Abstract
We conducted a comprehensive metabolic phenotyping of primary metabolism of photosynthetic tissue of Arabidopsis thaliana following spray treatment with a number of commercially used herbicides using a well established gas-chromatography mass-spectrometry profiling method. Applying this technique we were able to identify and quantify in excess of 80 polar metabolites and based on a combination of co-elution with standards and prediction from the mass spectra a similar number of lipophillic components within two chromatographic runs. The herbicides selected were glufosinate, sulcotrione, AE944 [N2-(1-ethyl-3-phenylpropyl)-6-(1-fluoro-1-methylethyl)-1,3,5-triazine-2,4-diamine], foramsulfuron, benfuresate and glyphosate. We determined causal changes in the metabolite profiles by following their time-dependent changes using a serial sampling strategy. The resultant profiles were compared both by looking at the largest changes in a metabolite by metabolite manner and by performance of statistical analyses. These data revealed that analysis of the polar metabolites allows clear separation of the compounds under test. This finding is discussed in the context of current strategies for agrochemical discovery.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-008-0149-8) contains supplementary material, which is available to authorized users.
Keywords: Herbicide, Mode of action, GC-MS, Metabolic profiling
Introduction
Agrochemicals are compounds that specifically control or arrest growth of weeds and pests. They have played an important role in the large increase of crop productivity achieved over the last 50 years (Lein et al. 2004). Until recently the first step in agrochemical discovery was to take a collection of chemicals, apply each to a small population of representative pests, and assess their efficacy by visual inspection. Testing of chemicals for efficacy on whole plants is direct and integrates several important attributes of a crop protectant including its uptake, transport, metabolism and ability to inhibit an important target protein (Lein et al. 2004). Commercially used herbicides act on a total of about 20 biochemically distinct and commercially relevant modes of action (Menne and Köcher 2007). Following the initial identification of lead chemicals, exploration and optimization of their structure is accompanied by research into their mode of action (Ott et al. 2003; Lein et al. 2004). In the search for more efficacious herbicides, knowledge concerning the biochemical mode of action is desirable for a number of reasons. Firstly, it facilitates access to advanced tools like such as ultra-high-throughput screening. Secondly, it can allow the determination of whether a novel analog shares the mode of action of its parent molecule. Thirdly, it is important in the classification and prioritization of the modes of action of novel leads found by biological screening. The commercial value of such findings is obviously far greater if the researcher avoids involving well-exploited targets for which novel compounds are not needed or targets historically associated with undesired side-effects (Petroff 1988; Aranibar et al. 2001; Fiehn et al. 2000; Sauter et al. 1991).
For these purposes investigations at the level of the metabolite have been demonstrated to represent effective diagnostic approaches. Indeed, one of the earliest applications of metabolic profiling in plants was the observation by Sauter and co-workers (Sauter et al. 1991), that barley plants treated with sub-lethal doses of various herbicides displayed different GC-MS metabolite profiles depending on the mode of action of the chemicals applied. In this study they compared the profiles of acetyl CoA carboxylase (ACC) and acetolactate synthase (ALS) inhibitors on the basis of visual comparison of metabolite profiles of plants treated with these herbicides. This study thus indicated the possibility that the mode of action of herbicides might be diagnosed by the analysis of reflective patterns of metabolite composition. Targetted GC-MS profiling of very long chain fatty acids in yeast expressing very long chain fatty acid elongases in the presence of, amongst others, the herbicides oxyacetamide and chloroacetanilide has also been used as a method of narrowing down the precise herbicidal mode of action (Trenkamp et al. 2004). Recently, a sophisticated broad range analysis of the mode of action has been carried out by Ott and colleagues, who used a combination of NMR-based metabolic profiling and bioinformatics to classify herbicides with unknown modes of action (Ott et al. 2003). This study illustrated a similar diagnostic power following the statistical analysis of proton NMR (1H-NMR) spectra to that which has been effectively used for non-invasive diagnosis of coronary disease (Brindle et al. 2002).
Another technology that has been successfully applied in the study of herbicide action is direct-infusion Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS), this technique was used by Oikawa et al. (2006) to profile metabolic changes following the growth of Arabidopsis plants for 1 week on agar containing sub-lethal levels of herbicides of varying mode of action. This study revealed that the herbicide treated samples were clearly distinguishable from non-treated samples and showed that in some instances the target site of inhibition could be retroactively located, but perhaps more importantly established an impressive bioinformatics resource to facilitate the use of FT-ICR-MS, which is by nature difficult to use (Iijima et al. 2008; Fernie et al. 2004; Aharoni et al. 2002), as a metabolomics tool. Here, we describe a similar set of experiments although our strategy differs somewhat in approach in two important ways. First, we utilized a well established GC-MS based metabolic profiling approach and secondly we applied herbicide by spraying and harvested plants at various time points post-treatment in order to evaluate the time-dependency of the metabolic response to inhibition of the herbicide targets. For this purpose we took a total of six partly commercial herbicides of known mode of action and compared the metabolic response of Arabidopsis to each of the chemicals. The relative efficacy of this method is discussed as is the possibility to adapt it into a high-throughput screening technology.
Methods
Chemicals
Herbicides were used from commercial sources (Riedel de Haen) or supplied by Bayer CropScience AG, Frankfurt with a minimum purity of 95%. The herbicides were dissolved in Dimethylsulfoxide (Sigma Aldrich) to 100 μg/μl to provide stock solutions. To provide spray solutions herbicide stock solutions were diluted in 800 μl 0.2% Agrotin supplemented with 16 μl EC-premixture/Diacetonalcohol 1:6 to the predefined desired concentrations (Eckes and Busch 2007; EC-premixture comprises 5% phenylsulfonate CA70, 7% Emulsogen EL360, 40% Isophorone and 48% methyloleate).
