Abstract
Objective
We explored automated concept-based indexing of unstructured figure captions to improve retrieval of images from radiology journals.
Design
The MetaMap Transfer program (MMTx) was used to map the text of 84,846 figure captions from 9,004 peer-reviewed, English-language articles to concepts in three controlled vocabularies from the UMLS Metathesaurus, version 2006AA. Sampling procedures were used to estimate the standard information-retrieval metrics of precision and recall, and to evaluate the degree to which concept-based retrieval improved image retrieval.
Measurements
Precision was estimated based on a sample of 250 concepts. Recall was estimated based on a sample of 40 concepts. The authors measured the impact of concept-based retrieval to improve upon keyword-based retrieval in a random sample of 10,000 search queries issued by users of a radiology image search engine.
Results
Estimated precision was 0.897 (95% confidence interval, 0.857–0.937). Estimated recall was 0.930 (95% confidence interval, 0.838–1.000). In 5,535 of 10,000 search queries (55%), concept-based retrieval found results not identified by simple keyword matching; in 2,086 searches (21%), more than 75% of the results were found by concept-based search alone.
Conclusion
Concept-based indexing of radiology journal figure captions achieved very high precision and recall, and significantly improved image retrieval.
Introduction
Images published in peer-reviewed journals provide valuable information for education and clinical decision support. Retrieval of images based on their visual properties and textual captions is an area of active research. 1 The articles in which the figures appear are indexed by Medical Subject Heading® (MeSH®) terms (U.S. National Library of Medicine, Washington, DC), 2 which enables users to find articles using medical concepts. Although MeSH-based search can help find journal articles, it is not well suited to the task of finding particular images in those articles. Such images generally have an associated figure caption. The caption's text provides more granular information, which can allow more robust search and retrieval of images. Searching the text within figure captions is plagued by the same challenges that are encountered when searching other clinical free-text information, such as radiology reports. We evaluated the impact of semantic indexing—the mapping of unstructured text to controlled terms—to improve retrieval of radiological images from journal articles.
Background
Semantic, or concept-based, indexing allows users to search for information using medical concepts. For example, concept-based searches recognize abbreviations, synonyms, and lexical variants. Most importantly, concept-based retrieval systems recognize subtypes of specific terms; for example, such systems understand that Parosteal Osteosarcoma is a type of Osteogenic Sarcoma, which is in turn a type of Bone Tumor. These systems require a robust model of medical knowledge to understand medical concepts and their interrelationships. Purely text-based retrieval systems are challenged by abbreviations and lexical variants, which stimulated our strategy to employ concept-based indexing.
To facilitate concept-based retrieval of images in articles, one could index the images using concepts extracted from the associated captions. However, it would be extremely laborious to perform this task manually. We explored an automated technique to map the unstructured (“free”) text of figure captions to concepts in a set of controlled vocabularies. Methods such as those described in this report can enable the radiology community to access more effectively the vast amounts of radiological image data being published online.
Several approaches have been explored for concept-based indexing of unstructured biomedical text. Systems such as MicroMeSH, 3 CHARTLINE, 4 CLARIT, 5 SAPHIRE, 6,7 Metaphrase, 8 and work by Nadkarni, et al 9 have been applied in a variety of applications to map unstructured text to the MeSH vocabulary and/or the UMLS Metathesaurus. The MetaMap program 10–12 offers a linguistically rigorous concept-discovery approach, and a version of the software can be obtained without cost. To improve retrieval of radiology images from the biomedical literature, we explored the use of MetaMap to index the text of radiology figure captions.
Methods
This work had two specific aims: (1) to evaluate the ability of a concept-mapping algorithm to correctly map free-text radiology figure captions to controlled vocabulary concepts, and (2) to measure the impact of concept-based searching on the performance of an image search engine. First, we used a concept-mapping algorithm to discover controlled-vocabulary terms in a collection of radiology figure captions and to index the captions accordingly. We applied standard information-retrieval performance metrics to measure the effectiveness of our semantic indexing process. Finally, we examined the effects of concept-based retrieval on real-life queries to a popular image search engine that uses this indexing approach. This investigation involved only analysis of information in the published literature, and did not involve any human subjects or protected health information; therefore, this study was exempt from Institutional Review Board review.
