Abstract
Maintenance of genome stability is essential for the accurate propagation of genetic information and cell growth and survival. Organisms have therefore developed efficient strategies to prevent DNA lesions and rearrangements. Much of the information concerning these strategies has been obtained through the study of bacterial and nuclear genomes. Comparatively, little is known about how organelle genomes maintain a stable structure. Here, we report that the plastid-localized Whirly ssDNA-binding proteins are required for plastid genome stability in Arabidopsis. We show that a double KO of the genes AtWhy1 and AtWhy3 leads to the appearance of plants with variegated green/white/yellow leaves, symptomatic of nonfunctional chloroplasts. This variegation is maternally inherited, indicating defects in the plastid genome. Indeed, in all variegated lines examined, reorganized regions of plastid DNA are amplified as circular and/or head-tail concatemers. All amplified regions are delimited by short direct repeats of 10–18 bp, strongly suggesting that these regions result from illegitimate recombination between repeated sequences. This type of recombination occurs frequently in plants lacking both Whirlies, to a lesser extent in single KO plants and rarely in WT individuals. Maize mutants for the ZmWhy1 Whirly protein also show an increase in the frequency of illegitimate recombination. We propose a model where Whirlies contribute to plastid genome stability by protecting against illegitimate repeat-mediated recombination.
Keywords: genome maintenance, microhomology, recombination
Plastids play diverse and essential roles in plants. Despite this central importance, surprisingly little is known about even the most basic aspects of the plastid genome structure, maintenance, and propagation. For example, while the textbook depiction of plastid DNA (ptDNA) is that of a genome-sized circular DNA molecule, recent evidence suggests instead that most of the ptDNA is organized in concatenated, branched linear forms with T4 phage-like features (1). This change of perception of plastid genome architecture requires a re-evaluation of the current rolling-circle model for plastid genome replication. It is now considered that a recombination-dependent replication process is responsible for the branched, multigenomic structures present in plastids (1). Recombination is also expected to play a crucial role in plastid genome maintenance. Indeed, because of its exposure to radiation and reactive oxygen species, the plastid genome is expected to accumulate mutations at a high rate. This situation stresses the importance of efficient DNA replication, recombination, and repair (DNA-RRR) pathways in these organelles (2). However, to date the mechanisms and enzymes involved in these pathways remain poorly characterized.
Evidence for recombination in plastid genomes abounds in the literature (2). An example is the recombination between the large inverted repeat sequences present in many plastid genomes (3). This flip-flop recombination is responsible for the 2 isomers of ptDNA, which differ only with respect to the orientation of the single-copy regions. More direct evidence of recombination comes from plastid transformation experiments, which demonstrate that foreign DNA is integrated into ptDNA by homologous recombination (4).
Homologues of bacterial genes involved in DNA-RRR are present in the nuclear genome of plants, and some of their encoded proteins are targeted to plastids. These include the recA homologs RECA1 (5) and RECA2, whose disruption is lethal in Arabidopsis (6), a Rec Q-like DNA helicase from rice (7), and genes for a gyrase A-like and 2 gyrase B-like subunits in Arabidopsis (8). Recently, 2 homologs of OSB1, a ssDNA-binding protein (SSB) that regulates recombination in mitochondria (9), were shown to localize to plastids. However, no role has yet been ascribed to these proteins. Finally, homologs of replication protein A (RPA), another ssDNA-binding protein family that plays an essential role in mammalian DNA-RRR, have recently been identified in plants. One member of this family is targeted to plastids (10).
Similar to many DNA-RRR proteins, Whirlies form a small family of ssDNA-binding proteins (11). They are involved in a variety of phenomena, ranging from pathogen defense (12) to telomeric homeostasis (13). In Arabidopsis, 3 Whirly genes are present and their proteins localize to organelles; AtWhy1 and AtWhy3 are targeted to plastids and AtWhy2 is targeted to mitochondria (14, 15).
