Abstract
Liver regeneration after two-thirds partial hepatectomy (2/3 PH) results in synchronized proliferation of hepatocytes and rapid restoration of liver mass. Understanding the mechanisms that regulate this process has both biological and clinical importance. Using cDNA microarray analysis, we investigated whether gene activation after 2/3 PH is specifically related to liver growth and hepatocyte proliferation. We generated gene expression profiles at 4, 12, 20, and 30 hours after 2/3 PH and compared them with profiles obtained at the same time points after 1/3 PH, a procedure that causes minimal DNA replication. Surprisingly, a significant number of genes whose expression is altered after 2/3 PH are similarly up- or down-regulated after 1/3 PH, particularly at 4 hours. We identified a number of genes and transcription factors that are more highly expressed (“preferential expression”) after 2/3 PH and show that a shift in transcriptional programs in the regenerating liver occurs between 4 and 12 hours after 2/3 PH, a time at which the decision to replicate appears to be made. These results show that the liver responds to PH with massive changes of gene expression, even in the absence of DNA replication. We suggest that the changes in gene expression during the first 4 to 6 hours after 2/3 PH may induce chromatin remodeling and facilitate the binding of new sets of transcription factors required for DNA replication.
Resection of hepatic tissue triggers a proliferative response known as liver regeneration in which quiescent hepatocytes enter the cell cycle and replicate. After 70% partial hepatectomy (2/3 PH) in mice, liver mass is fully restored during the second week after the operation. Restoration of mass is a consequence of a process of compensatory hyperplasia of cells of the liver remnant, as the liver lobes removed at the time of the operation do not re-grow.1,2,3,4 The human liver also has a high regenerative capacity, as shown by its growth in donors of right lobe grafts in living donor transplantation.5,6 Thus, the investigation of the cellular and molecular mechanisms of liver regeneration has biological and clinical implications, and is of fundamental importance.
Liver regeneration after 2/3 PH is a stepwise process that starts with an initiation phase, corresponding to the G0 to G1 transition, which primes hepatocytes to respond to growth signals. Primed hepatocytes enter the cell cycle, undergo one or two rounds of synchronous DNA replication followed by mitosis, and then return to a quiescent state. The regenerative process involves the activity of hundreds of genes and the activation of multiple pathways. However, despite the great progress achieved by the analyses of gene expression patterns in the regenerating liver,7,8,9,10,11 more information is still needed for a full understanding of the molecular mechanisms of liver regeneration.
An important question that has not been explored in detail is the extent to which the widespread changes in gene expression that occur during liver regeneration after 2/3 PH are linked to hepatocyte DNA replication. Designing studies to answer this question is made difficult for various reasons, particularly, the confounding factors created by surgical stress, the problems in choosing adequate controls (whether normal livers or liver of sham-operated mice) to measure relative changes in gene expression, and the variability of the data obtained from different animals.
Our approach to determine whether changes of gene expression in the regenerating liver are directly or indirectly linked to hepatocyte DNA replication has been to compare gene expression after 2/3 PH, the standard surgical procedure that produces robust DNA replication, with gene expression after 1/3 PH, a procedure that causes minimal replication. Previously, we showed that many proto-oncogenes and cytokines that are expressed early after 2/3 PH are also expressed after 1/3 PH, suggesting that some components of the immediate early gene response after 2/3 PH do not appear to be directly linked to the amount of tissue resected, and do not determine the magnitude of DNA replication after PH.12 This conclusion is similar to that presented by Lambotte et al in their studies of gene expression after a “temporary hepatectomy,” who indicated that the extent of DNA replication after PH is not determined at the initiating phase of liver regeneration, but may occur several hours later, possibly at a time when most hepatocytes reach the late G1 stage.13
To determine whether changes in gene expression after 2/3 PH occur even with a minimal replicative response, we have expanded our previous work, and performed a detailed analysis of global patterns of gene expression after 1/3 and 2/3 PH. Using an experimental design in which each mouse had its own normal liver as a control, thus reducing animal to animal variation, we analyzed gene expression profiles at 4, 12, 20 and 30 hours after both 1/3 and 2/3 PH, and identified transcription factors that may regulate genes that are preferentially expressed after 2/3 PH relative to 1/3 PH. We found that there are widespread changes of gene expression after 1/3 PH, a procedure that causes only minimal hepatocyte DNA replication, and that the expression of a large number of genes is similarly up- or down-regulated relative to normal liver after 1/3 or 2/3 PH. However, a group of genes showed a higher expression magnitude, up or down, after 2/3 PH relative to 1/3 PH (preferential expression). These genes contain binding sites for a small number of transcription factors whose profiles change drastically between 4 and 12 hours after 2/3 PH. We suggest that the change in the transcriptional program that occurs during this time may be associated with chromatin remodeling.
Materials and Methods
Surgical Procedures
Ten-week-old, male, wild-type C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) were kept on a 12 hours light/dark cycle with free access to food and water; 1/3 PH and 2/3 PH were performed as described.12 At the time of surgery, the resected left lobes were cut into approximately 5 mm3 cubes and stored in liquid nitrogen until use. Mice were sacrificed 4, 12, 20, or 30 hours later (6 mice/time point/type of surgery), and the regenerating lobes (right and caudate) were harvested as described.12 The experimental design is shown in Figure 1. All animal studies were performed under protocols approved by the Institutional Animal Care and Use Committee at the University of Washington.
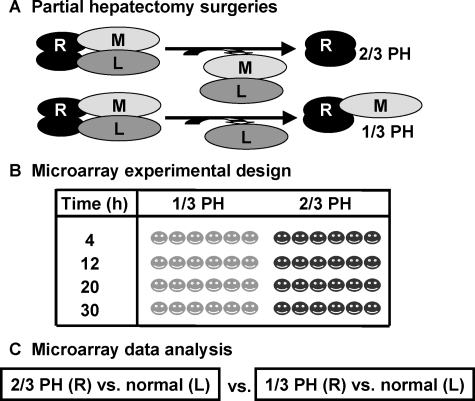
Figure 1.
Experimental design for 1/3 vs. 2/3 PH cDNA microarray analysis. A: Surgeries. One-third PH (1/3 PH) or two thirds PH (2/3/PH) were performed. The medial lobe (M) was not removed in 1/3PH. The left lobe (L) of every animal was snap frozen at the time of surgery. The mice were sacrificed and the right lobe (R) excised at each of the four time points. RNA was prepared from the R and L lobes of every animal. In all cases, the RNA from the L lobe was used as the normalizing RNA for microarray analysis. B: Microarray experimental design. 1/3 and 2/3 PH were performed (six mice at each type of surgery at each time point) and the livers were harvested at 4, 12, 20, and 30 hours. C: Microarray data analysis. We generated two groups of data from the microarray analysis by comparing 1/3 (R) or 2/3 (R) to its own normal (L) for each animal. At each time point 6 sets of data were generated. The two groups of microarray data were further compared using a two-class SAM method to identify preferentially expressed genes after 2/3 PH relative to 1/3 PH.
