Abstract
A conserved insulin-like pathway modulates both aging and pathogen resistance in Caenorhabditis elegans. However, the specific innate effector functions that mediate this pathogen resistance are largely unknown. Autophagy, a lysosomal degradation pathway, plays a role in controlling intracellular bacterial pathogen infections in cultured cells, but less is known about its role at the organismal level. We examined the effects of autophagy gene inactivation on Salmonella enterica Serovar Typhimurium (Salmonella typhimurium) infection in 2 model organisms, Caenorhabditis elegans and Dictyostelium discoideum. In both organisms, genetic inactivation of the autophagy pathway increases bacterial intracellular replication, decreases animal lifespan, and results in apoptotic-independent death. In C. elegans, genetic knockdown of autophagy genes abrogates pathogen resistance conferred by a loss-of-function mutation, daf-2(e1370), in the insulin-like tyrosine kinase receptor or by over-expression of the DAF-16 FOXO transcription factor. Thus, autophagy genes play an essential role in host defense in vivo against an intracellular bacterial pathogen and mediate pathogen resistance in long-lived mutant nematodes.
Immune functions decline with age (1), but the mechanisms underlying immunosenescence are not well understood. Because a conserved insulin-like signaling pathway controls both aging and pathogen resistance in Caenorhabditis elegans (2, 3), the identification of cellular innate immune functions regulated by this pathway may help elucidate the basis of age-related declines in immunity. One candidate target is autophagy, a lysosomal degradation pathway that decreases with age, is implicated in the degradation of intracellular bacteria, and is required for the lifespan extension of nematodes with mutation in the daf-2 insulin-like signaling pathway (4–7).
The autophagy pathway is mediated by evolutionarily conserved genes (called atg genes) and, in vitro, targets both extracellular bacteria that invade intracellularly and intracellular bacterial pathogens for lysosomal degradation (4). Autophagy limits the intracellular growth of Listeria monocytogenes in Drosophila (8), suggesting a role for autophagy in defense against intracellular bacteria in vivo. Besides promoting the direct degradation of intracellular pathogens, autophagy has other functions in immunity (4), including the delivery of microbial genetic material or peptides to endosomes or MHC Class II loading compartments, respectively, for activation of innate or adaptive immunity. Furthermore, a polymorphism in the atg gene, ATG16L1, is linked to genetic susceptibility to the inflammatory bowel disorder, Crohn's disease, which has led to the speculation that mutations in the autophagy pathway may alter the normal gut response to enteric bacterial pathogens (9). Indeed, loss of Atg16l1 in mouse models recapitulates certain aspects of the pathology of human Crohn's disease, and the Crohn's disease-associated ATG16L1 variant may have impaired antibacterial autophagy function in a human gut epithelial cell line (10).
The unicellular organism Dictyostelium discoideum and the multicellular organism C. elegans are useful models for studying interactions between hosts and pathogens (11, 12), including the gram-negative bacterium Salmonella typhimurium. Salmonella is a pathogen that causes human food-borne illness worldwide, infects both mammalian intestinal epithelial cells and macrophages, and has an increased propensity to cause invasive, extra-intestinal disease in the elderly (1, 13). In C. elegans, S. typhimurium establishes persistent infection in the intestinal lumen but does not replicate inside intestinal epithelial cells (14, 15), suggesting that host defense mechanisms to combat Salmonella infection may be more successful in nematode than in mammalian cells. In Dictyostelium, a soil amoeba that is highly susceptible to invasion by bacterial pathogens that commonly infect human macrophages (12), Salmonella is efficiently taken up but fails to multiply intracellularly. Therefore, the identification of factors that govern susceptibility to Salmonella infection in the nematode and soil amoeba may be relevant to understanding Salmonella host-pathogen interactions in mammalian intestinal epithelial cells and macrophages, respectively.
Long-lived mutant nematodes display resistance to Salmonella infection (2, 16). In C. elegans, the daf-2 insulin-like signaling pathway is a well-characterized regulator of pathogen resistance, longevity, and autophagy (2, 3, 5, 16). Nematodes with loss-of-function mutations in the insulin-like tyrosine kinase receptor daf-2 have extended lifespan and pathogen-resistant phenotypes (3, 16), including resistance to infection with S. typhimurium. The major target of the DAF-2 pathway is DAF-16, a forkhead transcription factor required for both longevity and pathogen resistance in daf-2 mutants (16). Moreoever, DAF-2 negatively regulates autophagy, and lifespan extension in daf-2 mutants requires atg genes (5–7). These findings, coupled with the role of autophagy in pathogen degradation, led us to postulate that autophagy may represent a mechanism of innate immunity against Salmonella infection in vivo that is required for the pathogen resistance of long-lived mutant nematodes.
To evaluate the role of autophagy in pathogen resistance, we used feeding RNAi to inactivate several C. elegans atg genes, including bec-1 and lgg-1, in S. typhimurium-infected wild-type, and long-lived daf-2-mutant and DAF-16 over-expressing nematodes. We also examined S. typhimurium infection in Dictyostelium with null mutations in three atg genes, ATG1, ATG6, and ATG7. Our results indicate that atg genes play a conserved role in antibacterial host defense in vivo and mediate pathogen resistance in long-lived mutant animals.
Results
Autophagy Mediates Host Defense Against Salmonella in C. elegans.
