Abstract
The type II restriction endonucleases are indispensible tools for molecular biology. Although enzymes recognizing nearly 300 unique sequences are known, the ability to engineer enzymes to recognize any sequence of choice would be valuable. However, previous attempts to engineer new recognition specificity have met limited success. Here we report the rational engineering of multiple new type II specificities. We recently identified a family of MmeI-like type II endonucleases that have highly similar protein sequences but different recognition specificity. We identified the amino-acid positions within these enzymes that determine position specific DNA base recognition at three positions within their recognition sequences through correlations between their aligned amino-acid residues and aligned recognition sequences. We then altered the amino acids at the identified positions to those correlated with recognition of a desired new base to create enzymes that recognize and cut at predictable new DNA sequences. The enzymes so altered have similar levels of endonuclease activity compared to the wild-type enzymes. Using simple and predictable mutagenesis in this family it is now possible to create hundreds of unique new type II restriction endonuclease specificities. The findings suggest a simple mechanism for the evolution of new DNA specificity in Nature.
INTRODUCTION
Discrete DNA binding is of fundamental biological importance, allowing the bound proteins to target their function to specific locations within the DNA. A more complete understanding of the molecular basis for specific DNA recognition is desirable, both to enable the engineering of proteins to bind at DNA sequences of choice, and to improve prediction of DNA specificity for the numerous uncharacterized DNA binding proteins increasingly available from next generation sequencing technologies. The type II restriction endonucleases (REs) and their companion DNA methyltransferases comprise two large, well-characterized families of DNA binding proteins that exhibit exquisite sequence specificity. REBASE currently reports that 274 unique sequences are recognized among the more than 4000 biochemically characterized type II REs and companion DNA methyltransferases (1). The REs have typically evolved very great discrimination between their recognition sequence and all other DNA sequences (2), though the companion DNA methyltransferases may be somewhat less precise, since there is not the same level of selection against modifying non-cognate sites as there is against cutting non-cognate sites in the host genome (3). Crystal structures are available for 31 of the REs and 13 of the DNA methyltransferases, many of which are co-crystals with the enzyme bound to its DNA target (1). These structural studies reveal that specific DNA recognition is generally quite complex, involving both direct and water-mediated contacts that typically saturate the available hydrogen bonding potential of the functional groups on the DNA bases recognized (4). The type II REs have diverged to the point that they generally share little amino-acid sequence similarity, except in the case of isoschizomers that recognize the same sequence or close homologs that recognize related sequences (5). This lack of sequence conservation and the complexity of specific base recognition have made it difficult to predict which amino-acid residues determine recognition, or to rationally alter the DNA specificity of type II endonucleases.
Although it would be desirable to engineer enzymes to bind at and act upon any DNA sequence of choice, to date attempts to alter DNA specificity in the type II REs have met with only limited success. Early on it was hoped that structural information would guide the rational mutagenesis of key residues to alter particular base recognition, for example in EcoRI (6), or to add contacts to increase the length of the recognition sequence, for example with EcoRV (7,8). However, efforts to rationally alter specificity in these enzymes were largely unsuccessful. A significant effort was made to alter BamHI recognition. Guided by the structure (9,10), residues making specific base contacts in BamHI were extensively mutagenized, and an in vivo selection for binding using a catalytically inactive BamHI was employed in an attempt to change the BamHI recognition sequence (11). No mutants recognizing a new sequence were isolated, though a mutant that preferentially cut at the same sequence but required that the adenine base be methylated was found (11). Comparison of the structures of BamHI (GGATCC) and BglII (AGATCT), which share similar recognition sequences, led to the conclusion that type II REs ‘are remarkably resilient to alterations in the binding specificity’ (12). BstYI (RGATCY) recognizes a degenerate sequence that includes BamHI and BglII sites, as well as AGATCC. Attempts were made to alter BstYI to recognize only AGATCT (BglII) by a directed evolution approach, taking advantage of the M.BglII DNA methyltransferase for host DNA protection (13). A variant enzyme that preferred AGATCT 12-fold over AGATCC, and that no longer cut GGATCC was obtained, but a complete change in specificity was not accomplished. Subsequently the structure of BstYI was determined (14). Analysis of the structures revealed that although BstYI, BglII and BamHI share many similarities and are ideal test cases for changing specificity, the enzymes use surprisingly distinct recognition strategies, leaving open the question whether it is possible to completely switch specificity, even for the relatively conservative change of reducing BstYI recognition to specificity for only the BglII sequence (15). In another study, an approach that used random mutagenesis coupled with a genetic screen was employed in an attempt to alter the specificity of NotI (GCGGCCGC). This approach successfully isolated variants that recognize the wild-type sequence plus several sequences that differ at one base (16), but was not successful in generating a completely new specificity. Overall, the lack of success in altering specificity in the orthodox type II REs has suggested the general rule that changing a contacting residue results in a drop in catalytic activity but not a change of specificity (17).
Somewhat more success in generating enzymes with new specificity has been achieved with the unorthodox type II REs. One such approach successfully generated new specificities by joining two existing half-site recognition domains into new combinations. It was observed that naturally occurring recombination could create new specificities in type I R-M systems, where each half site was derived from a different parental R-M system (18). This was exploited in vitro to generate type I R-M systems having new, hybrid specificities (19). This approach has recently been extended to type II REs that, like the type I enzymes, recognize split sequences, resulting in hybrid enzymes that recognize a new sequence consisting of one half site from each of two parental enzymes (20).
A different approach was applied to alter specificity for Eco57I [CTGAAG(16/14)] (21). This took advantage of the unorthodox nature of this type IIG enzyme, which is a fused endonuclease—DNA methyltransferase (22). In this approach, endonuclease activity was abolished while DNA methylation activity was retained, the enzyme was randomly mutated, then methylase selection (23) was performed using a separate enzyme that recognizes a different sequence to isolate variants that now recognized and modified this new sequence to protect against the selecting endonuclease. This yielded an Eco57I variant that changed recognition at the fourth position from A to R: CTGRAG. This approach successfully produced an enzyme with altered specificity; however, because the method requires an existing endonuclease for the methylase selection step, it is limited to the generation of variants having previously known recognition sequences or subsets thereof.