RNA Preparation
Total RNA was extracted from a matched pair of frozen liver tissues using Trizol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s directions. RNA obtained from the left lobe (L) of each mouse was used as a control for the regenerating right lobe (R). All RNA was quantified, assessed for quality, and amplified with a single round of Eberwine amplification as previously described.14
cDNA Microarrays
cDNA microarrays containing 13,425 mouse cDNAs from the NIA 15K collection (http://lgsun.grc.nia.nih.gov/cDNA/15k.html) were made at the Center for Expression Arrays at the University of Washington (http://www.expression.washington.edu) following established protocols that are described in detail elsewhere.14 We used two arrays for each animal—one array had the Cy3/Cy5 labels associated with L/R probes while the other was a dye reversal (ie, Cy3/Cy5 for R/L). There were two identical gene sets on each slide, which generated 2 × 2 = 4 technical replicates for each gene comparison per mouse. Array data have been deposited to Array Express, (http://www.ebi.ac.uk/microarray-as/ae, accession number E-MTAB-119).
Identification of Differentially Expressed Genes
We analyzed the normalized data using a custom version of TIGR’s MEV software15 that we have modified to connect to the Gene Traffic database. Several types of analyses were performed, as outlined in supplemental Table S1 (available at http://ajp.amjpathol.org): a) selection of genes that are differentially expressed (up or down) after 1/3 or 2/3 PH relative to normal liver, using a one-class significance analysis of microarray (SAM)16 with a false discovery rate of 5% for each type of surgery at each time point; b) selection of genes that are differentially expressed between 1/3 and 2/3 PH at all time points, using a two-class SAM with a false discovery rate of 20% at each time point; c) selection of genes that are differentially expressed after both 1/3 and 2/3 PH in relationship to normal liver in at least one time point; d) classification of genes preferentially expressed after 2/3 PH dependent on the type of surgery, the time after surgery, or dependent on both the type of surgery and the time after PH, using a two-factor analysis of variance using the type of surgery (1/3 or 2/3 PH) as one variable, and time (4, 12, 20, and 30 hours after PH) as the other. For this analysis a P value of <0.01 calculated by permutation testing was used, without further correction for multiple testing errors.
cDNA Data Pre-Processing and Pre-Filtering
After hybridization, washing and scanning, all of the data were transferred to a local GeneTraffic database (Iobion Software, http://www.iobion.com). All data were normalized using global LOWESS normalization.17 All gene annotations were updated in March 2006 using the SOURCE database.18 Out of 13,426 clones on the array, 10,240 have a UniGene cluster ID, and 7464 of these are unique. Duplicate UniGene IDs may not produce identical array data due to a variety of reasons, including the possible presence of alternative spliced versions of the same gene on the array.19 Duplicate UniGene IDs were treated the following way: we calculated the correlation coefficient of the expression values between duplicates using a tool built into the “R” package (http://www.r-project.org/), and treated the duplicates as one gene if the correlation coefficient was above 0.8 by averaging the ratios of those duplicates. For those duplicates having a correlation coefficient less than 0.8, we treated each individual clone as an independent gene associated with its own expression ratio. This approach generates a total of 10,143 clones used in the subsequent statistical analysis. Data from all technical replicates were averaged for subsequent analyses.
Gene Annotation and Identification of Over-Represented Biological Schemes
All of the gene functions are based on gene ontology (GO) terminology20 except for those specifically referenced. To identify biological schemes that are over-represented in each gene list, we used Expression Analysis Systematic Explorer (EASE).21,22 We used the unique UniGene cluster IDs as identifiers. All of the over-represented schemes are determined by an EASE score of less than 0.05.
Pathway Analysis
Pathway analyses were performed with GenMAPP 2 β.23,24 For gene selection, the threshold of ‘fold-change’ was set at 1.5 in creating the following 3 categories: 1) up-regulated (above 1.5), 2) down-regulated (below −1.5); and 3) criteria not met (values between −1.5 and +1.5). This criterion was chosen based on statistical calculations as described.25
Promoter Analysis
The Promoter Analysis and Interaction Network Toolset (PAINT) is a computational tool that can integrate functional genomics data with genomic sequence data to perform transcriptional regulatory network analysis.26 The differentially expressed genes at each time point were analyzed using this program to identify potential transcriptional regulatory elements (TREs) by the presence of transcription factor binding sites. The desired upstream sequences were set to 2000 bp, and the TRANSFAC public match program was applied. The Core similarity threshold was set to 1 and the P value was set to 0.05. The numbers of identified TREs present at each of the four time points were combined and re-scaled, with the total number of differentially expressed genes retrieved from the TRANSFAC database.
Real-Time RT-PCR and Immunohistochemistry
For validation of microarray results, 2 μg of RNA from 1/3 and 2/3 PH samples were reverse transcribed using the Invitrogen Retroscript kit, and real-time RT-PCR performed on the resultant cDNA amplicon using FAM-labeled primers for murine proliferating cell nuclear antigen (Pcna) and cyclin B1 (Ccnb1) (Applied Biosystems, Foster City, CA). Analyses were performed as previously described.12 For immunohistochemistry, the right lobes of each liver after 1/3 or 2/3 PH at 30 hours were fixed in 10% buffered formalin and embedded in paraffin. Specimens were stained with antibodies to PCNA (Transduction Laboratories, BD Biosciences, San Jose, CA) using the ABC method with diaminobenzidine as the chromogen. Hepatocytes with nuclear staining were considered PCNA-positive cells. For each sample, three high-power (×400) fields were counted, and differences between 1/3 and 2/3 PH were calculated using an unpaired t-test with Welch’s correction using GraphPad Prism (GraphPad Software, Inc., San Diego, CA).