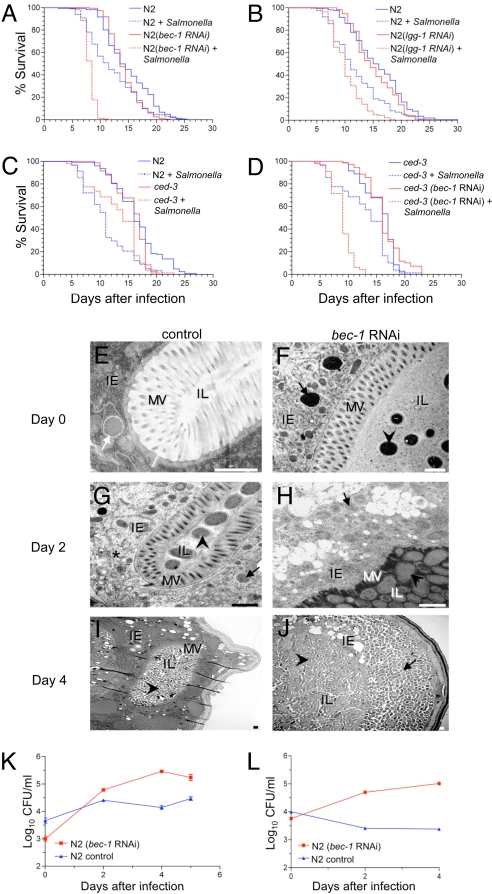
To evaluate whether atg genes function in host defense against Salmonella infection in C. elegans, we used feeding RNAi to silence two previously characterized C. elegans atg genes, bec-1 and lgg-1 (5), which are orthologs of yeast ATG6 and ATG8 and function in autophagic vesicle nucleation and autophagic vesicle expansion, respectively. The survival of Salmonella-infected animals treated with either bec-1 or lgg-1 RNAi was significantly shortened as compared to Salmonella-infected control vector-treated animals (Fig. 1 A and B, Table S1)]. Although a null mutation in bec-1 or high-dose atg gene RNAi injection is embryonically lethal (5, 17), feeding RNAi against bec-1 or lgg-1 did not shorten the lifespan of N2 (wild-type) animals exclusively fed nonpathogenic Escherichia coli (Fig. 1 A and B). We confirmed that feeding RNAi decreased the level of atg gene RNA by RT-PCR, and that bec-1 feeding-RNAi treatment exerted previously described effects of atg gene inhibition [e.g., abnormal dauer development in daf-2(e1370) mutants and autophagy inhibition in worms that transgenically express the autophagy marker GFP::LGG-1] (data not shown). We also knocked down the expression of atg-7 (involved in autophagic vesicle expansion) in N2 worms using RNAi. Similar to bec-1 and lgg-1 RNAi, the atg-7-RNAi animals were more susceptible to Salmonella infection than control animals (Fig. S1A, Table S2). Thus, inactivation of three different atg genes increased nematode susceptibility to lethal Salmonella infection. This is unlikely to reflect nonspecific or off-target effects of atg gene-RNAi treatment because no alterations in Salmonella susceptibility were observed in nematodes treated with feeding RNAi against two irrelevant control genes, unc-22 (required for normal muscle function) or him-3 (required for meiotic chromosome segregation) (Fig. S1 B and C, Table S2). Furthermore, neither bec-1-RNAi treatment [similar to a previous report by Hansen et al (7)] nor Salmonella infection altered pharyngeal pumping rates (Fig. S2), suggesting that the increased pathogen susceptibility in atg gene RNAi-treated animals is not because of alterations in food (bacterial) uptake.
Fig. 1.
Atg genes mediate host defense against Salmonella in C. elegans. (A and B) Survival curves of wild-type (N2) animals treated with either control vector or indicated atg gene RNAi-feeding plasmids following a 2-day exposure to S. typhimurium or normal food (i.e., nonpathogenic Escherichia coli) at 20 °C (see SI Materials and Methods for details). (C and D) Survival curves of wild-type (N2) and ced-3(n717)-mutant animals following a 2-day exposure to S. typhimurium or normal food without atg gene RNAi treatment (C) or with control vector or bec-1 RNAi treatment (D) at 20 °C. For (A) to (D), see Table S1 for statistical details. (E–J) Representative EMs of Salmonella-infected control N2 animals and bec-1-RNAi animals at day 0 (E and F), 2 (G and H), and 4 (I and J) after a 2-day Salmonella ingestion period. (F, G, I, and J) Arrowheads denote intraluminal bacteria. (E) White arrow denotes bacterial-containing early autophagosome. (G) Asterisk denotes autolysosome with partially degraded bacterial debris. (F–H and J) Black arrows denote SCVs inside intestinal epithelial cells. IE, intestinal epithelial cells; IL, intestinal lumen; MV, microvilli. (Scale bars, 1 μm.) (K and L) Growth curves of S. typhimurium in N2 animals treated with either control vector or bec-1-RNAi feeding plasmids in the absence (K) or presence (L) of 100 μg/ml gentamicin. For (K) and (L), values represent mean ± SEM for triplicate samples of ≈10 animals per treatment group per sample. Similar results were observed in two independent experiments.