Experimental design
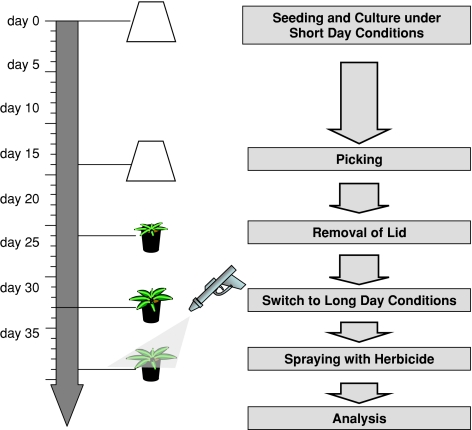
Fifteen Arabidopsis thaliana seeds (ecotype Col0 obtained from the Nottingham Arabidopsis Stock Center), were sown on soil using 11 cm plastic pots and grown under short day conditions (9 h 75 μE/m2s and 15 h darkness at 21°C) for 28 days. At day 14, plants were picked to result in ten evenly distributed plants. On day 28, plants were transferred to long-day conditions (16 h 75 μE/m2s at 21°C, 8 h darkness at 19°C). Following an adaptation period of 6 days, plants were evenly sprayed at a defined timepoint on day 34 with 800 μl of spray solutions containing herbicides as described above. Controls were treated similarly with spray solution devoid of herbicide. At timepoints 1, 3, 6, 12 and 24 h following spray applications rosettes of two pots each were harvested and pooled independently and immediately frozen in liquid nitrogen prior to storage at −70°C until further processing.
Metabolic profiling
Metabolite extraction was carried out on the exactly as described previously (Roessner et al. 2001; Schauer et al. 2006). 100 mg of Arabidopsis were homogenized by grinding in liquid nitrogen. Derivatization and GC-MS analysis were carried out as described previously (Lisec et al. 2006). The GC-MS system was comprised of a CTC CombiPAL autosampler, an Agilent 6890N gas chromatograph and a LECO Pegasus III TOF-MS running in EI+ mode. Metabolites were identified in comparison to database entries of authentic standards (Kopka et al. 2005; Schauer et al. 2005). Evaluation of non-polar metabolites was carried out following the method of Fiehn et al. (2000), since authentic chemical standards were not run for every metabolite the metabolite identity should be regarded as putative rather than exact for these metabolites.
Statistical analysis
The term significant is used in the text only when the change in question has been confirmed to be significant (P < 0.05) with the t-test using the algorithm embedded in Microsoft Excel. Principal component analysis (PCA) was performed on the data sets obtained from metabolite profiling with the software package TMEV (Saeed et al. 2003) using the default weighted covariance-estimation function. Further ICA analysis was performed using MetaGeneAlyse (Scholz et al. 2004). In this software (available at http://metagenealyse.mpimp-golm.mpg.de) ICA was performed following a PCA preprocessing step to optimize the outcome of the analysis. The data was log10-transformed and normalized to the mean of the entire sample set for each metabolite before the analysis. This transformation reduces the influence of rare high-measurement values, but does not change the discrimination in the data set.
Results
Experimental design
Herbicide treatments in the field are generally performed either by foliar spraying or by soil application. For the purposes of this study we decided to assess the effects of application by spraying since this represents the common method of application for the herbicides selected for these experiments. We decided to evaluate the effects of several herbicides of known mode of action, using a highly standardized growth and treatment cycle summarized in Fig. 1a. Given that we are looking at a short-term response to the herbicides we decided to adopt a serial sampling approach (harvesting plant material at 1, 3, 6, 12 and 24 h post-treatment), in order to follow the kinetics of the metabolic response to the herbicide. The herbicides we chose to study were glufosinate, sulcotrione, AE 944 [N2-(1-ethyl-3-phenylpropyl)-6-(1-fluoro-1-methylethyl)-1,3,5-triazine-2,4-diamine], foramsulfuron, benfuresate and glyphosate representing inhibitors of glutamine synthetase, 4-Hydroxyphenylpyruvate dioxygenase (4-HPPD), cellulose biosynthesis (exact target unknown; Kiedaisch et al. 2003) acetolactate synthase (ALS), very long chain fatty acid biosynthesis (VLCFA), and 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), respectively.
Fig. 1.
Overview of the standardized experimental treatment and growthconditions of the Arabidopsis thaliana (Col0) plants up until the herbicide application
Metabolic response of Arabidopsis to glufosinate treatment
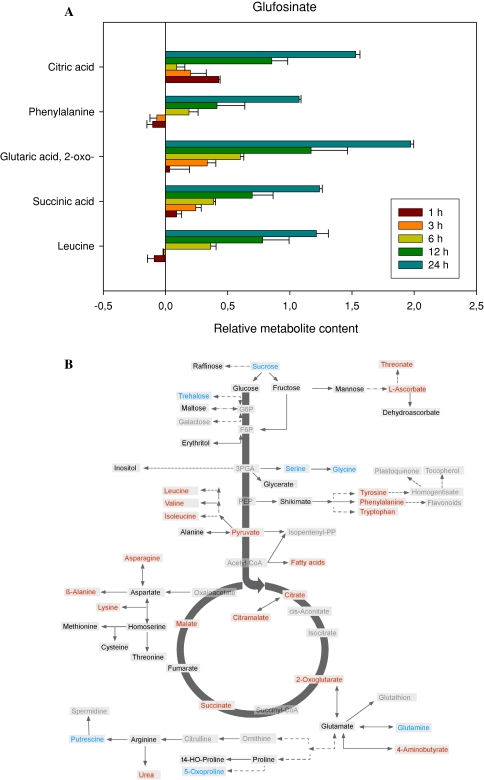
Treatment of Arabidopsis with the glutamine synthetase inhibitor glufosinate resulted in dramatic metabolic changes. The most prominent of these are represented, in log scale, in the histogram of Fig. 2a whilst the entire data set is available as Supplemental Table I. This study revealed that treatment with glufosinate resulted in a dramatic accumulation of the TCA cycle intermediate 2-oxoglutarate which reached 124% of the control level after 1 h, 222% after 3 h, 402% after 6 h, 2239% after 12 h and an incredible 9416% after 24 h. In addition, there were large increases in the concentrations of succinate and citrate, the branched chain amino acid leucine and the aromatic amino acid phenylalanine. Looking at the pathway map to identify the metabolites changing after 24 h revealed that the TCA cycle associated metabolites malate and citramalate also increased as did aspartate, β-alanine and lysine as well as other members of the branched chain and aromatic amino acid families, the fatty acids, metabolites of ascorbate metabolism and γ-aminobutyric acid (GABA) and urea. In contrast, sucrose and trehalose and serine and glycine were significantly lower 24 h after treatment as were putrescine, 5-oxoproline and glutamine (Fig. 2b).