The homing endonucleases have proved more amenable to rational specificity change than the type II REs, as evidenced by the recent successful alteration of a C:G to a G:C base pair through computational redesign of the specific contacts to this base pair (24). Additionally, significant progress has been made in engineering homing endonuclease DNA specificity, particularly with the enzyme I-CreI, through an approach of semi-rational mutagenesis and efficient screening for desired specificity changes (25–29). However, the type II REs have to date proved recalcitrant to engineering changes in recognition specificity.
MmeI is an unusual type II restriction enzyme that cuts DNA two turns of the helix away from its asymmetric recognition sequence (30–32). MmeI possesses both DNA methyltransferase and endonuclease activities in the same polypeptide. However, unlike previously described restriction-modification systems, MmeI relies on single strand modification for host protection (33). We have recently identified a family of REs highly similar to MmeI, which we proposed form a new subgroup within the type II endonucleases, the type IIL enzymes (34). The members of this family share significant primary amino-acid sequence similarity throughout their entire polypeptide chains. Their endonuclease and methyltransferase functions are highly conserved, however, their recognition sequences can be quite divergent. This overall conservation of function but diversification of substrate recognition indicates DNA specificity is undergoing rapid evolution. These proteins thus provide an excellent opportunity to study the determinants of DNA specificity. Currently no structure is available for any members of this family.
Here we report the rational engineering of new specificity variants within the MmeI family of type IIL REs. The engineered enzymes exhibit true conversion of specificity, and many exhibit specific activity comparable to the naturally occurring enzymes. The method described holds promise for the ability to engineer new type II REs, and also DNA methyltransferases, that specifically recognize more unique DNA sequences than have been isolated to date from natural sources.
MATERIALS AND METHODS
REs, S-adenosyl-l-methionine (AdoMet), T4 DNA Ligase, Phusion DNA polymerase (manufactured by Finnzymes Oy; Espoo, Finland), DNA size standards and competent cells were obtained from New England Biolabs (Ipswich, MA). DNA oligonucleotides were from New England Biolabs, Organic Synthesis Division (Ipswich, MA) or Integrated DNA Technologies (Coralville, IA). Plasmid preparation spin columns were from Qiagen (Valencia, CA). DNA purification spin columns were from Zymo Research (Orange, CA). Protein purification columns were from GE Healthcare (Piscataway, NJ).
Endonuclease assays
Endonuclease activity was assayed by incubating various amounts of enzyme in reaction buffer (NEBuffer 4: 20 mM Tris–acetate, pH 7.9, 10 mM magnesium acetate, 50 mM potassium acetate, 1 mM DTT, supplemented with 100 µg/ml BSA and 80 µM AdoMet) containing 1 µg substrate DNA per 50 µl for 1 h at 37°C. Reactions were terminated by addition of stop solution (50 mM EDTA, pH 8.0, 50% glycerol, 0.02% bromophenol blue), and reaction products were analyzed by electrophoresis in 1% LE agarose gels alongside DNA size standards: lambda-HindIII and PhiX174-HaeIII, or lambda-BstEII and pBR322-MspI.
Bioinformatics
The methodology to identify the particular amino acids that confer specific recognition depends on having a set of DNA binding proteins that have similar overall amino-acid sequences but that recognize differing DNA sequences. In this study, the MmeI family served as this input set of proteins (34). First, a multiple sequence alignment (MSA) of the amino-acid sequences was produced using the PROMALS web server (35). The default parameters were used, except that the ‘identity threshold above which fast alignment is applied’ was increased to 0.85. The alignment was used as produced. The MSA of the DNA recognition sequences was produced manually. Although the DNA recognized is double stranded and the enzymes make specific contacts to bases in both strands, for simplicity only the sequence of one strand was aligned, since the information in this strand determines the information in the complementary strand. The choice of strand to align was based on a functional attribute, which for the MmeI family enzymes was the direction that DNA cleavage occurs relative to the recognition sequence. The strand cut 3′ to the recognition site was aligned. Because the recognition sequences varied in length, the alignment was anchored about a common feature, the conserved adenine that is the target of modification, to put it in the proper register.
The aligned protein sequences were then grouped according to the DNA base recognized at the recognition alignment position under investigation and interrogated at each position within the protein MSA for correlations between the base recognized and the amino-acid residue present at that alignment position. Correlations in the current study were first identified by manual inspection, although subsequently the same positions have been identified using computer algorithms.
The aligned amino-acid positions that strongly correlated with DNA base recognition were selected for alteration. The residue at the correlated position was altered to an amino acid observed to correlate with recognition of the desired different DNA base. Alteration of recognition specificity typically required the coordinated alteration of a pair of amino-acid residues.
Mutagenesis
Mutagenesis was performed with the Phusion™ Site Directed Mutagenesis Kit protocol (New England Biolabs) using oligonucleotide primers that incorporated the desired amino-acid changes. The sequence of the mutagenic oligonucleotide primers used for the various amino-acid alterations are listed in Supplementary Table T1. The pairs of amino acids determining recognition at positions 3 or 6 were altered using a single pair of primers. The pair of amino acids specifying recognition at position 4 was altered sequentially using two separate pairs of primers, due to the distance separating them in the MmeI and homolog genes. The alteration at all three positions was also accomplished in two sequential steps using two separate pairs of primers (Supplementary Table T1).
Expression and characterization of altered enzymes
The altered enzymes were expressed, purified and their recognition sequence characterized as previously described (34). Briefly, plasmid carrying the altered enzyme gene was introduced into E. coli and isolates expressing active endonuclease were identified. Typically 0.5–4 g cells were grown, harvested, suspended in buffer, lysed by sonication and assayed for endonuclease activity. The altered endonuclease was partially purified over a 5 ml Heparin HiTrap column (GE Healthcare). Some enzymes were further purified using a second column, such as a 1 ml Q-HP or a 1 ml SP-HP (GE Healthcare). Column fractions containing the endonuclease were dialyzed against MmeI storage buffer (New England Biolabs) and used for subsequent analyses. Oligonucleotide primers used for cleavage position analysis are listed in Supplementary Table T1.
RESULTS
The DNA sequence specificity and respective amino-acid sequences for a family of enzymes remarkably similar to MmeI were recently described (34). The amino-acid MSA for the MmeI family members is shown in Supplementary Figure S1. The alignment of the DNA recognition sequences for these enzymes is shown in Supplementary Figure S2.