BEC-1 interacts with CED-9 (a Bcl-2 homolog) and disruption of bec-1 triggers apoptosis during embryo development (17). Therefore, we studied apoptosis-deficient nematodes with a loss-of-function mutation in the C. elegans caspase, ced-3(n717) to examine whether apoptosis is involved in the increased susceptibility of autophagy-deficient worms to Salmonella infection. Salmonella infection decreased the lifespan of wild-type worms more than that of ced-3(n717)-mutant animals (Fig. 1C and Table S1), indicating that the ced-3 mutation protects against Salmonella infection. [The basis for the difference between this finding and those of Aballay et al. (18) is unclear, but might reflect subtle differences in genetic strains]. Importantly, bec-1 RNAi significantly decreased the lifespan of Salmonella-infected ced-3-mutant animals (Fig. 1D and Table S1); these animals had similar mortality as bec-1 RNAi-treated Salmonella-infected wild-type animals (Fig. 1A). Therefore, the mechanism of increased Salmonella sensitivity because of atg gene inactivation does not involve caspase-dependent apoptosis.
bec-1 Restricts Bacterial Replication and Cytopathology in C. elegans Intestinal Epithelial Cells.
To examine the mechanism by which atg genes protect against Salmonella infection, we performed EM analyses of control vector- and bec-1-RNAi-treated animals (Fig. 1 E–J). Immediately after a 2-day Salmonella ingestion period, few organisms were observed in the intestinal lumen and some bacteria were observed inside the intestinal epithelial cells (Fig. 1 E and F). In the control group, intracellular bacteria were found primarily inside early intact autophagosomes or autolysosomes (Fig. 1E and data not shown), but rarely inside cytoplasmic Salmonella-containing vacuoles (SCVs). In contrast, in bec-1-RNAi animals, the cytoplasm contained intact SCVs (Fig. 1F). At day two after Salmonella ingestion, the number of organisms in the intestinal lumen increased in both groups, but more dramatically in the bec-1-RNAi animals (Fig. 1 G and H). Moreover, in control animals the intestinal epithelial cells contained rare, visible intact bacteria and numerous autolysosomes containing bacterial debris (Fig. 1G), whereas in bec-1-RNAi animals the intestinal epithelial cells contained numerous intact bacteria (which were similar in size and morphology to those found in the intestinal lumen) and very few autophagosomes or autolyososmes (Fig. 1H). At this stage, the epithelial cells in both groups had intact basement membranes and microvilli, although increased cytoplasmic vacuolization was observed in bec-1-RNAi animals. However, at day 4 after Salmonella ingestion, in the bec-1-RNAi group the intestinal epithelial cells were completely destroyed in most animals, and there were massive sheets of bacteria extending from the intestinal lumen to the body wall muscle (Fig. 1J). In contrast, the epithelial cells remained largely intact in the control animals (Fig. 1I), indicating that the atg gene bec-1 successfully protects the intestinal epithelium cells from overwhelming bacterial infection and cellular destruction.
These EM analyses suggest that Salmonella invades the intestinal epithelial cells in both control and bec-1-RNAi animals; however, in the former group the bacteria are efficiently degraded by the autophagy pathway, whereas in the latter group the intracellular bacterial population expands, leading to extensive cytoplasmic destruction and premature death of the organism. To confirm that Salmonella replicates intracellularly in bec-1-RNAi animals, we compared bacterial growth curves in animals cultured in the presence or absence of gentamicin, an antibiotic that inhibits extracellular but not intracellular Salmonella replication. In the absence of gentamicin, bacterial colony counts increased gradually over time in both control and bec-1-RNAi animals, with a larger magnitude of increase in the bec-1-RNAi animals (P < 0.0001, t-test) (Fig. 1K). In the presence of gentamicin, no increase was observed in bacterial colony counts in control animals (Fig. 1L). In contrast, in bec-1-RNAi animals, the Salmonella growth curve paralleled that observed in the absence of gentamicin, indicating that Salmonella replicates intracellularly in autophagy-deficient nematodes.
Atg Genes Restrict Intracellular Bacterial Replication and Host Cellular Destruction in Dictyostelium.
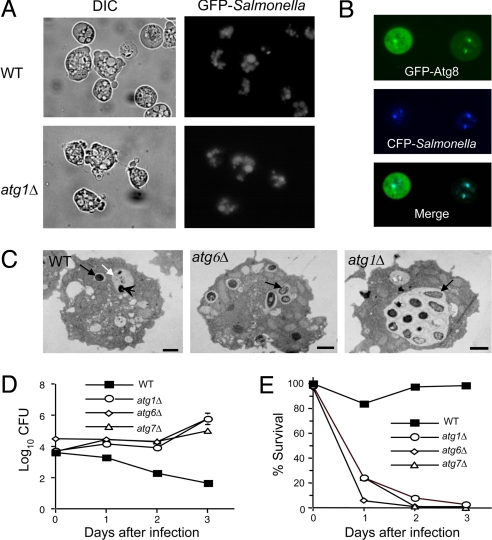
To further confirm that atg genes restrict intracellular Salmonella multiplication independently of bacterial invasion (which is difficult to assess in vivo in a multicellular organism), we used a unicellular model host organism, D. discoideum. Unlike many bacteria that replicate intracellularly in mammalian macrophages, Salmonella is rapidly degraded after internalization and is nonpathogenic in Dictyostelium (12). We found that GFP-labeled Salmonella invaded wild-type Dictyostelium and previously described Dictyostelium mutants (19) lacking the atg genes, ATG1 (a serine/threonine kinase involved in autophagy induction), ATG6 (the ortholog of bec-1), and ATG7 with similar kinetics and to a similar degree (Fig. 2A). In Dictyostelium that transgenically express a fluorescent autophagy marker protein, GFP-Atg8 (19), CFP-labeled Salmonella colocalized with GFP-Atg8 by 2-h postinfection (p.i.) (Fig. 2B), indicating that internalized bacteria are efficiently targeted to autophagosomes. Similarly, EM analysis revealed the degradation of Salmonella inside autolysosomes in wild-type Dictyostelium (Fig. 2C). In contrast, in atg1-, atg6-, and atg7-mutant Dicytostelium, no bacteria were observed inside autophagosomes or autolysosomes (Fig. 2C). Instead, the bacteria were observed exclusively in intact SCVs containing single bacteria or in larger vacuoles that contained multiple bacteria. Thus, the autophagic machinery is not necessary for bacterial invasion or formation of SCVs, but is required for the fusion of SCVs with lysosomal compartments. By 24-h p.i., Salmonella were rarely visible in wild-type Dictyostelium and the cytoplasm appeared normal, whereas atg1-, atg6-, and atg7-mutant Dictyostelium exhibited severe cytopathology, including extensive cytoplasmic vacuolization and disruption of plasma membrane integrity (data not shown).