Fig. 2.
Metabolic changes following treatment of Arabidopsis thaliana plants with the herbicide glufosinate. The graph shows the five most variant metabolites samples to the left of the line are increased in content following treatment, samples to the right are decreased in content following treatment. Data are presented on a logarithmic (base10) scale and are the mean of four independent samples. The full dataset is viewable in Supplemental Table I
Metabolic response of Arabidopsis to sulcotrione treatment
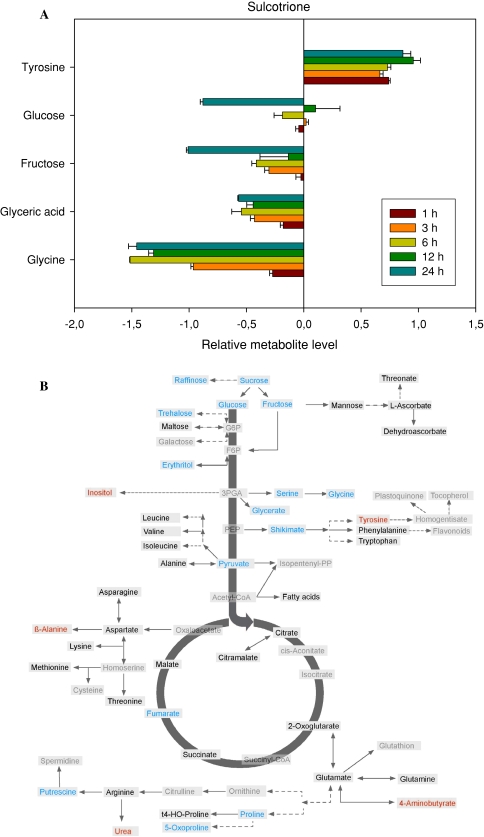
Application of the 4-HPPD inhibitor sulcotrione which is known to block plastoquinone and tocopherol biosynthesis (Schulz et al 1993; Pallett 2000), also resulted in large metabolic shifts, however, these were of a quite different nature to those described above for glufosinate. The most prominent of these are represented, in log scale, in the histogram of Fig. 3a (whilst the entire data set is available as Supplemental Table I). Intriguingly, the majority of changes observed were decreases with only tyrosine increasing whereas glucose, fructose, glycerate and glycine all decreased dramatically. The increase in tyrosine was very rapid with levels increasing mainly in the first hour following treatment and remaining relatively stable thereafter. Looking at the full data set in order to gain an overview of metabolites which display altered levels 24 h after sulcotrione treatment revealed that the trend of decreasing metabolite levels is reflected across the dataset with sucrose, raffinose, trehalose, erythritol, serine, shikimate, pyruvate, fumarate, putrescine, proline and 5-oxy-proline all being significantly lower, but only inositol, β-alanine, urea and GABA behaving similarly to Tyr, at this timepoint (Fig. 3b).
Fig. 3.
Metabolic changes following treatment of Arabidopsis thaliana plants with the herbicide sulcotrione. The graph shows the five most variant metabolites samples to the left of the line are increased in content following treatment, samples to the right are decreased in content following treatment. Data are presented on a logarithmic (base10) scale and are the mean of four independent samples. The full dataset is viewable in Supplemental Table I
Metabolic response of Arabidopsis to AE 944 treatment
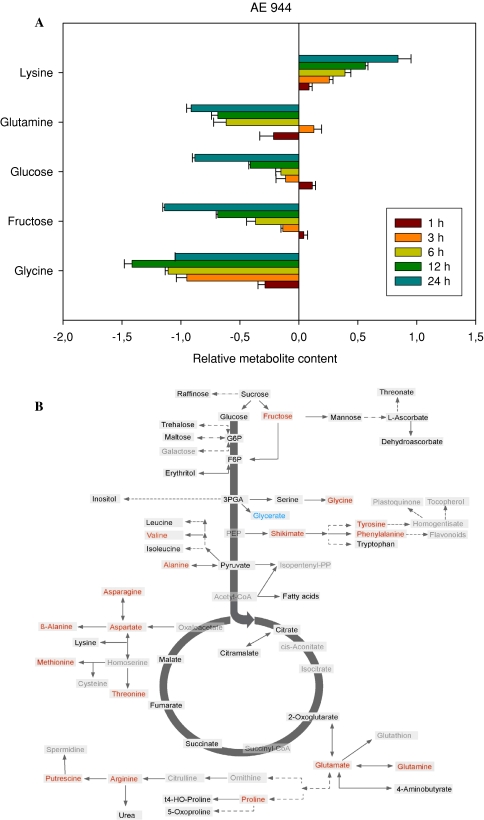
Application of the cell wall biosynthesis inhibitor AE 944 (Kiedaisch et al. 2003) also resulted in distinct shifts in metabolism. As was observed for the sulcotrione treatment the general trend was of decreased metabolite content with four of the five most variable metabolites displaying this behavior (see the histogram of Fig. 4a or the entire dataset—Supplemental Table I), namely glutamine, glucose, fructose and glycine, whilst the level of lysine was progressively increased over time. Analysis of the full data set revealed that after 24 h, the aspartate-derived β-alanine as well as the branched chain amino acids and the aromatic amino acids Tyr and Phe were additionally present at significantly elevated levels. In contrast, the levels of sucrose, raffinose, trehalose, mannose, pyruvate, citramalate, fumarate, succinate, putrescine, proline and glutamine were significant decreased at this timepoint (Fig. 4b).
Fig. 4.
