Abstract
Background
Cardiac hypertrophy is classically regarded as a compensatory response, yet the active tissue remodeling processes triggered by various types of mechanical stress can enhance or diminish the function of the heart. Despite the disparity in outcomes, there are similarities in the hypertrophic responses. We hypothesized that a generic genetic response that is not dependent upon the particular nature of the hypertrophic stimulus exists. To test our hypothesis, we compared the temporal evolution of transcriptomes measured in hearts subjected to either adaptive (exercise-induced) or maladaptive (aortic banding-induced) hypertrophy.
Methods and Results
Generic hypertrophy-associated genes were identified and distinguished from stimulus-dependent transcripts by coupling a metric of cardiac growth with a dynamic time warping algorithm to align transcriptome changes with respect to the hypertrophy response. The major differences in expression between the adaptive and maladaptive hypertrophy models were centered around genes involved in metabolism, fibrosis, and immune response. Conversely, transcripts with common expression patterns in both hypertrophy models were associated with signal transduction, cytoskeletal development and muscle contraction. Thus, despite the apparent differences in the expression response of the heart to either athletic conditioning or pressure overload, there is a set of genes that display similar expression profiles.
Conclusions
This finding lends support to the notion of a generalized cardiac growth mechanism that is activated in response to mechanical perturbation. The common and unique genetic signatures of adaptive and maladaptive hypertrophy may be useful in the diagnosis and treatment of pathological myocardial remodeling.
Keywords: heart, hypertrophy, gene expression, self-organizing map, GEDI, time warp
Introduction
Many risk factors associated with cardiovascular disease, such as high blood pressure, diabetes and obesity, increase the workload on the heart and can lead to gross and microscale morphological changes associated with a decrease in overall cardiac performance.1 Athletic conditioning also intensifies demand on the heart, but cardiac function is usually sustained or enhanced by this form of hypertrophic growth.2 At the micro-anatomical level, adaptive and maladaptive cardiac hypertrophy elicits distinct morphological changes in the cardiac tissue. Maladaptive cardiac remodeling is characterized by alterations in myocyte size, rarefaction in the myocardial vasculature, expression of fetal genes, and apoptotic loss of myocytes that leads to fibrosis.3 In contrast, adaptive hypertrophy entails sustained or increased capillary density in the myocardium, and an enhanced metabolic profile with no fetal gene expression and no sign of fibrosis.4 Nevertheless, the relative magnitude of overall myocardial growth observed in adaptive and maladaptive remodeling is remarkably similar.3 This suggests that at its core, cardiac growth may be driven in part by a generic mechanism, regardless of the nature of the hypertrophy stimulus.
Genome-scale analyses have provided “snapshots” of differential gene expression between normal and hypertrophied myocardium.5,6 In a detailed study, Izumo and colleagues looked at gene expression in mice subjected to either swim training as a model for exercise or transverse constriction of the aorta (“banding”) as a model for hemodynamic stress due to pressure overload.7 Mice subjected to regular bouts of swim exercise of increasing intensity over a period of four weeks experienced an increase in heart mass of up to 45%. Resting heart rate in these mice decreased with respect to non-exercised mice and no apoptosis or fibrosis was observed in the myocardium.7 Thus, changes in gene expression in these conditioned hearts were considered an “adaptive” hypertrophic response. In the same study, transverse aortic constriction induced pressure overload in the left ventricle that resulted in a subset of mice progressing to heart failure. Therefore, the changes in gene expression in this model represent a “maladaptive” hypertrophy response.
We hypothesized that a comparison of the respective gene expression patterns would differentiate genes whose expression was associated with a decrease in cardiac function from those associated with enhancement of cardiac performance, as well as identify non-specific transcriptional activity intrinsic to cardiac remodeling. The comparative dynamics of such an endeavor is complicated by the differences in the time courses of the different hypertrophy processes, where it is inappropriate to simply match up expression values collected at equal measurement intervals.8 However, a commonly varying feature between the two response profiles may serve as a reference frame for comparisons despite the differing time scales. Therefore, instead of the chronological time elapsed after stress application, we used the change in relative cardiac mass to register the genomic profiles of adaptive and maladaptive hypertrophy. This was achieved by employing dynamic time warping, an algorithm commonly used in studies of speech pattern recognition.9 In fact, the expression patterns of many genes activated in response to pressure overload hypertrophy were recently shown to exhibit a strong correlation to gross parameters of cardiac growth.10 Thus, we compared the two hypertrophy gene expression time courses based on heart weight to body weight ratio (HW/BW) as a surrogate time line. Despite the differences in the transcriptomes of these two hypertrophy responses, we uncovered several transcripts associated with cytoskeletal development and regulation of muscle contraction that displayed similar expression profiles when the profile data were compared relative to cardiac mass. In contrast, several fibrosis and immune response-related transcripts demonstrated significant changes in expression level only in the pressure overload model. Expression levels of transcripts associated with metabolism were significantly changed in the swim training model representing the adaptive response, but not in the pressure overload model representing the maladaptive response.