Fig. 2.
Atg genes restrict intracellular Salmonella replication and pathogenicity in D. discoideum. (A) Internalization and vacuolar localization of GFP-Salmonella in both wild-type (WT) and atg1 mutant Dictyostelium at 2-h postinfection (p.i.). Similar results were observed in atg6Δ and atg7Δ Dictyostelium (data not shown). (B) Colocalization of CFP-Salmonella and a transgenic autophagosomal marker, GFP-Atg8, at 2-h p.i. in wild-type Dictyostelium. (C) Representative EMs 12-h p.i. of wild-type Dictyostelium showing an intact SCV (black arrow) and an autolysosome (white arrow) containing a partially degraded Salmonella bacterieum (arrowhead), and of atg1Δ and atg6Δ Dictyostelium that lack autolyosomes but have SCVs (black arrows) that contain multiple organisms, indicative of active intracellular bacterial multiplication. (Scale bars, 1 μm.) (D) Growth of intracellular Salmonella in wild-type and atg gene-mutant Dictyostelium strains. (E) Survival of Dictyostelium infected in (D). For (D) and (E), results represent mean ± SEM for triplicate samples and similar results were observed in three independent experiments.
These findings are consistent with autophagic degradation of intracellular Salmonella in wild-type Dicytostelium and intracellular multiplication of Salmonella with resulting cytopathology in autophagy-deficient amoebae. To confirm this, we measured Salmonella replication (Fig. 2D) and cell survival (Fig. 2E) in the presence of gentamicin. In wild-type Dictyostelium, there was no bacterial growth (Fig. 2D), and the Dictyostelium remained healthy with a long-term survival curve that parallels that of amoebae grown in axenic media (Fig. 2E and data not shown). In contrast, in atg1-, atg6-, and atg7-mutant Dictyostelium, bacterial colony counts increased over time (Fig. 2D), indicating intracellular multiplication, and the Dictyostelium died within 24- to 72-h p.i. (Fig. 2E). Because Dictyostelium lack caspases or other essential components of the apoptotic machinery (20), this Salmonella-induced death of autophagy-deficient amoebae, like that of autophagy-deficient nematodes, does not involve caspase-dependent apoptosis. Taken together, these findings demonstrate that the autophagic machinery restricts intracellular bacterial multiplication and cytopathology in two model organisms, C. elegans and Dictyostelium.
Autophagy Deficiency Abrogates Pathogen Resistance of daf-2-Mutant Adults.
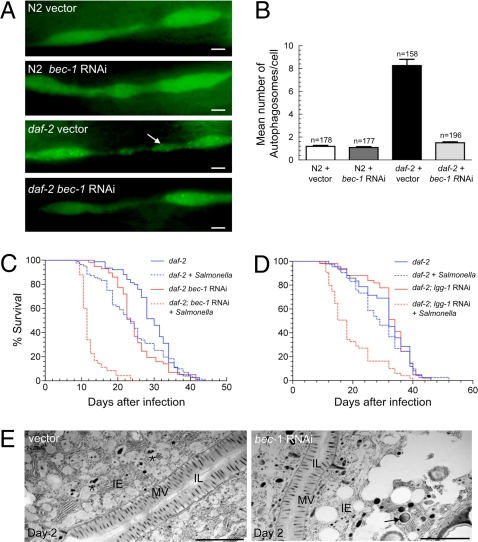
To evaluate whether autophagy is an effector mechanism of enhanced Salmonella resistance in long-lived C. elegans mutants, we first examined whether autophagy is induced in adult nematodes with a loss-of-function mutation in daf-2 (2). Using a previously described assay that measures autophagosomes in the seam cell, a type of specialized hypodermal cell that has detectable basal- and stimulus-induced autophagy (5), we observed a higher number of autophagosomes in daf-2(e1370)-mutant adults than in N2 wild-type worms (P < 0.0001, t-test), which was suppressed by bec-1-RNAi treatment (Fig. 3 A and B). Thus, the DAF-2 signaling pathway negatively regulates autophagy in the adult worm, consistent with the report by Hansen et al. (7).