Alteration of recognition specificity at position 6
Two positions in the aligned protein sequences were identified that had significant correlation to the DNA base recognized at position 6 in the recognition sequence alignment. Those enzymes recognizing ‘C’ at position 6 invariably had an arginine at the alignment position corresponding to MmeI R808, while those enzymes recognizing ‘G’ invariably had an aspartate residue at this position (Figure 1). A second amino-acid alignment position, corresponding to MmeI E806, had significant, though not invariant, correlation to the DNA base recognized at position 6. Here 9 of 10 enzymes recognizing ‘C’ had a glutamate at this position, while one had a threonine. For those enzymes recognizing ‘G’, 9 of 10 had a lysine at this position, while one enzyme had a glycine, but also had a two amino-acid insertion, ‘GV’, immediately preceding this glycine residue. Thus the enzymes recognizing the C:G base pair where ‘C’ is in the top strand have the amino-acid charge pattern negative–positive, that is ExR, while those recognizing the same base pair but with ‘G’ in the top strand have the pattern positive–negative, that is KxD. PspOMII, the lone enzyme not recognizing either ‘C’ or ‘G’ at this position but rather purine (‘R’), that is either ‘A’ or ‘G’, had the conserved aspartate of the ‘G’ recognizing enzymes paired with a second aspartate at the position corresponding to MmeI E806. However PspOMII also had a three amino-acid insertion, ‘GMY’, immediately preceding the two correlating amino-acid positions (Figure 1).
Figure 1.
A portion of the amino-acid multiple sequence alignment of the characterized MmeI family enzymes, grouped according to the DNA base recognized at position 6 of the recognition sequence alignment. The two amino-acid alignment positions correlated with base recognition are highlighted in magenta. ‘Consensus ss’ indicates predicted secondary structure, where ‘h’ indicates alpha-helix and ‘e’ indicates beta-strand.
In an initial attempt to change recognition specificity from ‘C’ to ‘G’ at this position, mutants were made in MmeI and NmeAIII that exchanged the completely conserved arginine to aspartate (MmeI) and vice versa (NmeAIII), however, both the MmeI R808D and NmeAIII D818R single mutants were inactive. Double mutants were then made that exchanged the amino acids at both highly correlated positions: MmeI E806K, R808D and NmeAIII K816E, D818R. The double mutant enzymes were active and now recognized the predicted new sequences: TCCRAG for the altered MmeI and GCCGAC for the altered NmeAIII (Figure 2, Supplementary Figures S3 and S4). Both of these new recognition sequences represent novel type II endonuclease specificities. The altered enzymes are specific for the new recognition sequence and no longer recognize the original recognition sequence. The amount of activity obtained from clones expressing the altered enzymes was equal to that obtained from the wild-type enzymes, indicating the engineered enzymes have comparable specific activities to the wild-type enzymes. The altered enzymes cut at the same distance relative to the new recognition sequence as the wild-type enzymes: Mme6GI cut at TCCRAG(20/18), and NmeA6CIII cut at GCCGAC(21/19) (Supplementary Figure S5).
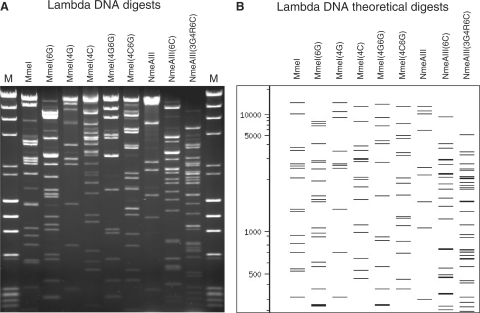
Figure 2.
Restriction fragment patterns for wild type and altered MmeI and NmeAIII variants. (A) Agarose gel showing endonuclease digestion fragment patterns of lambda DNA produced by the wild type and altered enzymes. MmeI cuts at TCCRAC(20/18); MmeI(6G) cuts at TCCRAG(20/18); MmeI(4G) cuts at TCCGAC(20/18); MmeI(4C) cuts at TCCCAC(20/18); MmeI(4G6G) cuts at TCCGAG(20/18); MmeI(4C6G) cuts at TCCCAG(20/18); NmeAIII cuts at GCCGAG(21/19); NmeAIII(6C) cuts at GCCGAC(21/19); NmeAIII(3G4R6C) cuts at GCGRAC(21/19). Size standards (M) are lambda-HindIII plus PhiX174-HaeIII. (B) Computer-generated restriction fragment patterns for cleavage of lambda DNA at the wild type and altered recognition sequences.
Alteration of recognition specificity at position 3
Two positions in the aligned protein sequences were identified that had significant correlation to the DNA base recognized at position 3 in the recognition sequence alignment. The protein MSA was grouped according to the DNA base recognized at position 3 in the recognition sequences alignment, where 7 enzymes recognize ‘C’, 10 recognize ‘G’, 3 recognize ‘T’ and 1 recognizes ‘Y’ (‘C’ or ‘T’) (Figure 3). The enzymes recognizing ‘G’ all had a positively charged amino acid (lysine or arginine) at the alignment position corresponding to MmeI E751, while those recognizing ‘C’ had a negatively charged amino acid, glutamate (5 of 7), or a serine (Figure 3). At a second alignment position corresponding to MmeI N773, the enzymes recognizing ‘G’ had either the negatively charged residue aspartate (7 of 10) or a serine. Enzymes recognizing ‘C’ had either an asparagine, a serine or a histidine at this position, but never an aspartate. The one enzyme recognizing ‘Y’ had the asparagine observed for those recognizing only ‘C’ at the MmeI N773 position, paired with a non-charged valine at the MmeI E751 position, suggesting the loss of discrimination between ‘C’ and ‘T’ may be due to the loss of a contact made by the glutamate or serine observed in the enzymes that recognize only ‘C’, coupled with flexibility for the asparagine residue to recognize either ‘A’ or ‘G’.
Figure 3.