Metabolic changes following treatment of Arabidopsis thaliana plants with the herbicide AE 944. The graph shows the five most variant metabolites samples to the left of the line are increased in content following treatment, samples to the right are decreased in content following treatment. Data are presented on a logarithmic (base10) scale and are the mean of four independent samples. The full dataset is viewable in Supplemental Table I
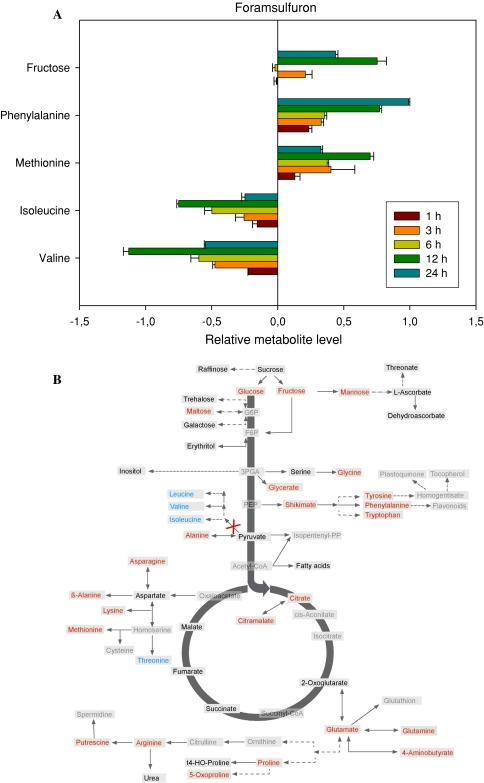
Metabolic response of Arabidopsis to foramsulfuron treatment
Next we evaluated the effect of treatment with the well characterized acetolactate synthase inhibitor foramsulfuron (Hacker et al. 2002). The pattern of metabolic change following this treatment was once again distinct from those following the other treatments described above. In this instance the changes were consistent in the first 12 h following treatment but appeared highly pleiotropic after this timepoint and we therefore focused on only the response up to 12 h following treatment with this chemical. The most dramatic changes after 12 h were seen in fructose, methionine and phenylalanine which increased and perhaps unsurprisingly given its mode of action the branched chain amino acids isoleucine and valine which were dramatically and rapidly depleted (leucine was the sixth most dramatically altered; see the histogram of Fig. 5a or the entire dataset—Supplemental Table I). When the full data set was analysed, again 12 h following treatment, all three branched chain amino acids are clearly dramatically decreased and there is a resultant build up of alanine, shikimate and all the aromatic amino acids, as well as a conserved increase in the sugars maltose, fructose, glucose and mannose, the amino acids glycine, asparagine, β-alanine, lysine, glutamine, glutamate, GABA, proline, 5-hydoxy-proline and arginine, putrescine, citrate and citramalate (Fig. 5b).
Fig. 5.
Metabolic changes following treatment of Arabidopsis thaliana plants with the herbicide formasulfuron. The graph shows the five most variant metabolites samples to the left of the line are increased in content following treatment, samples to the right are decreased in content following treatment. Data are presented on a logarithmic (base10) scale and are the mean of four independent samples. The full dataset is viewable in Supplemental Table I
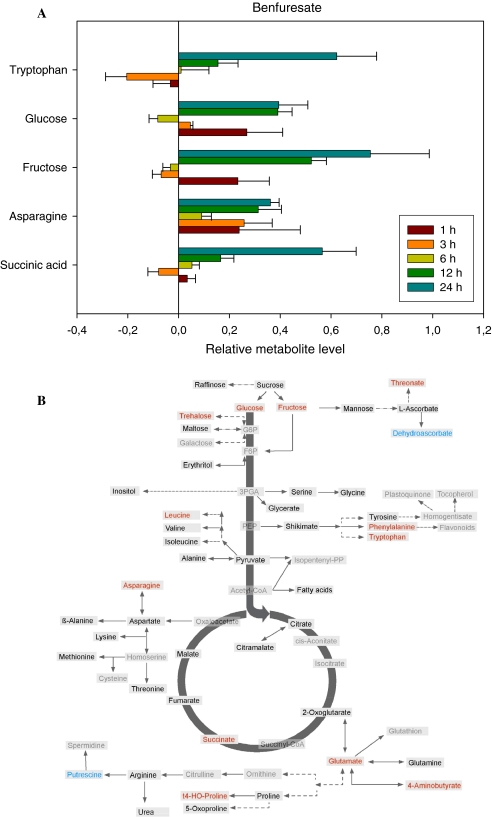
Metabolic response of Arabidopsis to benfuresate treatment
Treatment with the previously characterized very long chain fatty acid elongase inhibitor benfuresate (Trenkamp et al. 2004; Lechelt-Kunze et al. 2003) resulted in less dramatic changes in primary metabolites than has been described for the other herbicides (see Fig. 6a and Supplemental Table I). The most dramatically affected metabolites were tryptophan, aspartate, glucose, fructose and succinate, however, with the exception of asparagine (which increased in all timepoints following herbicide treatment), the directionality of the changes in these metabolites varied across the timecourse suggesting that these changes were more likely to be pleiotropic. Assessment of the polar metabolites 24 h after treatment, revealed conserved increases in trehalose, glucose, fructose threonate, leucine, phenylalanine, tryptophan, asparagine, succinate, glucose, GABA, 5-hydoxy-proline and asparagine and decreases in dehydroascorbate and putrescine (Fig. 6b). It should, however, be noted that given that these changes are inconsistent across the entire experiment, evaluation of the final changes are unlikely to be of high value in this instance. Since we knew this inhibitor affected fatty acid metabolism we also checked the non-polar profiles carefully, however, these also gave no clear indicators of the mode of action of the herbicide (data not shown).
Fig. 6.
