Abstract
Tnk1/Kos1 is a non-receptor protein tyrosine kinase found to be a tumor suppressor. It negatively regulates cell growth by indirectly suppressing Ras activity. We identified and characterized the critical cis-elements required for Tnk1/Kos1's promoter activity. Results indicate that the murine Tnk1 promoter lacks a conventional TATA, CAAT or initiator element (Inr) but contains multiple transcription start sites. Transcription is initiated by a TATA-like element composed of an AT rich sequence at -30 (30 bp upstream) from the major transcription start site and an Inr-like element that overlaps the multiple start sites. Deletion analysis of the m-Tnk1 promoter reveals the presence of both positive (-25 to -151) and negative (-151 to -1201) regulatory regions. The three GC boxes which bind Sp1 and Sp3 with high affinity, an AP2 site (that overlaps with an AML1 site) and a MED1 site comprise the necessary cis-elements of the proximal promoter required for both constitutive and inducible Tnk1/Kos1 expression. Importantly, results reveal that cellular stress reverses the repression of Tnk1/Kos1 and induces its expression through increased high affinity interactions between nuclear proteins Sp1, Sp3, AP2 and MED1 for the m-Tnk1 promoter. These findings provide a mechanism by which the m-Tnk1 promoter can be dynamically regulated during normal growth.
1. Introduction
We discovered through targeted disruption of the Tnk1 gene in mice that this ubiquitously expressed nonreceptor protein tyrosine kinase (NRPTK) possesses tumor suppressor activity since mice develop spontaneous tumors at a high rate (Hoehn et al., 1996; Hoare et al., 2003; Hoare et al., 2008). The human Tnk1 gene is located on chromosome 17p13.1, while the murine Tnk1 homolog is present on mouse chromosome 11, and composed of fourteen exons. Interestingly, the two gene products, Tnk1 and Tnk1/Kos1 (Thirty eight negative kinase1/Kinase of embryonic stem cell), are produced by alternative splicing (Hoare et al., 2008). While the deduced amino acid sequence of both the human and mouse Tnk1 transcript predict a 72 kDa protein, the 47 kDa Tnk1/Kos1 is produced when splicing from exons 8 to 9 fails to occur. Interestingly, in mice the 47kDa Tnk1/Kos1 is the predominant form. Mechanistically, both human Tnk1 and Tnk1/Kos1 have been found to negatively regulate cell growth by indirectly inhibiting Ras activity requiring the intrinsic kinase activity (Hoare et al., 2003; Hoare et al., 2008; Azoitei et al.). Furthermore, spontaneous tumors that develop in Tnk1 mice display up-regulated Ras activity (Hoare et al., 2008).
The expression of Tnk1/Kos1 may be dynamically regulated to maintain normal growth and development. For example, Tnk1/Kos1 may be involved in embryonic development because its expression is dramatically upregulated in a stage specific manner in developing mouse embryos and in murine embryonic stem cells in vitro following withdrawal of LIF (Hoare et al., 2003). The expression of Tnk1/Kos1 is also elevated in association with inhibition of cell growth and apoptosis following withdrawal of IL3 from factor dependent myeloid NSF.N1.H7 cells (Hoare et al., 2003). In contrast, epigenetic silencing of Tnk1/Kos1 expression has been observed in tumors that develop in the Tnk1/Kos1 heterozygous mice (Hoare et al., 2008). These findings indicate that Tnk1/Kos1 can be regulated at the transcriptional level but the mechanism is not yet clear. Here we identify and functionally characterize the murine Tnk1 promoter. Results indicate that Sp1, Sp3, AP2 and MED1 are necessary transcriptional regulators of Tnk1/Kos1 expression.
2. Materials and methods
2.1. Cell Culture
NIH3T3 cells (ATCC CRL1658) were grown in DMEM containing 10% Fetal Bovine Serum (FBS, Invitrogen) at 37°C, 5% CO2. The murine embryonic CCE stem cells were grown on feeder fibroblasts as described (Wiles and Keller, 1991). After two passages, the cells were grown without feeders. Drosophila Schneider 2 cells (ATCC CRL1963) were grown in Schneider's Drosophila medium containing 10% FBS. All media contained penicillin, streptomycin and L-glutamine.
2.2. Isolation of m-Tnk1 5' Flanking Region
A m-Tnk1 genomic clone (~ 8.5 kb) containing all the exons as well as the 5' and 3' flanking sequences was sub cloned into pZero 1.1 (Hoare et al., 2003). A HindIII-Nco1 fragment containing the entire 5' flanking region and the two non coding exons (-1202 to +621) was isolated and sub cloned into the pGL3 basic vector (Promega., Corp.) in the HindIII-Nco1 site so that the ATG of the luciferase cDNA overlaps with the Nco1 site. The -643Tnk deletion mutant was made by ligating a Sca1-Nco1 fragment to a Sma1-Nco1 digested pGL3 basic vector. The -487, -151, -89, -66 and -48 Tnk deletion mutants were created by performing a PCR reaction initially using forward primers (-487:5'GACTGTCTAGGACCAAAA CTGATGGTCGG 3'; -151: 5'CAAGTGGGAGGAGCTCCGCCACAAAG 3';-89: 5'GGCCAGCTTCCAGGATCCGCCCTC 3'; -66: 5'CCTCCACAGCCTGGGGTCCCG CCCTTTTTAG 3'; -48: 5'CCCTTTTTAGGATTTAATGCCCAGCC 3') and a common reverse primer corresponding to the region +611 to +636 (5'CCAGTAATGCC CATGGTCATGGCTTC 3') using Pfu DNA polymerase from Stratagene. The products were purified and digested with Nco1 and ligated to the Sma1-Nco1 digested pGL3 basic vector. The Hpa1-Nco1 fragment (-183Tnk), Bgl1-Nco1 fragment (-25Tnk), Dra3-Nco1 (-132Tnk) or Ase1-Nco1 (-113Tnk) were blunt ended at Dra3 and Ase1 sites and sub cloned at the Sma1-Nco1 site of the pGL3 basic vector. Digesting -1202Tnk Luc with Bgl2 and ligating the purified large fragment generated -247Tnk Luc. Site-specific mutations were created by introducing mutations in the PCR primers and performing an overlap PCR (Higuchi et al., 1988). The integrity of all PCR generated constructs were verified by DNA sequence analysis.