Results
Both 1/3 and 2/3 PH Cause Widespread Changes in Gene Expression
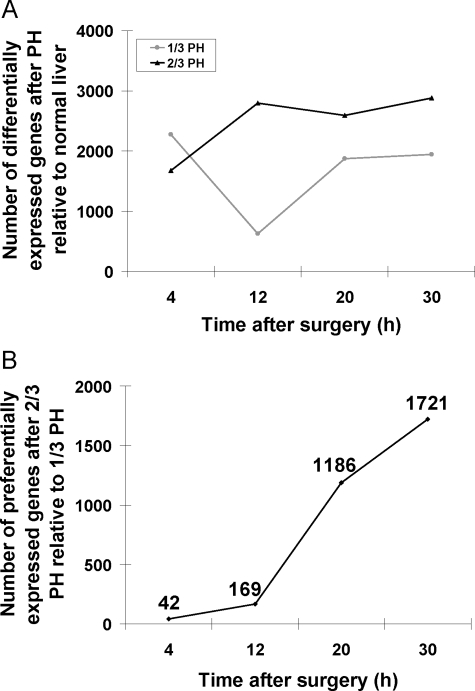
We examined gene expression profiles after 1/3 and 2/3 PH to study the similarities and differences in gene expression patterns after 2/3 PH, which leads to robust DNA replication, and 1/3 PH, in which the replicative response is minimal.12,13,27,28 For each animal, the left lobe resected at the time of the PH served as an internal control for its own right lobe, which was harvested 4 to 30 hours after the surgery (Figure 1, A–C). This design minimizes the variation inherent in comparing pre- and postoperative livers of different animals, and increases the statistical power for detecting differences in gene expression.29 Using a one-class SAM analysis, we first investigated the patterns of gene expression at 4, 12, 20, and 30 hours after 1/3 and 2/3 PH (see supplemental Tables S2 and S3 available at http://ajp.amjpathol.org) to select genes whose expression was either increased or decreased after PH, as compared with the normal liver from the same mouse (left lobe resected at the time of PH). Both 1/3 and 2/3 PH induce a large number of genes, ranging in number from 625 to 2877 at different time points (Figure 2A). Overall, the number of differentially expressed genes, after 2/3 PH relative to normal liver, increases from 1673 at 4 hours to 2877 at 30 hours. After 1/3 PH the largest number of differentially expressed genes relative to normal liver (ie, 2278) is detected at 4 hours. This number decreases to 625 at 12 hours, and then increases to 1945 from 20 to 30 hours. Genes that were differentially regulated after 1/3 or 2/3 PH compared with normal liver (ie, the left lobe from the same mouse) are listed in supplemental Tables S2 and S3 available at http://ajp.amjpathol.org, respectively. In summary, the results show that the expression of a large number of genes is altered, either up- or down-regulated, after 1/3 and 2/3 PH, and that between 4 and 12 hours, the number of differentially expressed genes relative to normal liver increases after 2/3 PH but decreases after 1/3 PH.
Figure 2.
Number of differentially expressed genes after PH relative to normal liver. A: The number of differentially expressed genes after 1/3 (gray line) and 2/3 PH (black line) relative to normal liver of the same animal. B: The number of preferentially expressed genes after 2/3 PH relative to 1/3 PH. RNA was prepared from liver samples taken at 4, 12, 20, and 30 hours post surgeries from both types of surgeries.
Preferential Expression of Genes after 2/3 PH as Compared with 1/3 PH
After compiling a group of genes that are up- or down-regulated after 1/3 or 2/3 PH relative to normal liver shown in Figure 2A, we selected genes that are preferentially expressed after 2/3 PH, as compared with 1/3 PH, using a two-class SAM with a false discovery rate of 20%. At 4 and 12 hours, only a few genes are preferentially expressed after 2/3 PH compared with 1/3 PH but the overall number of genes whose expression is preferentially modified after 2/3 PH increases significantly after 12 hours (Figure 2B). Supplemental Table S4 available at http://ajp.amjpathol.org lists genes that are preferentially regulated after 2/3 PH compared with 1/3 PH at all 4 time points (see supplemental Table S4 available at http://ajp.amjpathol.org). Given the importance of the hepatocyte growth factor/c-met system in liver regeneration,2,30,31,32 we examined the expression of c-met by real-time PCR because this gene was not among the genes in the microarray. There were no differences in the expression of c-met mRNA between 1/3 and 2/3 PH at 4, 12, and 30 hours. Previously we reported that hepatocyte growth factor mRNA levels did not differ between 1/3 and 2/3 PH.12
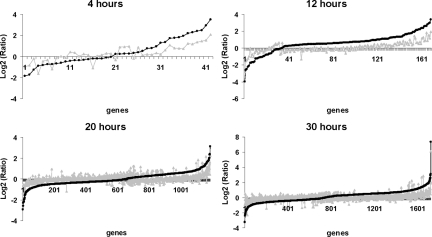
To learn more about gene expression differences between 1/3 and 2/3 PH, we plotted the log2 ratios of preferentially expressed genes in the regenerating liver relative to normal liver tissue at each time point after 1/3 or 2/3 PH (Figure 3). The data show that at each time point examined, the magnitude of expression of most genes is higher, either up or down, after 2/3 PH compared with 1/3 PH. Functional analysis by EASE of genes that are preferentially expressed after 2/3 PH (Table 1 and supplemental Table S5 available at http://ajp.amjpathol.org) shows an enrichment of transcriptional profiles at 12 hours after 2/3 PH of genes associated with “cell adhesion” and “blood vessel development.” At 20 hours the enrichment profiles shift to genes associated with “amine metabolism” and “amino acid metabolism,” and at 30 hours, there is enrichment of genes associated with “cell cycle,” “DNA replication,” and “S-phase of the cell cycle.” Surprisingly, our functional analysis of preferentially regulated genes at 4 hours after 2/3 PH relative to 1/3 PH did not reveal any enriched categories. We conclude that differences in gene expression between 1/3 and 2/3 PH are mostly due to the magnitude of gene expression, that is, changes in expression of individual genes, either up or down, are larger after 2/3 PH compared with 1/3 PH. Furthermore, enrichment for functional categories of expressed genes in 2/3 PH relative to 1/3 PH does not occur until 12 hours after the operation.
Figure 3.
The Log2(Ratio) of preferentially expressed genes after 1/3 (gray line) and 2/3 PH (black line) relative to normal. First, genes that are preferentially expressed after 2/3 PH relative to 1/3 PH were selected by a two-class SAM analysis. The selected genes were sorted by the Log2(Ratio) value (2/3 PH relative to normal). Next the Log2(Ratio) value of the same gene after 1/3 PH was plotted. The data shows that up- or down-regulation of gene expression is of greater magnitude after 2/3 PH.
Table 1.