Metabolic changes following treatment of Arabidopsis thaliana plants with the herbicide benfuresate. The graph shows the five most variant metabolites samples to the left of the line are increased in content following treatment, samples to the right are decreased in content following treatment. Data are presented on a logarithmic (base10) scale and are the mean of four independent samples. The full dataset is viewable in Supplemental Table I
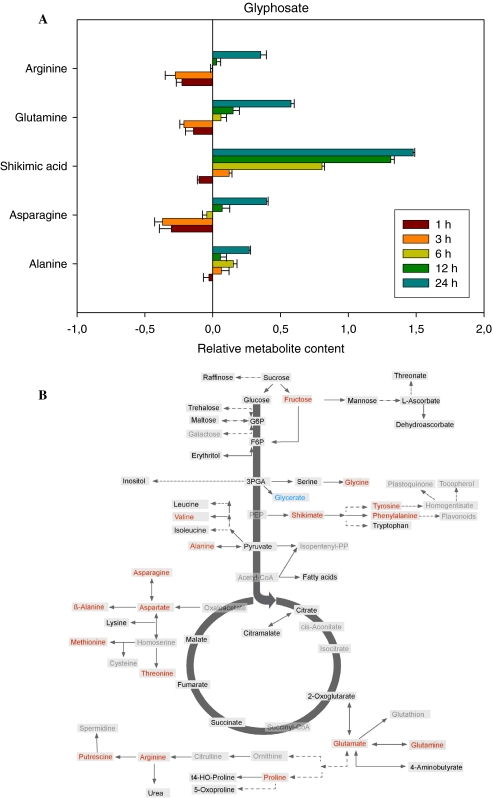
Metabolic response of Arabidopsis to glyphosate treatment
The final herbicide we evaluated was the EPSPS inhibitor glyphosate which has previously been well characterised at both the biochemical (Amrhein et al. 1980; DeMaria et al. 2006) and even the metabolomic level (Böttcher et al. 2008; Oikawa et al. 2006). The data collected here was largely in accordance with the earlier studies (see Fig. 7a and Supplemental Table I), with the outstanding metabolic change being the dramatic accumulation of shikimate as well as a general increase in Ala (although in both instances these metabolites were decreased 1 h after application of the herbicide) and large, albeit mixed, changes in the levels of Gln, Asp and Arg. When the full data set was evaluated, 24 h after treatment, additional increases were observed in levels of fructose, glycine, tyrosine, phenylalanine, valine, asparagine, aspartate, β-alanine, methionine, tyrosine, glutamate, glutamine, praline, arginine and putrescine. By contrast, the levels of glycerate were significantly decreased at this time point (Fig. 7b).
Fig. 7.
Metabolic changes following treatment of Arabidopsis thaliana plants with the herbicide glyphosate. The graph shows the five most variant metabolites samples to the left of the line are increased in content following treatment, samples to the right are decreased in content following treatment. Data are presented on a logarithmic (base10) scale and are the mean of four independent samples. The full dataset is viewable in Supplemental Table I
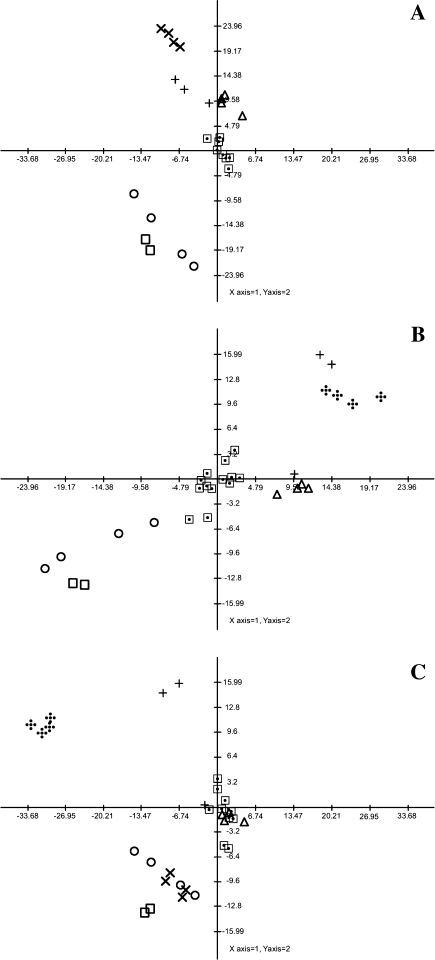
Statistical comparison of the datasets resulting from treatments with herbicides of different mode of action
In order to perform comparative analysis of the different herbicide treatments we chose to adopt principal component analysis (PCA). For this purpose we decided to restrict our analysis to the samples taken 24 h after treatment. The plots of the first three components, are presented in Fig. 8. As would perhaps be expected the clearest discrimination between the treatments was given by plotting principal component 1 against principal component 2 (Fig. 8a). Notably, the different treatments seem to be relatively easy to discriminate from one another but, reassuringly, the control treatments are indistinct from one another. From the perspective of this plot it would appear that the sulcotrione and AE 944 treatments were less easy to discriminate from one another than from the other treatment and the same holds true for the benfuresate and glyphosate treatments. That said, whilst these observations are conserved in the plot of component 2 against component 3 (Fig. 8b), the plot of component 1 against component 3 (Fig. 8c), clearly discrimates both sulcotrione from AE 944 and benfuresate from glyphosate treatments. We next performed an evaluation of the loadings, the parameters (in this case the metabolites) that exhibit the greatest influence on the discrimination. For principal component 1, the top five loadings (2-oxoglutarate, citrate, Tyr, Leu and Phe), include four metabolites thought to be diagnostic of the given mode of action with 2-oxoglutarate clearly indicative of inhibition of glutamine synthetase whilst Tyr and Phe are indicative of EPSPS inhibition and Leu is indicative of ALS inhibition. Similarly, Val and Iso, indicative of ALS inhibition, are amongst the top five loadings in principal component 1 and Tyr is again in the top five loadings of principal component 3.
Fig. 8.