Source Vocabularies
The Unified Medical Language System (UMLS®) Metathesaurus, licensed from the U.S. National Library of Medicine, served as the knowledge model for the image retrieval system. The Metathesaurus is a very large database of biomedical and health-related concepts, their various names, and the relationships among them. 13–15 It is built from the electronic versions of many different source vocabularies, such as classification schemes, thesauri, and lists of controlled terms used in patient care, health services billing, biomedical research, public health statistics, and biomedical literature indexing. To index the text corpus used in this study, we employed three source vocabularies from the UMLS, version 2006AA: Systematized Nomenclature of Medicine Clinical Terminology® (SNOMED-CT®; International Health Terminology Standards Development Organisation, Copenhagen, Denmark), 16,17 the Foundational Model of Anatomy, 18,19 and the MeSH vocabulary, as these are the dominant sources for terms relevant to our corpus. The aggregate vocabulary consisted of 1,735,102 terms representing 662,736 distinct concepts.
Concept Mapping Algorithm
We implemented the National Library of Medicine's MetaMap Transfer (MMTx) program to discover Metathesaurus concepts in unstructured (free-text) figure captions. MMTx employs a series of language-processing modules to map text to concepts in the UMLS Metathesaurus. 12,20 MMTx first parses text into components, including sentences, paragraphs, phrases, lexical elements, and tokens. Variants are generated from the resulting phrases. Candidate concepts from the UMLS Metathesaurus are retrieved and evaluated against the phrases. The best of the candidates are subsequently organized into a final mapping in such a way as to best cover the text. We employed MMTx's “strict” model of the UMLS Metathesaurus, version 2006AA. The strict filtering option limits the search to terms that are supported by both the MetaMap and PubMed Related Citations indexing methods. This approach tends to give a small list of very good candidate controlled terms, but may filter out some good recommendations as well.
Experimental Dataset
The ARRS GoldMiner® system (http://goldminer.arrs.org; American Roentgen Ray Society, Leesburg, VA) is a widely used image search engine that is freely available via the Internet. Goldminer uses both concept- and keyword-based search techniques to retrieve images from a large number of open-access, peer-reviewed journals. 21 To build the experimental dataset, we extracted 84,846 figure captions from the GoldMiner database. The figure captions, derived from GoldMiner's initial set of figures, were acquired from 9,004 articles published online from 1999 to 2006 in five peer-reviewed, English-language radiology journals: American Journal of Roentgenology (AJR), American Journal of Neuroradiology, British Journal of Radiology, RadioGraphics, and Radiology. All the articles from which the figures and captions are derived were available for open access. We created automated pattern-matching modules to remove hypertext mark up language (HTML) tags from the figure captions so that we could build a corpus containing only the text from the captions.
Information Retrieval Metrics
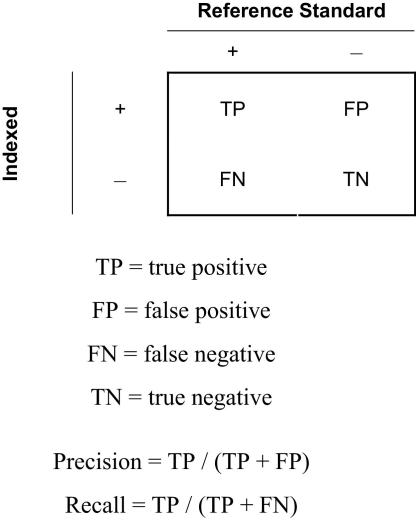
To assess the performance of our concept-mapping approach, we sought to evaluate the standard information-retrieval metrics of precision and recall (▶). 22,23 The Reference Standard is a Boolean value that indicates, based upon manual review, whether the specified concept is present in the figure caption. The Indexed variable indicates whether MMTx identified the concept in the figure caption.
Figure 1.
Contingency table, with variables used to compute precision and recall. “Reference Standard” indicates that a concept is present (+) or absent (−) in a figure caption, as determined by manual review. “Indexed” indicates whether the concept has been identified in the figure caption by the algorithm.
To compute precision and recall exactly, for each possible paring of concepts and captions one must compare whether the concept is truly present in the caption versus whether the algorithm has assigned it as present. However, for sets of c concepts and f figure captions, the cross-product—the set of all concept-caption pairs—has c × f elements. Here, the number of concept-caption pairs is 662,736 × 84,846, which exceeds 56 billion. Thus, we applied sampling strategies to estimate the precision and recall of the indexing technique.