Fig. 3.
Atg genes mediate insulin signaling-regulated resistance against Salmonella in C. elegans. (A) Representative images of autophagosomes (GFP::LGG-1 dots) in seam cells of N2 wild-type and daf-2-mutant animals treated either with RNAi control vector or bec-1 RNAi. The arrow denotes a representative autophagosome. (Scale bars, 2 μM.) (B) Quantification of autophagosomes per seam cell (mean ± SEM) for each genotype. n = number of seam cells per group in ≈20 animals. Similar results were obtained in two independent experiments. (C and D) Survival curves of daf-2(e1370)-mutant animals treated with either control or indicated atg gene-RNAi feeding plasmids following a 2-day exposure to S. typhimurium or normal food at 20 °C. See Table S1 for statistical details. (E) Representative EMs of Salmonella-infected control and bec-1-RNAi daf-2(e1370) animals two days after a 2-day Salmonella ingestion period. The asterisk denotes a representative autolysosome containing partially degraded bacteria. The arrow denotes representative SCV. (Scale bars, 2 μM.)
Next, we examined whether atg genes are required for the pathogen-resistance phenotype of daf-2(e1370)-mutant adults. Because atg genes have previously been shown to be required for the lifespan extension of daf-2 mutants (5, 6), we sought to avoid confounding effects of autophagy on lifespan regulation that occur independently of daf-2-mediated pathogen resistance. To do this, we used a low dose of the RNAi inducer isopropyl-β-d-thiogalactopyranoside (IPTG) (1 nM) that was titrated to identify a concentration at which bec-1 RNAi and lgg-1 RNAi did not have significant effects on the lifespans of uninfected daf-2(e1370)-mutant animals (Fig. 3 C and D and Table S1). Similar to previous reports with other bacterial pathogens (3), the control daf-2(e1370)-mutant animals survive longer than wild-type animals when infected with S. typhimurium (Fig. 3 compared to Fig. 1 and Table S1). Bec-1 and lgg-1 RNAi significantly shortened the lifespan of Salmonella-infected daf-2(e1370) animals at doses that only minimally shortened the lifespan of E. coli-fed control daf-2(1370) animals (Fig. 3 C and D, Table S1). Indeed, the mean lifespan of bec-1-RNAi or lgg-1-RNAi Salmonella-infected daf-2 animals was similar to that of control Salmonella-infected wild-type N2 animals (Fig. 1 A and B, Table S1), indicating that atg gene knockdown is sufficient to completely abrogate pathogen resistance conferred by a mutation in the insulin-like signaling pathway.
Similar to our observations in N2 animals, EM studies of Salmonella-infected daf-2(e1370) mutants demonstrated enhanced autophagic degradation of bacteria in control vs. bec-1-RNAi animals (Fig. 3E). Compared to N2 animals, there were fewer bacteria in the intestinal lumen of both control and bec-1-RNAi animals, and in control animals there was a marked increase in the number of autolysosomes containing partially degraded bacteria. Similar to bec-1-RNAi N2 animals, bec-1-RNAi daf-2(e1370) mutants displayed an increase in intact SCVs in intestinal epithelial cells, as well as increased intestinal epithelial cell pathologic changes, such as cytoplasmic vacuolization and destruction of the apical and basement membranes. These morphological observations suggest that increased intestinal epithelial cell autophagic activity may partially underlie the pathogen resistance of daf-2(e1370) mutants. Of note, pharyngeal pumping (and predicted bacterial uptake) was not altered in infected or uninfected daf-2(e1370) vs. N2 animals, nor in vector control vs. bec-1-RNAi daf-2(e1370) mutants (Fig. S2).
Autophagy Deficiency Blocks Pathogen Resistance Conferred by DAF-16 Over-Expression.
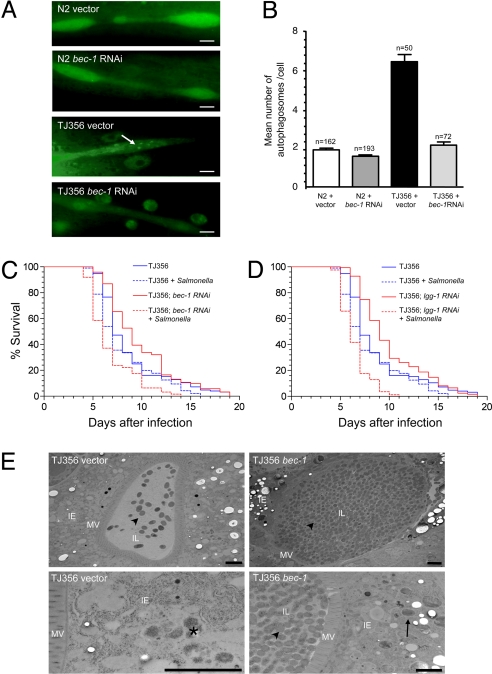
The daf-2 insulin-like signaling negatively regulates the activity of a forkhead transcription factor, DAF-16 (2), and daf-16 is required for the pathogen-resistance phenotype of daf-2 mutants (3). Thus, we postulated that DAF-16 over-expression might induce atg gene-dependent pathogen resistance. We introduced, by genetic crossing, the autophagy marker GFP::LGG-1 into TJ356 animals that carry additional daf-16 gene copies (21) and found that the number of autophagosomes per seam cell was significantly increased in TJ356 animals (P < 0.0001, t-test) in a manner that is suppressed by bec-1-RNAi treatment (Fig. 4 A and B). We also confirmed that seam cell autophagy was increased in another DAF-16 over-expressing strain, CF1139 (Fig. S3). The increased number of GFP::LGG-1 punctuate dots (autophagosomes) in the seam cells of TJ356 and CF1139 animals is unlikely to be a result of GFP over-expression per se (these strains express a DAF-16::GFP fusion protein) because we did not observe a similar increase in punctuate dots in GFP::LGG-1 animals crossed to another strain, JR667, that over-expresses GFP in seam cells (22) (Fig. S4). Furthermore, EM analyses revealed increased numbers of autolysosomes in the seam cells of TJ356 vs. N2 animals (see representative images in Fig. S5 A and B).