EASE Analysis of Preferentially Expressed Genes after 2/3 PH Relative to 1/3 PH
| System | Time (h) | Gene category | EASE score |
|---|---|---|---|
| GO_BP | 12 | Cell adhesion | 1.71E-02 |
| GO_BP | 12 | Angiogenesis | 2.48E-02 |
| GO_BP | 12 | Blood vessel development | 3.42E-02 |
| GO_BP | 20 | Amino acid and derivative metabolism | 1.89E-03 |
| GO_BP | 20 | Amino acid catabolism | 7.94E-03 |
| GO_BP | 20 | Amine metabolism | 9.81E-03 |
| GO_BP | 20 | Amine catabolism | 1.91E-02 |
| GO_BP | 20 | Amino acid metabolism | 2.22E-02 |
| GO_BP | 20 | Amino acid derivative metabolism | 4.81E-02 |
| GO_BP | 30 | Cell proliferation | 1.35E-03 |
| GO_BP | 30 | Cell cycle | 1.84E-03 |
| GO_BP | 30 | DNA replication and chromosome cycle | 2.80E-03 |
| GO_BP | 30 | Cell growth and/or maintenance | 9.22E-03 |
| GO_BP | 30 | DNA replication | 3.17E-02 |
| GO_BP | 30 | S phase of mitotic cell cycle | 3.17E-02 |
| GO_BP | 30 | DNA dependent DNA replication | 4.18E-02 |
Preferentially expressed genes after 2/3 PH relative to 1/3 PH at each time point were selected by two-class SAM analysis. Gene categories from the enriched biological schemes (GO_Biological Process; GO_BP) with an EASE score of less than 0.05 are shown. There were no enriched categories at 4 hours.
Non-Preferential Gene Activation after 2/3 Hepatectomy
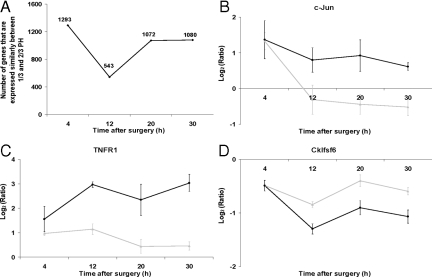
The lack of enriched categories at 4 hours after 2/3 PH relative to 1/3 PH suggested that at least at this time, there may be a large number of genes whose expression is similarly regulated in 2/3 and 1/3 PH. We then compiled a list of genes that showed expression changes of the same magnitude after 1/3 and 2/3 PH relative to their own internal normal liver control (see supplemental Table S6 available at http://ajp.amjpathol.org). At 4 hours, over 80% of the genes whose expression increases or decreases after 2/3 PH also showed changes of similar magnitude after 1/3 PH. The number of nonpreferentially expressed genes decreased at 12 hours, and remained at approximately 40% at 20 and 30 hours (Figure 4A). To explore the functional roles of genes whose expression showed similar alterations after 1/3 and 2/3 PH, we used the EASE program to identify enriched biological categories at each time point (Table 2 and supplemental Table S7 available at http://ajp.amjpathol.org). At 4 hours after either 1/3 or 2/3 PH the enriched categories predominantly included genes associated with lipid metabolism, which were all down-regulated (Table 2). The number of enriched biological categories was higher at 4 and 12 hours after 1/3 or 2/3 PH, and most of the enriched categories were down-regulated at 4 and 12 hours, but up-regulated at 20 and 30 hours. We were surprised at the number of genes that were activated at 4 hours after 1/3 PH in relationship to normal. To examine the expression of some of these genes at later time points after 1/3 and 2/3 PH, we plotted the log2 ratios for tumor necrosis factor receptor 1 (Tnfr1), c-Jun, and chemokine-like factor super family 6 (Cklfsf6) (Figure 4, B–D). Expression levels of c-Jun, Tnfr1, and Cklfsf6 are similar after both 1/3 and 2/3 PH at 4 hours, but the patterns diverge at later time points. For c-Jun there was a marked decrease in expression between 4 hours and 12 hours after 1/3 PH; for TNFR1, the expression increases after 2/3 PH, but remains constant or decreases after 1/3 PH; for Cklfsf6, the expression decreases between 4 and 12 hours, but the decrease is larger after 1/3 PH. In summary, while many genes showed preferential expression after 2/3 PH (preferentially expressed), there are a large number of genes, particularly at 4 hours, that are similarly regulated (nonpreferentially expression) after 1/3 and 2/3 PH. At 4 and 12 hours most changes in nonpreferentially expressed genes consisted in the down-regulation of gene expression, especially of genes associated with lipid metabolism.
Figure 4.
Genes that are nonpreferentially expressed in at least one time point in both 1/3 and 2/3 PH. A: Total number of nonpreferentially expressed genes after PH (black line) at each time point. B–D: Pattern of gene expression for genes that have similar expression at 4 hours after 2/3 or 1/3 PH, black lines and gray lines, respectively. Log2(Ratio) values are shown and plotted as described in Figure 2. TNFR1 (TNF receptor 1), and Cklfsf6 (Chemokine-like factor super family 6).
Table 2.
EASE Analysis of Nonpreferentially Expressed Genes after 1/3 or 2/3 PH at 4 Hours
| System | Time (h) | Direction | Gene category | EASE score | Bonferroni |
|---|---|---|---|---|---|
| GO_BP | 4 | Down | Steroid metabolism | 1.15E-06 | 1.24E-03 |
| GO_BP | 4 | Down | Steroid biosynthesis | 1.59E-05 | 1.72E-02 |
| GO_BP | 4 | Down | Sterol metabolism | 3.43E-05 | 3.70E-02 |
| GO_BP | 4 | Down | Lipid biosynthesis | 4.53E-05 | 4.88E-02 |
| GO_BP | 4 | Down | Cholesterol metabolism | 2.22E-04 | 2.39E-01 |
| GO_BP | 4 | Down | sterol biosynthesis | 2.79E-04 | 3.01E-01 |
| GO_BP | 4 | Down | Lipid metabolism | 3.16E-04 | 3.41E-01 |
| GO_BP | 4 | Down | Alcohol metabolism | 1.47E-03 | |
| GO_BP | 4 | Down | Cholesterol biosynthesis | 2.03E-03 | |
| GO_BP | 4 | Down | Electron transport | 3.64E-02 | |
| GO_BP | 4 | Down | Macromolecule biosynthesis | 4.89E-02 | |
| GO_BP | 4 | Up | Anion transport | 2.79E-02 | |
| GO_BP | 4 | Up | Cell-cell adhesion | 4.37E-02 | |
| KEGG | 4 | Down | Lipid metabolism | 8.43E-03 |
Nonpreferentially expressed genes (similarly regulated genes) after 1/3 or 2/3 PH relative to normal liver were selected by one class SAM analysis. Gene categories from the enriched biological schemes (GO_BP) with an EASE score less than 0.05 are shown. A KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis for lipid metabolism is also shown. A multiplicity correction of Bonferroni method is also shown for some categories, demonstrating the significance of the EASE selected categories. A complete list of EASE analysis with all time-point data is presented in Supplemental Table S7 available at http://ajp.amjpathol.org. GO_BP, GO_Biological_Process.
