Abstract
Background
From Paleo-Indian times to recent historical episodes, the Mesoamerican isthmus played an important role in the distribution and patterns of variability all around the double American continent. However, the amount of genetic information currently available on Central American continental populations is very scarce. In order to shed light on the role of Mesoamerica in the peopling of the New World, the present study focuses on the analysis of the mtDNA variation in a population sample from El Salvador.
Methodology/Principal Findings
We have carried out DNA sequencing of the entire control region of the mitochondrial DNA (mtDNA) genome in 90 individuals from El Salvador. We have also compiled more than 3,985 control region profiles from the public domain and the literature in order to carry out inter-population comparisons. The results reveal a predominant Native American component in this region: by far, the most prevalent mtDNA haplogroup in this country (at ∼90%) is A2, in contrast with other North, Meso- and South American populations. Haplogroup A2 shows a star-like phylogeny and is very diverse with a substantial proportion of mtDNAs (45%; sequence range 16090–16365) still unobserved in other American populations. Two different Bayesian approaches used to estimate admixture proportions in El Salvador shows that the majority of the mtDNAs observed come from North America. A preliminary founder analysis indicates that the settlement of El Salvador occurred about 13,400±5,200 Y.B.P.. The founder age of A2 in El Salvador is close to the overall age of A2 in America, which suggests that the colonization of this region occurred within a few thousand years of the initial expansion into the Americas.
Conclusions/Significance
As a whole, the results are compatible with the hypothesis that today's A2 variability in El Salvador represents to a large extent the indigenous component of the region. Concordant with this hypothesis is also the observation of a very limited contribution from European and African women (∼5%). This implies that the Atlantic slave trade had a very small demographic impact in El Salvador in contrast to its transformation of the gene pool in neighbouring populations from the Caribbean facade.
Introduction
El Salvador lies on the Pacific coast (without an Atlantic seaboard) and it is the smallest of the Central American countries. Most of the country rests on a fertile volcanic plateau. It is segmented by two volcanic ranges running roughly west to east, separated by broad, fertile valleys, such as that of the river Lempa. El Salvador was inhabited by Native American groups who were in part descendants of the Aztecs and Toltec of Mexico, such as the Pipil (a Nahua tribe) and the Lenca. These two Native American communities inhabited mainly the western regions, constituting about 60% of the population throughout the colonial era and into the early decades of independence [1], [2].
The development of coffee estates led to the slow but continuous dissolution of most of the communal lands of Native villages [1], [2]. Thus, the 1930 census, the last to contain the category, designated only 5.6% of the population as “Indian” – although it is not clear what criteria were used in arriving at this figure. Other independent estimates (considering religious activities, distinctive women's dress, language, and involvement in various handicrafts) placed the mid-twentieth-century Indian population at 20% (∼400,000 persons). The abandonment of Indian language and customs was hastened by political repression; most natives stopped wearing traditional dress, abandoned the Pipil language, and adopted ladino customs. By 1975 no more than ∼1% of the population wore distinctive Indian clothing or followed Indian customs. Nowadays, the official language in El Salvador is Spanish, although Nahua is still spoken among some natives.
Although the American continent has been the target of many forensic and population genetic studies, there are nevertheless many American regions, such as El Salvador, that remain genetically uncharacterized. The mtDNA molecule is commonly used in anthropological contexts because of particular features (maternal inheritance, lack of recombination and high average mutation rate) that confer great power for phylogenetic and phylogeographic inferences. Many mtDNA studies of Native Americans have, however, been limited to genotyping a handful of mtDNA coding region sites that simply distinguish the four major Native American mtDNA haplogroups, A2, B2, C1 and D1 (generally using RFLP typing); unfortunately, the information provided by these few SNPs is of limited value in forensic and population genetics.
Here we have sequenced the mtDNA control region in a sample from El Salvador in order to investigate to what extent the Native American component has survived the impact of European colonialism and the concomitant influx of African slaves to the Caribbean and Meso-America.
Materials and Methods
Sample collection and DNA extraction
A total of 90 saliva samples were collected from healthy unrelated individuals from El Salvador. DNA extraction was undertaken following standard phenol-chloroform protocol. DNA quantification was carried out using DyNA Quant 200 Fluorometer, Hoefer (APB, Uppsala, Sweden).
All the samples were collected anonymously by the Laboratorio de Genética Forense from the Instituto de Medicina Legal that belongs to the Corte Suprema de Justicia from San Salvador (El Salvador). Oral informed consent was required in all the cases. The study, including the oral informed consent protocol, was approved by the Ethical committee of the University of Santiago de Compostela, and it conforms to the Spanish Law for Biomedical Research (Law 14/2007- 3 of July).
PCR and sequence analysis
We analyzed the first and second hypervariable segments (HVS-I and HVS-II) of the mtDNA genome. We performed PCR amplifications using a 2700 Thermocycler (Applied Biosystems), using PCR and sequencing primers as reported in [3]. Cycling parameters were 95°C for 1 min, followed by 36 cycles of 95°C for 10 sec, 55°C for 30 sec and 72°C for 30 sec, and followed by 10 min at 15°C. We checked amplification products on a polyacrylamide gel visualized by silver staining and purified with Montage (Multiscreen PCR, Millipore Corporation, USA). We performed sequence reaction products on each strand by means of the ABI Prism dRhodamine Terminator cycle sequencing reaction kit (Applied Biosystems). DNA products were then purified by ethanol precipitation and sequence reaction products analyzed on the ABI Prism 3100 automatic sequencer (Applied Biosystems). We omitted population variation at the hypervariable sites (mainly related to the cytosine homopolymeric track around 310 and the CA-dinucleotide repeat around positions 522) from inter-population comparisons and phylogeographic analyses. We have used the same primers for amplification and sequencing described in [4].
Sequences were edited using the numbering system of the revised Cambridge Reference Sequence [5]. Most of the sequences could be read from np 16042–16569 and 21–550; for convenience, we will refer to these as HVS-I and HVS-II, although these sequence ranges encompass more than the canonical ranges of these control-region segments.
Quality checking
Problems with the quality of mtDNA data in forensic, clinical, and population genetic studies are unfortunately rather common; see, for instance, [6], [7], [8], [9], [10], [11]. In order to minimize the effects of potential laboratory and documentation errors, the data were read separately by two independent persons in the light of the known phylogeny. We checked phylogenetic inconsistencies by hand with special attention to private or unusual variants (e.g. rare transitions or indels). In some cases, we confirmed the sequences by repeated extraction and sequencing. In addition, to detect potential “phantom mutations” [7], we also checked the data using the computer program SPECTRA ([7], available at http://www.stats.gla.ac.uk/~vincent/fingerprint/index.html).
Statistical analysis and population comparison
Haplogroup nomenclature follows the most recently updated versions of the Native American phylogeny given in [12], [13], [14]. Diversity indices of HVS-I sequences (haplotype diversity, nucleotide diversity, average number of pairwise differences) were calculated using Arlequin 3.0 software [15]. Nucleotide and sequence diversity was computed as in the manner of Nei [16].
We estimated median-joining networks of HVS-I sequences using the Network 4.1.1.2 software [17], [18]. Coalescence times were calculated using the ρ statistic [19], [20] with an HVS-I mutation rate of one transition per 18,845 years applied for the sequence range 16090-16365 using the most recent estimates provided by Soares et al. [21].