Principal component analysis (PCA) of metabolite profiles following 24 h treatment with distinct herbicides. The PCA is presented as a combinations of the first three dimensions. Each data point represents an independent sample. The analysis of this data was performed using the TMEV software (Saeed et al. 2003). ⊡ untreated plants;  glufosinate; ◯ sulcotrione; □ AE 944; × foramsulfuron; + benfuresate; Δ glyphosate
glufosinate; ◯ sulcotrione; □ AE 944; × foramsulfuron; + benfuresate; Δ glyphosate
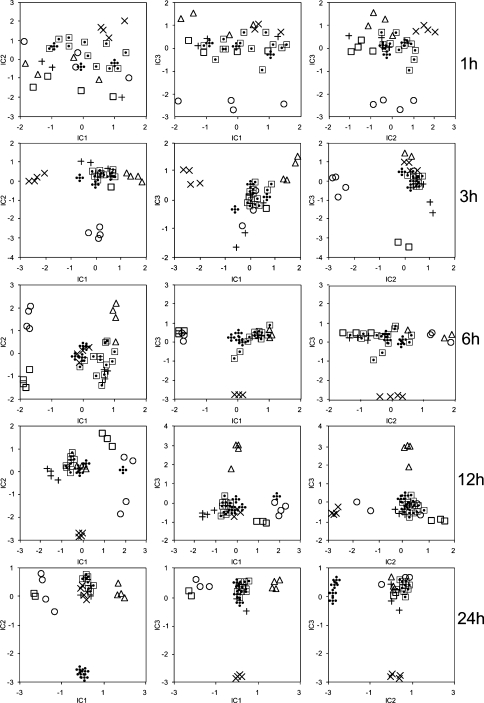
Given that both the point-by-point analysis and to a lesser extent the PCA analysis suggested that the effects of the herbicide treatments can be quite pleiotropic we decided to additionally evaluate the combined dataset by independent component analysis using the framework specifically adapted for metabolite profiling studies by Scholz et al. (2004). In this instance, rather than choosing a single sampling point to focus on we rather performed a comparative study across the kinetic of the experiment. The results of these analyses are presented in Fig. 9, for each time point three plots are shown which correspond to independent component (IC) 1C1 versus IC2, IC1 versus IC3 and IC2 versus IC1. In all instances a principal component analysis set to define four principal components was run as a preparatory analysis as detailed in Scholz et al. (2004). The discrimination afforded by this technique was largely as would be expected. At the first timepoint post treatment (1 h) there was essentially no discrimination between the treatments although it should be noted that the samples treated with AE 944, sulcotrione, foramsulfuron and glyphosate can be easily spotted in the Plot of IC2 versus IC3 at this timepoint. Three and six hours after treatment the separation of samples treated with these three herbicides was even clearer being obvious in all three plots. Twenty-four hours after treatment the glufosinate treated samples could additionally be clearly discriminated. The benfuresate samples were never clearly separable from the control samples—even when the analysis of lipophillic compounds was taken into consideration (data not shown).
Fig. 9.
Independent component analysis (ICA) of metabolite profiles of distinct herbicide treatments following different treatment timespans. Separate ICA are presented for each timepoint. The analysis of this data was performed on the basis of the first four principal components using the MetGenAlyse software (Scholz et al. 2004). ⊡ untreated plants;  glufosinate; ◯ sulcotrione; □ AE 944; × foramsulfuron; + benfuresate; Δ glyphosate
glufosinate; ◯ sulcotrione; □ AE 944; × foramsulfuron; + benfuresate; Δ glyphosate
Discussion
To combat crop losses to pests or weeds comprehensive strategies have recently been adopted including saturation transgenesis approaches to search for suitable herbicidal targets (Lein et al. 2004) as well as the application of genomics level screening methodologies such as transcript (Eckes nad Busch 2007; Yip et al. 1992; Raghavan et al. 2006), or metabolic profiling (Ott et al. 2003; Oikawa et al. 2006). Whilst the work described by Oikawa et al. observed the effects of herbicides supplied via the growth media over a longer time period, we favored a system by which plants were grown under very highly standardized conditions and then sprayed at a highly controlled developmental stage. Post-application plant material was also harvested at set times in a carefully controlled manner. The reasons for taking this approach were two-fold. Firstly, especially given our interest in changes at the level of the single metabolites themselves, we wanted to adopt as strict a procedure as possible in order to minimize pleiotropic or secondary effects that were not a direct consequence of the herbicide treatment. Secondly, application by spray was much more appropriate for the herbicides that we chose to work with here since this is the manner by which they are predominantly applied in the field. This approach allowed us to do two things. Firstly, we were able to demonstrate that simply measuring the polar phase of a simple ethanol extract by GC-MS produced data that enabled us to discriminate the herbicide treated samples from wild type in all the experiments. Intriguingly this was also true for the statistical tools PCA and ICA. That the treatments could be discriminated from one another using this approach was reassuring but perhaps not so surprising. Such discrimination had previously been demonstrated using similar statistical approaches following FT-ICR-MS analysis (Oikawa et al. 2006) and following machine learning approaches on the smaller metabolite data sets produced by NMR analysis (Ott et al. 2003). However, given the low unit cost of a standard GC-MS run (Stitt and Fernie 2003), these results highlight the potential of the approach taken here for high-throughput screening of novel agrochemicals.
The time-dependent sampling approach described here allowed us to identify metabolites that responded rapidly to the effects of the various herbicides. For several of the herbicides this strategy was highly effective. Of particular note are the five most effected metabolites following treatment with foramsulfuron, glufosinate and glyphosate with all three treatments showing metabolic changes previously characterized following these treatments (Gaston et al. 2003; Ray 1984; Servaites et al. 1987; Barsch et al. 2006; Böttcher et al. 2008), within a very short timeframe. Indeed after 24 h those plants treated with foramsulfuron displayed a metabolic profile that was not consistent with the mode of action—most probably due to a combination of pleiotropic and adaptive effects. Similarly the consistent increase in tryptophan in samples treated with sulcotrione is in accordance with its action as a 4-HPPD inhibitor (Pallett 2000) and likewise the accumulation of sugars following the treatment with AE 944 are what would be anticipated from a cell wall biosynthesis inhibitor (Kiedaisch et al. 2003). When taken together these five examples provide strong evidence that time-dependent metabolite profiling by GC-MS can aid in the identification of the mode of action of herbicides. The fact that many compounds with herbicidal activity exert their effects through the disruption of primary metabolism renders it likely that the coverage afforded by GC-MS approaches (Fernie et al. 2004) will be suitable for the detection of many different classes of herbicides. As a caveat to this statement it should be noted that the metabolite profiles following treatment with benfuresate provided little clue to its mode of action. Perhaps surprisingly, this was also true following GC-MS profiling of the lipophilic metabolites of samples collected following this treatment. That said further studies are probably required on compounds of similar mode of action to benfuresate in order to evaluate if there are metabolites diagnostic of very long chain fatty acid elongase inhibition present within the subset of metabolites accessible by this method.
In summary, in this article we have shown that a time dependent approach for GC-MS profiling based on serial sampling following herbicidal spray is likely to have high utility in aiding the process of mode of action identification of agrochemicals. The advantage of this approach is that it allows the discrimination of early and adaptive responses and is therefore more likely to facilitate a correct identification of the metabolic pathway or process that the herbicide is working on. Metabolic profiling of herbicide treated samples is also likely to be increasingly important given increasing public awareness with respect to crop safety issues (Catchpole et al. 2005; Shepherd et al. 2005). It should be noted that this approach has clear limitations, for example it was unable to provide useful clues to the mode of action of the very long chain fatty acid elongase inhibitor benfuresate and for certain other classes of herbicides—for example those that effect hormonal aspects of plant function. It, therefore, seems likely that for certain compound classes specific protocols, such as those used for determination of very long chain fatty acid elongase inhibitors (Trenkamp et al. 2004), are likely to retain their utility. That said, the fact that the identification of the mode of action of some of the inhibitors was facile on the basis of the behaviour of diagnostic metabolites suggests that this approach will likely play an important role in the acceleration of novel herbicide mode of action discoveries.