We used both “microaveraging” and “macroaveraging” to estimate these metrics. Microaveraging considers all concept-caption pairs as a single group. Macroaveraging computes the effectiveness measure separately for the set of captions associated with each concept, and then computes the mean of the results values. Macroaveraging is generally favored because it gives equal weight to each user query. 23
Reference Standard
To establish a reference standard, one of the authors (CEK) served as reviewer. The reviewer was presented sequentially with paired figure captions and concepts. For each concept-caption pair, the reviewer viewed the complete free-text figure caption, the UMLS concept unique identifier (CUI), and list of terms for that concept. The reviewer indicated whether the concept was present in the figure caption's text. To eliminate potential bias, the sequence of caption-concept pairs was randomized; the reviewer was blinded as whether one was determining if the concept might be present or absent within the figure caption.
Precision
Precision measures the fraction of retrieved documents that are relevant to a specific query, and is analogous to positive predictive value. To estimate the precision, we randomly selected 250 concepts among those that appeared in the collection. For each concept, we selected a random sample of up to five figure captions in which MMTx identified the concept as present. Those captions were reviewed manually to determine if the caption was indexed by specified concept correctly (true positive [TP]) or incorrectly (false positive [FP]). We computed the precision as the number of captions correctly indexed (TP) divided by the total number of captions indexed (TP + FP). We calculated the 95% confidence interval (CI95) for precision based on the size of the sample.
Recall
Recall measures the fraction of all the relevant documents in a collection that are retrieved by a specific query, and is akin to the concept of sensitivity. Here, recall is the number of figure captions that were indexed by a concept divided by the number of captions in which the concept was actually present. We estimated recall by sampling concepts and captions. We randomly selected 40 concepts, each of which MMTx had indexed in more than 10 figure captions. For each concept, the true positive (TP) value was estimated as the total number of “positive” captions (those indexed by that concept) multiplied by the overall precision value. Then, for each concept, we sampled 25 figure captions from among those that were not indexed by that concept and reviewed those concept-caption pairs. Those captions should be negative; the “Sample TN” is the number of true negative (TN) figures among the 25 sampled for each concept. Based on the Sample TN value, we extrapolated to the entire set of negative captions. Recall was computed as the number of correctly indexed captions (TP) divided by the number of captions that truly contained the concept (TP + TN).
We illustrate our estimation of recall with an example. Consider the concept Liver diseases (C0023895), which was identified in 90 figure captions (▶). Given an overall precision value of 90%, there are an estimated 81 “true positive” (TP) captions for this concept. Now we examine the sample of 25 captions not indexed by this concept. If one of the 25 sampled captions in fact contains the concept, then that caption is falsely negative; thus the false-negative fraction would be 1/25. To estimate the number of false negatives in the entire dataset, we multiply the false-negative fraction by the total number of negative captions (84,756 captions = 84,846 − 90) to yield 3,390. Thus, for Liver diseases, the estimated recall would be TP/(TP + FN) = 81/(81 + 3,390) = 0.023.
Table 2.
Table 2 Estimate of Recall from Sample of 40 Concepts
| CUI | Concept Name | Captions Indexed | Sample TN | Est. Recall |
|---|---|---|---|---|
| C0011304 | Demyelination | 51 | 25 | 1.000 |
| C0011331 | Dental Procedures | 18 | 25 | 1.000 |
| C0016911 | Gadolinium | 1,770 | 25 | 1.000 |
| C0018099 | Gout | 43 | 25 | 1.000 |
| C0020883 | Ileostomy | 30 | 25 | 1.000 |
| C0021925 | Intubation | 54 | 25 | 1.000 |
| C0023895 | Liver diseases | 90 | 24 | 0.023 |
| C0030424 | Paragonimiasis | 16 | 25 | 1.000 |
| C0032463 | Polycythemia Vera | 174 | 25 | 1.000 |