An mtDNA database of Native American populations was compiled for population comparisons: (i) from North America: Aleuts [22], Athapaskans, Inupiaq, Yakima [23], Chukchi and Siberian Eskimos [24], Bella Coola and Haida [25], Nuu-Chah-Nulth [26], Cheyenne [27], North Native Americans [various ethnic groups; 28], [29], [30], [31], [32], [33], Apache and Navajo [34], (ii) from Meso-America: Pima [27], Maya [33], Huetar [35], Kuna [36], Ngöbe [37], Quiché [38], Emberá and Wounan [39], Mexico [40], Central Native Americans [various ethnic groups] [28], El Salvador (present study), and (iii) from South America: Native Brazilians and Araucanians or Chileans [33], Equador [41], Embera and Gavião [42], Amazonas [43], Ayoreo [44], Chilean Mapuche and Pehueche, Yaghan [45], Argentinian Mapuche [46], Cayapas [41], Xavante, Zoró, and Gavião [42], Yanomami [47], [48], South Native Americans [various ethinic groups] [29], Tuacuarembó [49], Uruguay [50], Guahibo [51], Colombia [33], [52], Yuracaré, Trinitario, Movima, and Ignaciano [53], and 105 from Arequipa, Tayacaja and San Martin in Peru [54]. We also included the data collected from several studies of ancient DNA [55], [56], [57], [58], [59], [60], [61]. In addition, other datasets were additionally used for haplotype matching comparisons [34], [62], [63], [64], [65], [66]. In total, 3,843 mtDNAs profiles (mainly HVS-I segments) were compiled for comparison with our sample from El Salvador. Those population samples consisting of less than 15 individuals were only used for haplotype matching between populations. For comparison purposes the common reading frame 16090–16365 of the HVS-I was used.
Admixture analysis
We took two different approaches to carry out an admixture analysis.
The first model was applied as described in [67] although, instead of using haplogroup frequencies as variables, we used the frequencies of the shared haplotypes (matching haplotypes) between the source populations (North and South America) and El Salvador. The number of mtDNAs within each matching haplotype in El Salvador (ni: 1≤i≤C, the number of different matching haplotypes in the sample) was assumed to be a draw from a multinomial distribution with parameters 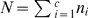 and
and 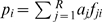 (1≤i≤C), where R is the number of source regions in America, fji is the frequency of the ith cluster in the jth source region (assumed to be known), and αj are the admixture coefficients. This model describes samples from an urn containing C different kinds of ball, where the urn has been created by mixing together R other urns in proportions given by the admixture coefficients. We chose to analyze this model in a Bayesian framework, which meant that we had to explore the distribution of the admixture coefficients, given the data. The prior distribution of the admixture coefficients was taken to be uninformative—namely, uniform on
(1≤i≤C), where R is the number of source regions in America, fji is the frequency of the ith cluster in the jth source region (assumed to be known), and αj are the admixture coefficients. This model describes samples from an urn containing C different kinds of ball, where the urn has been created by mixing together R other urns in proportions given by the admixture coefficients. We chose to analyze this model in a Bayesian framework, which meant that we had to explore the distribution of the admixture coefficients, given the data. The prior distribution of the admixture coefficients was taken to be uninformative—namely, uniform on 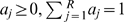 . The posterior distribution of the {αj} was explored with the Metropolis-Hastings algorithm, using a simple proposal, and was summarized by the posterior mean of each aj and its root-mean-square deviation about the mean. To assess model fit, we examined plots of standardized residuals.
. The posterior distribution of the {αj} was explored with the Metropolis-Hastings algorithm, using a simple proposal, and was summarized by the posterior mean of each aj and its root-mean-square deviation about the mean. To assess model fit, we examined plots of standardized residuals.
The second admixed model was applied as described [68]. The probability of origin of each of the sub-continental region was computed as 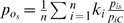 where, n is the number of El Salvador sequences with matches (≥1) in the whole continental dataset; ki, the number of times the sequence i is found in El Salvador; pis, the frequency of the sequence i in the sub-continental region dataset; and pic, the frequency of the sequence i in whole continental dataset.
where, n is the number of El Salvador sequences with matches (≥1) in the whole continental dataset; ki, the number of times the sequence i is found in El Salvador; pis, the frequency of the sequence i in the sub-continental region dataset; and pic, the frequency of the sequence i in whole continental dataset.
Founder analysis
The time to the most recent common ancestor (TMRCA) of haplogroup A2 in the phylogeny was estimated as described [19], [20].
In order to carry out a founder analysis [19], [69], we made some simplifying assumptions about the founding of El Salvador. We assumed (i) a single migration to El Salvador and (ii) that North America was the unique source population. Founder sequences were inferred as matches with samples from North America. An estimate of the time of the migration event was determined by averaging diversity over the clusters derived from each founder in El Salvador, as follows. Suppose there are r founder clusters. Let, ρi be the ρ value (average distance of the haplotypes of a clade from the respective root [19], [20]) for the it h founder cluster, σi be its estimated standard error [20] and ni be the number of sampled individuals in that cluster. Define,
Then,
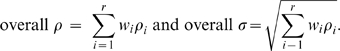 |
Values of ρ and σ were converted to age using the most recent mutation rate available for the HVS-I segment of 1 transition per 18,845 years (in the sequence range 16090–16365).
Results
Summary statistics
We observed a total of 55 different HVS-I, 40 different HVS-II, and 76 different combined HVSI/II mtDNA haplotypes in El Salvador (N = 90). Some HVS-I profiles are quite common, such as C16111T–T16223C–C16290T–G16319A–T16362T, appearing in 12 mtDNAs, and its one step-mutation ‘neighbour’ haplotype (16519 on top) appearing 12 times.
As shown in Table 1, El Salvador shows haplotype and nucleotide diversity values slightly lower than those observed in the continental North, South, and other Meso American populations, which is in part due to the fact that there is virtually only one Native American haplogroup (A2) represented in El Salvador sample. Note that these comparisons have to beviewed with care because the terms “North”, “South” and “Meso-American” refer to groups of population samples of different nature; some are Native American groups that have passed through severe prehistoric bottlenecks while others are at different levels of admixture with e.g. Europeans and Africans.
Table 1. Native American population mtDNA database considering the sequence range 16090–16365.
| n | H | D | π | M | |
| El Salvador | 90 | 49 | 0.917±0.025 | 0.013±0.002 | 3.5 |
| North America | 1215 | 243 | 0.950±0.003 | 0.020±0.000 | 5.1 |
| Meso America | 394 | 142 | 0.968±0.004 | 0.023±0.012 | 6.2 |
| South America | 1144 | 265 | 0.956±0.003 | 0.019±0.000 | 5.3 |
n = sample size; H = number of different haplotypes; D = haplotype diversity; π = nucleotide diversity; M = average number of nucleotide differences.
Phylogeography of Salvadorian Native American mtDNAs
Table 2 shows the full list of control region profiles from El Salvador and their haplogroup allocation. Frequencies of the typical Native American haplogroups A2, B2, and C1 are ∼91%, ∼2%, and ∼2%, respectively.
