentalization.
The tetracycline/doxycycline-regulatable TET-lys2ΔA746 frameshift allele detects net +1 mutations within an approximately 150 bp reversion window 4. Although high levels of transcription stimulate a wide variety of mutations in this system 2, only the complex mutations, in which the selected frameshift is accompanied by one or more nearby base substitutions, are relevant to the experiments reported here. Because complex mutations are completely dependent on the activity of the translesion synthesis (TLS) DNA polymerase zeta (Polζ), they likely arise during lesion bypass and provide a unique molecular signature of this TLS polymerase 5,6.
AP sites result either from spontaneous base loss or from the removal of a damaged base by the cognate DNA N-glycosylase, and their creation is the first step in the base excision repair (BER) pathway 7. The BER pathway usually proceeds by AP endonuclease-catalyzed cleavage of the sugar-phosphate backbone at the AP site, although some N-glycosylases have an associated AP lyase activity that also nicks the backbone. The subsequent steps in BER involve removal of blocked ends, gap filling by a DNA polymerase and ligation of the remaining nick. More than 95% of the AP endonuclease activity in S. cerevisiae is due to the Apn1 protein 8. When APN1 was deleted in a strain containing the TET-lys2ΔA746 allele, we found a 3.4-fold increase in the Lys+ rate under high-transcription conditions (WT and apn1; Table 1), but no significant increase under low-transcription conditions (WT+Dox and apn1+Dox; Table 1). Importantly, this increase was accompanied by a 20-fold elevation in the rate of complex mutations, with more than 50% occurring at a single site (Table 1 and Figure 1). This site is named according to the mononucleotide run where the selected frameshift occurs, and hence will be referred to as the 6A hotspot. When REV3, the gene encoding the catalytic subunit of Polζ, was deleted in addition to APN1, the Lys+ rate was reduced 2.2 fold and no complex mutations were observed among 82 revertants sequenced. A similar result was obtained when REV1 was deleted (data not shown).
Table 1.
Reversion of the TET-lys2ΔA746 allele
| Strain genotype | Lys+ rate ×10−8 | Complex mutations at the 6A hotspot | |
|---|---|---|---|
| (95% CI) | Number/total | Rate (×10−8)* | |
| WT | 4.28 (3.35−6.74) | 1/117 | 0.037 |
| WT + Dox | 0.584 (0.438−0.612) | 2/107 | 0.011 |
| apn1 | 14.5 (12.4−18.1) | 33/127 | 3.8 |
| apn1 + Dox | 0.646 (0.397−0.924) | 4/89 | 0.029 |
| apn1 rev3 | 6.54 (5.32−7.16) | 0/82 | <0.080 |
| apn1 ogg1 | 22.7 (17.7−42.8) | 24/93 | 5.9 |
| apn1 mag1 | 20.9 (16.2−25.1) | 31/93 | 7.0 |
| apn1 ung1 | 7.79 (5.69−13.1) | 8/92 | 0.68 |
| ntg1 ntg2 | 5.19 (4.28−7.16) | 2/93 | 0.11 |
| apn1 ntg1 ntg2 | 150 (125−192) | 43/94 | 69 |
| apn1 ntg1 ntg2 + Dox | 1.19 (0.949−1.32) | 4/89 | 0.054 |
| apn1 ntg1 ntg2 rev3 | 5.14 (2.65−10.2) | 0/92 | <0.057 |
| apn1 ntg1 ntg2 rev1 | 4.54 (2.64−6.91) | 1/88 | 0.052 |
| apn1 ntg1 ntg2 ung1 | 8.15 (6.52−14.8) | 4/89 | 0.37 |
Doxycycline (Dox) was added to repress transcription. CI = confidence interval.
Rates were calculated by multiplying the total Lys+ rate by the proportion of complex mutations at the 6A hotspot in the corresponding spectrum. When no such complex mutations were found, the rate was calculated assuming 1 event.
Figure 1.