Materials and Methods
Selection of Hypertrophy Model Datasets
A query of the Gene Expression Omnibus (GEO) revealed a pair of time course microarray gene expression datasets published by the CardioGenomics Program for Genomic Applications by Izumo and colleagues7 collected on mice subjected to swim training (GEO series GSE77) and pressure overload induced by transverse aortic constriction (GEO series GSE76) suitable for this study. In their study, FVB wild-type mice were subjected to a modified version of a previously described swim training protocol11 to stimulate cardiac hypertrophy with enhanced cardiac function and pressure overload was induced using a published transverse aortic constriction surgical procedure12 to represent cardiac hypertrophy associated with diminished cardiac function. Sedentary mice served as a control for exercise-induced hypertrophy. Mice that underwent a sham operation without aortic constriction served as the control condition for the pressure overload-induced hypertrophy model. Affymetrix mgu74A GeneChips® were used to measure gene expression levels in ventricular tissues isolated at ten minutes, two days, one week, two weeks, three weeks, and four weeks post-training in the exercise-induced hypertrophy model, and one hour, four hours, one day, two days, one week, and eight weeks post-surgery in the pressure overload-induced hypertrophy model. Gene expression measurements were collected in triplicate at each time point sampled in both models. To analyze this data, we downloaded normalized signal values and probe set “detection calls” computed by Affymetrix Microarray Suite (MAS) 5.0 software, as well as heart weight (HW) and body weight (BW) values made by the investigators and available from the CardioGenomics website.7
Pre-Processing of Gene Expression Data
Pre-processing of the data requires for both hypertrophy models to be evaluated relative to the sham and sedentary control conditions. Detection calls calculated by the Affymetrix MAS 5.0 software at the default cutoff of p < 0.05 were used to eliminate transcripts that were judged not significantly expressed at all time points in either of the time series being compared. Specifically, detection call scores from the three replicate arrays for each time point were evaluated and probe sets whose detection call was “marginal” or “absent” in two of the three replicate measurements at all time points were excluded from analysis. This resulted in a dataset with 5,153 transcripts. The MAS 5.0 signal values for these genes were averaged, log2-transformed, and analyzed with Significance Analysis of Microarrays (SAM) 3.00 software13 to identify the sets of these genes that demonstrated statistically different expression values between the band and sham (control) conditions in the banding model and between the exercise and sedentary conditions in the exercise model. The two-class, paired response format was used for the SAM analysis. The delta value for determining the false discovery rate cut-off was set at 0.379 for the exercise-induced hypertrophy model, resulting in a dataset of 511 genes with a false discovery rate (FDR) of 0.96%. For the banding-induced hypertrophy model, the delta value was set at 0.119, yielding 544 significant genes with an FDR of 31.63%. The delta values were chosen to create symmetrical data sets for the Gene Warp program, where the symmetry is defined by the same set of genes for both sets of experiments. In further analyses, the 511 significant genes from the exercise model and the 544 significant genes from the banding model were combined into a dataset consisting of 963 genes that were present in the SAM results for both hypertrophy models. The remaining 92 transcripts consisted of genes that were present in the SAM results for the maladaptive hypertrophy model, but not the adaptive model, and vice versa.
Analysis of Cardiac Growth
Post-mortem heart weight and body weight values for the set of mice used at each measurement interval in the time series were averaged and used to compute the HW/BW ratio representative of each time point. Linear regression analysis was used to compare the HW/BW ratios calculated for the adaptive and maladaptive time courses.
Alignment of Time Course Expression Data
Log2-transformed signal ratios (SLR) were calculated for the transcripts whose expression levels were significantly changed in response to the adaptive and maladaptive hypertrophy stimuli. The time course expression data for the adaptive and maladaptive models were then aligned based on the similarity of the HW/BW associated with each time point. Finally, the aligned expression data were input into genewarp, a program implementing the classic dynamic time warping algorithm modified for analyzing global gene expression9, to assess the alignment. The quality of the alignment was determined by measuring the weighted Euclidean distance D(a,b) along the warping path between the global expression profiles of the two models. For comparison, the hypertrophy expression data was also evaluated with the genewarp program with the expression data aligned with respect to the order of the measurement intervals rather than HW/BW. All analyses were conducted with the program's parameters set to their default values.
Global Analysis of Differences in Gene Expression
The adaptive and maladaptive models were evaluated relative to each other to identify their differences. We used SAM to identify the set of transcripts among the 963 transcripts that were found to be significantly expressed in both the exercise and banding hypertrophy models whose expression significantly differed between the exercise and banding hypertrophy models. For this analysis, the SLR values from the exercise and banding datasets were input into SAM, and the two-class, unpaired response format was used. The delta threshold was set at 0.424, yielding an FDR of 5.28%. This analysis resulted in a list of 314 genes with significantly different expression between the exercise and banding models. The expression values for these differentially expressed genes were then subjected to cluster analysis and visualization using Gene Expression Dynamics Inspector (GEDI) 2.1 software.14 SLR values for these transcripts aligned based on change in HW/BW were input into GEDI. Grid size was set to nine by eight and all other parameters were set to their default values.