Recent evidence indicates that Whirlies bind organelle DNA without apparent sequence specificity in vivo. In Arabidopsis, AtWhy2 binds to many regions of the mitochondrial genome with no obvious sequence consensus (15). Similarly, in maize, the plastid-localized ZmWhy1 interacts with DNA from throughout the plastid genome (16). Knockdown mutations of ZmWhy1 lead to ivory or pale green plants, indicating that this Whirly is involved in chloroplast biogenesis. This phenotype was attributed to a defect in the maturation of the atpF and 23S ribosomal RNAs, but the participation of ZmWhy1 in DNA recombination or repair has not been ruled out.
To better understand the role of plastid-targeted Whirlies (ptWhirlies), we characterized an Arabidopsis double KO line of the AtWhy1 and AtWhy3 genes (KO1/3). Variegation patterns, which appear on leaves in ≈4.6% of the progeny, correlated with the selective rearrangement and amplification of large regions of the plastid genome. We show that the rearrangements are produced by illegitimate recombination at short direct repeats that border the amplified regions in intact ptDNA. We suggest that AtWhy1 and AtWhy3 function as antirecombination proteins, contributing to safeguard plastid genome integrity.
Results
Arabidopsis ptWhirlies Are Involved in Chloroplast Biogenesis.
To investigate the role of Whirlies in plastids, we obtained mutant plants that no longer produce the AtWhy1 (KO1) and/or AtWhy3 (KO3) proteins (Fig. 1 A and B). Only one Whirly is detected in the single KOs, and no Whirlies are detected in KO1/3 extracts. We then monitored the ssDNA-binding activity of Whirlies by EMSA. As shown in Fig. S1 a strong DNA-binding signal is observed with Col-0 (WT) proteins, whereas less intense signals are detected with KO1 and KO3, respectively. No signal was observed in KO1/3, confirming the absence of ptWhirlies in these plants.
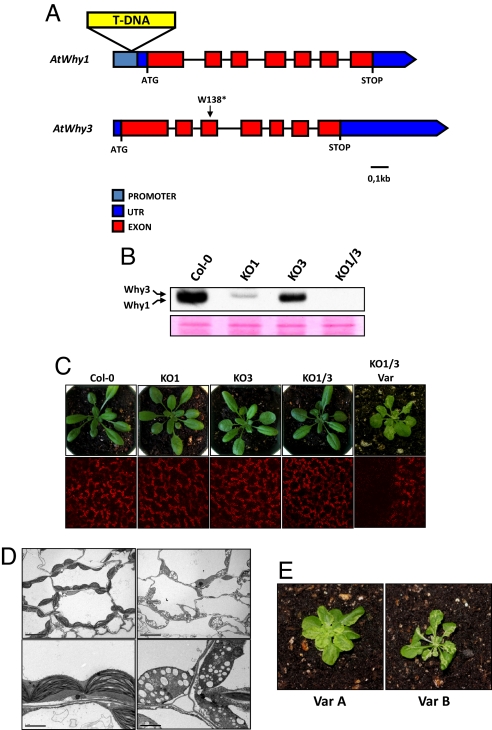
Fig. 1.
AtWhy1 and AtWhy3 are involved in the biogenesis of chloroplasts. (A) Physical maps of the AtWhy1 (AT1G14410) and AtWhy3 (AT2G02740) genes. The position of the T-DNA insertion in the KO1 line is indicated. KO3 is a TILLING line with a mutation that changes a TGG codon to a TGA stop codon in the AtWhy3 gene. An asterisk indicates the position of the mutation. (B) Protein gel blot analysis of simple and double ptWhirlies KO plants. Crude plastid proteins were separated by SDS/PAGE on a 15% polyacrylamide gel. Whirlies were detected by using an anti-AtWhy1/3 antibody. A section of the blot stained with Ponceau red is presented below as a loading control. (C) (Upper) Four-week-old individuals of the indicated genotypes are shown. (Lower) Fluorescence of chlorophyll was visualized by confocal microscopy. (D) Transmission electron microscopy of sections from green (Left) and white (Right) sectors of variegated leaves of KO1/3 plants. (Scale bars: 10 μm in Upper; 2 μm in Lower.) (E) Variegation phenotype varies between independent lines. Four-week-old individuals from the 2 variegated lines Var A (Left) and Var B (Right) are shown.