Electronic supplementary material
MOESM1 [INSERT CAPTION HERE] (XLS 64 kb)
MOESM2 [INSERT CAPTION HERE] (XLS 63 kb)
MOESM3 [INSERT CAPTION HERE] (XLS 53 kb)
MOESM4 [INSERT CAPTION HERE] (XLS 64 kb)
MOESM5 [INSERT CAPTION HERE] (XLS 56 kb)
MOESM6 [INSERT CAPTION HERE] (XLS 64 kb)
Acknowledgement
We thank Nicolas Schauer for help in the early stages of this project, Sonia Osario-Algar for help with the figures and Corinna van Almsick and Franka Hübner for plant cultivation and spray application
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-008-0149-8) contains supplementary material, which is available to authorized users.
References
- Aharoni, A., de Vos, R. C. H., Verhoeven, H. A., Maliepaard, C. A., Kruppa, G., Bino, R., et al. (2002). Non-targeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS,6, 217–234. doi:10.1089/15362310260256882. [DOI] [PubMed]
- Amrhein, N., Schab, J., & Steinrucken, H. C. (1980). The mode of action of glyphosate. Naturwissenschaft,67, 356–357. doi:10.1007/BF01106593. [DOI]
- Aranibar, N., Singh, B. K., Stockton, G. W., & Ott, K. H. (2001). Automated mode of action detection by metabolic profiling. Biochemical and Biophysical Research Communications,286, 150–155. doi:10.1006/bbrc.2001.5350. [DOI] [PubMed]
- Barsch, A., Carvalho, H. G., Cullimore, J. V., & Niehaus, K. (2006). GC-MS profiling implies three independent ways of ammonium assimilation in Medicago truncatula root nodules. Journal of Biotechnology,127, 79–83. doi:10.1016/j.jbiotec.2006.06.007. [DOI] [PubMed]
- Böttcher, C., Centeno, D., Freitag, J., Höfgen, R., Köhl, K., Kopka, J., et al. (2008). Teaching (and learning from) metabolomics: The 2006 PlantMetaNet ETNA Metabolomics research school. Physiologia Plantarum,132, 136–144. [DOI] [PubMed]
- Brindle, J. T., Antti, H., Holmes, E., Tranter, G., Nicholson, J. K., Bethell, H. W. L., et al. (2002). Rapid and non-invasive diagnosis of the presence and severity of coronary heart disease using H-1-NMR-based metabonomics. Nature Medicine,8, 1439–1444. doi:10.1038/nm802. [DOI] [PubMed]
- Catchpole, G. S., Beckmann, M., Enot, D. P., Mondhe, M., Zywicki, B., Taylor, J., et al. (2005). Hierarchical metabolomics demonstrates substantial composition similarity between genetically modified and conventional potato crops. Proceedings of the National Academy of Sciences of the United States of America,102, 14458–14462. doi:10.1073/pnas.0503955102. [DOI] [PMC free article] [PubMed]
- DeMaria, N., Becerril, J. M., Garcia-Plazaola, J. I., Hernandez, A., De Felipe, M. R., & Fernandez-Pascual, M. (2006). New insights on glyphosate mode of action in nodular metabolism: role of shikimate accumulation. Journal of Agricultural and Food Chemistry,54, 2621–2628. doi:10.1021/jf058166c. [DOI] [PubMed]
- Eckes, P., & Busch, M. (2007). Fast identification of the mode of action of herbicides by DNA chips. In W. Krämer & U. Schirmer (Eds.), Modern crop protection compounds (pp. 1161–1174). Weinheim, Germany: Wiley.
- Fernie, A. R., Trethewey, R. N., Krotzky, A., & Willmitzer, L. (2004). Metabolite profiling: From diagnostics to systems biology. Nature Reviews. Molecular Cell Biology,5, 763–769. doi:10.1038/nrm1451. [DOI] [PubMed]
- Fiehn, O., Kopka, J., Dörmann, P., Altmann, T., Trethewey, R. N., & Willmitzer, L. (2000). Metabolic profiling for plant functional genomics. Nature Biotechnology,18, 1157–1161. doi:10.1038/81137. [DOI] [PubMed]
- Gaston, S., Ribas-Carbo, M., Busquets, S., Berry, J. A., Zabalaza, A., & Royuela, M. (2003). Changes in mitochondrial electron partitioning in response to herbicides inhibiting branched chain amino acid biosynthesis in soybean. Plant Physiology,133, 1351–1359. doi:10.1104/pp.103.027805. [DOI] [PMC free article] [PubMed]
- Hacker, E., Bieringer, H., Willms, L., Schnabel, G., Koecher, H., Hagenteister, H., et al. (2002). Foramsulfuron plus safener—a new technology for weed control in maize. Journal of Plant Diseases and Protection,18(Special Issue), 747–756.
- Iijima, Y., Nakamura, Y., Ogata, Y., Tanaka, K., Sakurai, N., Suda, K., et al. (2008). Metabolite annotations based on the integration of mass spectral information. The Plant Journal,54, 949–962. doi:10.1111/j.1365-313X.2008.03434.x. [DOI] [PMC free article] [PubMed]
- Kiedaisch, B. M., Blanton, R. L., & Haigler, C. H. (2003). Characterisation of a novel cellulose synthesis inhibitor. Planta,217, 922–930. doi:10.1007/s00425-003-1071-y. [DOI] [PubMed]
- Kopka, J., Schauer, N., Krueger, S., Birkmeyer, C., Usadel, B., Bergmuller, E., et al. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics (Oxford, England),21, 1635–1638. doi:10.1093/bioinformatics/bti236. [DOI] [PubMed]
- Lechelt-Kunze, C., Meissner, R. C., Drewes, M., & Tietjen, K. (2003). Flufenacet herbicide treatment phenocopies the fiddlehead mutant in Arabidopsis thaliana. Pest Management Science,59, 847–856. doi:10.1002/ps.714. [DOI] [PubMed]
- Lein, W., Bornke, F., Reindl, A., Erhardt, T., Stitt, M., & Sonnewald, U. (2004). Target-based discovery of novel herbicides. Current Opinion in Plant Biology,7, 219–225. doi:10.1016/j.pbi.2004.01.001. [DOI] [PubMed]
- Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., & Fernie, A. R. (2006). Gas chromatrography mass spectrometry-based metabolite profiling in plants. Nature Protocols,1, 387–396. doi:10.1038/nprot.2006.59. [DOI] [PubMed]
- Menne, H., & Köcher, H. (2007). HRAC classification of herbicides and resistance development. In W. Krämer & U. Schirmer (Eds.), Modern crop protection compounds (pp. 5–26). NY: Wiley.