Global Analysis of Commonalities in Gene Expression
The adaptive and maladaptive models were evaluated relative to each other to identify their commonalities. Transcripts with similar expression profiles in the adaptive and maladaptive hypertrophy models were identified with a novel analysis program designated Statistical Investigator of Transcriptional Homology (SITH). This software utilizes a modified form of the dynamic time warping algorithm used in the global analysis to identify individual genes within a pair of genes in the expression datasets that exhibit comparable temporal expression patterns. A score (di) for gene i is computed by summing the Euclidean distances calculated for the expression measures taken at each pair of aligned time points multiplied by a time weight that adjusts for the length of the path through the distance table. This distance score indicates how closely the temporal expression profiles of gene i in the two experiments match. A cutoff value can then be set to limit the range of distance scores that represent similarly expressed transcripts. The distance score di was set to 0.45 in this study, meaning those transcripts that received distance scores less than or equal to 0.45 were considered to have matching temporal expression profiles. Transcripts determined to have matching expression profiles were visualized using Cluster and Treeview software.15 Functional classification and enrichment analysis of the set of commonly expressed genes was conducted using the web-based DAVID bioinformatics resources.16 Enrichment was assessed using a “heuristic fuzzy multiple linkage” algorithm that assigns enrichment scores to functional groups based on the geometric mean of a modified Fisher Exact statistical test.16
Statistics
Data are presented as mean±SEM. Pearson product moment correlation was used to test for similarity between the HW/BW values observed over time in the adaptive and maladaptive models. Significant differences were revealed to exist at p < 0.05.
Results
Exercise and Banding Stimuli Elicit a Similar Magnitude of Myocardial Growth
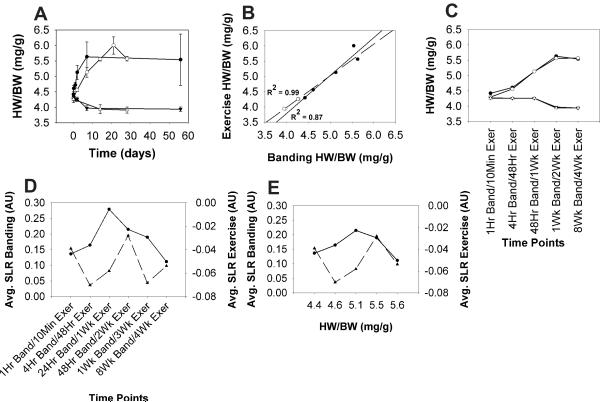
To achieve time warping based on relative progression of hypertrophy, we first plotted the average HW/BW for each time point in the exercise and aortic banding hypertrophy data sets to compare the changes in cardiac mass over time (Fig. 1A). While the rate of change in cardiac mass differed between the two hypertrophy models, the magnitude of overall growth appeared to be similar. This was compared quantitatively with linear correlation analysis of the change in HW/BW observed in the adaptive (exercise) and maladaptive (aortic banding) models (Fig. 1B), which showed a strong correlation between the control (r2=0.97, p<0.01) and hypertrophy (r2=0.87, p<0.01) conditions. This analysis indicated that the exercise training and aortic banding stimuli elicited an analogous increase in cardiac mass albeit with disparate characteristic time scales. Closer inspection of the temporal evolution of HW/BW in response to the hypertrophy stimuli identified the time points in the adaptive and maladaptive hypertrophy models with nearly identical HW/BW values (Fig. 1C). Since alignment of these time points via HW/BW could be achieved without violating their temporal order, it was postulated that this metric of cardiac growth might serve as a common feature to facilitate comparison between the time series gene expression data of the adaptive and maladaptive hypertrophy models. In other words, the gene expression values could be mapped from the time space into the cardiac growth (HW/BW) domain, or mass space, as a new reference system for comparing the hypertrophy models. When the data sets are examined in mass space, this registration results in five different mass points, versus the six time points in the traditional comparison. This does not imply a loss of fidelity, but rather a shift of the data points over a defined mass range for comparison.
Fig. 1.
(A) Average heart weight to body weight ratios (HW/BW) for the adaptive hypertrophy experimental (Exercise, ◯) and control (Sedentary, Δ) conditions, as well as the maladaptive hypertrophy experimental (Banding, ●) and control (Sham, ▲) conditions plotted against time. Error bars represent standard error of the mean (SEM). (B) HW/BW for the adaptive and maladaptive hypertrophy experimental (●) and control (◯) conditions were plotted against each other and subjected to Pearson correlation analysis (C) HW/BW from selected time points in the adaptive and maladaptive hypertrophy models were plotted to visualize their similarity over the time courses (D) Average Log2 signal ratios (SLR) for the distinct set of significantly expressed genes from the adaptive (▲) and maladaptive (●) models were calculated at each time point in the time series and aligned based on the order of the measurement intervals or (E) based on heart weight to body weight ratio. AU = arbitrary units.