A portion of the amino-acid multiple sequence alignment of the characterized MmeI family enzymes, grouped according to the DNA base recognized at position 3 of the recognition sequence alignment. The two amino-acid alignment positions correlated with base recognition are highlighted in orange. The boxes indicate the amino-acid pairs observed at the two correlated positions for each DNA base recognized. ‘Consensus ss’ indicates predicted secondary structure, where ‘h’ indicates alpha-helix and ‘e’ indicates beta-strand.
To alter recognition at position 3, an MmeI single position variant was made where the asparagine at position 773 was changed to aspartate: N773D; however, this variant was inactive for both endonuclease and DNA methyltransferase activities (33). A double mutant was then made that changed both correlated positions: MmeI E751R, N773D. This altered enzyme was active and now recognized the predicted new sequence in which the ‘C’ recognized by wild-type MmeI at position 3 is changed to ‘G’: TCGRAC (Supplementary Figure S6). This altered Mme3GI enzyme exhibited reduced activity compared to the wild-type enzyme such that complete digests were not obtained. Alteration of the glutamate at position 751 to lysine in the double mutant, E751K, N773D, resulted in the same specificity change from ‘C’ to ‘G’ at position 3 and similarly reduced overall enzyme activity. Mutation of the equivalent positions in NmeAIII, S761R and N783D, resulted in the same conversion of recognition from ‘C’ to ‘G’ at position 3, though again with significant reduction in overall activity (data not shown). However, the NmeAIII variant that combined the S761R and N783D mutations with alterations at positions 4 and 6 produced enough activity to achieve complete digestions and demonstrate the change at position 3 from recognition of ‘C’ to ‘G’ (Figure 2, Supplementary Figures S3 and S4).
Alteration of recognition specificity at position 4
The protein MSA was grouped according to the recognized base at position 4 of the recognition sequence alignment (Figure 4). At this position one enzyme recognizes ‘A’, five recognize ‘C’, nine recognize ‘G’, four recognize ‘R’, that is either ‘A’ or ‘G’, and two recognize any base, ‘N’. Those that recognize a purine base, that is only ‘A’, only ‘G,’ or ‘R’, were observed to have an arginine conserved at the position in the MSA corresponding to MmeI R810, while no enzyme that recognized ‘C’ or ‘N’ had an arginine at this position. A second position appeared necessary, in conjunction with the arginine, to distinguish between ‘A’, ‘G’ or ‘R’. A second position was also necessary to distinguish between ‘C’ and ‘N’, since one enzyme recognizing each had valine or methionine at the MmeI R810 position. The amino-acid residues at a second alignment position, corresponding to MmeI A774, were also found to correlate with recognition at position 4. Here enzymes recognizing ‘C’ had a lysine residue in four of five cases, while the fifth had a serine, while none of the enzymes recognizing bases other than ‘C’ had lysine at this position. The two enzymes not specifically recognizing any base at position 4 had glycine at this position. Since glycine has no side chain to specifically interact with any DNA base, this is consistent with the observed lack of specific recognition. The distinction between recognition of only ‘G’ and ‘R’ was postulated to occur based on the size of the residue at position A774. Those enzymes recognizing ‘R’ had a small residue at the A774 position, while the majority of the enzymes recognizing only ‘G’ had a residue with a larger side chain at this position that might preclude recognition of the A:T base pair. To test this, a single point mutation was made in MmeI: A774L. The MmeI A774L enzyme was active at the same level as wild-type MmeI but recognized only ‘G’ at position 4, whereas wild-type MmeI recognizes both ‘A’ and ‘G’, confirming that this amino-acid position participates in specificity determination for position 4 (Figure 2, Supplementary Figures S3 and S4). The reverse mutation was made in NmeAIII, where changing the wild-type leucine to alanine, L784A, resulted in recognition of ‘R’ at position 4 in the altered enzyme, versus recognition of only ‘G’ in the wild-type NmeAIII (Figure 2, Supplementary Figures S3 and S4).
Figure 4.
A portion of the amino-acid multiple sequence alignment of the characterized MmeI family enzymes, grouped according to the DNA base recognized at position 4 of the recognition sequence alignment. The two amino-acid alignment positions correlated with base recognition at position 4 are highlighted in green. The boxes indicate the amino-acid pairs observed at the two correlated positions for each DNA base recognized. ‘Consensus ss’ indicates predicted secondary structure, where ‘h’ indicates alpha-helix and ‘e’ indicates beta-strand.
Confirmation that both positions A774 and R810 confer recognition for position 4 was obtained by converting MmeI from recognition of ‘R’ to ‘C’ at this position. An MmeI variant was created where both positions where changed: MmeI A774K, R810S. This pair of mutations altered MmeI recognition from ‘R’ to ‘C’ at the fourth position, resulting in the new recognition specificity: 5′-TCCCAC-3′ (Figure 2, Supplementary Figures S3 and S4). The MmeI A774K, R810S mutant was specific for TCCCAC and had a similar level of activity compared to wild-type MmeI, however this mutant cut DNA nearly, but not quite to completion (Figure 2, Supplementary Figures S3 and S4).
Methylation specificity
The specificity of DNA methylation produced in vivo by the altered enzymes was tested. Plasmid DNAs of constructs expressing several of the altered enzymes were digested with wild-type MmeI or the altered enzymes. The plasmid DNAs were not cut by the enzyme variant they expressed, indicating they modified the same sequence at which they cut DNA. The plasmid DNAs that expressed the altered enzymes were cut by wild-type MmeI, indicating these enzymes no longer modified the wild-type MmeI recognition sequence. The altered endonucleases cut the wild-type MmeI plasmid DNA, indicating wild-type MmeI does not modify the sequences recognized by the altered enzymes (Supplementary Figure S7). The altered enzymes are thus specific for modification, as well as endonuclease activity, at their new, altered recognition sequence.