2.3. Primer Extension Analysis
An antisense primer from exon 2 (5'GAGTTGGTC ACCCCAGGCGGCCTGAG 3'; located between +220 and +245) was end labeled (0.8 × 109 CPM) using [γ 32P] ATP (Amersham Pharmacia Biotech.) and T4 Poly Nucleotide Kinase (New England Biolabs, Inc.). The labeled primer was annealed to 1 μg of mouse liver poly A+RNA in 50 mM Tris-HCl (pH 8.3) containing 50 mM KCl, 10 mM MgCl2, 10 mM DTT, 1mM each dNTP and 0.5 mM spermidine at 50°C for 1 hour and slowly cooled. Primer extension was carried out using AMV Reverse Transcriptase (Promega, Corp.) for 1 hour at 40°C in the presence of 0.7 mM Sodium Pyrophosphate and 6 μg Actinomycin D. The reaction products were digested with RNase A (20 μg/ml) for 30 min at 37°C followed by treatment with Proteinase K (200 mg/ml, Promega, Corp.) and finally purified by phenol-chloroform extraction/precipitation. The extended products were analyzed on a 6% acrylamide, 8 M urea gel along with an end labeled ϕX 174 Hinif 1 DNA marker and chain termination sequencing reaction (USB) performed using the same primer and m-Tnk1 genomic clone containing exons 1, 2 and the 5' flanking region. Total RNA was prepared using TRIZOL reagent (Life Technology) and PolyA+RNA was purified by the PolyAT tract mRNA isolation system (Promega, Corp.).
2.4. Transfection, Reporter Assays and Immunoblotting
The mammalian expression plasmids Sp1, Sp3 and AP2α were purified by the Qiagen plasmid purification system (Qiagen Inc.). Transient transfections using the plasmids were done by calcium phosphate co-precipitation (Promega, Corp.) in triplicate in 35 mm plates using 2 μg of the promoter construct and 300 ng of CMVβGal (Promega, Corp.). Transfections were also performed using Lipofectamine™ (Invitrogen). Murine stem cells (CCE) were transfected by electroporation (Gene Pulser set at 250 volts, 500 μF, 5-15 millisecond range, Bio-Rad Laboratories). Forty-eight hours following transfection, cells were washed with PBS three times and lysed in 200 μl of lysis buffer (Luciferase Assay System, Promega, Corp.). Firefly Luciferase light units were measured in a BD Monolight™ 3010C Luminometer (BD PharMingen) using 20 μl of cleared lysate. To study the effect of serum starvation, transfected cells were grown in medium containing 0.5% BSA for 24 hours before measurement of enzyme activity. The reporter construct (-151Tnk Luc) was co-transfected with Sp1, Sp3 or AP2 expression plasmids. Cotransfection of the reporter construct with pCDNA (Invitrogen) served as the control. Western analysis was performed by lyzing cells in RIPA lysis buffer and subjecting the clarified cell lysate (50 -100 μg) to 10% SDS PAGE (Hoare et al., 2003). The resolved protein bands were transferred on to nitrocellulose membrane and immuno-blotted with α-Tnk1/Kos1 (Hoare et al., 2003), α-Sp1, α-Sp3, α-AP2 and α-Actin (Santa Cruz Biotechnology). Protein was estimated using the Bradford reagent (BioRad Laboratories, CA).
2.5. Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared from NIH3T3 cells as described (Shapiro et al., 1988). Protein content was measured using a protein assay reagent from Bio-Rad. Oligonucleotides corresponding to the regions -39 to -69 (Tnk30): 5'CCTCCACAGCCTGGGGTCCCGCCCTTTTTAG 3'; -63 to -88 (Tnk25): 5'GCCAGCTTCCAGGATCCGCCCTCCACAGC 3'; -39 to -76 (Tnk38): 5'GATCCGCCCTCCACAGCCTGGGGTCCCGCCCTTTTTA 3'; -1 to -24 (Tnk24) 5' CCACTTGGGCGGCCCCCGTAGCTG 3' and +54 to +80 (Tnk-MED1) 5' TGGGACCAGCGGGCTCCCTTTGGGGT 3' were synthesized, annealed with complementary sequences and end labeled with [γ]32P ATP and T4 Poly Nucleotide Kinase (New England Biolabs, Inc.). The probes were purified using MicroSpin G50 columns (Amersham Pharmacia Biotech.). The nuclear proteins (10 μg) extracted from cells were incubated at room temperature for 20 min with 1 ng probe in 10 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 4 % glycerol and 1 μg poly dI-dC. In the competition assays, non-radioactive competitor (unlabeled) oligonucleotides were incubated with the nuclear extract for 10 min before addition of the probe(s). Oligonucleotides containing consensus binding sites for Sp1, AP2 and MED1 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and the AML1 consensus site (5'GGATATTTGCGGTTAGCA 3') was synthesized. The samples were analyzed on a 4 % denaturing polyacrylamide gel (Hoare et al., 1999). For the super-shift assay, 1μl of antibody specific for Sp1, Sp3, or AP2α (Santa Cruz Biotechnology Inc.) was added to the nuclear extract and incubated for 20 minutes before addition of the labeled probe(s).