Alignment of Hypertrophy Gene Expression Time Series Using HW/BW
Gene expression data sets downloaded from the Cardiogenomics website7 were subjected to low-level processing to remove genes that were not considered to be expressed significantly above noise (see MATERIAL AND METHODS). This produced a dataset of 963 genes that were present in both hypertrophy models after the SAM analyses. To assess the benefit of using HW/BW as an alignment system to study the gene expression profiles we compared the distance scores D(a,b) [= a high-dimensional distance in gene expression space between two profiles at (warped) time t ] between the profiles of adaptive and maladaptive hypertrophy time series microarray data aligned based on temporal progression alone to that computed for time series data aligned based on HW/BW. The alignment based solely on temporal progression resulted in a high distance score of 1001.25, indicating that the global expression profiles were not well aligned between the hypertrophy model datasets (Fig. 1D). In contrast, the alignment that utilized HW/BW rather than time as an alignment parameter produced a much lower distance score of 31.34, demonstrating a markedly better alignment of the datasets (Fig. 1E). Thus, the alignment based on the common feature of HW/BW drastically reduced the distance between gene expression profiles and hence removed noise in gene expression differences due to the disparity of the characteristic time scales.
Differential Gene Expression between Exercise and Banding Models
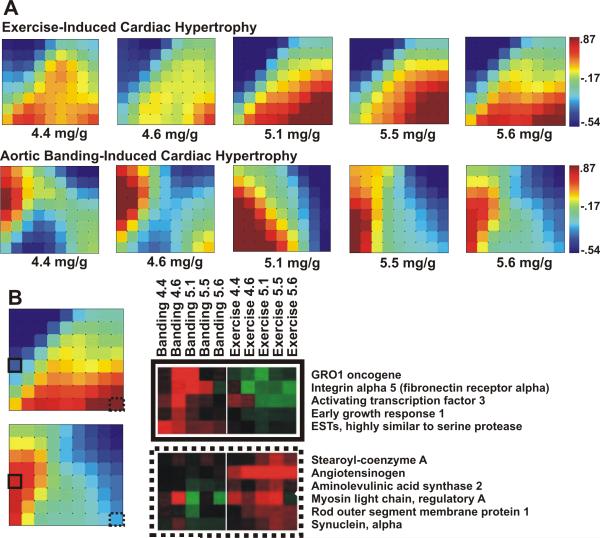
The mass aligned hypertrophy gene expression data was subjected to SAM13 analysis to identify transcripts that exhibited differential expression between the adaptive and maladaptive hypertrophy models. This yielded 314 significantly differentially expressed genes. To assess global patterns in the transcriptome, this set of differentially expressed genes was analyzed and visualized using the program Gene Expression Dynamics Inspector (GEDI).14 The GEDI mosaics representing these transcripts indicate that as the HW/BW surpasses 5 mg/g, the global expression profiles in each hypertrophy model approach a steady state. They also reveal that these transcripts cluster into two large, oppositely-regulated groups over the course of the growth processes initiated in the compared hypertrophy models (Fig. 2A). Closer inspection of the most distinctive mini-clusters in the GEDI mosaics showed that transcripts up-regulated in response to aortic banding pertain primarily to cell adhesion, immune response, and transcriptional regulation processes (Fig. 2B). In contrast, genes that were up-regulated primarily in response to the exercise stimulus were associated with metabolism and regulation of muscle contraction.
Fig. 2.
(A) Genes expressed primarily in either the adaptive or maladaptive model were input into GEDI to produce mosaics of global gene expression with respect to HW/BW. Scale bars indicate the centroid SLR value associated with each tile color in the mosaics. (B) Inspection of the gene clusters comprising the GEDI mosaics reveals the identities of genes differentially expressed in the hypertrophy models. Heat maps of gene clusters from mosaic tiles with solid and dashed outlines provide examples of genes demonstrating this expression pattern.