| C0037939 | Spinal Neoplasms | 291 | 25 | 1.000 |
| C0038895 | Surgical Aspects | 2,121 | 22 | 0.159 |
| C0040132 | Thyroid Gland | 334 | 25 | 1.000 |
| C0042382 | Vascularization | 53 | 25 | 1.000 |
| C0085406 | Anisotropy | 186 | 25 | 1.000 |
| C0149554 | Frontal Horn | 79 | 25 | 1.000 |
| C0179376 | Bottle, device | 15 | 25 | 1.000 |
| C0185792 | Incision of sternum | 16 | 25 | 1.000 |
| C0205556 | Qualitative | 58 | 25 | 1.000 |
| C0225897 | Left ventricular structure | 648 | 25 | 1.000 |
| C0226862 | Structure of straight sinus | 53 | 25 | 1.000 |
| C0280100 | Solid tumor | 67 | 20 | 0.003 |
| C0332218 | Difficult | 396 | 25 | 1.000 |
| C0332272 | Better | 1,567 | 25 | 1.000 |
| C0428772 | Left ventricular ejection fraction | 23 | 25 | 1.000 |
| C0443343 | Unstable status | 80 | 25 | 1.000 |
| C0449379 | Connection | 186 | 25 | 1.000 |
| C0450195 | Cervicothoracic | 27 | 25 | 1.000 |
| C0489800 | Left Calf | 59 | 25 | 1.000 |
| C0521104 | Permission | 947 | 25 | 1.000 |
| C0522537 | Xenograft type of graft | 11 | 25 | 1.000 |
| C0560737 | Bone structure of hamate | 28 | 25 | 1.000 |
| C0600080 | Stretching exercises | 65 | 25 | 1.000 |
| C1269584 | Entire posterior semicircular canal | 11 | 25 | 1.000 |
| C1278929 | Entire liver | 4,283 | 25 | 1.000 |
| C1280264 | Entire pterygoid muscle | 25 | 25 | 1.000 |
| C1280605 | Entire infratemporal fossa | 19 | 25 | 1.000 |
| C1280839 | Entire incus | 53 | 25 | 1.000 |
| C1305627 | Entire superior ramus of pubis | 11 | 25 | 1.000 |
| C1446409 | Positive | 1,371 | 25 | 1.000 |
| C1457873 | Os trigonum disorder | 19 | 25 | 1.000 |
| MACRO-AVERAGE | 0.930 |
For each concept, the table lists the UMLS concept unique identifier (CUI), the concept name, and the number of “positive” captions (indexed by that concept). For each concept, 25 “negative” figure captions (those not indexed by the concept) were sampled. The number of true negatives in that sample (sample TN) is indicated, and the estimated recall value is computed.
F 1 Value
For values of precision, P, and recall, R, we computed the F 1 score, the harmonic mean of precision and recall, as:
Impact on Search Engine Performance
To evaluate how semantic indexing enhances search, we obtained a set of 10,000 randomly selected entries from the ARRS GoldMiner search engine's log file. Each log-file entry included the total number of images (N) retrieved, the number of images found by concept-based search alone (C), and the number found by keyword-based search alone (K). Because the total search result is the union of the concept- and keyword-based searches, N ≤ C + K. We computed the fraction of results that were contributed by concept-based search alone—that is, (N–K) / N—to assess the extent to which concept-based searching increased the number of total results. Keyword-based search used the MySQL database management system's case-insensitive, whole-word “FULLTEXT” indexing method.
Results
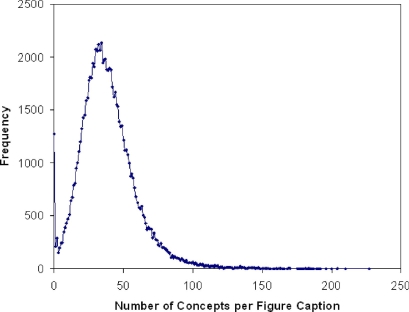
The MMTx program identified 31,108 unique concepts in the radiology figure captions. A figure caption with its indexing terms is shown as an example in ▶. The number of concepts found per figure caption ranged from 0 to 227 (median, 36; mean ± SD, 38.6 ± 20.1). The distribution of the number of concepts per caption is shown in ▶.
Table 1.
Table 1 Concepts Identified in Figure Caption Number 54194 48 : “Portal vein gas. Contrast material-enhanced CT scans obtained at the top of the liver show tubular areas of decreased attenuation in the periphery of the liver (arrows), findings that are consistent with gas in the intrahepatic portal veins”
| CUI | Concept Name |
|---|---|
| C1305775 | Entire portal vein |
| C0017110 | Gases |
| C0596601 | Gastrointestinal gas |
| C0032718 | Portal vein structure |
| C0205054 | Hepatic |
| C1278960 | Entire vein |
| C0042449 | Veins |
| C0009924 | Contrast Media |
| C0040405 | X-ray computed tomography |
| C0441633 | Scanning |
| C0520510 | Materials |
| C1301820 | Obtained |
| C0000811 | Termination of pregnancy |
| C1278929 | Entire liver |
| C0023884 | Liver |
| C0151747 | Renal tubular disorder |
| C0332208 | Tubular formation |
| C0205216 | Decreased |
| C0205100 | Peripheral |
| C0336721 | Arrow |
| C0243095 | Finding |
| C0582254 | Intrahepatic portal vein |
| C1512948 | Intrahepatic |
Figure 2.