3. Results
3.1. Transcription start site of murine Tnk1 gene
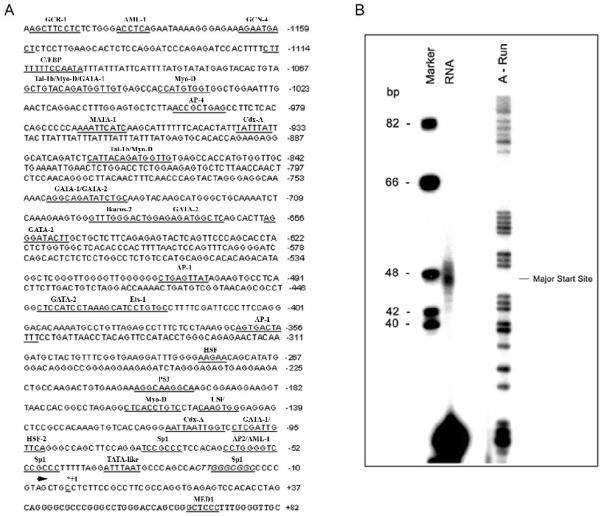
The murine Tnk1 (m-Tnk1) gene located on chromosome 11 is organized into 14 exons, of which exons 1, 2 and part of exon 3 (50 bp) are non-coding sequences (1, 3). To identify the nucleotide(s) from which transcription is initiated in the promoter, primer extension analysis was initially performed. An antisense primer to the start of exon 2 was annealed to mouse poly-A+RNA and then extended using AMV reverse transcriptase as described in materials and methods. Results reveal the presence of extended products of 46, 47, 49 and 50 bp, indicating that multiple start sites exist for the initiation of Tnk1 transcription (Figure 1B). The major transcription start site is assigned at the C (C+1 in Figure 1A), corresponding to the major band that appears at the 47 bp from the primer (Figure 1B). Interestingly, the Tnk1 gene lacks a conventional TATA, CAAT or Inr element, however, a TATA-like box is present consisting of an AT rich region (ATTTAAT) found 30 bp (-30) upstream of the major transcription start site. Also a “loose” consensus for an Inr, GTAGCTGCC (+2 to -8) is shown that overlaps with the multiple transcription start sites identified (Figure 1).
Figure 1. m-Tnk1 promoter region.
A, Putative binding sites for transcription factors in the 5' flanking region are underlined and indicated above the sequence. The nucleotide corresponding to the major transcription site is shown in bold as (+1). Filled arrowhead shows the start of exon1. B, The Tnk1/Kos1 transcription start site was determined by primer extension analysis. Labeled size marker (ϕX 174 Hinif1, lane 1) and sequencing ladder for A (lane 3) were run in parallel to the reaction products (lane2). The indicated major band is 47 bases away from the primer.
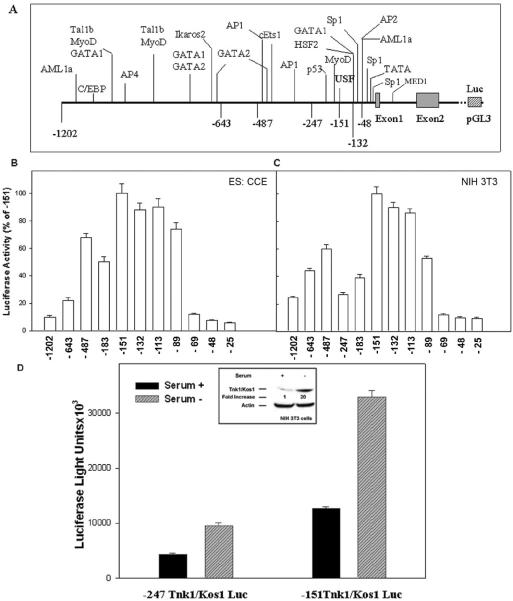
3.2. The 5' Flanking Promoter Sequence of Tnk1 Contains both Positive and Negative Regulatory Regions
In silico analysis of the m-Tnk1 promoter revealed a number of putative sites that may represent binding by lineage-specific transcription factors including GATA 1 & 2, Tal1b, Ikaros, AML1 and C/EBP (Figures 1A and 2A). However, the proximal promoter region contains consensus sites for Sp1, AP2, p53, USF, MyoD and MED1 (Figures 1A and 2A). There are three GC boxes that harbor the Sp1 sites. To determine the region(s) required for m-Tnk1 gene expression, we created a series of 5' deletions starting from the -1202/+621 to -25/+621 sites and ligated them in the pGL3 Basic Luciferase reporter (Luc) vector. Murine CCE embryonic stem cells or NIH3T3 fibroblasts were transfected using the reporter constructs (Figures 2B and 2C). Results reveal that the -151Tnk Luc demonstrates the highest level of luciferase activity in both cell lines. In addition, a 10-fold reduction in luciferase activity is observed for the -1202Tnk Luc compared to -151Tnk Luc which indicates a repression of the promoter activity. The promoter activity also remains repressed when the -643Tnk Luc, -487Tnk Luc and -183Tnk Luc are tested. Therefore, we can conclude that silencer-binding sites exist at locations between -151 and -1202 in the m-Tnk1 promoter. While progressive deletion from -151 to -89 results in only a marginal reduction of promoter activity, a major reduction (~90%) occurs when the region between -89 and -69 is deleted. In addition, further reduction in the promoter activity is observed with the -48 and -25 deletion mutants, indicating that the -89 to -25 region of the promoter is necessary for m-Tnk1 gene expression. Collectively, these data suggest that the -151 deletion mutant (-151Tnk) may represent the minimal m-Tnk1 promoter region.
Figure 2. Deletion analysis of m-Tnk1 promoter.
A, Potential regulatory elements and the deletion sites are indicated. B and C, Full length or progressively deleted regions of m-Tnk1 promoter fused to luciferase reporter plasmid pGL3 (2 μg) were transfected into stem cells (B, CCE) or NIH3T3 cells (C) and luciferase activities were determined. Luciferase activity is normalized to β-galactosidase activity from a co-transfected CMV-βGal plasmid (300ng). Activities are expressed as a percentage (± S.D) of three replicate determinations relative to the construct expressing highest activity. D, Inset shows a 20 fold increase in Tnk1/Kos1 protein expression in NIH 3T3 cells following withdrawal of serum for 24 hours. The level of Actin served as the loading control. Histogram shows the relative luciferase activities of the deletion mutants-247Tnk Luc or -151Tnk Luc in the presence or absence of serum. NIH 3T3 cells transfected with deletion mutants (2 μg) along with CMV-βGal plasmid were grown in the presence or absence of serum for 24 hours. Cells were lysed and luciferase activities were measured. The data represent the average of triplicate (±S.D.) determinations normalized to β-galactosidase activity.