Commonalities in Gene Expression between Exercise and Banding Models
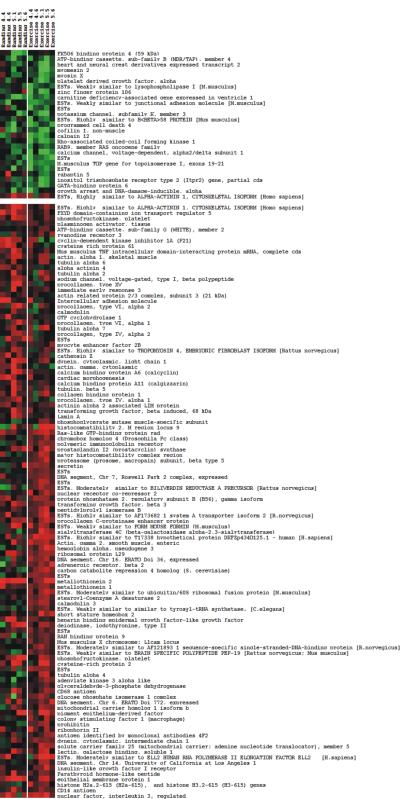
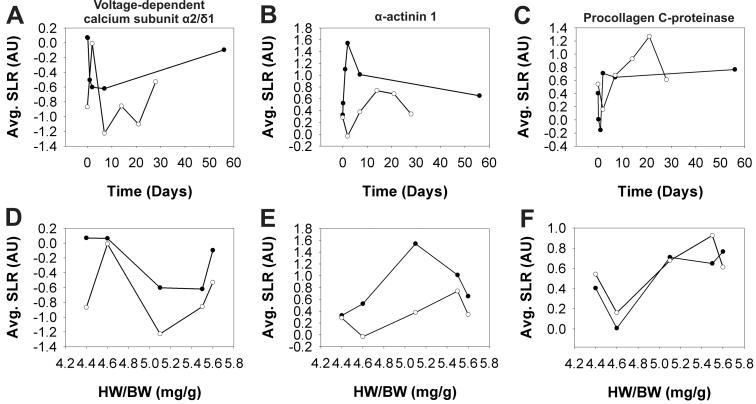
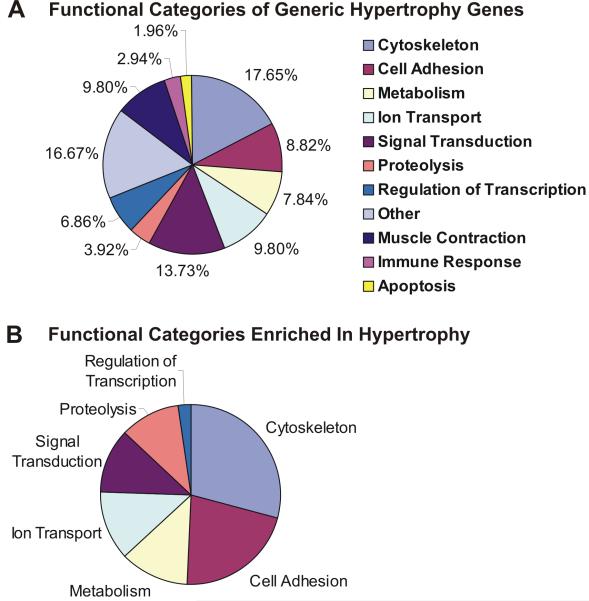
To identify genes that may be involved in a generic hypertrophy, we next examined the HW/BW aligned exercise and banding gene expression data to find individual genes that exhibited similar expression profiles in both datasets after application of the dynamic time warping algorithm (Fig. 3). The similarity of the temporal expression patterns of individual transcripts in the two mass-aligned profiles was determined using SITH (see MATERIALS and METHODS) which computes a distance scores di for gene i. Values for di ranged from di 0.089 for i = tubulin α6 to 2.65 for i = connective tissue growth factor, with lower scores di representing a greater degree of similarity between the temporal expression profiles of a gene i observed in the hypertrophy models. A cut-off score of 0.45 was chosen to distinguish transcripts with similar expression profiles in the exercise and banding time courses. That is, all genes with di < 0.45 were considered to have similar temporal patterns of expression in both types of hypertrophy. Between the HW/BW aligned hypertrophy models, 136 transcripts (14.5 %) displayed distance scores less than 0.45. The vastly different expression profiles for the connective tissue growth factor gene observed in the adaptive and maladaptive responses, coupled with the previous association of increased connective tissue growth factor expression with heart failure17, suggest that the SITH analysis correctly distinguished transcripts with common expression patterns in response to the adaptive and maladaptive stimuli from those that embody a stimulus-specific response. Additionally, comparison of the time-aligned and mass-aligned profiles for genes with distance scores less than the selected cut-off value, such as voltage-gated calcium channel subunit α2/δ1 (Fig. 4A, D), α-actinin 1 (Fig. 4B, E), and procollagen C-proteinase (Fig. 4C, F), showed that the similarities in the expression profiles revealed by the SITH analysis for the mass-aligned hypertrophy models was not evident when they were aligned based on time. The transcripts identified as having common temporal expression profiles (low di) were subjected to functional enrichment analysis to identify the cellular processes they participate in (Fig. 5). The results suggest that the functional categories to which the generic hypertrophy associated genes belong include primarily cytoskeletal organization, extracellular matrix, metabolism, ion transport, cell adhesion, proteolysis, and regulation of transcription.
Fig. 3.
Transcripts with comparable expression profiles between the adaptive and maladaptive hypertrophy models were subjected to 2 group K-means clustering and their expression profiles visualized using a heat map.
Fig. 4.
Average Log2 signal ratios (SLR) were plotted against time (A, B, C) and heart weight to body weight (HW/BW) (D, E, F) for the adaptive (exercise, ◯) and maladaptive (banding, ●) hypertrophy experimental conditions to compare the gene expression profiles for voltage-dependent calcium channel α2/δ1 (A, D), α-actinin 1 (B, E), and procollagen C-proteinase enhancer (C, F) before and after mass alignment. AU = arbitrary units.
Fig. 5.