Histogram showing the number of concepts identified per figure caption. The text corpus included 1,273 figure captions (1.5%) in which no concept was identified.
At least one concept was discovered in 83,573 (99.95%) of the 83,615 nonempty figure captions. The five most common concepts appeared in 41–62% of all captions, whereas 4,035 concepts appeared only once. The 50 most common concepts (0.2% of all concepts identified) accounted for 25% of references to concepts in the entire collection.
Precision
By selecting up to five figure captions indexed for each of 250 randomly selected concepts, 890 figure captions were identified. Of these captions, 784 were indexed correctly; microaveraging yielded an estimated precision of 0.881 (CI95, 0.859–0.903). By macroaveraging on the 250 concepts, the mean precision was 0.897 (CI95, 0.857–0.937).
Recall
Review of the sample of 25 figure captions classed as “negative” for each of the 40 randomly sampled concepts (1,000 figure captions) found nine figure captions to be classified incorrectly (▶). Macroaveraging on the 40 concepts yielded an estimated recall of 0.930 (CI95, 0.838–1.000). The F 1 value based on macroaveraging was 0.913.
Search Results
In 5,535 queries (55%) of the random sample of 10,000 ARRS GoldMiner search queries, concept-based retrieval found figure captions that were not found by searching only for keywords. In 2,086 searches (21%), concept-based search accounted for more than 75% of the total search results.
Discussion
Indexing is critical to rapid and accurate retrieval of pertinent information from large databases. Typical indexing techniques are based on keywords, for which exact (letter-by-letter) matches are sought within the text corpus. Conceptual indexing differs from keyword indexing in that the text is labeled with descriptive terms, usually taken from a controlled terminology. Concept-based indices often possess taxonomical structure—i.e., relationships among concepts—which enable applications to use the term hierarchy to expand or generalize the search. The concept hierarchy also often contains terminological information such as lexical variants, abbreviations, and synonyms that can be exploited in searching the raw text.
Conceptual indexing offers several advantages: for one, it allows the recognition of a term's lexical variants, semantic variants, synonyms, and abbreviations. For example, the term “esophagus” has the lexical variant “oesophagus” and the adjectival form “esophageal”. The term “hepatocellular carcinoma” has the synonym “hepatoma” and the abbreviation “HCC”. Conceptual indexing allows search engines to intelligently unify such lexical variants, as well as to expand queries to retrieve information based on the meaning of the concept and its relationships to other concepts.
In radiology figure captions, descriptions tend to focus on anatomy, diseases, radiological findings, and imaging techniques—a subset of general language which is much more varied. The focused scope of radiology language may account for the high performance of our approach.
Our concept-based indexing approach has been incorporated into GoldMiner to improve retrieval for user searches. The results for the 10,000 search queries suggests that concept-based indexing substantially increases the number of images retrieved; in fact, our methods have been adopted in the current release of GoldMiner. Given the high precision and recall of the concept-based index, the images retrieved should be highly relevant to the query terms. Identification of age, sex, and imaging-modality metadata in radiology figure captions also can be accomplished with high recall and precision. 24
Concept-based indexing of text has been undertaken in earlier work. For example, in using the SAPHIRE system to index concepts in radiology reports, the researchers found recall of 63% but a precision of only 30%. 25,26 The precision and recall found in this study were very high. Some of the differences in our results from that of prior work may relate to the methods for concept recognition and differences in the domain of the text being indexed.