Since the expression of Tnk1/Kos1 is induced under stress following withdrawal of serum (Figure 2D inset; Hoare et al., 2003), we next compared the promoter activity of the -151Tnk Luc with the -241Tnk Luc constructs in NIH 3T3 cells. In brief, cells were transiently transfected and serum was either removed or not in order to measure reporter activity. Results show that an increase in promoter activity is observed but the -151Tnk Luc demonstrates a 3.5 fold higher level in luciferase activity compared to the -247Tnk Luc in the absence of serum (Figure 2D). These data confirm that the deletion mutant -151Tnk represents the minimal m-Tnk1 promoter.
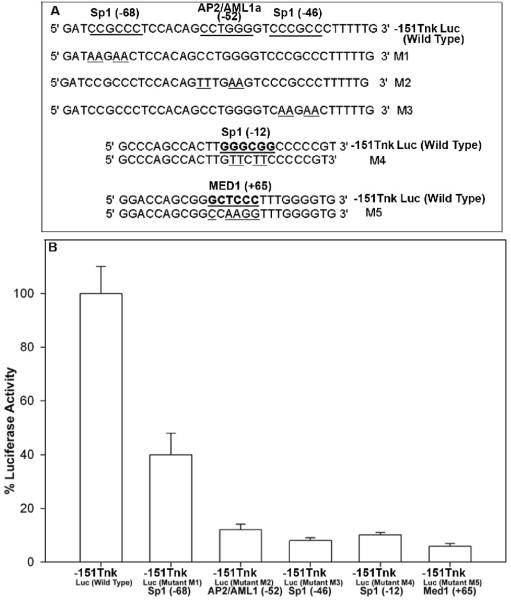
3.3. Functional Analysis of the minimal m-Tnk1 promoter
Potential regulatory elements within the minimal m-Tnk1 promoter include three Sp1 consensus binding sites (i.e. at -68, -42 and -12), an AP2 site (that overlaps with an AML1 site at -52) and a MED1 site (at +66; Figs. 1, 3). To test the functionality of each of these cis-elements, mutations were introduced in the minimal promoter construct, -151Tnk Luc as indicated (Figure 3A). The mutants were transiently transfected in NIH3T3 cells and luciferase activities were measured under serum starvation condition. Results indicate that a mutation of the Sp1 site at -68 drastically reduces the promoter activity by about 60%. However, mutations of the Sp1 at -46 and -12 sites, the AP2/AML1 site at -52 and the MED1 site at +66, all further decrease the promoter activity by 80%-90%, indicating that the integrity of these transcription factors putative binding sites are required for Tnk1/Kos1 expression (Figure 3B).
Figure 3. Effect of mutation of Sp1, AP2/AML1 and MED1 binding sites on the minimal m-Tnk1 promoter activity.
A, Specific mutations at the three Sp1 sites, the AP2/AML1 site and the MED1 site are created. Mutated nucleotides at the sites are underlined. B, NIH3T3 cells were transiently transfected with the wild type and the mutated -151Tnk Luc constructs and luciferase activities were determined. The data represent the average of triplicate (+ S.D.) determinations normalized to β-galactosidase activity.
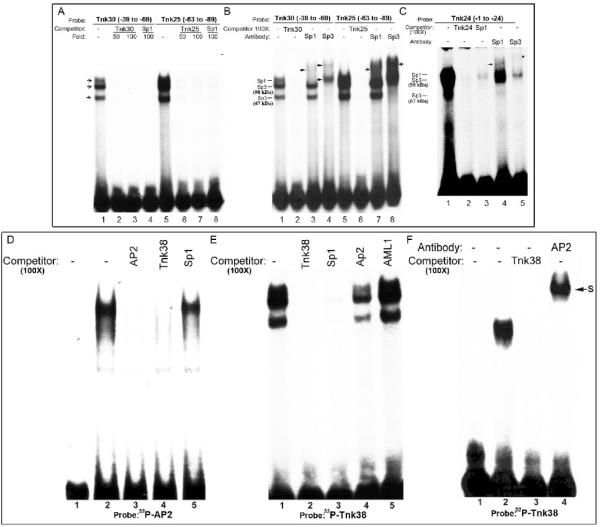
3.4. Both Sp1 and Sp3 bind to GC boxes located in the m-Tnk1 promoter
The Sp1 family of transcription factor is known to bind to the GC box that contains its consensus binding site (Wierstra 2008). To determine Sp1's specificity for binding to the m-Tnk1 proximal promoter, three oligonucleotide probes - Tnk30 (-39 to -69), Tnk25 (-63 to -88) and Tnk24 (-1 to -24) representing the three GC boxes were synthesized. The oligonucleotide probes were end labeled with [γ]32P ATP and incubated with nuclear extracts prepared from NIH3T3 cells to perform an EMSA. Results reveal the presence of three retarded mobility bands that can be competed with a 100 fold molar excess of the unlabeled probe, demonstrating specificity for the interaction (Figures 4A, lanes 1-3, 5-7; 4B, lanes 1, 2, 5, 6; 4C, lanes 1, 2). Further, these three bands are competed using a 100 fold molar excess of the unlabeled oligonucleotide that contains a bona fide Sp1 binding site, indicating that Sp1 and Sp1-like proteins specifically bind to these three GC boxes in the m-Tnk1 promoter (Figures 4A, lanes 4,8; 4C lane 3). A super-shift assay using antibodies against Sp1 and Sp3 verified the identity of the retarded bands based on the molecular weights of Sp1 (~105 kDa) and Sp3 (~98 kDa and ~67 kDa). Thus results indicate that the Sp1 antibody shifts the lower mobility band to a higher position while the Sp3 antibodyx retards the other two bands (Figures 4 B and 4C). We can then conclude that Sp1 and Sp3 are functionally part of the protein-DNA complexes associated with the GC boxes present in m-Tnk1 promoter that regulate its expression (Figures 4B, lanes 1, 3, 4, 5, 7, 8; 4C, lanes 1, 4, 5).
Figure 4. Identification of nuclear proteins that interact with Sp1 and AP2 binding sites in the m-Tnk1 promoter.