(A) Gene ontology biological process terms were used to identify the cellular processes represented in the list of similarly expressed genes and the proportion of genes that fell into each category was plotted. (B) Functional enrichment analysis provided by the NIH's DAVID Bioinformatics Resources (see MATERIALS & METHODS) identified the biological processes terms that were over-represented in the list of similarly expressed genes. Segment sizes represent the enrichment scores with geometric means > 0.05, with larger segments indicating higher enrichment.
Discussion
Previous studies have used expression profiling to identify genes contributing to hypertrophic growth. Friddle, et al., did expression profiling of pharmacological models of cardiac hypertrophy in mice and reported groups of known and novel genes whose expression was transiently regulated distinctly during induction and regression of hypertrophy.18 Several models of experimentally induced-hypertrophy in rats have identified a group of 139 genes with consistent differential expression, suggesting a common genetic program that underlies pathogenic hypertrophic growth.19 Zhao, et al., used aortic constriction to induce hypertrophic growth in the hearts of mice to identify individual genes that were either uniquely up-, or down-regulated, over the course of days at three unique time points.20 In an examination of adaptive and maladaptive hypertrophy, Kong et al., conducted microarray profiling of physiological and pathological hypertrophic growth in Dahl salt-sensitive rats.21 This study reported increases in stress response and inflammation-related genes in pathological processes but not in physiological hypertrophy. In the case of the latter, differential expression of genes associated with metabolic function and protein synthesis were observed. These studies are important, because they documented changes in expression in a variety of experimentally-induced cardiac hypertrophy, however, they illustrate the lack of analytical strategies and tools for looking at hypertrophic growth as a dynamic event and the means of comparing and contrasting adaptive versus maladaptive growth.
Our study differed from previous work because our comparison of adaptive and maladaptive hypertrophic growth was in mass space, thus allowing us to compare disparate timescales of different studies as the growth occurred. We hypothesized that the concordant change in cardiac mass observed in mouse models of these hypertrophic stimuli was in part the product of a generic tissue remodeling program that is activated regardless of the nature of the inciting stimulus. The average HW/BW values associated with each time point in the hypertrophy time courses revealed that certain time points exhibited similar values, and hence could be used to align the time courses without violating the temporal order of the time points. We postulated that it may be possible to use this metric of cardiac growth as a common feature between these dissimilar hypertrophy models to facilitate comparison of the gene expression changes associated with each. This comparison allowed us to distinguish between stimulus-dependent and -independent hypertrophic gene expression.
In spite of the pronounced differences in the stimulus-dependent responses of the heart to adaptive and maladaptive stimuli, several transcripts had similar expression profiles between the contrasting hypertrophy models. Functional enrichment analysis revealed that transcripts coding for components of the sarcomere, such as α-actinin, were observed to have similar expression patterns in response to the adaptive and maladaptive stimuli. Likewise, transcripts coding for ion transport-related transcripts, particularly one associated with L-type calcium channels, were also commonly expressed in the adaptive and maladaptive models. Also of note, an enrichment of proteolysis related transcripts was also observed in the list of commonly expressed genes, such as the transcript for procollagen C-proteinase enhancer protein. Taken together, the cellular processes represented in the list of genes found to have similar expression patterns in the exercise and banding hypertrophy models suggest that a baseline level of remodeling occurs in the myocytes and extracellular matrix comprising the myocardium to enhance its performance in response to hypertrophic stimuli.
Genes whose expression was perturbed primarily by the exercise stimulus were transcripts associated with metabolic processes and muscle contraction. Notably the expression of myosin light chain, regulatory 7 and angiotensinogen transcripts were significantly up-regulated in response to exercise training. Angiotensinogen plays a role in the renin-angiotensin system as the substrate that angiotensin converting enzyme (ACE) converts into angiotensin I during the process of blood pressure regulation. Studies have shown that the heart possesses an intra-cardiac renin-angiotensin system associated with various initiators of cardiac hypertrophy.22 Additionally, enhanced expression of transcripts coding for proteins involved in lipid and glucose metabolism observed in the adaptive hypertrophy model corresponded well to those reported in the literature.4,23 As expected, these transcripts expressed specifically in response to the adaptive stimulus contrast sharply with the genes expressed exclusively in response to the maladaptive stimulus.
Fibrosis and immune response related transcripts dominated the list of genes whose expression was explicitly modulated in response to the maladaptive stimulus, consistent with previous reports.20,24,25 Among the transcripts that were highly expressed only in response to aortic banding were several well characterized clinical biomarkers for heart failure, such as brain derived neutrophic factor, natriuretic peptide precursor type A, and natriuretic peptide precursor type B.26 Several isoforms of collagen, such as procollagen type I, α1 and procollagen type III, α1, as well as the genes for secreted phosphoprotein 1 (a.k.a. osteopontin), fibronectin 1, and the fibronectin receptor integrin α5, were also highly up-regulated exclusively in response to the maladaptive stimulus. It is not surprising that the expression of these particular transcripts is significantly altered exclusively in response to the maladaptive stimulus because they are involved in augmenting the extracellular matrix of the myocardium, a hallmark of fibrosis.3 The cell surface antigen CD44 and the fibronectin receptor integrin α5 were both highly expressed with similar expression profiles in response to the maladaptive stimulus and are associated with osteopontin. It has been hypothesized that osteonpontin is a component of the mechanism that regulates the cardiac response to increased pressure or volume load on the heart because cardiac fibroblasts and/or cardiac myocytes demonstrate radically increased osteopontin expression in response to angiotensin II stimulation associated with the onset of heart failure.27 As shown in this study and widely reported in the literature, fibrosis plays a major role in the heart's response to maladaptive, but not adaptive stimuli.