Table 2. MtDNA haplotypes in El Salvador.
| ID# | HVS-I (minus 16000) | HVS-I reading range (minus 16000) | HVS-II | HVS-II reading range | HG |
| 1 | 42 111 223 244 290 319 362 519 | 024-569 | 64 73 146 152 153 154 235 263 309+C 315+C 523-524del | 021-540 | A2 |
| 2 | 51 111 223 290 299 319 362 | 024-569 | 64 73 146 153 235 263 315+C 523-524del | 021-589 | A2 |
| 90 | 111 136 153 223 290 311 319 362 | 024-560 | 64 73 146 153 235 263 309+CC 315+C 523-524del | 021-595 | A2 |
| 10 | 111 136 153 223 290 319 362 | 024-520 | 64 73 146 153 235 263 309+CC 315+C 523-524del | 021-590 | A2 |
| 8 | 111 136 223 290 311 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-560 | A2 |
| 43 | 111 136 223 290 311 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 55 | 111 136 223 290 319 362 | 024-560 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 62 | 111 136 223 290 319 362 | 024-560 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-540 | A2 |
| 93 | 111 136 223 290 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 61 | 111 172 223 290 319 362 519 | 024-589 | 64 73 146 153 195 235 263 309+CC 315+C | 021-590 | A2 |
| 83 | 111 175 223 290 300 319 362 | 024-569 | 64 73 153 235 263 309+C 315+C | 021-440 | A2 |
| 77 | 111 175 290 300 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-560 | A2 |
| 35 | 111 181 187 223 290 304 319 362 | 024-564 | 64 73 146 153 207 235 263 309+C 315+C | 021-320 | A2 |
| 64 | 111 182C 183C 189 223 290 319 362 | 024-549 | 64 73 146 153 235 263 309+CC 315+C | 021-320 | A2 |
| 67 | 111 182C 183C 189 223 290 319 362 | 024-560 | 64 73 146 153 235 263 315+C 523-524del | 021-600 | A2 |
| 48 | 111 183C 189 223 290 319 362 381 519 | 024-560 | 64 73 146 153 235 263 309+C 315+C | 021-310 | A2 |
| 46 | 111 187 209 223 290 319 362 371 519 | 024-560 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-550 | A2 |
| 40 | 111 187 223 234 290 319 362 390 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-570 | A2 |
| 54 | 111 187 223 290 319 362 | 024-589 | 64 73 146 153 235 263 309+C 315+C | 021-410 | A2 |
| 6 | 111 187 223 290 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-560 | A2 |
| 13 | 111 187 223 290 319 362 | 024-520 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-535 | A2 |
| 17 | 111 187 223 290 319 362 | 024-530 | 64 73 146 153 235 263 315+C 523-524del | 021-569 | A2 |
| 47 | 111 189 223 290 319 362 | 024-530 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-550 | A2 |
| 89 | 111 189 223 274 290 319 362 | 024-540 | 64 73 146 153 235 263 309+CC 315+C | 021-510 | A2 |
| 87 | 111 189 223 290 311 319 362 | 024-550 | 64 73 146 153 235 263 292 309+C 315+C 523-524del | 021-600 | A2 |
| 24 | 111 189 223 290 319 324 362 | 034-569 | 64 73 146 153 235 263 309+CC 315+C 523-524del | 021-535 | A2 |
| 32 | 111 189 223 290 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-535 | A2 |
| 57 | 111 189 223 290 319 362 | 024-560 | 73 146 152 153 235 263 309+CC 315+C | 021-320 | A2 |
| 75 | 111 209 223 290 291 319 362 477 | 024-569 | 64 73 146 152 153 235 263 315+C 523-524del | 021-600 | A2 |
| 74 | 111 209 223 290 293C 319 362 519 | 024-569 | 64 73 146 153 235 263 309+CC 315+C4 523-524del | 021-550 | A2 |
| 28 | 111 209 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 31 | 111 209 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-595 | A2 |
| 99 | 111 223 234 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 356+C 523-524del | 021-600 | A2 |
| 21 | 111 223 243 290 299 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 7 | 111 223 290 299 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-560 | A2 |
| 37 | 111 223 290 299 319 362 | 024-560 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-535 | A2 |
| 44 | 111 223 290 299 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 49 | 111 223 290 299 319 362 | 024-520 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-570 | A2 |
| 82 | 111 223 290 300 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 23 | 111 223 290 311 319 360 362 | 024-569 | 146 153 235 263 309+CC 315+C 523-524del | 021-539 | A2 |
| 26 | 111 223 290 311 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 4 | 111 223 290 311 319 362 | 024-569 | 64 73 146 153 235 263 315+C 523-524del | 021-600 | A2 |
| 3 | 111 223 290 319 360 362 | 024-569 | 143 146 152 153 204 235 263 309+CC 315+C 523-524del | 021-570 | A2 |
| 59 | 111 223 290 319 362 | 024-560 | 64 73 146 152 153 215 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 92 | 111 223 290 319 362 | 024-569 | 64 73 146 152 153 235 263 315+C | 021-320 | A2 |
| 38 | 111 223 290 319 362 | 024-560 | 64 73 146 153 235 263 309+C 315+C | 021-535 | A2 |
| 78 | 111 223 290 319 362 | 024-520 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 79 | 111 223 290 319 362 | 024-530 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 84 | 111 223 290 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-530 | A2 |
| 20 | 111 223 290 319 362 | 024-549 | 64 73 146 153 235 263 309+CC 315+C | 021-560 | A2 |
| 29 | 111 223 290 319 362 | 024-500 | 64 73 146 153 235 263 309+CC 315+C 523-524del | 021-570 | A2 |
| 36 | 111 223 290 319 362 | 024-560 | 64 73 153 214 235 263 315+C 523-524del | 021-600 | A2 |
| 63 | 111 223 290 319 362 | 024-525 | 73 146 150 153 235 263 315+C 523-524del | 021-560 | A2 |
| 68 | 111 223 290 319 362 | 024-550 | n.d | – | A2 |
| 42 | 111 223 290 319 362 391 | 024-569 | 64 73 146 153 235 263 309+CC 315+C 523-524del | 021-600 | A2 |
| 103 | 111 223 290 319 362 518 519 | 024-569 | 64 73 146 153 235 263 315+C 523-524del | 021-550 | A2 |
| 91 | 111 223 290 319 362 519 | 024-560 | 64 73 146 150 153 174+C 235 263 309+CC 315+C 523-524del | 021-320 | A2 |
| 53 | 111 223 290 319 362 519 | 024-569 | 64 73 146 150 153 235 263 315+C 523-524del | 021-600 | A2 |