Retrieval of images based on their visual content and textual annotations is an area of active research. The ImageCLEFmed 2008 medical image retrieval task, part of the Cross-Language Evaluation Forum (CLEF) information retrieval challenge, employed a subset of images that have been indexed by ARRS GoldMiner. 1 Yu and colleagues have explored the analysis of figure captions and associated text from journal articles to answer biological questions. 27,28
Because of the rich interconnections among its component vocabularies, the UMLS Metathesaurus is an important source of medical knowledge. 13–15 Indexing of unstructured text to standardized vocabularies—similar to that done in this study—has improved information retrieval in several other biomedical domains. The KnowledgeMap system has been used to identify Metathesaurus concepts in the impression text of electrocardiogram reports. 29 Dermatlas, a Web-based collection of dermatology cases, was indexed to MeSH terms using the National Library of Medicine's Medical Text Indexer (MTI). 30 Ontology-based indexing has been shown to aid retrieval and extraction of information from the biomedical literature. 31,32 Shah, et al developed and applied techniques to map free-text annotations of tissue microarray data to structured vocabularies. 33 We chose the approach described here because MMTx offered high-quality indexing, was readily available, and was integrated with the UMLS Metathesaurus. Lexical expansions and exploitation of knowledge in UMLS make this approach particularly advantageous in the radiology domain to improve recall of matching concepts. Preliminary analysis of GoldMiner's performance showed that this indexing approach functioned well in our domain.
A limitation of our work is that we did not measure recall for all concepts, but estimated it by sampling. To measure recall most accurately, one would have to determine how many relevant documents are retrieved for each search concept. Given the number of concepts and the size of the database, such measurement would have been prohibitive. We believe our estimated recall based on sampling figure captions and concepts is a reasonable approach to this limitation.
Although widely used, MMTx, which is incorporated into our system, has several limitations: it is relatively slow, it is limited to UMLS vocabularies, and it is unable to process negation. Because figure captions from journal articles are processed as a “background” task, MMTx's processing speed was not detrimental to our project. Investigators have developed a new MetaMap module that identified 91% of the concepts found by MMTx in 14% of the time taken by MMTx. 34 Alternative algorithms, such as MGREP 35 or MTag, 36 may provide sufficient speed to allow real-time mapping of clinical text to controlled vocabularies. 37 Such systems would allow flexibility to use vocabularies, such as RadLex, which are not yet part of the UMLS. RadLex offers terms for radiology-specific observations that are not found in other terminologies. One goal is to integrate semantic indexing of clinical radiology reports in real time. Real-time indexing could allow integration of clinical systems with ontology-based knowledge resources.
Another limitation of MMTx is that it depends on UMLS for its source terminologies, and UMLS lacks terminologies specific to radiology. RadLex, a unified vocabulary for radiology that is being transformed into an ontology of radiology knowledge 38–40 may help improve our concept-based image retrieval method. Until RadLex is incorporated into the UMLS Metathesaurus, other tools must be used to map text to terms in that lexicon. Another limitation is that MMTx lacks negation detection, so that both positive and negative statements are indexed equivalently. Although satisfactory for figure captions (which generally mention negative concepts only if relevant, e.g., “no evidence of appendicitis”), such an approach likely would retrieve too many false-positive results when dealing with clinical text such as radiology reports.
Concept-based indexing of clinical documents is an area of active investigation. 41 Although text mining and semantic indexing have been applied successfully to molecular biology and the biomedical literature, relatively few studies have explored their application to clinical content. 42 In radiology, automated techniques have been used to code findings in cancer-related radiology reports, 43 to identify findings of congestive heart failure, 44 and to identify clinically important findings. 45 Semantic indexing has improved noun phrase identification 46 and overall precision of information retrieval 47 in radiology reports. Real-time semantic indexing of the content of radiology reports creates opportunities to integrate the reporting process with clinical decision support and point-of-care learning, and may improve the quality of radiology practice and learning.
Conclusions
Our goal was to assess the performance of the MMTx system for concept-based indexing of radiology figure captions. In our study, MMTx demonstrated precision of 0.897 and estimated recall of 0.930. This indexing approach has been incorporated into the ARRS GoldMiner Web-based image search engine. Concept-based indexing allowed retrieval of results not identified by keyword-based retrieval in more than half of all actual search queries, based on a large sample. Concept-based indexing can achieve high precision and recall, and can improve retrieval of radiology images and their textual captions.
Footnotes
This study was supported in part by the American Roentgen Ray Society. The work also was supported in part by the National Center for Biomedical Ontology under roadmap-initiative grant U54 HG004028 from the NIH.
References
- 1.Müller H, Kalpathy-Cramer J, Kahn Jr CE, et al. Overview of the ImageCLEFmed 2008 medical image retrieval taskIn: Peters C, Giampiccolo D, Ferro N, et al. editors. Evaluating Systems for Multilingual and Multimodal Information Access—. Denmark: 9th Workshop of the Cross-Language Evaluation Forum Aarhus; 2009.