Traditionally, the genetic differences between the diseased and healthy heart are mined for therapeutic opportunities. In this study, we have identified similarities between the expression profiles of adaptive and maladaptive hypertrophy which may be generally involved in cardiac growth. Most of these commonly expressed genes code for proteins which support the structure and function of cardiomyocytes, yet, the architectural differences between the conditions are profound. This suggests that overlapping molecular signatures in the responses to beneficial and pathological stimuli may result in disparate outcomes, This offers a starting point to investigate the cause of the divergent development to better understand the disease course and how it might be reversed.
Limitations of this Study
While the vast majority of gene expression studies focus on finding genes differentially expressed in various pathological situations we used a new analysis technique to identify common elements in two distinct models of cardiac hypertrophy, for commonalities can provide a deeper understanding of fundamental processes that do not depend on specific mechanistic aspects.
In doing so we first uncovered a relationship between HW/BW and the gene expression profiles in adaptive or maladaptive hypertrophy. However, HW/BW may not be the best physiological indicator of cardiac hypertrophy. HW/BW was chosen for this study because the values for these physiological parameters are commonly reported in studies of cardiac hypertrophy. Nonetheless, body weight can fluctuate independently of heart weight due to perturbations in diet and/or exercise, leading to false-positive or false-negative reporting of hypertrophy. A more stable parameter, such as tibia length instead of BW may provide a more reliable comparison against heart weight for determining the incidence of cardiac hypertrophy.
In addition, artifactual differences in the experimental protocols often hard to avoid when comparing physiological and pathological manifestations of the same process. The sham operation and sedentary control groups against which the aortic banding- and exercise-induced hypertrophy groups were compared consisted of mice with slight differences in age, weight, and the manner in which they were treated. This may introduce unspecific differences in the transcriptomes of the mice. However, since our emphasis is on the characterization of commonalities between two hypertrophy models, technical (non-relevant) differences will not have affected the specificity of our findings albeit the sensitivity could have been reduced (omission of interesting, commonly altered genes).
Finally, the analysis algorithms utilized in this study required that the gene expression datasets be “symmetrical”, that is for every transcript in the banding dataset, the same transcript had to be present in the exercise dataset as well. This symmetry was created by using different stringency criteria in the SAM analyses initially conducted on the banding and exercise-induced hypertrophy expression datasets. While we attempted to account for this discrepancy by applying more restrictive standards in our characterization of the differences and similarities between the banding and exercise datasets, the criteria used for the selection of the genes were established subjectively. Therefore, the results presented in this study provide what we feel is a representative, but incomplete sampling of the differences and commonalities in gene expression between these models of hypertrophy.
Acknowledgments
Sources of Funding Funding for this work was provided by the Harvard University Joseph H. Clark research support fund and NIH R01 grant# HL079126-01A2 awarded to K.K. Parker.
Footnotes
Disclosures None.
References
- 1.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Eng. J. Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 2.Fagard RH. Impact of different sports and training on cardiac structure and function. Cardiol. Clin. 1997;15:397–412. doi: 10.1016/s0733-8651(05)70348-9. [DOI] [PubMed] [Google Scholar]
- 3.Wakatsuki T, Schlessinger J, Elson EL. The biochemical response of the heart to hypertension and exercise. TRENDS Biochem. Sci. 2004;29:609–617. doi: 10.1016/j.tibs.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Strom CC, Aplin M, Ploug T, Christoffersen TEH, Langfort J, Viese M, Galbo H, Haunso S, Sheikh SP. Expression profiling reveals differences in metabolic gene expression between exercise-induced cardiac effects and maladaptive cardiac hypertrophy. FEBS Journal. 2005;272:2684–2695. doi: 10.1111/j.1742-4658.2005.04684.x. [DOI] [PubMed] [Google Scholar]