Combinatorial alteration of recognition specificity at multiple positions
Having identified the positions and amino-acid residues conferring recognition at single positions, a combinatorial approach was taken to generate enzymes that recognize new DNA bases of choice at multiple positions within the recognition sequence. Several enzymes were formed that changed recognition at two positions within their recognition sequence. MmeI was altered from ‘R’ to ‘G’ at position 4 and ‘C’ to ‘G’ at position 6 by combining three mutations: A774L, E806K and R808D. The enzyme so altered now specifically recognized 5′-TCCGAG-3′ and exhibited similar specific activity as wild-type MmeI (Figure 2, Supplementary Figures S3 and S4). Similarly an altered enzyme that recognized ‘C’ at position 4 along with ‘G’ at position 6, 5′-TCCCAG-3′, was formed from the quadruple mutant MmeI A774K, E806K, R808D, R810S (Figure 2, Supplementary Figures S3 and S4). This enzyme had activity comparable to that of the position 4C altered MmeI. An enzyme altered at positions 3 and 6 was made from MmeI E751R, N773D, E806K, R808D, which recognized the predicted new sequence 5′-TCGRAG-3′ (data not shown). Like the MmeI position 3G variant, this construct was specific for the new sequence but had a reduced level of endonuclease activity. An enzyme altered at positions 3 and 4 was formed by changing three amino acids in NmeAIII: S761R, N783D and L784A. This variant altered recognition from ‘C’ to ‘G’ at position 3 and from ‘G’ to ‘R’ at position 4, resulting in an enzyme that recognized 5′-GCGRAG-3′ (data not shown). A variant altered at all three positions was created. Recognition in NmeAIII was changed from 5′-GCCGAG-3′ to 5′-GCGRAC-3′ by changing five amino-acid residues: S761R, N783D, L784A, K816E and D818R (Figure 2, Supplementary Figures S3 and S4). However for MmeI, alteration at each of the three pairs of amino acids conferring recognition to a set of residues that gave active enzyme when altered singly and in pairs, that is to E751R, N773D, A774K, E806K, R808D and R810S, resulted in an inactive enzyme, indicating that not every combination of changes results in an active enzyme.
DISCUSSION
We have rationally created new specificity at three separate positions within the recognition sequences of a family of type IIL REs. It is remarkable that simple mutations result in predictable, authentic alteration of DNA specificity in the MmeI family enzymes. These results are quite different than those encountered in previous efforts to alter the more canonical type IIP restriction enzymes, such as BamHI or EcoRI. Because the MmeI family has retained highly similar protein sequences while evolving different recognition specificity, we were able to develop and apply a methodology that used correlations between position specific base recognition and position-specific amino-acid residues to identify the particular amino-acid residues within the proteins that recognize given DNA base pairs. We have demonstrated the utility of this methodology in one family of type IIL REs, and are exploring whether this approach will be applicable to other sets of proteins.
The MmeI family is adapted for specificity change
The MmeI family enzymes have features that make them naturally adapted to generate recognition sequence diversity. The endonuclease domain is separate from the target recognition domain (TRD), and thus catalysis need not be affected by alterations to DNA binding, as evidenced by the fact that the enzymes remain bound to and modify their recognition site following DNA cleavage. The potential toxicity of an endonuclease that acquires new specificity in the absence of corresponding modification imposes a significant barrier to endonuclease specificity change. Because the MmeI family enzymes require only the one strand modification they themselves produce, their single DNA binding domain is able to direct recognition for both their endonuclease and protective modification activities. Any alteration of this lone TRD thus results in coordinated change of host protection and endonuclease specificity, requiring only that a naive host survive any endonucleolytic damage sustained before modification at the new specificity is established. This eliminates the need to alter two DNA TRDs simultaneously to the same new specificity, as would be required to change specificity in most type II REs that require the presence of a separate DNA methyltransferase for host protection. The MmeI family thus represents a form of R-M system naturally optimized to allow specificity change. This is evidenced by the broad diversity of DNA specificity observed in these otherwise highly similar proteins. This biology greatly simplifies the task of forming new specificity in vitro, and represents a simple evolutionary mechanism for forming new specificities in Nature.
Predicted amino-acid contacts to the DNA bases
The MmeI family enzymes use discrete pairs of amino-acid residues to accomplish recognition of the DNA base pairs at positions 3, 4 and 6 of their recognition sequences. Analysis of the amino-acid residues that correlate with recognition of each base pair strongly suggests which DNA strand is contacted by each residue position. At recognition position 6, the arginine at position R808 is well suited to form a bidentate hydrogen bond to the N7 and O6 positions of the guanine base in the major groove, while the glutamate at position E806 could accept a hydrogen bond from the N4 amino group of cytosine (36,37). Such a pattern is observed in the structures of other REs, such as SfiI, where E106 and R220 specify recognition of the C3:G3 base pair (38). Conversely for enzymes recognizing G:C at position 6, the lysine at alignment position E806 could hydrogen bond to the top strand guanine N7 or O6 positions or both, while the aspartate at alignment position R808 could accept a hydrogen bond from the bottom strand cytosine N4 moiety. This suggests the amino acid corresponding to position MmeI E806 contacts the DNA base in the top strand at position 6, while the residue corresponding to MmeI R808 contacts the DNA base in the bottom strand (Figure 5).
Figure 5.
Predicted amino acid–DNA base contacts for MmeI. The MmeI amino-acid residue predicted to contact each individual base at positions 3, 4 and 6 of the double-stranded recognition sequence is shown. ‘Mtase’ indicates the methyltransferase binding pocket for the adenine that is flipped out of the helix and modified by the methyltransferase activity of the enzyme.
Similarly for recognition sequence position 4, enzymes that recognize a purine in the top strand have an arginine residue at the protein alignment position corresponding to MmeI R810, while those that recognize a guanine in the bottom strand most often have a lysine at the alignment position corresponding to MmeI A774. The R810 position arginine could form a bond to the N7 position of either purine base in the top strand. The lysine at the A774 alignment position of the enzymes recognizing ‘C’ in the top strand is well suited to bond to the guanine in the bottom strand, while glutamine or serine at the R810 position could accept a hydrogen bond from the cytosine N4 moeity. A hydrogen bond to a side chain is apparently not needed for recognition of the top strand C, however, as valine or methionine are observed to pair with the lysine to specify the C:G pair. Likewise, the alanine in MmeI at position A774 is unlikely to make specific contact to either pyrimidine base in the bottom strand, which is consistent with the observed degenerate recognition; however, the glutamate present in RpaB5I at this position could contact the bottom strand cytosine and, in concert with the R810 arginine, specify recognition of the G:C base pair in the same manner observed at position 6. These results suggest the amino-acid position corresponding to MmeI R810 contacts the base in the top strand at position 4, while the amino acid at the position corresponding to MmeI A774 interacts with the base in the bottom strand (Figure 5). The spacing between the MmeI R808 and R810 residues and their predicted contacts, that is, the bottom strand guanine at position 6 (R808) and top strand purine at position 4 (R810), appears strikingly similar to recognition in SfiI, where R218 and R220 each contact a guanine base that are two base pairs apart and located in opposite strands [R218 contacts (C1)-G1 and R220 contacts G3-(C3)] (38). The finding that MmeI can use 774K+810S to specify a C:G base pair, while 774L+810R specifies a G:C base pair is also remarkably similar to the computational redesign solution developed to change a C:G base pair to a G:C base pair in the homing endonuclease I-MsoI by the double mutation of K28L+T83R (24).