- Oikawa, A., Nakamura, Y., Ogura, T., Kimura, A., Suzuki, H., Sakarai, N., et al. (2006). Clarification of pathway-specific inhibition by Fourier transform ion cyclotron resonance/mass spectrometry-based metabolic phenotyping studies. Plant Physiology,142, 398–413. doi:10.1104/pp.106.080317. [DOI] [PMC free article] [PubMed]
- Ott, K. H., Aranibar, N., Singh, B. J., & Stockton, G. W. (2003). Metabononics classifies pathways affected by bioactive compounds. Artificial neural network classification of NMR spectra of plant extracts. Phytochemistry,62, 971–985. doi:10.1016/S0031-9422(02)00717-3. [DOI] [PubMed]
- Pallett, K. E. (2000). Mode of action of isoxaflutole—a case study of an emerging target site. In A. H. Cobb & R. C. Kirkwood (Eds.), Herbicides and their mechanisms of action (pp. 215–238). Sheffield: Sheffield Academic Press.
- Petroff, O. A. C. (1988). Biological 1HNMR spectroscopy. Comparative Biochemistry and Physiology,90B, 249–260. [DOI] [PubMed]
- Raghavan, C., Ong, E. K., Dalling, M. J., & Stevenson, T. W. (2006). Regulation of genes associated with auxin, ethylene and ABA pathways by 2-4-dichlorophenoxyacetic acid in Arabidopsis. Functional and Integrative Genomics,6, 60–70. doi:10.1007/s10142-005-0012-1. [DOI] [PubMed]
- Ray, T. B. (1984). Site of action of chlorsulfon. Plant Physiology,133, 1351–1359.
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., et al. (2001). Metabolite profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. The Plant Cell,13, 11–29. [DOI] [PMC free article] [PubMed]
- Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: A free open-source system for microarray data management and analysis. BioTechniques,34, 374–378. [DOI] [PubMed]
- Sauter, H., Lauer, M., & Fritsch, H. (1991). Metabolic profiling of plants—a new diagnostic technique. In D. R. Baker, J. G. Fenyes, & M. K. Moberg (Eds.), Synthesis and chemistry of Agrochemicals II. ACS Symposium Series 443 (pp. 288–299). Washington, DC: American Chemical Society.
- Schauer, N., Semel, Y., Roessner, U., Gur, A., Balbo, I., Carrari, F., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nature Biotechnology,24, 447–454. doi:10.1038/nbt1192. [DOI] [PubMed]
- Schauer, N., Steinhauser, D., Strelkov, S., Schomburg, D., Allison, G., Moritz, T., et al. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters,579, 1332–1337. doi:10.1016/j.febslet.2005.01.029. [DOI] [PubMed]
- Scholz, M., Gatzek, S., Sterling, A., Fiehn, O., & Selbig, J. (2004). Metabolic fingerprinting: Detecting biological features by independent component analysis. Bioinformatics, 20, 2447–2454. [DOI] [PubMed]
- Schulz, A., Ort, O., Beyer, P., & Kleinig, H. (1993). SC-0051, a 2-benzoyl-cyclohexane-1, 3-dione bleaching herbicide, is a potent inhibitor of the enzyme 4-hydroxyphenylpyruvate dioxygenase. FEBS Letters,319, 162–166. doi:10.1016/0014-5793(93)80013-K. [DOI] [PubMed]
- Servaites, J. C., Tucci, M. A., & Geiger, D. R. (1987). Glyphosate effects on carbon assimilation, ribulose bisphosphate carboxylase activity, and metabolite levels in sugar beet leaves. Plant Physiology,85, 370–374. [DOI] [PMC free article] [PubMed]
- Stitt, M., & Fernie, A. R. (2003). From measurements of metabolites to metabolomics an “on the fly” perspective illustrated by recent studies of carbon-nitrogen interactions. Current Opinion in Biotechnology,14, 136–144. doi:10.1016/S0958-1669(03)00023-5. [DOI] [PubMed]
- Shepherd, L. V. T., McNicol, J. W., Razzo, R., Taylor, M. A., Davies, H. V., & McNicol, J. W. (2005). Assessing the potential for unintended effects in genetically modified potatoes perturbed in metabolic and developmental processes. Targeted analaysis of key nutrients and anti-nutrients. Transgenic Research,15, 409–425. doi:10.1007/s11248-006-0012-5. [DOI] [PubMed]
- Trenkamp, S., Martin, W., & Tietjen, K. (2004). Specific and differential inhibition of very-long chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proceedings of the National Academy of Sciences of the United States of America,101, 11903–11908. doi:10.1073/pnas.0404600101. [DOI] [PMC free article] [PubMed]
- Yip, W. K., Moore, T., & Yang, S. F. (1992). Differential accumulation of transcripts for four tomato 1-aminocyclopropane-1-carboxylate synthase homologs under various conditions. Proceedings of the National Academy of Sciences of the United States of America,89, 2475–2479. doi:10.1073/pnas.89.6.2475. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MOESM1 [INSERT CAPTION HERE] (XLS 64 kb)
MOESM2 [INSERT CAPTION HERE] (XLS 63 kb)
MOESM3 [INSERT CAPTION HERE] (XLS 53 kb)
MOESM4 [INSERT CAPTION HERE] (XLS 64 kb)
MOESM5 [INSERT CAPTION HERE] (XLS 56 kb)
MOESM6 [INSERT CAPTION HERE] (XLS 64 kb)