Identification of nuclear proteins that bind to GC boxes (A, B and C). EMSA was performed by incubating nuclear extract (10μg) prepared from NIH3T3 cells with 32P labeled Tnk oligonucleotides: Tnk30 (A, lane1; B, lane1), Tnk25 (A, lane5; B, lane5) and Tnk24 (C, lane1). Competition assays were carried out in the presence of 50 or 100 fold molar excess of cold Tnk30 (A, lanes 2, 3; B, lane 2), Tnk25 (A, lanes 6,7; B, lane 6) and Tnk24 (C, lane 2) or Sp1 consensus oligonucleotide (A, lanes 4,8; C, lane3). Super-shift assays were performed using antibodies against Sp1 (B, lanes 3, 7; C, lane 4) and Sp3 (B, lanes 4, 8; C, lane5). Arrows indicate supershifted complexes. Identification of nuclear proteins that bind to the AP2 consensus site (D). Gel shift assay for nuclear proteins prepared from NIH 3T3 that complex with 32P labeled AP2 oligonucleotide (lane 2). The radio-labeled band was competed with 100 fold molar excess of unlabeled AP2 oligonucleotide (lane 3), Tnk38 oligonucleotide (lane 4) or Sp1 oligonucleotide (lane 5). Lane 1 represents reaction mixture without nuclear extract. Identification of nuclear proteins that bind to the AP2/AML1 consensus site (E). The protein DNA complex formed between 32P labeled Tnk38 (-40 to -76) and NIH3T3 nuclear extract (lane1) is tested for competition with 100 fold molar excess of unlabeled oligonucleotides Tnk38 (lane 2), Sp1 (lane 3), AP2 (lane 4) and AML1a (lane 5). AP2 binds the AP2/AML1 consensus site (F). Purified AP2 recombinant protein (50ng) was allowed to react with 32P labeled Tnk38 (lane 2) in the presence of 100 fold molar excess of unlabeled Tnk38 (lane 3) or an antibody for AP2α (lane 4). S indicates the position of the super-shifted complex. Lane 1 shows reaction mixture without protein.
3.5.. AP2α binds selectively to a site that overlaps with the AP2 and AML1 consensus sequences
The loss of m-Tnk1 minimal promoter activity due to mutation in the AP2/AML1 site at -52 indicates that this site is essential for transcription (Figures 4D, 4E and 4F). This loss in activity may be due to the inability of AP2 or AML1 to bind the promoter. To test whether AP2 activity is present in a nuclear extract prepared from NIH3T3 cells, a consensus AP2 oligonucleotide was radio-labeled and used for an EMSA. Results indicate that the radiolabeled AP2-DNA complex is competed with a 100 fold molar excess of either an unlabeled AP2 oligonucleotide or a Tnk38 oligonucleotide that contains the AP2/AML1 overlapping binding site (Figure 4D, lanes 2-4). These data demonstrate that AP2 specifically binds to its cognate site in the m-Tnk1 proximal promoter. Importantly, under similar conditions, a 100 fold molar excess of unlabeled Tnk38 oligonucleotide fails to compete the radiolabeled AML1-DNA complex, indicating that AML1 does not bind at this site (data not shown). In contrast, the AP2-DNA complex can be partially inhibited (by ~50%) using an unlabeled Sp1 consensus oligonucleotide, indicating that Sp1 may be a partner of AP2 through protein-protein interaction (Figure 4D, lane5). This data raises the possibility of a potential “cross talk” between the respective transcription factors in the transcriptional regulation of m-Tnk1 promoter.
To further characterize binding of AP2 to the proximal m-Tnk1 promoter, an EMSA was performed using radio-labeled Tnk38 and nuclear extract prepared from NIH 3T3 cells. Results indicate that formation of protein-DNA complexes observed are specifically competed with a 100 fold molar excess of unlabeled Tnk38 (Figure 4E, lane2) or Sp1 oligonucleotide (Figure 4E, lane 3). However, even a 100 fold molar excess of the unlabeled AML1 oligonucleotide fails to compete the radiolabeled complex (Figure 4E, lane 5), confirming that Sp1 but not AML1 binds to the proximal promoter. However, unlike the AML1 oligonucleotide, a 100 fold molar excess of the unlabeled AP2 consensus oligonucleotide does displace approximately 50% of the complex, indicating that AP2 may be part of this protein-DNA complex through its interaction with Sp1 (Figure 4E, lane 4). To further test the binding of AP2, purified recombinant AP2 was incubated with radiolabeled Tnk38 oligonucleotide and an EMSA was performed (Figure 4F). Results clearly show formation of an AP2-DNA complex that is competed with a 100 fold molar excess of unlabeled Tnk38 and is super-shifted by the AP2α antibody (Figure 4F, lanes 2-4). These data indicate that AP2 binding is indispensible for m-Tnk1 promoter activity.
3.6. Effect of exogenous Sp1, Sp3 and AP2 expression on m-Tnk1 Promoter Activity and endogenous Tnk1/Kos1 expression
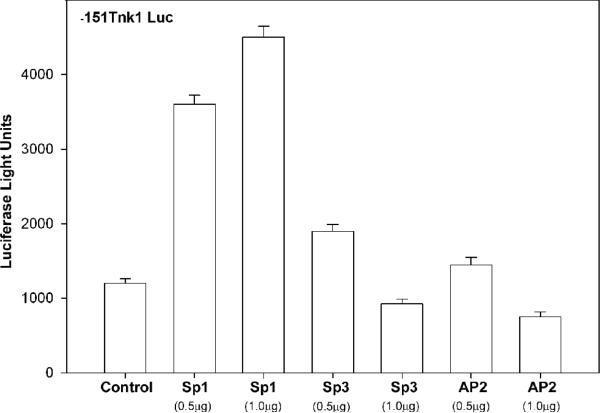
Since the m-Tnk1 minimal promoter binds Sp1, Sp3 and AP2, we tested the effect of expression of these individual proteins in the absence of serum on m-Tnk1 promoter activity by measuring the luciferase activity of -151Tnk Luc reporter. Results indicate that ectopic expression of Sp1 in NIH 3T3 cells when transfected with 0.5 μg or 1.0 μg of plasmid enhances m-Tnk1 minimal promoter activity by 3-4 fold, which results in a 2-3 fold increase in endogenous Tnk1/Kos1 expression versus vector-only transfected control cells (Figures 5 and 5-inset). In contrast, we observe only a marginal increase in promoter activity (1.25-1.5 fold) with either Sp3 or AP2 when cells were transfected with 0.5 μg of their respective expression plasmids (Figure 5). However, when cells are transfected with 1.0 μg of the Sp3 or AP2 plasmid m-Tnk1 minimal promoter activity is repressed as is endogenous Tnk1/Kos1 expression (Figures 5 and 5-inset). These findings suggest that Sp3 and AP2, unlike Sp1 can function as activators or repressors of m-Tnk1 promoter activity in a mechanism dependent upon their relative expression. In support of this, it has been reported that Sp3 can compete with Sp1 to block promoter activity and AP2 may negatively regulate transcription through self inhibition at higher expression levels depending on the cell type or context (Wiestra, 2008; Hilger-Eversheim et al., 2000; Buettner et al., 1993). It is relevant that Sp1 and Sp3 are abundantly expressed in NIH 3T3 cells, while AP2 is bearly detectable, which may explain the observed dose dependent inhibition by both Sp3 and AP2 on mTnk1 promoter activity (Figure 5 inset, control lanes). Thus, our findings suggest a direct transcriptional link between Sp1, Sp3, AP2 and m-Tnk1 promoter activity and protein expression.