| 34 | 111 223 290 319 362 519 | 024-569 | 64 73 146 150 153 235 263 315+C 523-524del | 021-600 | A2 |
| 76 | 111 223 290 319 362 519 | 024-550 | 64 73 146 152 153 235 263 309+C 315+C | 021-320 | A2 |
| 27 | 111 223 290 319 362 519 | 024-569 | 64 73 146 152 153 235 263 309+C 315+C 523-524del | 021-589 | A2 |
| 14 | 111 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 51 | 111 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 86 | 111 223 290 319 362 519 | 021-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 88 | 111 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 11 | 111 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 315+C 523-524del | 021-570 | A2 |
| 9 | 111 223 290 319 362 519 | 024-569 | 73 146 152 153 197 235 263 309+C 315+C 523-524del | 021-590 | A2 |
| 15 | 111 223 290 319 362 519 | 024-569 | 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 25 | 111 223 290 319 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-544 | A2 |
| 97 | 111 261 290 319 362 519 | 024-550 | 73 146 152 153 235 263 315+C 523-524del | 021-600 | A2 |
| 45 | 111 290 311 319 362 391 | 024-569 | 64 73 146 153 235 263 315+C 523-524del | 021-590 | A2 |
| 50 | 111 290 311 319 362 391 | 024-569 | 64 73 146 153 235 263 315+C 523-524del | 021-589 | A2 |
| 19 | 111 290 319 362 391 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 58 | 153 223 240 290 319 362 | 024-560 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-590 | A2 |
| 18 | 189 223 290 319 362 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-585 | A2 |
| 56 | 223 290 311 319 362 | 024-560 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-530 | A2 |
| 5 | 223 290 316 319 362 | 024-569 | 64 73 146 153 182 235 263 309+C 315+C 523-524del | 021-560 | A2 |
| 60 | 223 290 319 352 362 | 024-560 | 64 73 146 153 182 235 263 309+C 315+C 523-524del | 021-589 | A2 |
| 70 | 223 290 319 362 | 024-545 | 64 73 146 153 235 263 315+C 523-524del | 021-600 | A2 |
| 100 | 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-600 | A2 |
| 16 | 92 111 189 223 290 319 362 519 | 024-569 | 64 73 146 153 235 263 309+C 315+C 523-524del | 021-570 | A2 |
| 81 | 93 111 136 223 290 319 324 362 | 024-569 | 73 146 235 263 309+C 315+C 523-524del | 021-580 | A2 |
| 33 | 129 183C 189 217 269 519 | 024-569 | 73 146 152 195 234 263 309+CC 315+C 499 | 021-570 | B2 |
| 22 | 183C 189 217 519 | 024-569 | 73 263 309+C 315+C 499 | 021-570 | B2 |
| 41 | 183C 189 223 256 298 325 327 | 024-560 | 73 249del 263 290-291del 309+C 315+C 489 523-524del | 021-555 | C1 |
| 39 | 189 223 298 325 327 362 519 | 024-560 | 73 195 249del 263 290-291del 315+C 489 | 021-580 | C1 |
| 12 | 519 | 024-569 | 153 263 315+C 523-524del | 021-600 | H? |
| 52 | 129 148 168 172 187 188G 189 223 230 278 293 311 320 | 024-560 | 93 95C 185 189 236 247 263 315+C 523-524del | 021-560 | L0a1a |
| 30 | 126 271 294 296 304 519 | 024-569 | 73 263 315+C | 021-580 | T2 |
| 96 | 51 129C 189 362 | 024-540 | 73 152 217 263 309+C 315+C 340 508 | 021-600 | U2e |
HG = haplogroup.
n.d. = non determined.
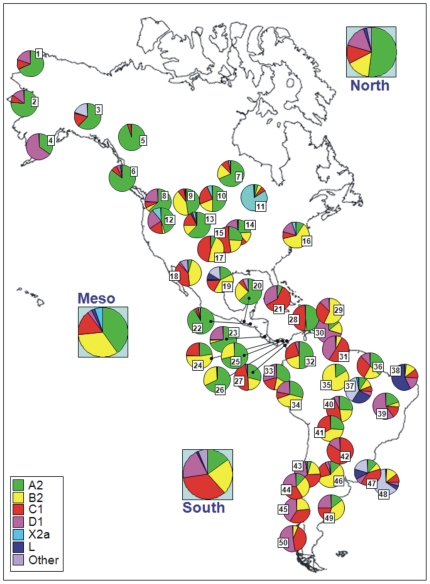
Figure 1 shows the frequency distribution of the main mtDNA American haplogroups in Native American populations. Although haplogroup A2 is at high frequencies in Meso America, El Salvador is particularly distinct from the other populations by its extremely high A2 haplogroup frequency. Note also that there exists substantial heterogeneity of haplogroup frequency patterns in America (even between neighbouring populations).
Figure 1. Haplogroup patterns in America.
Codes for populations are as follow: North America: 1 = Chukchy, 2 = Eskimos [24]; 3 = Inuit (collected from the HvrBase database [84]; 4 = Aleuts [22]; 5 = Athapaskan [23]; 6 = Haida [25]; 7 = Apache [85], 8 = Bella Coola [25]; 9 = Navajo [85]; 10 = Sioux, 11 = Chippewa [66], 12 = Nuu-Chah-Nult [26]; 13 = Cheyenne [66]; 14 = Muskogean populations [31]; 15 = Cheyenne-Arapaho [30]; 16 = Yakima [23]; 17 = Stillwell Cherokee [30]; Meso-America: 18 = Pima [27]; 19 = Mexico [40]; 20 = Quiche [38]; 21 = Cuba [38]; 22 = El Salvador (present study); 23 = Huetar [35]; 24 = Emberá [39]; 25 = Kuna [36]; 26 = Ngöbé [37]; 27 = Wounan [37]; South America: 28 = Guahibo [51]; 29 = Yanomamo from Venezuela [48]; 30 = Gaviao [42]; 31 = Yanomamo from Venezuela and Brazil [86]; 32 = Colombia [52]; 33 = Ecuador (general population), 34 = Cayapa [41]; 35 = Xavante [42]; 36 = North Brazil [43]; 37 = Brazil [64]; 38 = Curiau [60]; 39 = Zoró [42]; 40 = Ignaciano, 41 = Yuracare [53]; 42 = Ayoreo [44]; 43 = Araucarians [33]; 44 = Pehuenche, 45 = Mapuche from Chile [45]; 46 = Coyas [4]; 47 = Tacuarembó [49]; 48 = Uruguay [50]; 49 = Mapuches from Argentina [46]; 50 = Yaghan [59].
The phylogeny of A2 in El Salvador is clearly star-like (Figure 2); its root is, identified by the diagnostic sites C16111T–T16223C–C16290T–G16319A–T16362C in HVS-I, and C64T–A73G–T146C–A153G –A235G–A263G–315+C in HVS-II. There are no very solid diagnostic sites in the control region that would allow us to classify A2 sub-lineages from El Salvador [12], [14]. Moreover, several control-region variants regarded as haplogroup diagnostic, such as C64T and A153G, show reversions: complete genome sequence data confirm the existence of multiple back and parallel mutations within haplogroup A2 [12], [14]. Although many of them are well-known hotspots (e.g. T146C), others such as position 64, seem to behave as hotspots only within A2 (see e.g. [12]). Other Native American lineages, like D1, D4h3 and X2a [13] are absent from our sample from El Salvador.
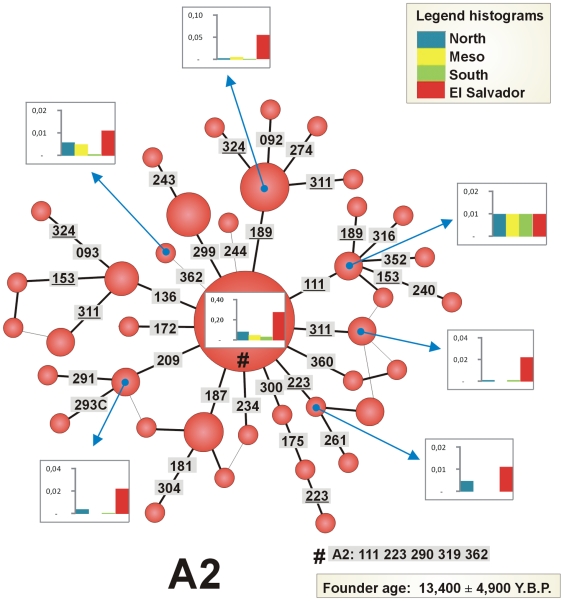
Figure 2. Phylogenetic network of haplogroup A2 mtDNA sequences from El Salvador.