At position 3, a similar pattern occurs where enzymes recognizing guanine in the top strand have lysine or arginine at the position corresponding to MmeI E751, paired with either a negatively charged aspartate or a polar serine residue at the position corresponding to MmeI N773. Enzymes recognizing cytosine in the top strand have a negatively charged glutamate or a polar serine residue at the MmeI E751 position, paired with an asparagine, a polar serine, or in one case a histidine at the MmeI N773 position. The asparagine residue could form two hydrogen bonds to either purine; by donating bonds from the amine group to the O6 and N7 of guanine, or by donating and accepting a bond from the N6 and N7 of adenine, as occurs in the structure of PvuII (39). Serine or histidine could likewise form hydrogen bond to guanine. Recognition of the G:C base pair is analogous to SfiI, which recognizes the G1:C1 base pair with an arginine and a serine (R218, S210) (38), or BglI, which recognizes the same G:C base pair with arginine and aspartate (R277, D268) (40). In PspPRI the uncharged, relatively small valine residue present at position E751 may permit the presence of either pyrimidine base in the top strand, while the asparagine at position N773 could contact either purine base in the bottom strand to generate the observed pyrimidine: purine specificity. These results suggest the amino-acid position corresponding to MmeI E751 contacts the base in the top strand at position 3, while the amino acid at the position corresponding to MmeI N773 contacts the base in the bottom strand (Figure 5).
Taken together the recognition results imply these enzymes do not completely saturate the potential for hydrogen bonds with the bases in their target sequence. This lack of a requirement to saturate the potential base contacts likely gives these enzymes greater potential to change their specificity. The trade off may be that these enzymes also exhibit less discrimination between their specific and similar sites than typical type IIP REs. However, since any non-cognate sites bound can also be modified, such ‘star activity’ is likely to be less toxic to the host cell in MmeI-like R-M systems.
Mechanism for evolution of new DNA specificity
The finding that conserved amino-acid positions determine specificity in the MmeI family enzymes opens a window on the evolution of new DNA recognition specificities. The predicted secondary structural features observed in the MSA are remarkably conserved among these enzymes. A number of these secondary structure features are also conserved within the TRDs of the N6-adenine methyltransferases in general, and it has been suggested many methylases, including 5mC producing methyltransferases such as M.HhaI, may share a common structural core for recognition (41). It thus appears that many DNA methyltransferases, and perhaps other DNA binding proteins, have diverged from a common ancestor to the point that sequence conservation is not readily observed, but have retained a common structural core that allows specific recognition of short DNA sequences. In the MmeI family, these shared structural elements are able to present different amino-acid residues at the proper surface of the protein to specifically interact with different combinations of the DNA bases. This represents a ‘plug and play’ approach to recognition, where a given pair of amino acids determines the particular base recognized at a given position in the DNA recognition sequence.
It is remarkable that the proteins described are able to accommodate very different amino-acid residues, both in physical properties and size, at the conserved positions and still present these residues at the DNA interface such that they are able to form functional base-specific contacts. This implies flexibility in the proteins in order to position the different sized residues correctly to contact the DNA bases. For example, the residues in MmeI that recognize the C:G base pair at position 6, glutamate and arginine, are both larger than the residues that specify the same bases in the inverse strands, lysine and aspartate. MmeI can accommodate either lysine or arginine at position 806, in combination with aspartate at 808, to recognize the G:C base pair, and similarly can use either lysine or arginine at position 751, paired with aspartate at 773, to recognize G:C at position 3 (data not shown). This remarkable flexibility allows the enzymes to adopt new specificity from very simple mutations yet remain functional. While some changes resulted in lowered specific activity, in nature selection of secondary mutations could operate to restore any loss in activity occasioned by the change in specificity. The ability to alter multiple positions in these enzymes further demonstrates the flexible nature of the recognition domain.
The common structure and high degree of overall protein sequence similarity in this family makes it likely that new specificities might also be generated in Nature through recombination events that exchange all or part of the TRD region. A number of strains harbor multiple MmeI family enzymes, increasing the opportunity for such recombination to occur (for example, Agmenellum quadruplicatum PR-6 has three systems: AquII, AquIII and AquIV). An example of a possible exchange of TRDs can be seen in Microcystis aeruginosa PCC7806. This strain harbors two putative MmeI homologs, genes IPF4888 and IPF5080. These proteins have nearly identical amino-acid sequences (98.5% identities) everywhere except within their TRD region, where the level of identities drops to 55%, suggesting these enzymes derived from a recent parental system that acquired two different TRDs (Supplementary Figure S8).
The flexible nature of these proteins also means there are often multiple amino-acid combinations that can recognize a given base. Such natural flexibility is a boon to engineering the enzymes but complicates the bioinformatic identification of the recognition determinants.
Biological systems contain noise in correlations
Bioinformatic identification of amino-acid positions that determine recognition can be complicated by ‘noise’ in the correlations between DNA base recognition and amino-acid residues that results from the diversity in amino acid–base interactions. The amino acids present at the two positions that determine recognition for position 4 illustrate this difficulty. The same pair of amino-acid residues, threonine and arginine, specify ‘A’ or ‘G’ in one enzyme (SdeAI), but only ‘G’ in two others (DrdIV, MaqI). This conflict indicates there are additional factors affecting recognition to be discovered, such as structural influences that position or limit the movement of the residues in the identified positions to interact with the DNA to determine recognition. The short insertion of three amino acids in PspOMIII immediately preceding the amino acids determining recognition of position 6 may represent such a structural influence. The residues at position 4 demonstrate how the diversity and flexibility these proteins exhibit in achieving specific base recognition complicates the bioinformatic task of identifying correlations between position specific amino-acid residues and DNA base recognition. Bioinformatic approaches are therefore likely to benefit from a fuzzy approach that allows some ambiguity or even a small degree of outright conflict when assessing correlations.