Fig. 4.
Atg genes are required for DAF-16-mediated resistance to Salmonella infection. (A) Representative pictures of autophagosomes in seam cells of N2 wild-type and TJ356 animals treated either with RNAi control vector or bec-1 RNAi. TJ356 is heterozygous for both gfp::lgg-1 and daf-16::gfp. The oval-shaped green staining near the seam cells in TJ356 animals represent nuclei of intestinal cells that express DAF-16::GFP. The arrow denotes a representative autophagosome. (Scale bars, 2 μM.) (B) Quantification of autophagosomes per seam cell (mean ± SEM) for each genotype. n = number of seam cells per group in ≈20 animals. Similar results were obtained in two independent experiments. (C and D) Survival curves of TJ356 animals treated with either control or indicated atg gene RNAi-feeding plasmids following exposure to S. typhimurium or normal food at 25 °C. n = number of animals per treatment group. See Table S1 for statistical details. (E) Representative EMs of Salmonella-infected control and bec-1-RNAi TJ356 animals two days after a 2-day Salmonella ingestion period. The asterisk denotes a representative autolysosome containing partially degraded bacteria. Arrowheads represent intestinal luminal bacteria and the arrow denotes representative SCV-containing bacteria. (Scale bars, 2 μM.)
Next, we examined whether autophagy induction is required for the pathogen resistance of TJ356 animals (23). The Salmonella infection survival assay was performed at 25 °C to reproduce published experimental conditions in which pathogen resistance was observed (23). Salmonella-infected TJ356 animals lived significantly longer than control N2 wild-type worms (Fig. S5C and Table S2), indicating that DAF-16 over-expression confers resistance to Salmonella infection. This resistance to Salmonella infection was blocked by atg gene knockdown; bec-1 and lgg-1 RNAi significantly shortened the lifespan of Salmonella-infected TJ356 animals but did not shorten the lifespan of E. coli-fed control TJ356 animals (Fig. 4 C and D and Table S1). Similar to N2 animals, EM analyses of Salmonella-infected TJ356 animals revealed increased luminal bacteria and decreased lysosomal bacterial degradation in intestinal epithelial cells in bec-1-RNAi vs. control animals (Fig. 4E). We also examined the pathogen-resistance phenotype of CF1139 animals. Salmonella-infected CF1139 animals lived significantly longer than control animals, although the resistance was not as strong as in TJ356 animals, and this pathogen resistance was blocked by both bec-1 and lgg-1 RNAi (Table S2). Together, these data indicate that atg genes are required for resistance to Salmonella both in worms with a loss-of-function mutation in the DAF-2 insulin-like tyrosine kinase receptor or with over-expression of the DAF-16 forkhead transcription factor.
Discussion
Our findings demonstrate that atg genes play an evolutionarily conserved role in host defense against the mammalian intracellular bacterial pathogen, S. typhimurium. In C. elegans and Dictyostelium, the genetic knockdown or knockout of essential atg genes dramatically alters the fate of the invading bacterium and of the host organism. The invading bacterium, rather than being targeted for lysosomal degradation, establishes an intracellular replicative niche, which leads to cellular destruction and premature organismal death. Thus, in these two model organisms, the presence or absence of atg genes is sufficient to determine whether Salmonella is a successful intracellular pathogen.
In mammals, Salmonella is phagocytosed by macrophages and can invade nonphagocytic cells through the activity of the type-III secretion system (24). Salmonella replicates intracellularly within a specialized compartment, the SCV, and to be a successful intracellular pathogen, Salmonella must avoid fusion of the SCV with the lysosome (24). Our results indicate that in both Dictyostelium and C. elegans, Salmonella invades intracellularly, resides in SCVs, and is targeted to lysosomes for degradation in wild-type but not autophagy-deficient organisms. Therefore, we propose that the autophagic machinery is used by these organisms to deliver Salmonella to the lysosome. Autophagy likely plays a similar role in mammalian cells, as previous in vitro studies have shown that autophagy inactivation enhances Salmonella replication in macrophages (25). Perhaps a major difference in Salmonella infection in mammalian cells (where it replicates intracellularly) versus that in lower eukaryotic organisms (where it cannot replicate intracellularly) may relate to the ability of the bacterium to partially antagonize the host autophagic machinery. This could explain why genetic inactivation of the autophagic machinery in lower eukaryotes (such as C. elegans or Dictyostelium) mimics Salmonella pathogenesis in mammalian cells, in that the bacteria can replicate inside and destroy intestinal epithelial cells and phagocytic cells.