Only the variation contained in the HVS-I segment (range 16090–16365) was used. Positions are referred to the rCRS minus 16000; a transversion is specifically indicated as a suffix. The areas of the circles are proportional to the number of individuals bearing the corresponding haplotype in the El Salvador sample. The histograms indicate the frequency of the El Salvadorian mtDNA founder clusters in other continental regions (North, Meso, and South America). To compute the TMRCA and founder ages, we resolved the network to a tree (shown by the bold connections between haplotypes), using the positional mutation rates reported in [21]. Resolved parallel mutations are shown underlined. The ancestral haplotype is indicated by a hash.
The sub-clade of A2 carrying C16360T is particularly prevalent in Meso America, especially in the Huetar (12 matches; ∼44% of the Huetar sample) from Costa Rica [35] and the Ngöbé (three matches; ∼7% of the Ngöbe sample) from Panama [37]; in El Salvador this variant was also present in two individuals. The haplotype C16111T–C16187T–T16223C–C16290T–G16319A–T16362C is virtually only shared with the Ngöbe (19 matches that make up ∼41% of the Ngöbe sample) but was also detected in one Uruguayan [50]. El Salvador shares a higher number of haplotypes with North America (19), followed by Meso-America (10), and then South America (8); note however that the database for Meso-America (n = 395) is of a much lower sample size than the one from North (n = 2,010) and South (n = 1,596). These results roughly indicate a clear imprint of North in Meso-America and also the existence of lineages that are mainly concentrated in Meso-America (probably due to the fact that these were founders in the region and experienced posterior expansion); in some instances, some of these South mtDNAs could have been carried from Meso-America in some wave of migration towards the South (such as the one indicated above observed in Uruguay, or e.g. C16111T–T16189C–T16223C–C16290T–16311–G16319A–T16362C, which was also found in three Brazilians [64]).
We found only four Native American mtDNAs not belonging to haplogroup A2: two haplogroup B2 and two haplogroup C1 mtDNAs. We did not find any exact match amongst published data for the B2 sequence #33 that carries the distinctive variant A16269G. The haplogroup C1 sequence #41 carries C16256T; this uncommon variant within haplogroup C1 has been also observed in the Yanomama from Venezuela [47] and the Zoró from Brazil [42]. Haplotype C1 #39 was only observed in one Brazilian [43] and one Guahibo from Venezuela [51], but also in two ancient Taino samples from the Caribbean [57].
Non-Native American haplotypes in El Salvador
Signals of a European contribution to our sample from El Salvador are limited to three haplotypes (see Table 1): haplotype #96 belongs to haplogroup U2e, with exact matches in several West European locations (e.g. Northwest Spain and Portugal [70]); in Madeira [71], etc. Haplotype #12 can be assigned most plausibly to haplogroup H, while #30 probably belongs to haplogroup T2; this sequence curiously matches published sequences only observed in Portugal and Brazil [64], [72] but also a single hit in Poland [73].
We detected only one sequence belonging to a typical sub-Saharan haplogroup in El Salvador. It belongs to L0a1a, a sub-clade highly prevalent in southeast Africa [67], [74], [75], where we find exact matches in HVS-I and HVS-II. Exact matches are also found in, for example, the Atlantic African southwest coast, in Cabinda [76], and in the Tongas [77]. Although it is not possible to determine with precision the African origin of this haplotype, southeast Africa (Mozambique) is probably the best candidate population source.
Admixture analysis
The admixture analysis carried out as in [67] indicated that North America accounts for ∼92% of the lineages in El Salvador, the remaining ∼8% coming from South America. The method described in [68] indicated that North America contributed to El Salvador ∼76% of the mtDNA lineages, in contrast to the ∼24% coming from South America.
Founder analysis
We inferred seven founders in our Salvadorian sample, all present in North American populations. Some sequence matches were not considered founders because they were detected only in Mexico and not in North American populations; they are more likely the result of recent gene flow between El Salvador and neighbouring populations. Some other potential founders were also rejected because they were present mostly as singletons analyzed in North American laboratories but belonging to e.g. ‘Hispanic’ or other sample populations without information about their ethnic affiliation. All the founders are indicated in Figure 1. The founder age of haplogroup A2 in El Salvador was estimated as 13,400±5,200 Y.B.P.
Discussion
El Salvador is the smallest Latin American republic and also the most densely populated. Although historically El Salvador has been home to a culturally diverse mix of peoples, including Native Americans, Africans, and west Europeans, by the 1980s the population of the country was essentially considered to be homogeneous in terms of ethnicity and basic cultural identity. Virtually all Salvadorans speak Spanish, the official language, as their mother tongue, and the vast majority are generally characterized as “mestizos” (or “ladinos”, a term more commonly used in Central America), popularly used to refer to those persons of mosaic geographic ancestry who follow a wide variety of indigenous and “hispanic” customs and habits that over the centuries have come to constitute Spanish-American cultural patterns. In the late 1980s, the ethnic composition of the population was estimated as 89% “mestizo”, 10% Native American, and 1% “white” [78]. Therefore, in contrast to most other Central American countries, El Salvador no longer possessed an ethnically or linguistically distinct Native American population, although persons of native-like ethnicity or cultural heritage still lived in the western parts of the country. Similarly, there was no ethnically or culturally distinct African-American population as there is in neighbouring populations [79]. However, there is a general belief that much of the Salvadorian population in the 1980s had a predominantly Native American ancestry[1].
The results of the present study have shown that, in contrast to the cultural patterns observed in the today's El Salvador population, most of the mtDNA profiles found are typically Native American; haplogroup A2 account for ∼90% of the Salvadorian sample. Correspondingly, the impact of Europeans on the mtDNA pool of El Salvador is very low (∼2%). It seems that the Spanish conquerors and more recent European demographic influences did not contribute significantly to the today's genetic composition of El Salvador in the maternal side. This contrasts with the European Y-chromosome contribution to the El Salvador gene pool. According to [80] about one half in metropolitan areas and two thirds in rural populations of El Salvador belong to non-Native American haplogroups; for instance, the most common Y-chromosome haplogroup in Europe (namely, R1b) is present in El Salvador at 24% in metropolitan areas and 43% in rural regions. Concomitantly, the Native American Y-chromosome proportion in El Salvador (represented by haplogroup Q3) is about 31–49%.
Therefore, the mtDNA and Y-chromosome variation in El Salvador displays an extreme version of a pattern that was also observed in other American populations [81], [82]: the indigenous female contribution is much higher than the indigenous male contribution.
Our results show that the impact of African-American lineages on the mtDNA pool of El Salvador was very low, as indicated by the presence of only one mtDNA of sub-Saharan origin in our sample. The scarcity of the sub-Saharan component strikingly contrasts with the situation on the Caribbean coast, where (as a consequence of the Atlantic slave trade) it is clearly predominant [67], [74], [75], [79]. The Y-chromosome variation shows a similar pattern: no lineages of African ancestry have been detected in El Salvador [80].