- 2. Medical subject headings, National Library of Medicinehttp://www.nlm.nih.gov/mesh/ 2009. Accessed: Oct 3, 2007.
- 3.Elkin PL, Cimino JJ, Lowe HJ, et al. Mapping to MeSH: The art of trapping MeSH equivalence from within narrative text Proc Annu Symp Comput Appl Med Care 1988:185-190.
- 4.Miller RA, Gieszczykiewicz FM, Vries JK, Cooper GF, CHARTLINE Providing bibliographic references relevant to patient charts using the UMLS Metathesaurus knowledge sources Proc Annu Symp Comput Appl Med Care 1992:86-90. [PMC free article] [PubMed]
- 5.Evans DA, Hersh WR, Monarch IA, Lefferts RG, Handerson SK. Automatic indexing of abstracts via natural-language processing using a simple thesaurus Med Decis Mak 1991;11:S108-S115. [PubMed] [Google Scholar]
- 6.Hersh W, Hickam DH, Haynes RB, McKibbon KA. Evaluation of SAPHIRE: An automated approach to indexing and retrieving medical literature Proc Annu Symp Comput Appl Med Care 1991:808-812. [PMC free article] [PubMed]
- 7.Hersh WR, Hickam DH, Haynes RB, McKibbon KA. A performance and failure analysis of SAPHIRE with a MEDLINE test collection J Am Med Inform Assoc 1994;1:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuttle MS, Olson NE, Keck KD, et al. Metaphrase: An aid to the clinical conceptualization and formalization of patient problems in healthcare enterprises Methods Inf Med 1998;37:373-383. [PubMed] [Google Scholar]
- 9.Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: A feasibility study J Am Med Inform Assoc 2001;8:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rindflesch TC, Aronson AR. Semantic processing in information retrieval Proc Annu Symp Comput Appl Med Care 1993:611-615. [PMC free article] [PubMed]
- 11.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus Proc Annu Symp Comput Appl Med Care 1994:240-244. [PMC free article] [PubMed]
- 12.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program Proc AMIA Symp 2001:17-21. [PMC free article] [PubMed]
- 13.Lindberg DAB, Humphreys BL, McCray AT. The unified medical language system Methods Inf Med 1993;32:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The unified medical language system: An informatics research collaboration J Am Med Inform Assoc 1998;5:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodenreider O. The unified medical language system (UMLS): Integrating biomedical terminology Nucleic Acids Res 2004;32:D267-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.In: Coté RA, Rothwell DJ, Beckette R, Palotay J, editors. SNOMED International: The Systematized Nomenclature of Human and Veterinary Medicine. Northfield, IL: College of American Pathologists; 1993.
- 17.SNOMED International SNOMED CT. College of American Pathologists. http://www.snomed.org/snomedct/ 1993. Accessed: Jul 14, 2005.
- 18.Rosse C, Mejino Jr JL. A reference ontology for biomedical informatics: The foundational model of anatomy J Biomed Inform 2003;36:478-500. [DOI] [PubMed] [Google Scholar]
- 19. Foundational model of anatomy, Structural Informatics Group; University of Washingtonhttp://sig.biostr.washington.edu/projects/fm/ 2003. Accessed: Jan 16, 2006.
- 20.Browne AC, Divita G, Aronson AR, McCray AT. UMLS language and vocabulary tools AMIA Annu Symp Proc 2003:798. [PMC free article] [PubMed]
- 21.Kahn CE, Thao C. GoldMiner: A radiology image search engine AJR Am J Roentgenol 2007;188:1475-1478. [DOI] [PubMed] [Google Scholar]
- 22.Lewis DD. Evaluating text categorizationIn: Asilomar CA, editor. Proceedings of the Speech and Natural Langugage Workshop. Morgan Kaufmann; 1991. pp. 312-318.