Our findings clearly indicate that atg genes function in a cell-autonomous manner, in intestinal epithelial cells in the nematode or in the unicellular organism, Dictyostelium, to limit intracellular bacterial replication and cellular destruction. In the presence of gentamicin that kills extracellular bacteria, Salmonella only multiplies successfully and only causes cytopathology when atg genes are inactivated. However, in C. elegans we cannot exclude multifactorial bases for increased bacterial multiplication or for increased animal mortality in autophagy-deficient organisms. We observed a defect in intestinal epithelial cell bacterial lysosomal degradation in atg gene RNAi-treated animals, but increased bacterial replication might also result from enhanced epithelial cell bacterial uptake or invasion or impaired intestinal epithelial cell secretion of antimicrobial peptides. Indeed, in autophagy-deficient organisms, we observed an increase in luminal bacteria, and in daf-2-mutant animals (which have higher levels of basal autophagy), a decrease in luminal bacteria. Recently, mice defective in intestinal expression of the atg genes, Atg16L1 or Atg5, were found to have defects in the secretory function of the Paneth cell, a key cell type involved in gut antimicrobial peptide secretion (10). Interestingly, several genes encoding antibacterial lysozymes were induced in daf-2 mutants, including 2 intestinally expressed genes, lys-7 and lys-8 (26). Furthermore, genes encoding C. elegans antimicrobial peptides, abf-2 and spp-1, are induced during Salmonella infection and required to limit Salmonella infection in the intestinal lumen (27). Thus, it is possible that, in addition to restricting intracellular bacterial multiplication, autophagy is also involved in the production and secretion of antimicrobial peptides that limit intraluminal replication.
The mechanisms by which atg gene inactivation increases susceptibility to lethal Salmonella infection are not completely understood. Ultrastructural analyses of both Salmonella-infected nematodes and slime molds at later time points after infection reveal virtually complete cellular effacement by bacteria. Thus, one speculation is that the high bacterial burden or harmful products secreted by the bacteria leads to cytotoxicity. Although Salmonella has been reported to cause apoptotic death of intestinal epithelial cells (28), we did not observe a role for apoptosis in the increased lethality of Salmonella-infected autophagy-deficient nematodes. Another possibility is that, in addition to its role in decreasing the intracellular bacterial burden, autophagy, as a prosurvival mechanism, functions to “buffer” the stress imposed by intracellular bacterial infection.
Our findings not only identify an important role for atg genes in antibacterial host defense in vivo, they also identify atg genes as mediators of the pathogen-resistance phenotype of long-lived mutant nematodes. Pathogen resistance in daf-2-mutant animals requires at least two essential atg genes: bec-1 and lgg-1. Thus, autophagy is likely a specific innate effector that mediates not only lifespan extension (5–7), but also pathogen resistance in nematodes with mutation in the insulin-like signaling pathway. This connection between autophagy-mediated lifespan extension and pathogen resistance may be evolutionarily conserved. Perhaps age-related decreases in autophagy that occur in mammals may be mechanistically linked to age-related increases in susceptibility to certain infectious diseases, including those caused by intracellular bacterial pathogens.
One apparent discrepancy between our findings and a previous report (7) relates to the role of DAF-16 in autophagy regulation. In the present study, we found that two different DAF-16 over-expressing strains had increased levels of autophagy. However, Hansen et al. (7) found that a daf-16 null mutation did not block autophagy induction in daf-2 mutants. One potential explanation for this discrepancy is that another unidentified protein may function redundantly with DAF-16 in autophagy regulation. This might explain why a daf-16 null mutation has no effect on autophagy, whereas DAF-16 over-expression induces autophagy. Further studies are required to more clearly delineate the role of DAF-16, and other potential transcription factors downstream of DAF-2, in mediating autophagy and pathogen resistance in C. elegans. However, of note, forkhead transcription factors seem to play an evolutionarily conserved role in autophagy induction; FOXO over-expression induces autophagy in Drosophila (29) and Foxo3 regulates autophagy in mouse muscle cells (30, 31).
In conclusion, our data demonstrate that autophagy is a critical host-defense mechanism that limits intracellular infection with the bacterial pathogen S. typhimurium. Based upon our findings in nematode intestinal epithelial cells and in Dictyostelium, we speculate that the autophagic machinery may play a conserved role in protecting mammalian epithelial cells and phagocytes from bacterial attack. Furthermore, our data demonstrate a critical role for autophagy in mediating insulin-like signaling-regulated pathogen resistance in long-lived mutant nematodes. Thus, human atg gene polymorphisms (e.g., the Crohn's ATG16L variant) or age-related changes that reduce autophagy may contribute to impaired intestinal immunity to bacterial pathogens.
Materials and Methods
C. elegans, Dictyostelium, and Salmonella Strains.
Wild-type strains were the C. elegans Bristol strain N2, the D. discoideum strain DH1, and Salmonella enterica Serovar Typhimurium ATCC14028s (S. typhimurium). All mutant strains have been previously published and are described in the SI Materials and Methods. Strains TJ356 and CF1139 were out-crossed with our laboratory strain of N2 before Salmonella infection experiments. See details in the SI Materials and Methods.
RNAi Methods.