There are no clear signals of recent genetic drift events in the general population from El Salvador, as observed in, for instance, neighbouring but isolated Native American populations such as the Ngöbé from Panamá [37] which shows extremely reduced levels of mtDNA diversity (reflecting passage through post-conquest population bottlenecks). Haplogroup A2 is at high frequency in El Salvador (∼90% of the sample) and a high percent of the lineages (45%; computed using the sequence range 16090–16365) remain unobserved in other American populations. Admixture analysis indicates that the main mtDNA influence in El Salvador can be attributed to North America. The phylogeny of A2 is rather star-like and the founder age was 12,600±4,900 years. The shape of this phylogeny points to the existence of a prehistoric demographic expansion. Considering the most recently estimated age of A2 in the American continent as a whole of 13,400±5,200 [21] (largely determined from North American samples) as a proxy for the time of the expansion into the Americas, it can be tentatively suggested that the initial settlement of El Salvador occurred rather soon after the initial colonization of the American continent, and that El Salvador largely contains the descendants of the mtDNAs in that original pool with scarce subsequent demographic influence from other American or non-American populations. Indeed, since we have genotyped samples collected in urban areas we would expect to have an even higher prevalence of the Native American component in more isolated groups from the country, as is in fact observed on the Y-chromosome side where the Native American component is higher in rural than in metropolitan areas [80].
In contrast to the high impact of the Atlantic slave trade on the Central American Caribbean coast [79], the Pacific side (at least for El Salvador) appears to have preserved its Native American mtDNA heritage intact to the present day. At the same time, this study has also shown that El Salvador harbours haplogroup frequency patterns quite different from other modern Native American communities. At the individual haplotype level, El Salvador shows numerous mtDNAs that have never been observed in other American regions, even within Central America. These features provide little support to those that assume (or claim) that “Hispanics” or Native American communities are sufficiently homogeneous to justify the portability of forensic databases from one country to another (e.g. SWGDAM; [34]); see [83] for a discussion.
Acknowledgments
We would like to thank Vilma de Aguilar (Ministerio de Salud Pública de El Salvador) for helping with the sample collection.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was partially supported by grants from the Xunta de Galicia (Grupos Emerxentes; 2008/037), Ministerio de Ciencia e Innovacion (SAF2008-02971), and Fundacion de Investigacion Medica Mutua Madrilena (2008/CL444) given to AS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ministerio de Educación ES. Historia de El Salvador 1994.
- 2.Lardé y Larín J. 2000. El Salvador: descubrimiento, conquista y colonización: Dirección de Publicaciones e Impresos, El Salvador.
- 3.Álvarez-Iglesias V, Salas A, Cerezo M, Ramos-Luis E, Jaime JC, et al. Genotyping coding region mtDNA SNPs for Asian and Native American haplogroup assignation. Int Congress Series. 2006;11:4–6. [Google Scholar]
- 4.Álvarez-Iglesias V, Jaime JC, Carracedo Á, Salas A. Coding region mitochondrial DNA SNPs: targeting East Asian and Native American haplogroups. Forensic Sci Int: Genet. 2007;1:44–55. doi: 10.1016/j.fsigen.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 6.Bandelt H-J, Salas A, Bravi CM. Problems in FBI mtDNA database. Science. 2004;305:1402–1404. doi: 10.1126/science.305.5689.1402b. [DOI] [PubMed] [Google Scholar]
- 7.Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salas A, Carracedo Á, Macaulay V, Richards M, Bandelt H-J. A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem Biophys Res Commun. 2005;335:891–899. doi: 10.1016/j.bbrc.2005.07.161. [DOI] [PubMed] [Google Scholar]
- 9.Salas A, Yao Y-G, Macaulay V, Vega A, Carracedo Á, et al. A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005;2:e296. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Y-G, Salas A, Bravi CM, Bandelt H-J. A reappraisal of complete mtDNA variation in East Asian families with hearing impairment. Hum Genet. 2006;119:505–515. doi: 10.1007/s00439-006-0154-9. [DOI] [PubMed] [Google Scholar]
- 11.Bandelt HJ, Salas A. Contamination and sample mix-up can best explain some patterns of mtDNA instabilities in buccal cells and oral squamous cell carcinoma. BMC Cancer. 2009;16:113. doi: 10.1186/1471-2407-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Achilli A, Perego UA, Bravi CM, Coble MD, Kong QP, et al. The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies. PLoS ONE. 2008;3:e1764. doi: 10.1371/journal.pone.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, et al. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 14.Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, et al. Beringian standstill and spread of Native American founders. PLoS ONE. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 16.Nei N. New York: Columbi University Press; 1987. Molecular evolutionary genetics: [Google Scholar]
- 17.Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 18.Bandelt H-J, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forster P, Harding R, Torroni A, Bandelt H-J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 20.Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soares P, Ermini L, Thomson N, Mormina M, Rito T, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet in press. 2009. [DOI] [PMC free article] [PubMed]
- 22.Rubicz R, Schurr TG, Babb PL, Crawford MH. Mitochondrial DNA variation and the origins of the Aleuts. Hum Biol. 2003;75:809–835. doi: 10.1353/hub.2004.0009. [DOI] [PubMed] [Google Scholar]
- 23.Shields GF, Schmiechen AM, Frazier BL, Redd A, Voevoda MI, et al. mtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am J Hum Genet. 1993;53:549–562. [PMC free article] [PubMed] [Google Scholar]
- 24.Starikovskaya YB, Sukernik RI, Schurr TG, Kogelnik AM, Wallace DC. mtDNA diversity in Chukchi and Siberian Eskimos: implications for the genetic history of ancient Beringia and the peopling of the New World. Am J Hum Genet. 1998;63:1473–1491. doi: 10.1086/302087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward RH, Redd A, Valencia D, Frazier B, Pääbo S. Genetic and linguistic differentiation in the Americas. Proc Natl Acad Sci U S A. 1993;90:10663–10667. doi: 10.1073/pnas.90.22.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward RH, Frazier BL, Dew-Jager K, Pääbo S. Extensive mitochondrial diversity within a single Amerindian tribe. Proc Natl Acad Sci U S A. 1991;88:8720–8724. doi: 10.1073/pnas.88.19.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kittles RA, Bergen AW, Urbanek M, Virkkunen M, Linnoila M, et al. Autosomal, mitochondrial, and Y chromosome DNA variation in Finland: evidence for a male-specific bottleneck. Am J Phys Anthropol. 1999;108:381–399. doi: 10.1002/(SICI)1096-8644(199904)108:4<381::AID-AJPA1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, et al. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- 29.Torroni A, Sukernik RI, Schurr TG, Starikovskaya YB, Cabell MF, et al. mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet. 1993;53:591–608. [PMC free article] [PubMed] [Google Scholar]