TET-lys2ΔA746 reversion spectra. Only the first 130 nt of the 150 nt reversion window are shown. The position of the A deletion that characterizes the allele is indicated by a dash within the sequence. Most simple insertions and deletions are indicated below the sequence by +, −2, −5, etc.; complex insertions and deletions (cins and cdel, respectively) are indicated above the sequence. Complex mutations at the 6A hotspot that are associated with a T to G transversion are highlighted in dark gray; insertions of G into a 2G run are highlighted in light gray. N = the number of independent revertants sequenced.
Ntg1 and Ntg2 are functionally-related proteins that possess AP lyase as well as N-glycosylase activity 9, and genetic data indicate that they can substitute for Apn1 in AP site processing 10 11. Under high-transcription conditions, the overall reversion rate and the rate of complex mutations at the 6A hotspot were synergistically elevated in an apn1 ntg1 ntg2 triple mutant (10- and 18- fold, respectively) relative to the apn1 single and ntg1 ntg2 double mutant rates (Table 1 and Figure 1). Of particular significance, 42 of 43 complex mutations at the 6A hotspot in the triple mutant contained, in addition to the selected 6A to 7A frameshift mutation, a T to G transversion at one of the two T residues immediately 3’ of the 6A run (highlighted in dark gray in Figure 1; Table 2). As in the apn1 single mutant, the elevation in total Lys+ rate and the rate of complex mutations at the 6A hotspot in the triple mutant were dependent on highly-elevated transcription and on the presence of Rev3 and Rev1.
Table 2.
Complex mutations at the 6A hotspot (AGCTGAAAAAATTC)
| Sequence | Number of events in mutant background | |
|---|---|---|
| apn1 | apn1 ntg1 ntg2 | |
| AGCTGAAAAAAATgC | 10 | 26 |
| AGCTGAAAAAAAgTC | 4 | 16 |
| AGCTGCAAAAAATTC | 4 | 0 |
| AGCTGACAAAAATTC | 5 | 0 |
| AGCTGAACAAAATTC | 2 | 0 |
| AGCTcAAAAAAATTC | 3 | 0 |
| AGCTaAAAAAAATTC | 2 | 0 |
| AGCaGAAAAAAATTC | 1 | 0 |
| AGCTGAAAAAAAaTC | 1 | 0 |
| AGCTGAAAAgTC | 1 | 0 |
| AGCTGAAAAAAAAAATgC | 0 | 1 |
| Total | 33 | 43 |
Insertions are underlined and base substitutions are in lowercase.
The synergism observed between apn1 and ntg1 ntg2 suggests that a major lesion responsible for TAM in this system is an AP site. In addition to Ntg1 and Ntg2, there are three other N-glycosylases in yeast: Ogg1, Mag1 and Ung1, which primarily remove 8-oxoguanine, methylated purines and uracil from DNA, respectively 7. To determine if any of these glycosylases might be responsible for generating the AP sites that accumulate under high-transcription conditions, each of the corresponding genes was deleted from the apn1 background. The total reversion rate and the rate of complex mutations at the 6A hotspot were reduced only in the apn1 ung1 double mutant. This effect was much more pronounced when UNG1 was deleted in the apn1 ntg1 ntg2 background, with the overall reversion rate decreasing over 10-fold, and the rate of complex mutations at the 6A hotspot decreasing more than 100-fold (Table 1).
The striking Ung1-dependence of TAM in BER-deficient (apn1 and apn1 ntg1 ntg2) strains demonstrates that uracil removal is the major source of AP sites in highly-transcribed DNA. There are two known sources of uracil in DNA: (1) spontaneous deamination of cytosine to uracil and (2) direct incorporation of dUTP during DNA synthesis. The incorporation of dUTP into genomic DNA is normally suppressed by the essential enzyme Dut1, which converts dUTP to dUMP 12. To examine whether incorporation of dUTP is responsible for TAM in the apn1 ntg1 ntg2 mutant, the DUT1-encoded dUTPase was overexpressed by transforming mutant strains with a high-copy plasmid containing galactose-inducible DUT1 13. Transcription-associated and spontaneous mutagenesis were monitored in parallel by measuring TET-lys2ΔA746 reversion and forward mutation at the CAN1 locus, respectively. While Dut1 overexpression had little effect on the CAN1 forward mutation rate, it was associated with a 10-fold reduction in the lys2ΔA746 reversion rate (Supplementary Table 2) as well as a proportional decrease in complex insertions at the 6A hotspot (data not shown). Most of the uracil present in highly-transcribed DNA is thus derived from direct dUTP incorporation rather than from cytosine deamination.