Patterns of Preferential Gene Activation after 2/3 PH
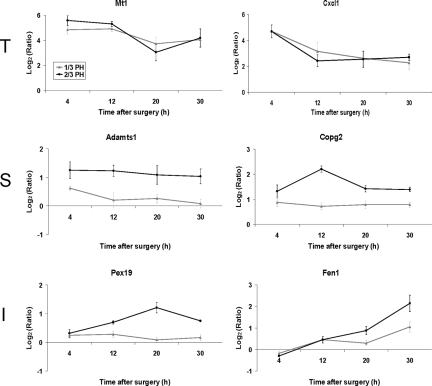
We next performed an independent statistical analysis of our dataset using a two-factor analysis of variance to identify general patterns of gene expression and identify genes whose expression levels are dependent on the time after surgery, type of surgery, or both (referred to as interaction). Examples of genes that represent these different patterns of expression are shown in Figure 5, in which the expression patterns of six genes are shown: 1) metallothioniein 1 (Mt1) and chemokine ligand 1 (Cxcl1), whose expression depend on the time after surgery; 2) A disintegrin-like and metalloprotease with thrombospondin type 1 motif (Adamts 1), and coatomer protein complex, subunit gamma 2 (Copg2), which depends on the type of surgery, and 3) peroxisome biogenesis factor 19 (Pex19) and Flap structure specific endonuclease 1 (Fen1), whose expression depends both on the time after surgery and the type of surgery. For each of these three categories of gene expression we performed a functional analysis using the EASE program (see supplemental Table S8 available at http://ajp.amjpathol.org). The highest enriched categories were: cell proliferation genes for time dependent expression, cell organization and biogenesis genes for surgery dependent expression, and cell growth and maintenance genes for the interaction category.
Figure 5.
Pattern of gene expression for genes selected by two-factor analysis of variance. The expression pattern of two-factor analysis of variance selected genes are shown, with genes that are time dependent (T), surgery dependent (S), or interaction dependent (I). Expression of T genes may vary with time but are similar after 2/3 or 1/3 PH; expression of S genes varies depending on the type of surgery, but relatively little regarding time; expression of I genes depends both on the type of surgery and time after surgery (2/3 PH black line; 1/3 PH gray line). Typical patterns are shown. Abbreviations: Mt1, Metallothionein 1; Cxcl1, Chemokine (C-X-C motif) ligand 1; Adamts1, A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 1; Copg2, Coatomer protein complex, subunit gamma 2; Pex19, Peroxisome biogenesis factor 19; Fen1, Flap structure specific endonuclease 1.
To gain a global view of the patterns of genes preferentially expressed after 2/3 PH compared with 1/3 PH, we performed a hierarchical clustering analysis of genes dependent on both time and surgery type (interaction category). Six gene expression patterns were observed, but approximately 80% of the genes fit into two expression patterns corresponding to patterns 1 and 2 (see supplemental Figure S1 available at http://ajp.amjpathol.org). Essentially, gene expression was not significantly different at 4 or 12 hours, but was different at 20 and 30 hours after 2/3 PH. The expression of the genes that fit patterns 1 and 2 were significantly up- or down-regulated only at 20 and 30 hours after 2/3 PH.
Expression of Genes Associated with Proliferative Pathways
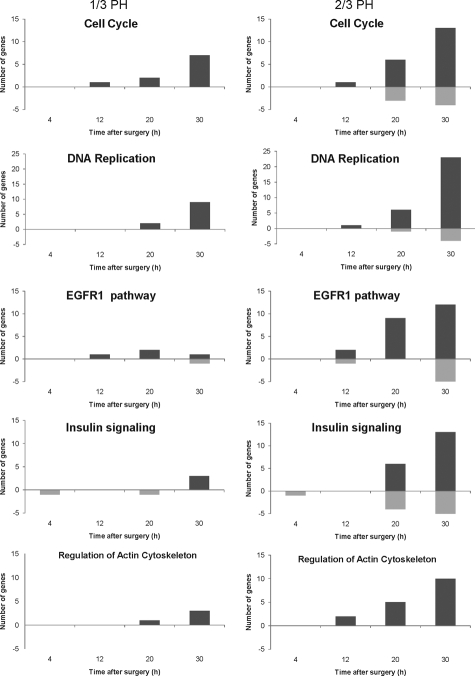
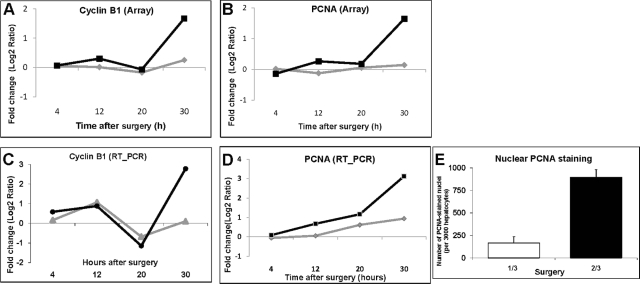
We used the GenMAPP program to compare the number of genes involved in the cell cycle, DNA replication, Epidermal growth receptor 1 and insulin signaling, and the regulation of actin cytoskeleton after 1/3 and 2/3 PH (Figure 6). The number of genes activated in each of these pathways increases significantly from 20 to 30 hours after 2/3 PH. In contrast, there are only minimal changes in the number of genes activated in each of the pathways after 1/3 PH. A list of the genes for each category and surgery type can be found in supplemental Table S9 (available at http://ajp.amjpathol.org). The main findings are summarized below. Among cell cycle and DNA replication genes, there were increases in cyclin A and the exonuclease Xrn1 at 12 hours after 2/3 PH, while cyclins A, B1 (Figure 7, A and C), and E increased at 30 hours after 2/3 PH, but showed no change or smaller increases after 1/3 PH . At 20 and 30 hours after 2/3 PH, the expression of many genes involved in DNA replication increased in agreement with the data of Otu et al.9 These genes included minichromosome maintenance complex components 3 and 5 (Mcm3, Mcm5), Orc2, Cdc6, Cdc2, Mad2, Cdc25a, and Cdt1 (chromatic licensing and DNA replication factor aka Ris2). In contrast, expression of these genes after 1/3 PH was either not changed or was increased at a lesser magnitude than after 2/3 PH. The microarray analysis showed expression of PCNA after 2/3 PH but not 1/3 PH (however PCNA was detectable by real-time PCR and immunohistochemistry (Figure 7, B, D and E). DNA dependent ligase 1 (Lig1) and flap structure specific endonuclease 1 (Fen1) encode proteins that form complexes with PCNA. Lig1 mRNA was not detectable after 1/3 PH and Fen1 was detected at 30 hours, but not at 20 hours after PH.