C. elegans feeding-RNAi experiments were performed as described (32), with the exception that a lower dose (1 nM instead of 1 mM) of IPTG was used to induce expression of bec-1 RNAi in daf-2(e1370)-mutant animals and DAF-16 over-expression strains (TJ356 and CF1139) to avoid decreases in lifespan in uninfected animals. The construction of feeding-RNAi plasmids is described in the SI Materials and Methods.
Salmonella Infection and Survival Studies.
All C. elegans RNAi treatment, Salmonella infection, and lifespan experiments were performed at 20 °C unless otherwise indicated, as described in detail in the SI Materials and Methods. All survival experiments were repeated at least two times, and the results shown represent an analysis of the combined data for the total number of animals per experimental group in all experiments. The results of all individual experiments were similar to that of the combined data. The mean lifespans, total number of worms per group, total censored animals, and survival statistical analyses are listed in Tables S1 and S2. See the SI Materials and Methods for details of statistical methods. Salmonella infection and survival studies in Dictyostelium were performed as described in the SI Materials and Methods.
Measurement of Bacterial Growth.
See the SI Materials and Methods for further details of bacterial growth measurement.
Microscopic Analyses.
For transmission EM of C. elegans, ≈100 adult nematodes per experimental group were collected and processed as previously described (5). Transmission EM analysis of Dictyostelium was performed as previously described (33). Light microscopic analyses of fluorescent nematodes and amoebae were performed using a Zeiss Axioplan2 Imaging microscope.
Autophagy Induction Analysis.
N2- and daf-2(e1370)-transgenic strains carrying the gfp::lgg-1 autophagy marker were described previously (5) and similar methods were used to measure seam cell autophagy in young adults (i.e., within 12 h beyond the L4 stage). The generation of TJ356 and CF1139 animals carrying the gfp::lgg-1 autophagy marker is described in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Alejandro Aballay, Leon Avery, Scott Cameron, Simon Daefler, and Richard Kessin for providing reagents, Tom Januszewski for assistance with electron microscopy, and the Caenorhabditis Genetics Center for C. elegans strains used in this study. This work was supported by National Institutes of Health Grant RO1 AI051367 (to B.L.), National Institutes of Health Clinical Translational Science Award Grant UL1 RRO24982 (to B.A.-H.), and an Ellison Medical Foundation Senior Scholars Award in Infectious Diseases (to B.L.) and a Ellison Medical Foundation New Scholars Award in Aging (to K.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813319106/DCSupplemental.
References
- 1.Gavazzi G, Krause KH. Ageing and infection. Lancet Infect Dis. 2002;2:659–666. doi: 10.1016/s1473-3099(02)00437-1. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Garsin D, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melendez A, et al. Autophagy genes are essential for dauer development and lifespan extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 6.Hars ES, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 7.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massey DC, Parkes M. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn's disease. Autophagy. 2007;3:649–651. doi: 10.4161/auto.5075. [DOI] [PubMed] [Google Scholar]
- 10.Deretic V, Master S, Singh S. Autophagy gives a nod and a wink to the inflammasome and Paneth cells in Crohn's disease. Dev Cell. 2008;15:641–642. doi: 10.1016/j.devcel.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millet AC, Ewbank JJ. Immunity in Caenorhabditis elegans. Curr Opin Immunol. 2004;16:4–9. doi: 10.1016/j.coi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Skriwan C, et al. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol. 2002;291:615–624. doi: 10.1078/1438-4221-00177. [DOI] [PubMed] [Google Scholar]
- 13.Voetsch AC, et al. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38(Suppl 3):S127–S134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- 14.Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 15.Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol. 2000;10:1543–1545. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 16.Kurz CL, Tan MW. Regulation of aging and innate immunity in C. elegans. Aging Cell. 2004;3:185–193. doi: 10.1111/j.1474-9728.2004.00108.x. [DOI] [PubMed] [Google Scholar]
- 17.Takacs-Vellai K, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 18.Aballay A, Ausubel FM. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc Natl Acad Sci USA. 2001;98(5):2735–2739. doi: 10.1073/pnas.041613098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto GP, et al. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol Microbiol. 2004;51:63–72. doi: 10.1046/j.1365-2958.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- 20.Golstein P, Aubry L, Levraud JP. Cell-death alternative model organisms: Why and which? Nat Rev Mol Cell Biol. 2003;4:798–807. doi: 10.1038/nrm1224. [DOI] [PubMed] [Google Scholar]
- 21.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 22.Terns RM, Kroll-Conner P, Zhu J, Chung S, Rothman JH. A deficiency screen for zygotic loci required for establishment and patterning of the epidermis in Caenorhabditis elegans. Genetics. 1997;146:185–206. doi: 10.1093/genetics/146.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci USA. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finlay BB, Brumell JH. Salmonella interactions with host cells: in vitro to in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355:623–631. doi: 10.1098/rstb.2000.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 26.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 27.Alegado RA, Tan MW. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol. 2008;10:1259–1273. doi: 10.1111/j.1462-5822.2008.01124.x. [DOI] [PubMed] [Google Scholar]
- 28.Guiney DG. The role of host cell death in Salmonella infections. Curr Top Microbiol Immunol. 2005;289:131–150. doi: 10.1007/3-540-27320-4_6. [DOI] [PubMed] [Google Scholar]
- 29.Juhasz G, et al. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14:1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001 doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon JM, Rupper A, Cardelli JA, Isberg RR. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun. 2000;68:2939–2947. doi: 10.1128/iai.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.