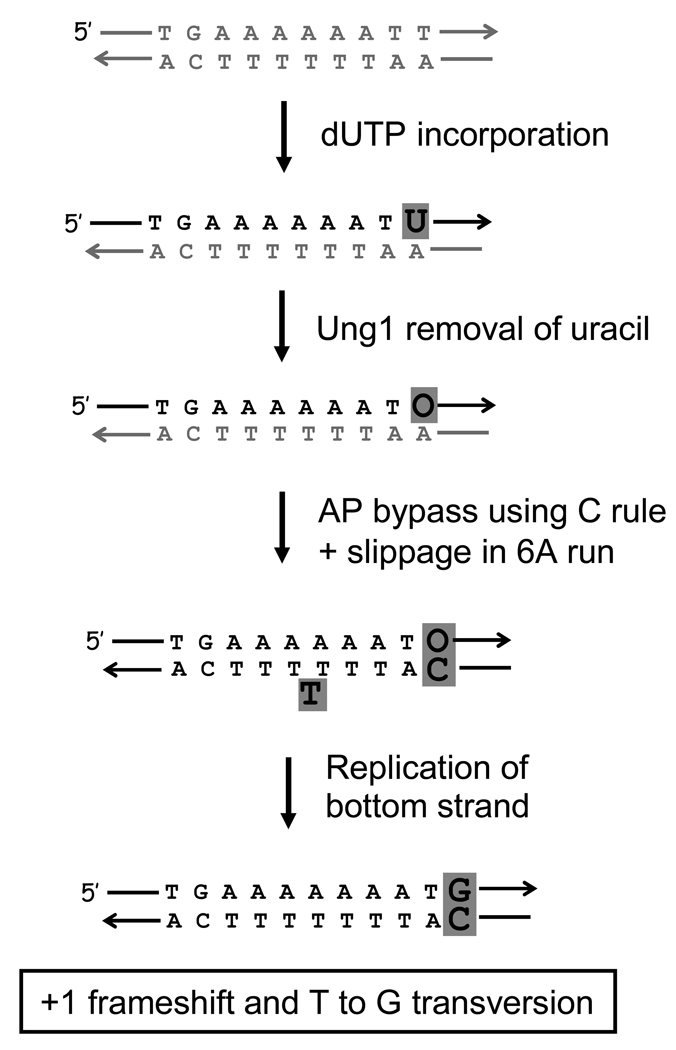
Previous analyses of complex mutation hotspots led us to suggest that these events result from misincorporation opposite a discrete lesion, followed by slippage in a short homopolymer run 5. The most relevant features of the misincorporation-slippage model are that (1) the site of the most frequent hotspot-associated base substitution marks the site of the initiating lesion and (2) the nature of the most frequent base substitution reflects the identity of the lesion. The distinctive base substitution pattern at the 6A hotspot (AAAAAATT to AAAAAAAgT or AAAAAAATg; Table 2) is also more consistent with direct dUTP incorporation than with cytosine deamination, which would specifically result in base substitutions at cytosines. We suggest that dUTP is incorporated into the nontranscribed strand of the mutational target in place of one of the thymines immediately downstream of the 6A run (Figure 2). Following removal of the uracil by Ung1, specific insertion of dCTP during Polζ-assisted bypass of the resulting AP site will generate a T to G transversion. If bypass is followed by slippage in the 6A run that immediately follows, a complex mutation of the type observed will result. In agreement with the specificity of AP site bypass proposed here, it should be noted that a reduction in Dut1 activity has been associated with an Ung1-dependent increase in T to G transversions 13. Although it is formally possible that the complex events at the 6A hotspot reflect a unique structural feature rather a more general, transcription-associated increase in dUTP incorporation, the presence of an additional hotspot in the apn1 single and apn1 ntg1 ntg2 triple mutants (TTTTTGG to TTTTTGGG; highlighted in light gray in Figure 1) makes this unlikely. Of particular significance, the second hotspot has the same genetic requirements as the 6A hotspot (Supplemental Table 3) and similarly can be explained as a complex mutation resulting from cytosine insertion during AP site bypass coupled with slippage in a short homopolymer run (Supplemental Figure 1).