- 30.Malhi RS, Schultz BA, Smith DG. Distribution of mitochondrial DNA lineages among Native American tribes of Northeastern North America. Hum Biol. 2001;73:17–55. doi: 10.1353/hub.2001.0008. [DOI] [PubMed] [Google Scholar]
- 31.Bolnick DA, Smith DG. Unexpected patterns of mitochondrial DNA variation among Native Americans from the southeastern United States. Am J Phys Anthropol. 2003;122:336–354. doi: 10.1002/ajpa.10284. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz JG, Smith DG. Distribution of the 9-bp mitochondrial DNA region V deletion among North American Indians. Hum Biol. 1994;66:777–788. [PubMed] [Google Scholar]
- 33.Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, et al. Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol. 1993;10:23–47. doi: 10.1093/oxfordjournals.molbev.a039987. [DOI] [PubMed] [Google Scholar]
- 34.Monson KL, Miller KWP, Wilson MR, DiZinno JA, Budowle B. The mtDNA Population Database: an integrated software and database resource for forensic comparison. Forensic Sci Commun. 2002;4:no 2. [Google Scholar]
- 35.Santos M, Ward RH, Barrantes R. mtDNA variation in the Chibcha Amerindian Huetar from Costa Rica. Hum Biol. 1994;66:963–977. [PubMed] [Google Scholar]
- 36.Batista O, Kolman CJ, Bermingham E. Mitochondrial DNA diversity in the Kuna Amerinds of Panama. Hum Mol Genet. 1995;4:921–929. doi: 10.1093/hmg/4.5.921. [DOI] [PubMed] [Google Scholar]
- 37.Kolman CJ, Bermingham E, Cooke R, Ward RH, Arias TD, et al. Reduced mtDNA diversity in the Ngöbé Amerinds of Panamá. Genetics. 1995;140:275–283. doi: 10.1093/genetics/140.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boles TC, Snow CC, Stover E. Forensic DNA testing on skeletal remains from mass graves: a pilot project in Guatemala. J Forensic Sci. 1995;40:349–355. [PubMed] [Google Scholar]
- 39.Kolman CJ, Bermingham E. Mitochondrial and nuclear DNA diversity in the Choco and Chibcha Amerinds of Panama. Genetics. 1997;147:1289–1302. doi: 10.1093/genetics/147.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green LD, Derr JN, Knight A. mtDNA affinities of the peoples of North-Central Mexico. Am J Hum Genet. 2000;66:989–998. doi: 10.1086/302801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickards O, Martinez-Labarga C, Lum JK, De Stefano GF, Cann RL. mtDNA history of the Cayapa Amerinds of Ecuador: detection of additional founding lineages for the Native American populations. Am J Hum Genet. 1999;65:519–530. doi: 10.1086/302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward RH, Salzano FM, Bonatto SL, Hutz MH, Coimbra CEA, et al. Mitochondrial DNA polymorphism in 3 Brazilian Indian tribes. Am J Hum Biol. 1996;8:317–323. doi: 10.1002/(SICI)1520-6300(1996)8:3<317::AID-AJHB2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 43.Santos SE, Ribeiro-Dos-Santos AK, Meyer D, Zago MA. Multiple founder haplotypes of mitochondrial DNA in Amerindians revealed by RFLP and sequencing. Ann Hum Genet. 1996;60 (Pt 4):305–319. doi: 10.1111/j.1469-1809.1996.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 44.Dornelles CL, Battilana J, Fagundes NJ, Freitas LB, Bonatto SL, et al. Mitochondrial DNA and Alu insertions in a genetically peculiar population: the Ayoreo Indians of Bolivia and Paraguay. Am J Hum Biol. 2004;16:479–488. doi: 10.1002/ajhb.20038. [DOI] [PubMed] [Google Scholar]
- 45.Moraga ML, Rocco P, Miquel JF, Nervi F, Llop E, et al. Mitochondrial DNA polymorphisms in Chilean aboriginal populations: implications for the peopling of the southern cone of the continent. Am J Phys Anthropol. 2000;113:19–29. doi: 10.1002/1096-8644(200009)113:1<19::AID-AJPA3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 46.Ginther C, Corach D, Penacino GA, Rey JA, Carnese FR, et al. Genetic variation among the Mapuche Indians from the Patagonian region of Argentina: mitochondrial DNA sequence variation and allele frequencies of several nuclear genes. Exs. 1993;67:211–219. doi: 10.1007/978-3-0348-8583-6_17. [DOI] [PubMed] [Google Scholar]
- 47.Merriwether DA, Kemp BM, Crews DE, Neel JV. Gene flow and genetic variation in the Yanomama as revealed by mitochondrial DNA. In: Renfrew C, editor. America Past, America Present: Genes and Languages in the Americas and Beyond. Cambridge: McDonald Institute for Archaeological Research; 2000. pp. 89–124. [Google Scholar]
- 48.Williams SR, Chagnon NA, Spielman RS. Nuclear and mitochondrial genetic variation in the Yanomamo: a test case for ancient DNA studies of prehistoric populations. Am J Phys Anthropol. 2002;117:246–259. doi: 10.1002/ajpa.10035. [DOI] [PubMed] [Google Scholar]
- 49.Bonilla C, Bertoni B, Gonzalez S, Cardoso H, Brum-Zorrilla N, et al. Substantial Native American female contribution to the population of Tacuarembo, Uruguay, reveals past episodes of sex-biased gene flow. Am J Hum Biol. 2004;16:289–297. doi: 10.1002/ajhb.20025. [DOI] [PubMed] [Google Scholar]
- 50.Pagano S, Sans M, Pimenoff V, Cantera AM, Alvarez JC, et al. Assessment of HV1 and HV2 mtDNA variation for forensic purposes in an Uruguayan population sample. J Forensic Sci. 2005;50:1239–1242. [PubMed] [Google Scholar]
- 51.Vona G, Falchi A, Moral P, Calo CM, Varesi L. Mitochondrial sequence variation in the Guahibo Amerindian population from Venezuela. Am J Phys Anthropol. 2005;127:361–369. doi: 10.1002/ajpa.20070. [DOI] [PubMed] [Google Scholar]
- 52.Torres MM, Bravi CM, Bortolini MC, Duque C, Callegari-Jacques S, et al. A revertant of the major founder Native American haplogroup C common in populations from northern South America. Am J Hum Biol. 2006;18:59–65. doi: 10.1002/ajhb.20461. [DOI] [PubMed] [Google Scholar]
- 53.Bert F, Corella A, Gene M, Perez-Perez A, Turbon D. Mitochondrial DNA diversity in the Llanos de Moxos: Moxo, Movima and Yuracare Amerindian populations from Bolivia lowlands. Ann Hum Biol. 2004;31:9–28. doi: 10.1080/03014460310001616464. [DOI] [PubMed] [Google Scholar]
- 54.Vernesi C, Fuselli S, Castri L, Bertorelle G, Barbujani G. Mitochondrial diversity in linguistic isolates of the Alps: a reappraisal. Hum Biol. 2002;74:725–730. doi: 10.1353/hub.2002.0060. [DOI] [PubMed] [Google Scholar]