- 5.MacLellan WR, Schneider MD. Genetic dissection of cardiac growth control pathways. Annu. Rev. Physiol. 2000;62:289–319. doi: 10.1146/annurev.physiol.62.1.289. [DOI] [PubMed] [Google Scholar]
- 6.Mirotsou M, Watanabe CMH, Schultz PG, Pratt RE, Dzau VJ. Elucidating the molecular mechanism of cardiac remodeling using a comparative genomic approach. Physiol. Genomics. 2003;15:115–126. doi: 10.1152/physiolgenomics.00071.2003. [DOI] [PubMed] [Google Scholar]
- 7.Genomics of Cardiovascular Development, Adaptation, and Remodeling. NHLBI Program for Genomic Applications, Harvard Medical School. URL: http://www.cardiogenomics.org [October, 2006]
- 8.Bar-Joseph Z. Analyzing time series gene expression data. Bioinformatics. 2004;20:2493–2503. doi: 10.1093/bioinformatics/bth283. [DOI] [PubMed] [Google Scholar]
- 9.Aach J, Church GM. Aligning gene expression time series with time warping algorithms. Bioinformatics. 2001;17:495–508. doi: 10.1093/bioinformatics/17.6.495. [DOI] [PubMed] [Google Scholar]
- 10.Mirotsou M, Dzau VJ, Pratt RE, Weinberg EO. Physiological genomics of cardiac disease: quantitative relationships between gene expression and left ventricular hypertrophy. Physiol. Genomics. 2006;27:86–94. doi: 10.1152/physiolgenomics.00028.2006. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan ML, Cheslow Y, Vikstrom K, Malhotra A, Geenen DL, Nakouzi A, Leinwand LA, Buttrick PM. Cardiac adaptations to chronic exercise in mice. Am. J. Physiol. 1994;267:H1167–1173. doi: 10.1152/ajpheart.1994.267.3.H1167. [DOI] [PubMed] [Google Scholar]
- 12.Tarnavski O, McMullen JR, Schinke M, Nie Q, Kong S, Izumo S. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genomics. 2004;16:349–360. doi: 10.1152/physiolgenomics.00041.2003. [DOI] [PubMed] [Google Scholar]
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 17.Weinberg EO, Mirotsou M, Gannon J, Dzau VJ, Lee RT, Pratt RT. Sex dependence and temporal dependence of the left ventricular genomic response to pressure overload. Physiol. Genomics. 2003;12:113–127. doi: 10.1152/physiolgenomics.00046.2002. [DOI] [PubMed] [Google Scholar]
- 18.Friddle CJ, Koga T, Rubin EM, Bristow J. Expression profiling reveals distinct sets of genes altered during induction and regression of cardiac hypertrophy. Proc Natl Acad Sci U S A. 2000;97:6745–50. doi: 10.1073/pnas.100127897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strøm CC, Kruhøffer M, Knudsen S, Stensgaard-Hansen F, Jonassen TE, Orntoft TF, Haunsø S, Sheikh SP. Identification of a core set of genes that signifies pathways underlying cardiac hypertrophy. Comp Funct Genomics. 2004;5(67):459–70. doi: 10.1002/cfg.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao M, Chow A, Powers J, Fajardo G, Bernstein D. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol. Genomics. 2004;19:93–105. doi: 10.1152/physiolgenomics.00040.2004. [DOI] [PubMed] [Google Scholar]
- 21.Kong SW, Bodyak N, Yue P, Liu Z, Brown J, Izumo S, Kang PM. Genetic expression profiles during physiological and pathological cardiac hypertrophy and heart failure in rats. Physiol Genomics. 2005;21:34–42. doi: 10.1152/physiolgenomics.00226.2004. [DOI] [PubMed] [Google Scholar]
- 22.Raman VK, Lee YA, Lindpaintner K. The cardiac rennin-angiotensin-aldosterone system and hypertensive cardiac hypertrophy. Am. J. Cardiol. 1995;76:18D–23D. doi: 10.1016/s0002-9149(99)80487-1. [DOI] [PubMed] [Google Scholar]
- 23.Selvetella G, Hirsch E, Notte A, Tarone G, Lembo G. Adaptive and maladaptive hypertrophic pathways: points of convergence and divergence. Cardiovasc. Res. 2004;63:373–380. doi: 10.1016/j.cardiores.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Johnatty SE, Dyck JRB, Michael LH, Olsen EN, Abdellatif M. Identification of genes regulated during mechanical load-induced cardiac hypertrophy. J. Mol. Cell. Cardiol. 2000;32:805–815. doi: 10.1006/jmcc.2000.1122. [DOI] [PubMed] [Google Scholar]
- 25.Van den Bosch BJC, Lindsey PJ, Van den Burg CMM, Van der Vlies SA, Lips DJ, Van der Vusse GJ, Ayoubi TA, Doevendans PA, Smeets HJM. Early and transient gene expression changes in pressure overload-induced cardiac hypertrophy in mice. Genomics. 2006;88:480–488. doi: 10.1016/j.ygeno.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Maisel AS, Bhalla V, Braunwald E. Cardiac biomarkers: a contemporary status report. Nat. Clin. Pract. Cardiovasc. Med. 2006;3:24–34. doi: 10.1038/ncpcardio0405. [DOI] [PubMed] [Google Scholar]
- 27.Graf K, Do YS, Ashizawa N, Meehan WP, Giachelli CM, Marboe CC, Fleck E, Hsueh WH. Myocardial osteopontin expression is associated with left ventricular hypertrophy. Circulation. 1997;96:3063–3071. doi: 10.1161/01.cir.96.9.3063. [DOI] [PubMed] [Google Scholar]