Figure 2.
Model for complex insertions at the 6A hotspot. Black lines and letters correspond to newly-synthesized DNA. O = AP site.
Given the highly conserved nature of DNA structure and basic metabolic processes, it seems likely that TAM will be a general phenomenon affecting mutation accumulation in all organisms. The significance of the results reported here is that they directly link the availability of dUTP for DNA synthesis to the process of transcription. One intriguing possibility is that a locally high concentration of UTP is required to support efficient transcription and that this concomitantly elevates the dUTP:dTTP ratio within the dNTP pool available to the replicative DNA polymerases. Such dUTP incorporation into transcriptionally-active DNA could occur during normal DNA replication and/or during repair synthesis. These observations not only have broad implications for gene-specific mutagenesis and evolution, but also provide indirect evidence for nucleotide pool compartmentalization within the nucleus.
METHODS SUMMARY
The WT strain containing the TET-lys2ΔA746 reporter on Chromosome III near ARS306 was previously described 2. In all strains used here, transcription occurs in the same direction as replication fork movement. A PvuII fragment of p424-GAL1-DUT1 13 was cloned into PvuII-digested pRS426 14 to create p426-GAL1-DUT1. A complete strain list is given in Supplemental Table 1.
For mutation rates and spectra, 1 ml cultures (YEP supplemented with 2% glycerol plus 2% ethanol) with or without added doxycycline hyclate (2 µg/ml; Sigma) were inoculated with ~250,000 cells grown in the same medium. After 3 days, Lys+ revertants were selected on synthetic 2% dextrose plates deficient in lysine. Mutation rates were determined by the method of the median and 95% confidence intervals were calculated as described previously 15 using data from 10 to 24 cultures. Sequencing of a PCR fragment amplified from independent revertants was done by the High Throughput Genomics Unit at the University of Washington (Seattle, WA).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank S. Boiteux for providing the p424-GAL-DUT1 plasmid. We thank members of the lab and Tom Petes for discussions and comments on the manuscript. This work was supported by a grant from the National Institutes of Health (R01 GM038464).
REFERENCES
- 1.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair. 2007;6:1285. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippert MJ, Freedman JA, Barber MA, Jinks-Robertson S. Identification of a distinctive mutation spectrum associated with high levels of transcription in yeast. Mol. Cell. Biol. 2004;24:4801. doi: 10.1128/MCB.24.11.4801-4809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harfe BD, Jinks-Robertson S. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4766. doi: 10.1128/mcb.19.7.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harfe BD, Jinks-Robertson S. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell. 2000;6:1491. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 6.Minesinger BK, Jinks-Robertson S. Roles of RAD6 epistasis group members in spontaneous Polζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2005;169:1939. doi: 10.1534/genetics.104.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Popoff SC, Spira AS, Johnson AW, Demple B. The yeast structural gene (APN1) for the major apurinic endonuclease: homology to E. coli endonucelase IV. Proc. Natl. Acad. Sci. USA. 1985;82:3182. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senturker S, et al. Substrate specificities of ntg1 and ntg2 proteins of Saccharomyces cerevisiaefor oxidized DNA bases are not identical. Nucl. Acids Res. 1998;26:5270. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2929. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You HJ, et al. Saccharomyces cerevisiaeNtg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 12.Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillet M, Van Der Kemp PA, Boiteux S. dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:2056. doi: 10.1093/nar/gkl139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy number vectors. Gene. 1992;119:122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 15.Spell RM, Jinks-Robertson S. In: Genetic Recombination: Reviews and Protocols. Waldman AS, editor. Vol. 262. Totowa, NJ: Humana Press; 2004. p. 3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.