- 55.Monsalve MV, Stone AC, Lewis CM, Rempel A, Richards M, et al. Brief communication: molecular analysis of the Kwaday Dan Ts'finchi ancient remains found in a glacier in Canada. Am J Phys Anthropol. 2002;119:288–291. doi: 10.1002/ajpa.10116. [DOI] [PubMed] [Google Scholar]
- 56.Stone AC, Stoneking M. Ancient DNA from a pre-Columbian Amerindian population. Am J Phys Anthropol. 1993;92:463–471. doi: 10.1002/ajpa.1330920405. [DOI] [PubMed] [Google Scholar]
- 57.Lalueza-Fox C, Calderon FL, Calafell F, Morera B, Bertranpetit J. MtDNA from extinct Tainos and the peopling of the Caribbean. Ann Hum Genet. 2001;65:137–151. doi: 10.1017/S0003480001008533. [DOI] [PubMed] [Google Scholar]
- 58.Lalueza-Fox C, Gilbert MT, Martinez-Fuentes AJ, Calafell F, Bertranpetit J. Mitochondrial DNA from pre-Columbian Ciboneys from Cuba and the prehistoric colonization of the Caribbean. Am J Phys Anthropol. 2003;121:97–108. doi: 10.1002/ajpa.10236. [DOI] [PubMed] [Google Scholar]
- 59.Moraga M, Santoro CM, Standen VG, Carvallo P, Rothhammer F. Microevolution in prehistoric Andean populations: chronologic mtDNA variation in the desert valleys of northern Chile. Am J Phys Anthropol. 2005;127:170–181. doi: 10.1002/ajpa.10438. [DOI] [PubMed] [Google Scholar]
- 60.Ribeiro-Dos-Santos AK, Santos SE, Machado AL, Guapindaia V, Zago MA. Heterogeneity of mitochondrial DNA haplotypes in Pre-Columbian Natives of the Amazon region. Am J Phys Anthropol. 1996;101:29–37. doi: 10.1002/(SICI)1096-8644(199609)101:1<29::AID-AJPA3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 61.García-Bour J, Pérez-Pérez A, Álvarez S, Fernández E, López-Parra AM, et al. Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: ancient mtDNA sequences and Y-chromosome STR characterization. Am J Phys Anthropol. 2004;123:361–370. doi: 10.1002/ajpa.10337. [DOI] [PubMed] [Google Scholar]
- 62.Brown MD, Hosseini SH, Torroni A, Bandelt H-J, Allen JC, et al. mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am J Hum Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dornelles CL, Bonatto SL, De Freitas LB, Salzano FM. Is haplogroup X present in extant South American Indians? Am J Phys Anthropol. 2005;127:439–448. doi: 10.1002/ajpa.20103. [DOI] [PubMed] [Google Scholar]
- 64.Alves-Silva J, da Silva Santos M, Guimaraes PE, Ferreira AC, Bandelt H-J, et al. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ribeiro-Dos-Santos AK, Carvalho BM, Feio-Dos-Santos AC, Santos SE. Nucleotide variability of HV-I in Afro-descendents populations of the Brazilian Amazon Region. Forensic Sci Int. 2006 doi: 10.1016/j.forsciint.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 66.Smith DG, Malhi RS, Eshleman J, Lorenz JG, Kaestle FA. Distribution of mtDNA haplogroup X among Native North Americans. Am J Phys Anthropol. 1999;110:271–284. doi: 10.1002/(SICI)1096-8644(199911)110:3<271::AID-AJPA2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 67.Salas A, Richards M, Lareu MV, Scozzari R, Coppa A, et al. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet. 2004;74:454–465. doi: 10.1086/382194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mendizabal I, Sandoval K, Berniell-Lee G, Calafell F, Salas A, et al. Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evol Biol. 2008;8:213. doi: 10.1186/1471-2148-8-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 70.González AM, Brehm A, Pérez JA, Maca-Meyer N, Flores C, et al. Mitochondrial DNA affinities at the Atlantic fringe of Europe. Am J Phys Anthropol. 2003;120:391–404. doi: 10.1002/ajpa.10168. [DOI] [PubMed] [Google Scholar]
- 71.Brehm A, Pereira L, Kivisild T, Amorim A. Mitochondrial portraits of the Madeira and Acores archipelagos witness different genetic pools of its settlers. Hum Genet. 2003;114:77–86. doi: 10.1007/s00439-003-1024-3. [DOI] [PubMed] [Google Scholar]
- 72.Pereira L, Prata MJ, Amorim A. Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation. Ann Hum Genet. 2000;64:491–506. doi: 10.1046/j.1469-1809.2000.6460491.x. [DOI] [PubMed] [Google Scholar]
- 73.Malyarchuk BA, Grzybowski T, Derenko MV, Czarny J, Wozniak M, et al. Mitochondrial DNA variability in Poles and Russians. Ann Hum Genet. 2002;66:261–283. doi: 10.1017/S0003480002001161. [DOI] [PubMed] [Google Scholar]
- 74.Salas A, Richards M, De la Fé T, Lareu MV, Sobrino B, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salas A, Carracedo Á, Richards M, Macaulay V. Charting the Ancestry of African Americans. Am J Hum Genet. 2005;77:676–680. doi: 10.1086/491675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beleza S, Gusmão L, Amorim A, Carracedo Á, Salas A. The genetic legacy of western Bantu migrations. Hum Genet. 2005;117:366–375. doi: 10.1007/s00439-005-1290-3. [DOI] [PubMed] [Google Scholar]
- 77.Trovoada MJ, Pereira L, Gusmao L, Abade A, Amorim A, et al. Pattern of mtDNA variation in three populations from Sao Tome e Principe. Ann Hum Genet. 2004;68:40–54. doi: 10.1046/j.1529-8817.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- 78.CONCULTURA. Biblioteca de Historia Salvadoreña, El Salvador, Historia de sus pueblos, villas y ciudades. Consejo Nacional para la Cultura y el Arte 2000 [Google Scholar]
- 79.Salas A, Richards M, Lareu MV, Sobrino B, Silva S, et al. Shipwrecks and founder effects: Divergent demographic histories reflected in Caribbean mtDNA. Am J Phys Anthropol. 2005;128:855–860. doi: 10.1002/ajpa.20117. [DOI] [PubMed] [Google Scholar]
- 80.Lovo-Gómez J, Blanco-Verea A, Lareu MV, Brion M, Carracedo Á. The genetic male legacy from El Salvador. Forensic Sci Int. 2007;171:198–203. doi: 10.1016/j.forsciint.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 81.Dipierri JE, Alfaro E, Martínez-Marignac VL, Bailliet G, Bravi CM, et al. Paternal directional mating in two Amerindian subpopulations located at different altitudes in northwestern Argentina. Hum Biol. 1998;70:1001–1010. [PubMed] [Google Scholar]
- 82.Salas A, Jaime JC, Álvarez-Iglesias V, Carracedo Á. Gender bias in the multi-ethnic genetic composition of Central Argentina. J Hum Genet. 2008;53:662–674. doi: 10.1007/s10038-008-0297-8. [DOI] [PubMed] [Google Scholar]
- 83.Salas A, Bandelt H-J, Macaulay V, Richards MB. Phylogeographic investigations: The role of trees in forensic genetics. Forensic Sci Int. 2007;168:1–13. doi: 10.1016/j.forsciint.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 84.Burckhardt F, von Haeseler A, Meyer S. HvrBase: compilation of mtDNA control region sequences from primates. Nucleic Acids Res. 1999;27:138–142. doi: 10.1093/nar/27.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Budowle B, Allard MW, Fisher CL, Isenberg AR, Monson KL, et al. HVI and HVII mitochondrial DNA data in Apaches and Navajos. Int J Legal Med. 2002;116:212–215. doi: 10.1007/s00414-001-0283-6. [DOI] [PubMed] [Google Scholar]
- 86.Merriwether DA, Kemp BM, Crews DE, Neel JV. Cambridge: McDonald Institute for Archaeological Research; 2002. Gene flow and genetic variation in the Yanomama as revealed by mitochondrial DNA. pp. 89–124. [Google Scholar]