Figure 6.
Preferentially expressed genes in multiple signaling pathways after 1/3 and 2/3 PH. The preferentially expressed genes after 1/3 (gray boxes) or 2/3 PH (black boxes) relative to normal liver at each time point were imported into GenMAPP with UniGene cluster IDs as the identifier. The number of genes on Gen MAPP, either up- or down-regulated in 1/3 and 2/3 PH relative to normal were counted and plotted as a function of time.
Figure 7.
Comparison of PCNA and Cyclin B1 expression by microarray analysis and real-time PCR. (A) and (C) show expression profiles for Cyclin B1 and PCNA from microarray analysis, respectively; (B) and (D), profiles from real-time PCR, where 2/3 PH is represented by black lines, and 1/3 PH represented by gray lines. E: Immunohistochemistry for nuclear PCNA protein at 30 hours after surgeries as total number of PCA staining nuclei s/3000 hepatocytes. PCNA; proliferating cell nuclear antigen.
Among the genes included in the Epidermal growth factor receptor 1 and insulin signaling categories, increases in expression occurred only after 2/3 PH, with the exception of Git1 and Egr1, which were highly expressed after 1/3PH. In contrast, genes in the category of regulation of actin cytoskeleton increased after both 1/3 and 2/3 PH (Figure 6), although the changes after 1/3PH were generally of lower magnitude.
The Differentially Expressed Genes after 2/3 PH Are Regulated by a Small Group of Transcription Factors
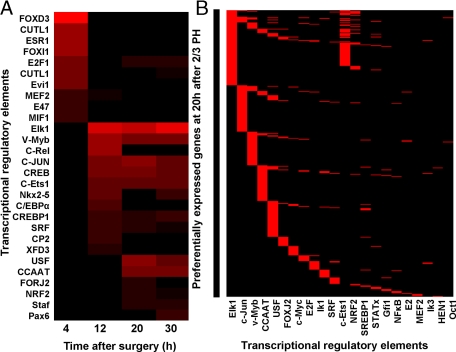
To identify transcriptional regulators that might function as inducers of genes that are differentially regulated after 2/3 PH (Figure 2B), we imported the gene list at each time point into the Promoter Analysis and Interaction Network Toolset (PAINT) program to retrieve TREs. To reveal the correlations between identified transcription factors, we normalized the number of identified TREs to the total number of genes retrieved at each time point, and calculated the percentage of genes with each specific TREs on their 5′ end sequences (Figure 8A). The differentially regulated genes at 4 hours have some unique transcription factor binding sites compared with the rest of the time points. At this time, FoxD3 binding sites are present in 25% of retrieved genes. A striking transition occurs at 12 hours, as the TREs detected at 4 hours were not detected at later time points. The differentially expressed genes at 12, 20 and 30 hours share various transcription factor binding sites including those for Elk-1, Myb, c-jun, CREB, and ETS-1 (see supplemental Table S10 available at http://ajp.amjpathol.org).
Figure 8.
Transcriptional regulation of preferentially expressed genes after 2/3 PH. The preferentially expressed genes after 2/3 PH relative to 1/3 PH (supplemental Table S4 available at http://ajp.amjpathol.org) were subjected to Promoter Analysis and Interaction Network Toolset analysis. A: Pattern of identified Transcription Regulatory Elements (TREs) at 4, 12, 20, and 30 hours. The number of TRE at each time point was normalized by the total number of preferentially expressed genes retrieved from the TRANSFAC database. A higher percentage of genes containing the specific TRE is represented by red color. The two CUTL1 represent TRE ids of CDP CR1 and CDP CR3 in TRANSFAC database respectively, which encode the same gene of Cut-like 1 (CUTL1). B: TREs associated with genes preferentially expressed after 2/3 PH relative to 1/3 PH at 20 hours after 2/3 PH. The gene symbols associated with specific TREs on their 5′ end are shown to the left of the panel and appears as a solid line.
In Figure 8B, we show the predicted TREs for genes that are preferentially up-regulated at 20 hours after 2/3 PH relative to 1/3 PH. We found that more than 80% of the genes preferentially up-regulated at 20 hours after 2/3 PH have at least one binding site for one or more of the 5 transcription factors: c-Jun, CEBP-β, Myb, Ets1, and Elk1. Of these, c-jun and CEBP β box have been studied in mouse liver, and been proposed to play critical roles in liver regeneration.33,34 The pattern of identified TREs for c-Jun in Figure 8A is consistent with the array expression data (Figure 4B) showing that c-Jun was differentially expressed from 12 to 30 hours after 2/3 PH relative to 1/3 PH, but the expression after 1/3 or 2/3 PH was not significantly different at 4 hours. The data in Figure 8B also show that there are genes that have more than one transcription factor binding site, indicating that redundant regulatory mechanisms exist at the transcription factor level.
Discussion
To determine whether changes of gene expression in the regenerating liver are directly or indirectly linked to hepatocyte DNA replication, we compared the patterns of gene expression after 2/3 PH that produces robust DNA replication, with those after 1/3 PH, a procedure that causes minimal replication. Previously, we showed that many of the proto-oncogenes and cytokines that are expressed after 2/3 PH are also expressed after 1/3 PH, indicating that these genes are activated regardless of the biological outcome of the operation. In the present work, we greatly expanded the original studies by doing an extensive analysis of the global patterns of gene expression after 1/3 and 2/3 PH. To obtain reliable results, we extracted RNA from the right lobe after either 2/3 or 1/3 PH, and compared the right lobe gene expression patterns with those obtained from the left lobe removed from the same animal at the time of the operation. Thus, for each mouse, its own normal hepatic tissue served as the control sample. Moreover, we used 12 mice per time point (six for each PH procedure) to permit a meaningful statistical analysis of the data. We have not compared RNA expression patterns of liver resected at the time of PH with un-operated livers. We assumed that whatever differences may exist in the expression patterns between normal liver resected at the time of the operation and from un-operated mice reflect the operative procedures including anesthesia, manipulation of the liver, bleeding, etc. For these reasons, we feel that resected normal tissues are a better baseline to evaluate changes in gene expression in our experiments.