Figure 5. Effect of expression of Sp1, Sp3 and AP2 on m-Tnk1 promoter activity and endogenous level of Tnk1/Kos1.
NIH3T3 cells were transfected with the minimal promoter, -151Tnk Luc (2μg) along with either the control, pCDNA, CMV-Sp1, CMVSp3 or CMV-AP2. CMV-βGal was co-transfected in all cases for normalizing transfection efficiency. The transfected cells were deprived of serum for 24 hours prior to the determination of luciferase activity. The data represent the average of triplicate (± S. D.) determinations normalized to β-galactosidase activity. Inset shows the ectopic expression of Sp1, Sp3 and AP2 in the transfected cells. The endogenous expression level of Tnk1/Kos1 in the transfected cells is indicated. The level of Actin served as the loading control.
3.7. MED1 selectively binds to its consensus site in the m-Tnk1 promoter
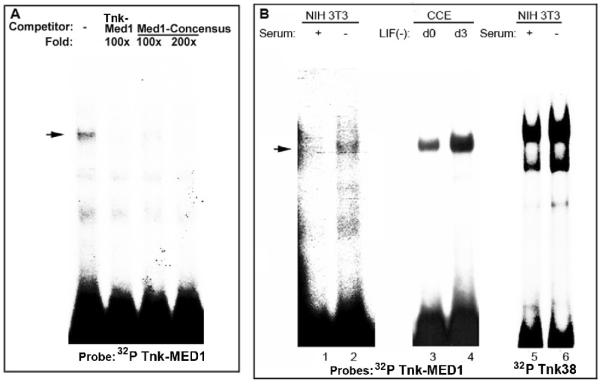
MED1 (Multiple start site element downstream) is considered a master switch that can regulate utilization of multiple transcription start sites to enhance efficient transcription from TATA-less promoters (Ince and Scotto, 1995). In support of this notion, mutation of the MED1 site at +66 results in the complete loss of the m-Tnk1 minimal promoter activity (Figure 3). To test whether MED1 can bind to this site, the Tnk-MED1 (+54 to +80) oligonucleotide was radiolabeled and incubated with nuclear extract prepared from NIH 3T3 cells (Figure 6A). An EMSA reveals the formation of a protein-DNA complex, which is competed with a 100 fold molar excess of unlabeled Tnk-MED1 or a 100 fold molar excess of the MED1 consensus oligonucleotide (Figure 6A, lanes 1-4). These data indicate that MED1 can selectively bind to regulate the transcriptional activity of the m-Tnk1 promoter.
Figure 6. MED1 positively regulates Tnk1/Kos1 transcription.
Identification of the nuclear proteins that bind to the MED1 consensus binding site (A). The protein-DNA complex formed between 32P labeled Tnk-MED1 and NIH3T3 nuclear extract (lane1) is competed with 100 to 200 fold molar excess of unlabeled Tnk-MED1 or an oligonucleotide containing the MED1 consensus site. Effect of serum or Leukemia inhibitory factor (LIF) withdrawal on the protein-DNA complex formation (B). 32P labeled Tnk-MED1 or 32P labeled Tnk38 was incubated with nuclear extract prepared from NIH 3T3 cells in the presence of serum (lanes 1,5) and in the absence of serum (lane 2,6) or nuclear extract prepared from CCE cells d0 (lane3) and d3 (lane 4) of LIF withdrawal.
3.8. Cellular stress differentially regulates m-Tnk1 promoter activity in a Sp1/AP2 and MED1 dependent manner
Since cellular stress induces the up-regulation of Tnk1/Kos1 expression through transcriptional activation (Figure 2D), we tested the effect of serum or Leukemia inhibitory factor (LIF) withdrawal on the MED1 or Sp1/AP2-DNA interactions in NIH 3T3 and murine embryonic CCE stem cells. Radiolabeled Tnk oligonucleotides containing either MED1 (Tnk-MED1) or both Sp1 and AP2 sites (Tnk38) were incubated with nuclear extract prepared from either NIH 3T3 or CCE cells to perform an EMSA (Figure 6B). Results show a dramatic increase in the high affinity binding of Sp1/AP2 and MED1 in response to serum or LIF withdrawal compared to binding in the presence of serum or LIF. These findings indicate that loss of Sp1/AP2 and MED1 binding to the m-Tnk1 promoter results in repression of Tnk1/Kos1, which is reversed under cellular stress and may occur by induction/activation of Sp1, AP2 and MED1.