- 23.Hersh W. Evaluation of biomedical text-mining systems: Lessons learned from information retrieval Brief Bioinform 2005;6:344-356. [DOI] [PubMed] [Google Scholar]
- 24.Kahn CE. Effective metadata discovery for dynamic filtering of queries to a radiology image search engine J Digit Imaging 2008;21:269-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe HJ, Antipov I, Hersh W, Smith CA, Mailhot M. Automated semantic indexing of imaging reports to support retrieval of medical images in the multimedia electronic medical record Methods Inf Med 1999;38:303-307. [PubMed] [Google Scholar]
- 26.Hersh W, Mailhot M, Arnott-Smith C, Lowe H. Selective automated indexing of findings and diagnoses in radiology reports J Biomed Inform 2001;34:262-273. [DOI] [PubMed] [Google Scholar]
- 27.Yu H. Towards answering biological questions with experimental evidence: Automatically identifying text that summarize image content in full-text articles Proc AMIA Annu Fall Symp 2006:834-838. [PMC free article] [PubMed]
- 28.Yu H, Lee M. Accessing Bioscience Images from Abstract Sentences. vol 22. BioInform 2006;22:e547-e556. [DOI] [PubMed] [Google Scholar]
- 29.Joubert M, Fieschi M, Robert JJ, Volot F, Fieschi D. UMLS-based conceptual queries to biomedical information databases: An overview of the project Ariane Unified medical language system J Am Med Informatics Assoc 1998;5:52-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim GR, Aronson AR, Mork JG, Cohen BA, Lehmann CU. Application of a medical text indexer to an online dermatology atlas Medinfo 2004:287-291. [PubMed]
- 31.Müller HM, Kenny EE, Sternberg PW, Textpresso An ontology-based information retrieval and extraction system for biological literature PLoS Biol 2004;2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller HM, Rangarajan A, Teal TK, Sternberg PW. Textpresso for neuroscience: Searching the full text of thousands of neuroscience research papers Neuroinform 2008;6:195-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah NH, Rubin DL, Supekar KS, Musen MA. Ontology-based annotation and query of tissue microarray data AMIA Annu Symp Proc 2006:709-713. [PMC free article] [PubMed]
- 34.Bashyam V, Divita G, Bennett DB, Browne AC, Taira RK. A normalized lexical lookup approach to identifying UMLS concepts in free text Stud Health Technol Inform 2007;129:545-549. [PubMed] [Google Scholar]
- 35.Dai M, Shah NH, Xuan W, et al. An efficient solution for mapping free text to ontology terms AMIA Summit on Translational BioInformatics, San Francisco CA. 2008.
- 36.Jin Y, McDonald RT, Lerman K, et al. Automated Recognition of Malignancy Mentions in Biomedical Literature BMC BioInform 2006;7:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia N, Shah NH, Rubin DL, Chiang AP, Musen MA. Technical report: Comparing concept recognizers for ontology-based indexing: MGREP v MetaMap, Stanford Universityhttp://bmir.stanford.edu/file_asset/index.php/1349/BMIR-2008-1332.pdf 2006. Accessed: Nov 5, 2008.
- 38.Langlotz CP. RadLex: A new method for indexing online educational materials Radiographics 2006;26:1595-1597. [DOI] [PubMed] [Google Scholar]
- 39. RadLex: A lexicon for uniform indexing and retrieval of radiology information resources, Radiological Society Of North Americahttp://www.rsna.org/Radlex/ 2006. Accessed: Jan 20, 2006.
- 40.Rubin DL. Creating and curating a terminology for radiology: Ontology modeling and analysis J Digit Imaging 2008;21:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uzuner Ö. Second i2b2 workshop on natural language processing challenges for clinical records AMIA Annu Symp Proc 2008:1252-1253. [PubMed]
- 42.Collier N, Nazarenko A, Baud R, Ruch P. Recent advances in natural language processing for biomedical applications Int J Med Inform 2006;75:413-417. [DOI] [PubMed] [Google Scholar]
- 43.Mamlin BW, Heinze DT, McDonald CJ. Automated extraction and normalization of findings from cancer-related free-text radiology reports AMIA Annu Symp Proc 2003:420-424. [PMC free article] [PubMed]
- 44.Friedlin J, McDonald CJ. A natural language processing system to extract and code concepts relating to congestive heart failure from chest radiology reports AMIA Annu Symp Proc 2006:269-273. [PMC free article] [PubMed]
- 45.Dreyer KJ, Kalra MK, Maher MM, et al. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: Validation study Radiology 2005;234:323-329. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon J Am Med Inform Assoc 2005;12:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Lowe HJ, Hersh WR. A pilot study of contextual UMLS indexing to improve the precision of concept-based representation in XML-structured clinical radiology reports J Am Med Inform Assoc 2003;10:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebastia C, Quiroga S, Espin E, et al. Portomesenteric vein gas: Pathologic mechanisms, CT findings, and prognosis Radiographics 2000;20:1213-1224Discussion:1224–16. [DOI] [PubMed] [Google Scholar]