The main findings of this study can be summarized as follows: 1) despite the lack of significant cell replication, 1/3 PH causes widespread changes in gene expression; 2) comparison of gene expression between 1/3 and 2/3 PH showed two patterns; a) genes whose expression increased or decreased after both 1/3 and 2/3 PH, but the magnitude of the changes were higher after 2/3 PH (referred to as preferentially expressed genes); and b) genes whose expression also changed after both operations, but and the magnitude of the changes was similar in 1/3 and 2/3 PH (referred to as nonpreferentially expressed genes); 3) the main differences in gene expression between 2/3 and 1/3 PH reflected changes (increases or decreases) in the magnitude of gene expression (preferential expression) rather than the activation of new sets of genes after 2/3 PH; 4) at 4 hours there is a large overlap between genes expressed in 1/3 and 2/3 PH, but the overlap decreases after 12 hours, and the preferentially expressed genes increases; 5) genes involved in the assembly and activation of DNA pre-replicative complexes and for cyclin A, E and B1 are highly expressed after 2/3 PH, but are not changed or show small changes in magnitude after 1/3 PH; 6) more than 80% of the genes preferentially expressed after 2/3 PH relative to 1/3 PH at 12 hours or later have at least one binding site for six transcription factors: Elk-1, c-Jun, CCAAT CEBP-β, Myb, Ets-1, and USF (upstream stimulatory factor, a member of the basic helix-loop-helix leucine zipper family). It is surprising that we did not find an increase in cyclin D1 mRNA as Mullany et al reported that this cyclin has proliferative and transcriptional effects during liver regeneration.35
Our initial hypothesis was that gene expression changes after 1/3 PH would be relatively small and very different from those occurring after 2/3 PH. We found instead that the changes in gene expression after 1/3 PH relative to control liver of the same animal were very large, and that at 4 hours there was little preferential gene expression in 2/3 PH compared with 1/3 PH. More than 1000 genes were similarly expressed after 2/3 and 1/3 PH. The most salient changes among these genes were the marked down-regulation of genes associated with lipid biosynthesis and metabolism, and the up-regulation of genes associated with cell–cell adhesion. The decrease in the expression of genes related to lipid metabolism after 2/3 PH has been demonstrated by White et al,7 Our results indicate that a similar decrease occurs after 1/3 PH. Thus, in the first 4 hours after PH similar changes in gene expression occur after 1/3 and 2/3 PH, indicating that the liver initially responds to a partial hepatectomy with massive changes in gene expression, even if the operation does not result in DNA replication. This conclusion does not imply that changes in gene expression that occur in both 1/3 and 2/3 PH are not important for liver regeneration, but it does suggest that the great majority of changes in gene expression that occur during the first four hours after 2/3 PH may be a “wake up” call for quiescent hepatocytes, while the decision about a replicative response is done at later times. Specificity toward liver growth and DNA replication seems to occur at about 12 hours or later after 2/3 PH, and is associated with a change in the transcription program.
DNA replication starts at origins marked by the origin recognition complex, which associates with Cdc6 and Cdt1 (Ris2). Mcm proteins are recruited to these sites forming pre-replication complexes.36 Most of the genes involved in the initiation and elongation of DNA replication are highly expressed at 30 hours after 2/3 PH, but have lower expression or are not detected after 1/3 PH. It is interesting to note that Chk1, a check point kinase, that inactivates Cdc25 and stops cell cycle progression after DNA damage is also increased after 2/3 PH. Changes in the expression of this gene, as well as the down-regulation of Mcm10 may prevent unscheduled DNA replication after 2/3 PH.37 In addition, PCNA expression and its association with Fen-1 and Lig1 might be a key regulatory step for controlling DNA replication during liver regeneration.
The examination of transcription factors associated with genes that are preferentially expressed after 2/3 PH, showed that between 4 and 12 hours after 2/3 PH there is a major change in the liver transcriptional program. Although, as discussed above, in the first 4 hours after 2/3 PH the pattern of gene expression does not appear to be linked to DNA replication, specificity toward replication occurs 12 hours or later after PH. At 4 hours the TREs for the transcription factors encoded by FOXD3, FOXI1 (FhK10), CDPCR1(CCAAT displacement protein/cut homeobox, CUTL1), ER, and E2F-1 were highly represented. The TRE binding patterns of preferentially expressed genes after 2/3 PH shifts at 12 hours with activation of genes that have TREs for c-jun, CCAAT CEBP-β box, Myb, Ets-1, Elk1, and USF. Expression of these factors during liver regeneration have also been reported by Juskeviciute and co-workers.38 At 12 hours there is a preferential expression of genes associated with cell adhesion and angiogenesis, and at later times, preferentially expressed genes are mostly associated with amino acid metabolism and protein synthesis at 20 hours, and with cell growth and DNA replication at 30 hours. We do not know the mechanisms that regulate the transition between the nonspecific priming period of the first 4 hours after 2/3 PH and the preferential expression of genes associated with growth and replication that starts at 12 hours, but our previous data indicated that Heparin binding epidermal growth factor (HB-EGF) expression plays a crucial role in this transition.12
Several reports in the literature are relevant to the present studies. Lambotte et al showed that in rats, the changes in the expression of some individual genes occurring shortly after PH are not related to the amount of the functional mass of the liver, and that the extent of the proliferative response is controlled at later times corresponding to mid to late G1.13 These and other data,39 as well as our own results suggest that the mechanisms of control of gene expression at the priming phase may differ from those at the cell cycle progression stage. We hypothesize that the activation of forkhead genes Foxd3 and Foxl1 in the first 4 hours after PH may remodel chromatin,40 as forkhead proteins have a DNA binding domain that is quite similar to the winged-helix structures of histones H1 and H5.41 The early remodeling of chromatin may create new binding sites, which, at 12 hours and later times after 2/3 PH, are occupied by transcription factors associated with cell growth and DNA replication. In interpreting our results, it should be taken into account that the gene expression experiments were performed with whole liver RNA, precluding an analysis of the localization of genes and transcription factors among liver cells.
In summary, our results suggest that the large gene response in the first four hours after 2/3 PH does not predict whether DNA replication is going to occur. However, the early response primes or “prepares” the cell for the later changes, and appears to open new sites in the genome that, starting at 12 hours, are occupied by transcription factors required for DNA replication. The mechanisms that account for the shifts in transcription factor binding during liver regeneration remain to be established.
Supplementary Material
Acknowledgments
We thank Dr. Alison Crowe and Dr. Shelley Barton for helpful discussions, Cynthia Yost for qPCR analysis, and Stephanie Brown and Greg Lawrence for assistance in preparing the manuscript.