4. Discussion
We recently discovered that Tnk1/Kos1, a novel NRPTK, is a tumor suppressor (Hoare et al., 2008), which is consistent with our earlier report that Tnk1/Kos1 is a negative regulator of cell growth (Hoare et al., 2003; Azoitei et al., 2007). Tnk1/Kos1 is ubiquitously expressed (albeit at low levels) and cellular stress can upregulate expression, suggesting that Tnk1/Kos1 may be regulated at the transcriptional level (Hoare et al., 2003). Therefore, to determine whether and how Tnk1/Kos1 may be transcriptionally regulated, we identified and characterized the murine Tnk1/Kos1 promoter (5' flanking region from -1202 to +621), and determined and tested the functionality of cis-elements that regulate its activity. Mapping the transcription initiation site revealed the presence of multiple start sites clustered within a five-nucleotide long stretch in exon1 (Figure 1). While no conventional TATA, CAAT, or Inr element was found that could initiate transcription, importantly an AT rich region (ATTAAT) does exist that may function as a TATA-like box at 30 bp upstream of the major transcription start site. Consistent with this possibility, TFIID is known to bind and stimulate transcription from a variety of such AT rich sequences (Hahn et al., 1989; Singer et al., 1990). In silico analysis has also revealed the presence of a potential Inr-like consensus sequence CGTAGCTGCC (+2 to -8) that overlaps the multiple transcription start sites in the m-Tnk1 promoter (shown in bold with major start site underlined, Figure 1). Interestingly, a similar functional Inr-like sequence has been reported for the Ha-Ras (Lu et al., 1994) and Msx1 (Takahashi et al., 1997) promoters. Therefore, we propose that the Inr-like sequence contained in the m-Tnk1 core promoter region may represent such a functional element. In support of this, multiple transcription start sites in several promoters that lack a consensus Inr-binding site have been shown to possess Inr activity, even though the activity does not depend on initiation of transcription at an adenosine residue (Lu et al., 1994; O'Shea-Greenfield and Smale, 1992). In addition, the presence of a regulatory element at +66 for MED1 (Multiple start site Element Downstream; consensus sequence, GCTCCC), downstream of the start sites, suggests that MED1 may also act as a selector or activator of the major start site for the efficient initiation of transcription of m-Tnk1 (Figure 1). In support of this notion, the loss of MED1 binding at the +66 site results in the complete loss of minimal promoter activity when mutant -151Tnk Luc (M5) is expressed (Figure 3). Therefore, we propose that MED1, in concert with the Inr-like element identified, may regulate initiation of transcription of the m-Tnk1 promoter.
Characterization of the m-Tnk1 promoter was also carried out by generating a series of 5' deletion mutants (from -1202/+621 to -25/+621). Analyzing of the promoter activity of the mutant reporter constructs was performed in either murine embryonic stem (CCE) or NIH3T3 cells (Figure 2). Results demonstrate the presence of both silencer (-1202 to -151) and activator (-151 to -25) elements in the promoter sequence identified (Figures 2 and 3). Thus, the loss of silencer and activator elements can lead to similar luciferase activities for the -487Tnk Luc and -89Tnk Luc reporter plasmids respectively (Figure 2B). Further, the presence of three GC boxes in the activator region suggests the potential involvement of the Sp1 and Sp3 transcription factors in the regulation of Tnk1/Kos1expression. Interestingly, the Sp family is comprised of nine structurally related members that can bind to the same consensus sequence and may play different functional roles (Wierstra, 2008). The founder member, Sp1, is a ubiquitously expressed C2H2-type zinc-finger transcription factor that can directly stabilize the binding of TFIID to the core promoter elements (Wierstra, 2008; Smale et al., 1990) by physically interacting with the TATA binding proteins (TBP), hTAFII130 and hTAFII55 (Emili et al., 1994; Tanese et al., 1996). Sp1 was initially considered to be simply a constitutive activator of housekeeping or TATA-less genes. However, more recently it has become clear that Sp1 is involved in the regulation of cell growth and tumorigenesis (Wierstra, 2008; Safe and Abdelrahim, 2005). In addition, a functional interplay between the ubiquitously expressed family members Sp1 and Sp3 can occur in the regulation of transcription of a number of genes (Wierstra, 2008). For example, while Sp1 is primarily described as an activator of gene transcription, Sp3 may function as either an activator (Wierstra, 2008; Ihn and Trojanowska, 1997) or a repressor (Wierstra, 2008; Hagen et al., 1994), depending on the cell type and the promoter tested. While our data indicate that Sp1 and Sp3 are required for m-Tnk1 gene expression, they may also function to either stimulate or suppress Tnk1/Kos1 expression depending upon their expression relative to one another (Figures 3, 4, 5). Our data suggests that AP2, a cell type specific transcription factor (Hilger-Eversheim et al., 2000; Moser et al., 1995), may also play a role in the transcriptional activation of Tnk1/Kos1. Furthermore, like Sp3, AP2 can also suppress Tnk1/Kos1 expression dependent upon its expression level. (Figures 3, 4, 5). It should be noted that Sp1 may be a versatile partner of AP2 and their interaction can be highly cell type/context-dependent (Wierstra, 2008; Safe and Abdelrahim, 2005; Mitchell et al., 2006). Therefore, our findings indicate that Sp1, Sp3, AP2 and MED1 may potentially regulate the m-Tnk1 promoter in a combinatorial manner, which could account for any differential expression of Tnk1/Kos1 observed in various tissues. Furthermore, any enhancement of binding of transcription factors like Sp1, Sp3, AP2 and MED1 under stress conditions may result in enhanced m-Tnk1 promoter activation and induction of Tnk1/Kos1.
Methylation of promoter CpG sites is now well characterized in the mechanism of regulation of gene transcription, most notably for tumor suppressors (Jones and Baylin, 2002; Liu et al., 2008). Unlike human Tnk1, the m-Tnk1 promoter lacks a classical CpG “Island” but does contain multiple CpG sites in the proximal promoter region and may potentially regulate of Tnk1/Kos1 expression (Figure 1). In support of this mechanism, we discovered that Tnk1/Kos1 expression is silenced in tumor tissue but not in the adjacent, uninvolved tissue from the same heterozygous Tnk1+/- mice (Hoare et al., 2008). This likely explains why tumors develop in these mice. Further, mapping of the Tnk1/Kos1 promoter CpG sites of tumors derived from the Tnk1+/- mice reveal that all 7 CpG sites are methylated but none of the same sites are methylated in the adjacent, uninvolved tissue (Figure 1; -100 to +22; 3). Importantly, the CpG sites are located within the Tnk1/Kos1 core promoter region that contains the GC boxes. Therefore, it is possible that methylation will prevent Sp/MED1 proteins from binding to and activating the m-Tnk1 promoter (Wierstra, 2008; Liu et al., 2008). Thus, methylation of the CpG sites in the m-Tnk1 promoter is a likely mechanism for the negative regulation of Tnk1/Kos1 expression (Hoare et al., 2008).