Naming
The potential to generate new specificities by rational engineering creates the question of how to name these newly engineered enzymes. Naming the prototype enzymes has followed the accepted convention (42). We propose a naming scheme for the altered enzymes that uses the name of the prototype enzyme altered, followed by the position(s) altered listed by an Arabic numeral and the new base recognized at that position in parentheses. For example, an MmeI variant that recognizes at position 6 a G would be named MmeI(6G), while an NmeAIII variant that recognized at position 3 a G, position 4 an R and position 6 a C would be named: NmeAIII(3G4R6C). This is a functional naming system that describes the recognition of the enzymes. To distinguish different combinations of amino acids that produce the same recognition sequence, an additional tag consisting of an Arabic numeral following the name would be added in the order such enzymes were formed, for example MmeI(6G)-2 to distinguish the MmeI(6G) E806R, R808D variant from the original MmeI(6G) E806K, R808D enzyme.
The MmeI family offers an attractive system to study the determinants of base specific recognition. These enzymes offer a readily altered DNA recognition domain coupled with an efficient means to characterize any change in DNA-binding specificity through the location of endonuclease cleavage. Saturation mutagenesis to explore the possibilities for base recognition is tractable for each base position, given that only a pair of amino-acid residues determines recognition, and such studies are underway. As the repertoire of amino-acid combinations that specify a given base pattern is determined, it will be increasingly possible to engineer enzymes with desired specificities, and also to predict the recognition specificity for the many uncharacterized homologs increasingly available from genome and environmental sequencing projects. Observed homologs whose sequences do not match a known pattern become high priority targets for expression and characterization, as these are most likely to yield new recognition specificities.
Designer enzymes
Using the enzymes and method described it is now possible to rationally engineer hundreds of new type II RE (and DNA methyltransferase) specificities. At present we have rationally altered position 3 to two bases (C and G), position 4 to four base patterns (C, G, R and N), and position 6 to three base patterns (C, G and N). Among the enzymes characterized to date there are eight unique patterns for the first two positions in those enzymes recognizing six base pairs, and also eight unique patterns for the first three positions among the enzymes recognizing seven base pairs. Implementation of the demonstrated alterations would provide 192 unique specificities recognizing six base pairs, and a further 192 unique specificities recognizing seven base pairs: {[8 (6 base pair)+8 (7 base pair)] unique first position patterns known × 2 (position 3) ×4 (position 4) × 3 (position 6) = 384}. We have demonstrated that many of the identified alterations produce fully functional enzymes having the targeted specificity change. Although we have not yet experimentally demonstrated alteration at positions 0, 1 and 2, we anticipate that this is likely to be possible. Establishing changes to any or all of these recognition positions would greatly increase the number of recognition sequences that can be engineered. The ability to change all six recognition positions to just those bases observed in the currently characterized enzymes would result in enzymes with (2 × 5 × 5 × 4 × 5 × 1 × 3) = 3000 different recognition specificities. This represents a significant increase from the 274 unique type II RE recognition sequences currently reported in REBASE (1). These results move us closer to the goal of creating ‘designer REs’ that recognize and cleave at any desired DNA sequence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: New England Biolabs, Inc. 240 County Road Ipswich, MA 01938 USA.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Roberts for his valuable mentoring, and for extensive discussion and critique of the manuscript. We thank Yu Zheng and Chandra Pedamallu for implementing algorithms to identify correlations. We thank Geoffrey Wilson, Jim Samuelson, Iain Murray, Harriet Strimpel and William Jack for critical reading of the manuscript and discussion.
REFERENCES
- 1.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halford S.E. The specificity of the EcoRI restriction endonuclease. Biochem. Soc. Trans. 1980;8:399–400. doi: 10.1042/bst0080399. [DOI] [PubMed] [Google Scholar]
- 3.Heitman J., Ivanenko T., Kiss A. DNA nicks inflicted by restriction endonucleases are repaired by a RecA- and RecB-dependent pathway in Escherichia coli. Mol. Microbiol. 1999;33:1141–1151. doi: 10.1046/j.1365-2958.1999.01556.x. [DOI] [PubMed] [Google Scholar]
- 4.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki J.M. Crystallographic and bioinformatic studies on restriction endonucleases: Inference of evolutionary relationships in the ‘midnight zone' of homology. Curr. Protein Pept. Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg J., Wang B., Frederick C., Reich N., Greene P., Grable J., McClarin J. Development of a protein design strategy for EcoRI endonuclease. In: Oxender D.L., Fox F., editors. Protein Eng. NY: Alan R. Liss; 1987. pp. 237–250. [Google Scholar]
- 7.Lanio T., Jeltsch A., Pingoud A. On the possibilities and limitations of rational protein design to expand the specificity of restriction enzymes: a case study employing EcoRV as the target. Protein Eng. 2000;13:275–281. doi: 10.1093/protein/13.4.275. [DOI] [PubMed] [Google Scholar]
- 8.Jeltsch A., Wenz C., Wende W., Selent U., Pingoud A. Engineering novel restriction endonucleases: principles and applications. Trends Biotechnol. 1996;14:235–238. doi: 10.1016/0167-7799(96)10030-5. [DOI] [PubMed] [Google Scholar]
- 9.Newman M., Strzelecka T., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of restriction endonuclease BamHI phased at 1.95 A resolution by MAD analysis. Structure. 1994;2:439–452. doi: 10.1016/s0969-2126(00)00045-9. [DOI] [PubMed] [Google Scholar]
- 10.Newman M., Strzelecka T., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 11.Dorner L.F., Bitinaite J., Whitaker R.D., Schildkraut I. Genetic analysis of the base-specific contacts of BamHI restriction endonuclease. J. Mol. Biol. 1999;285:1515–1523. doi: 10.1006/jmbi.1998.2408. [DOI] [PubMed] [Google Scholar]