Footnotes
Address reprint requests to Nelson Fausto, Department of Pathology, University of Washington, Box 357740, Seattle, WA 98195. nfausto@u.washington.edu.
Supported by NIH grants, CA-23226 and CA-74131 (N.F.), CA-127228 (J.S.C.), NIAID 5P01AI052106 and NCRR 1S10RR01942 (R.E.B.), and the EPT Training Program (NIEHS 2 T32 ES007032 to R.S.M.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Current address of C.M.: Institut Cochin, Université Paris Descartes, Inserm (U567), 75014 Paris, France.
References
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- Haga J, Shimazu M, Wakabayashi G, Tanabe M, Kawachi S, Fuchimoto Y, Hoshino K, Morikawa Y, Kitajima M, Kitagawa Y. Liver regeneration in donors and adult recipients after living donor liver transplantation. Liver Transpl. 2008;14:1718–1724. doi: 10.1002/lt.21622. [DOI] [PubMed] [Google Scholar]
- Pomfret EA, Pomposelli JJ, Gordon FD, Erbay N, Lyn Price L, Lewis WD, Jenkins RL. Liver regeneration and surgical outcome in donors of right-lobe liver grafts. Transplantation. 2003;76:5–10. doi: 10.1097/01.TP.0000079064.08263.8E. [DOI] [PubMed] [Google Scholar]
- White P, Brestelli JE, Kaestner KH, Greenbaum LE. Identification of transcriptional networks during liver regeneration. J Biol Chem. 2004 doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci USA. 2002;99:11181–11186. doi: 10.1073/pnas.122359899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, Libermann TA, Karp SJ. Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem. 2007;282:11197–11204. doi: 10.1074/jbc.M608441200. [DOI] [PubMed] [Google Scholar]
- Fukuhara Y, Hirasawa A, Li XK, Kawasaki M, Fujino M, Funeshima N, Katsuma S, Shiojima S, Yamada M, Okuyama T, Suzuki S, Tsujimoto G. Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol. 2003;38:784–792. doi: 10.1016/s0168-8278(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Kelley-Loughnane N, Sabla GE, Ley-Ebert C, Aronow BJ, Bezerra JA. Independent and overlapping transcriptional activation during liver development and regeneration in mice. Hepatology. 2002;35:525–534. doi: 10.1053/jhep.2002.31351. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Nivison M, Jackson LF, Fox R, Lee DC, Campbell JS, Fausto N. Heparin-binding epidermal growth factor-like growth factor links hepatocyte priming with cell cycle progression during liver regeneration. J Biol Chem. 2005;280:2562–2568. doi: 10.1074/jbc.M412372200. [DOI] [PubMed] [Google Scholar]
- Lambotte L, Saliez A, Triest S, Tagliaferri EM, Barker AP, Baranski AG. Control of rate and extent of the proliferative response after partial hepatectomy. Am J Physiol. 1997;273:G905–G912. doi: 10.1152/ajpgi.1997.273.4.G905. [DOI] [PubMed] [Google Scholar]
- Li J, Adams L, Schwartz SM, Bumgamer RE. RNA amplification, fidelity and reproducibility of expression profiling. C R Biol. 2003;326:1021–1030. doi: 10.1016/j.crvi.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland W. Robust locally weighted regression and smoothing scatter plots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Wei C, Li J, Bumgarner RE. Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics. 2004;5:87. doi: 10.1186/1471-2164-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- Bucher NL, Swaffield MN. The rate of incorporation of labeled thymidine into the deoxyribonucleic acid of regenerating rat liver in relation to the amount of liver excised. Cancer Res. 1964;24:1611–1625. [PubMed] [Google Scholar]
- Inderbitzin D, Studer P, Sidler D, Beldi G, Djonov V, Keogh A, Candinas D. Regenerative capacity of individual liver lobes in the microsurgical mouse model. Microsurgery. 2006;26:465–469. doi: 10.1002/micr.20271. [DOI] [PubMed] [Google Scholar]
- Li J, Pritchard DK, Wang X, Park DR, Bumgarner RE, Schwartz SM, Liles WC. cDNA microarray analysis reveals fundamental differences in the expression profiles of primary human monocytes, monocyte-derived macrophages, and alveolar macrophages. J Leukoc Biol. 2007;81:328–335. doi: 10.1189/jlb.0206124. [DOI] [PubMed] [Google Scholar]
- Borowiak M, Garratt AN, Wustefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci USA. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh CG, Factor VM, Sanchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci USA. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe S, Bowen WC, Bell AW, Nejak-Bowen K, Luo JH, Michalopoulos GK. Cell cycle effects resulting from inhibition of hepatocyte growth factor and its receptor c-Met in regenerating rat livers by RNA interference. Hepatology. 2007;45:1471–1477. doi: 10.1002/hep.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepniak E, Ricci R, Eferl R, Sumara G, Sumara I, Rath M, Hui L, Wagner EF. C-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20:2306–2314. doi: 10.1101/gad.390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum LE, Li W, Cressman DE, Peng Y, Ciliberto G, Poli V, Taub R. CCAAT enhancer-binding protein beta is required for normal hepatocyte proliferation in mice after partial hepatectomy. J Clin Invest. 1998;102:996–1007. doi: 10.1172/JCI3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullany LK, White P, Hanse EA, Nelsen CJ, Goggin MM, Mullany JE, Anttila CK, Greenbaum LE, Kaestner KH, Albrecht JH. Distinct proliferative and transcriptional effects of the D-type cyclins in vivo. Cell Cycle. 2008;7:2215–2224. doi: 10.4161/cc.7.14.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwani W, Alexandrow M, Chellappan S. Prohibitin physically interacts with MCM proteins and inhibits mammalian DNA replication. Cell Cycle. 2009;8:1621–1629. doi: 10.4161/cc.8.10.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell. 2007;18:4085–4095. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juskeviciute E, Vadigepalli R, Hoek JB. Temporal and functional profile of the transcriptional regulatory network in the early regenerative response to partial hepatectomy in the rat. BMC Genomics. 2008;9:527. doi: 10.1186/1471-2164-9-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JL, Boukaba A, Sandoval J, Georgieva EI, Latasa MU, Garcia-Trevijano ER, Serviddio G, Nakamura T, Avila MA, Sastre J, Torres L, Mato JM, Lopez-Rodas G. Transcription of the MAT2A gene, coding for methionine adenosyltransferase, is up-regulated by E2F and Sp1 at a chromatin level during proliferation of liver cells. Int J Biochem Cell Biol. 2007;39:842–850. doi: 10.1016/j.biocel.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci USA. 2002;99:3663–3667. doi: 10.1073/pnas.062041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26:155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.