5. Conclusion
We have identified and characterized the m-Tnk1 promoter. Functional analysis indicates that Sp1, Sp3, AP2 and MED1 may regulate differential expression of Tnk1/Kos1. Induction of Tnk1/Kos1 occurs when the m-Tnk1 promoter is functionally activated during stress in a mechanism that results from high affinity Sp1/Sp3/AP2/Med1 binding to the promoter and the loss or lower affinity binding to the promoter can result in repression. This may explain, at least in part, how Tnk1/Kos1 is dynamically regulated during normal growth.
Acknowledgments
We thank R. Tjian, G. Suski and R. Buettner for providing the expression plasmids Sp1, Sp3 and AP2, respectively. This work is supported by a grant from National Cancer Institute CA109150.
Abbreviations used
- Tnk1/Kos1
Thirty eight negative kinase1/Kinase of stem cell 1
- PCR
Polymerase chain sreaction
- RACE
Rapid amplification of cDNA ends
- EMSA
Electrophoretic gel mobility shift assay
- bp
Base pair
- MED1
Multiple start site element downstream
- LIF
Leukemia Inhibitory Factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Hoare K, Hoare S, Smith OM, et al. Kos1, a nonreceptor tyrosine kinase that suppresses Ras signaling. Oncogene. 2003;22:3562–77. doi: 10.1038/sj.onc.1206480. [DOI] [PubMed] [Google Scholar]
- Hoare S, Hoare K, Reinhard MK, et al. Tnk1/Kos1 knockout mice develop spontaneous tumors. Cancer Res. 2008;68:8723–32. doi: 10.1158/0008-5472.CAN-08-1467. [DOI] [PubMed] [Google Scholar]
- Hoehn GT, Stokland T, Amin S, et al. Tnk1: a novel intracellular tyrosine kinase gene isolated from human umbilical cord blood CD34+/Lin-/CD38- stem/progenitor cells. Oncogene. 1996;12:903–13. [PubMed] [Google Scholar]
- Azoitei N, Brey A, Busch T, Fulda S, et al. Thirty-eight-negative kinase1 (TNK1) facilitates TNFα-induced apoptosis by blocking NF-κB activation. Oncogene. 2007;26:6536–45. doi: 10.1038/sj.onc.1210476. [DOI] [PubMed] [Google Scholar]
- Wiles MV, Keller G. Multiple hematopoietic lineages develop from embryonic stem (ES) cells in culture. Development. 1991;111:259–269. doi: 10.1242/dev.111.2.259. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro DJ, Sharp PA, Wahli WW. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- Hoare S, Copland JA, Wood TG, et al. Identification of a GABP alpha/beta binding site involved in the induction of oxytocin receptor gene expression in human breast cells, potentiation by c-Fos/c-Jun. Endocrinology. 1999;140:2268–2279. doi: 10.1210/endo.140.5.6710. [DOI] [PubMed] [Google Scholar]
- Wierstra I. Sp1:Emerging roles-Beyond constitutive activation of TATA-less housekeeping genes. BBRC. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, et al. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. 2000;260:1–12. doi: 10.1016/s0378-1119(00)00454-6. [DOI] [PubMed] [Google Scholar]
- Buettner R, Kannan P, Imhof A, et al. An alternatively spliced mRNA from the AP-2 gene encodes a negative regulator of transcriptional activation by AP2. Mol. Cell Biol. 1993;13:4174–4. doi: 10.1128/mcb.13.7.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince TA, Scotto KW. A conserved downstream element defines a new class of RNA polymerase II promoters. J. Biol. Chem. 1995;270:30249–30252. doi: 10.1074/jbc.270.51.30249. [DOI] [PubMed] [Google Scholar]
- Hahn S, Buratowski S, Sharp PA. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer VL, Wobbe CR, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- Lu J, Lee W, Jiang C. Start site selection by Sp1 in the TATA-less human Haras promoter. J. Biol. Chem. 1994;269:5391–5402. [PubMed] [Google Scholar]
- Takahashi T, Guron C, Shetty S, et al. A minimal murine MSX-1 gene promoter. Organization of its cis-regulatory motifs and their role in transcriptional activation in cells in culture and in transgenic mice. J. Biol. Chem. 1997;272:22667–22678. doi: 10.1074/jbc.272.36.22667. [DOI] [PubMed] [Google Scholar]
- O'Shea-Greenfield A, Smale ST. Roles of TATA and initiator elements in determining the start location and direction of RNA polymerase II transcription. J. Biol. Chem. 1992;267:1391–1402. [PubMed] [Google Scholar]
- Smale ST, Schmidt MC, Berk AJ. Transcriptional activation by Sp1 as directed through TATA or initiator: Specific requirement for mammalian transcription factor IID. Proc. Natl. Acad. Sci. USA. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Greenblat J, Ingles CJ. Spieces-specific interaction of the glutamine-rich activation domain of Sp1 with the TATA box-binding protein. Mol. Cell. Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N, Saluja D, Vassallo MF, Chen JL, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc. Natl. Acad. Sci. USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur.J. Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Ihn H, Trojanowska M. Sp3 is a transcriptional activator of the human alpha2(I) collagen gene. Nucleic Acids Res. 1997;25:3712–3717. doi: 10.1093/nar/25.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Pscherer A, Imhof A, Bauer R, et al. Cloning and characterization of a second AP-2 transcription factor:AP-2 beta. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Abdelrahim M, Weng J, et al. Regulation of KiSS-1 metasis suppressor gene expression in breast cancer cells by direct interaction of transcription factors activator pritein-2α and specificity protein-1. J.Biol.Chem. 2006;281:51–58. doi: 10.1074/jbc.M506245200. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang L, Niu Z, et al. Promoter methylation inhibits BRD7 expression in human nasopharyngeal carcinoma cells. BMC Cancer. 2008;8:253–66. doi: 10.1186/1471-2407-8-253. [DOI] [PMC free article] [PubMed] [Google Scholar]