- 12.Lukacs C.M., Kucera R., Schildkraut I., Aggarwal A.K. Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution. Nat. Struct. Biol. 2000;7:134–140. doi: 10.1038/72405. [DOI] [PubMed] [Google Scholar]
- 13.Samuelson J.C., Xu S.Y. Directed evolution of restriction endonuclease BstYI to achieve increased substrate specificity. J. Mol. Biol. 2002;319:673–683. doi: 10.1016/S0022-2836(02)00343-1. [DOI] [PubMed] [Google Scholar]
- 14.Townson S.A., Samuelson J.C., Vanamee E.S., Edwards T.A., Escalante C.R., Xu S.Y., Aggarwal A.K. Crystal structure of BstYI at 1.85. ANG. resolution: a thermophilic restriction endonuclease with overlapping specificities to BamHI and BglII. J. Mol. Biol. 2004;338:725–733. doi: 10.1016/j.jmb.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 15.Townson S.A., Samuelson J.C., Xu S.Y., Aggarwal A.K. Implications for switching restriction enzyme specificities from the structure of BstYI bound to a BglII DNA sequence. Structure. 2005;13:791–801. doi: 10.1016/j.str.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Samuelson J.C., Morgan R.D., Benner J.S., Claus T.E., Packard S.L., Xu S.Y. Engineering a rare-cutting restriction enzyme: genetic screening and selection of NotI variants. Nucleic Acids Res. 2006;34:796–805. doi: 10.1093/nar/gkj483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves J., Vennekohl P. Protein engineering of restriction enzymes. Nucleic Acids Mol. Biol. 2004;14:393–411. [Google Scholar]
- 18.Fuller-Pace F.V., Bullas L.R., Delius H., Murray N.E. Genetic recombination can generate altered restriction specificity. Proc. Natl Acad. Sci. USA. 1984;81:6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubler M., Braguglia D., Meyer J., Piekarowicz A., Bickle T.A. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992;11:233–240. doi: 10.1002/j.1460-2075.1992.tb05046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurenaite-Urbanaviciene S., Serksnaite J., Kriukiene E., Giedriene J., Venclovas C., Lubys A. Generation of DNA cleavage specificities of type II restriction endonucleases by reassortment of target recognition domains. Proc. Natl Acad. Sci. USA. 2007;104:10358–10363. doi: 10.1073/pnas.0610365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rimseliene R., Maneliene Z., Lubys A., Janulaitis A. Engineering of restriction endonucleases: using methylation activity of the bifunctional endonuclease Eco571 to select the mutant with a novel sequence specificity. J. Mol. Biol. 2003;327:383–391. doi: 10.1016/s0022-2836(03)00142-6. [DOI] [PubMed] [Google Scholar]
- 22.Janulaitis A., Vaisvila R., Timinskas A., Klimasauskas S., Butkus V. Cloning and sequence analysis of the genes coding for Eco57I type IV restriction-modification enzymes. Nucleic Acids Res. 1992;20:6051–6056. doi: 10.1093/nar/20.22.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szomolanyi I., Kiss A., Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980;10:219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- 24.Ashworth J., Havranek J.J., Duarte C.M., Sussman D., Monnat R.J., Stoddard B.L., Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnould S., Chames P., Perez C., Lacroix E., Duclert A., Epinat J.C., Stricher F., Petit A.S., Patin A., Guillier S., et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2005;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 26.Chames P., Epinat J.C., Guillier S., Patin A., Lacroix E., Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith J., Grizot S., Arnould S., Duclert A., Epinat J.C., Prieto P.C., Redondo P., Blanco F.J., Bravo J., Montoya G., et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fajardo-Sanchez E., Stricher F., Paques F., Isalan M., Serrano L. Computer design of obligate heterodimer meganucleases allows efficient cutting of custom DNA sequences. Nucleic Acids Res. 2008;36:2163–2173. doi: 10.1093/nar/gkn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redondo P., Prieto J., Munoz I.G., Alibes A., Stricher F., Serrano L., Cabaniols J.P., Daboussi F., Arnould S., Perez C., et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 30.Boyd A.C., Charles I.G., Keyte J.W., Brammar W.J. Isolation and computer-aided characterization of MmeI, a type II restriction endonuclease from Methylophilus methylotrophus. Nucleic Acids Res. 1986;14:5255–5274. doi: 10.1093/nar/14.13.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucholski J., Zmijewski J.W., Podhajska A.J. Two intertwined methylation activities of the MmeI restriction-modification class-IIS system from Methylophilus methylotrophus. Gene. 1998;223:293–302. doi: 10.1016/s0378-1119(98)00450-8. [DOI] [PubMed] [Google Scholar]
- 32.Nakonieczna J., Kaczorowski T., Obarska-Kosinska A., Bujnicki J.M. Functional analysis of MmeI from methanol utilizer Methylophilus methylotrophus, a subtype IIC restriction-modification enzyme related to type I enzymes. Appl. Environ. Microbiol. 2009;75:212–223. doi: 10.1128/AEM.01322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan R.D., Bhatia T.K., Lovasco L., Davis T.B. MmeI: a minimal type II restriction-modification system that only modifies one DNA strand for host protection. Nucleic Acids Res. 2008;36:6558–6570. doi: 10.1093/nar/gkn711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan R.D., Dwinell E.A., Bhatia T.K., Lang E.M., Luyten Y.A. The MmeI family: type II restriction-modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res. 2009;37:5208–5221. doi: 10.1093/nar/gkp534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pei J., Kim B.H., Tang M., Grishin N.V. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res. 2007;23:802–808. doi: 10.1093/nar/gkm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeman N.C., Rosenberg J.M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanamee E.S., Viadiu H., Kucera R., Dorner L., Picone S., Schildkraut I., Aggarwal A.K. A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. EMBO J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng X., Balendiran K., Schildkraut I., Anderson J.E. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman M., Lunnen K., Wilson G., Greci J., Schildkraut I., Phillips S.E.V. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J. 1998;17:5466–5476. doi: 10.1093/emboj/17.18.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturrock S.S., Dryden D.T.F. A prediction of the amino acids and structures involved in DNA recognition by type I DNA restriction and modification enzymes. Nucleic Acids Res. 1997;25:3408–3414. doi: 10.1093/nar/25.17.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S.K., Dryden D.T., Dybvig K., et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.