The KO1 and KO3 plants have no apparent phenotype. Interestingly, although most KO1/3 plants have a WT appearance, some individuals have a smaller size and a variegated phenotype with white/yellow sectors on some leaves (Fig. 1C Upper). In these sectors, a strong diminution in chlorophyll autofluorescence is observed (Fig. 1C Lower). Examination of plastids by electron microscopy reveals that in the white sectors thylakoid stacks are replaced by large round vesicles, indicating that plastid development is compromised (Fig. 1D Right). By contrast, plastids from green sectors of the same leaf appear normal (Fig. 1D Left). We then evaluated the frequency of sectored individuals in large populations of Col-0 and Whirly mutant plants (Table S1). Although no variegated individuals were recovered from Col-0, KO1, and KO3 populations, 4.6% of KO1/3 plants had at least one variegated leaf sector (Table S1). These data indicate that elimination of both ptWhirlies triggers changes that ultimately lead to strong interference with chloroplast development and function.
The Variegated Phenotype of KO1/3 Plants Is Maternally Inherited.
The severity of the variegation phenotype in the KO1/3 population is variable. Although some plants exhibit chlorosis on a single leaf, others have most of their leaves covered by yellow/white sectors (data not shown and Fig. 1E). Phenotypic differences were also observed between different variegated individuals, suggesting that the defects responsible for variegation differ from one plant to another (Fig. 1E). From this first generation of variegated KO1/3 plants, we selected 2 lines with a strong variegation phenotype (Var A and Var B) and set out to define the molecular basis of impaired plastid development.
Maternal inheritance of variegation is often linked to modifications of organelle genomes (9, 17, 18). Crosses were performed between variegated line Var B and Col-0 plants to determine the inheritance mode of the variegation. When variegated plants were used as male parents, no variegation was observed in the heterozygous F1 progeny. However, when Var B plants were used as female in the same cross, variegation was found in 46% of the progeny (47 of 103 F1 plants). This finding indicates that variegation is maternally inherited and that reintroduction of Whirlies is unable to completely rescue the plastid defect, suggesting that the plastids in variegated sectors are irreversibly damaged, most likely at the genetic level.
Amplification of Reorganized ptDNA Regions in Variegated Plants.
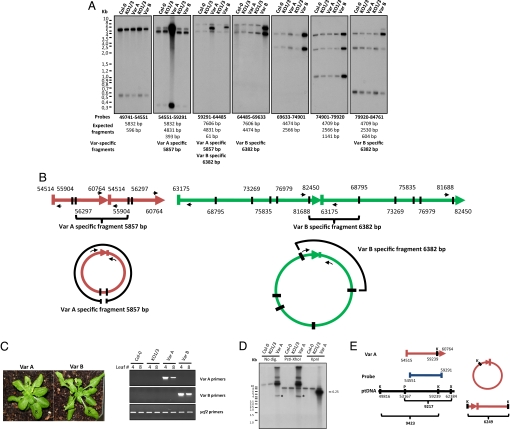
Because Whirlies bind DNA in organelles (refs. 15 and 16 and Figs. S2 and S3), it is likely that variegation in KO plants results from mutations in their ptDNA. We thus searched for ptDNA rearrangements by DNA hybridization. Although no change was detected when comparing HindIII-digested Col-0 to nonvariegated (green) KO1/3 DNA, unique amplified regions were identified in digested ptDNA of the Var A and Var B lines (Figs. 2A and S4). Amplified DNA was estimated to be 10–25 times more abundant than WT ptDNA. In addition, for both variegated lines, new HindIII fragments (≈5.8 kb in Var A and ≈6.3 kb in Var B) were detected by using probes located at the extremities of the amplified regions. These new bands indicate that the amplified regions are reorganized either as circular molecules and/or head-tail concatemers (Fig. 2B).
Fig. 2.
Variegated plants contain rearranged amplified ptDNA regions. (A) DNA gel blot (10 μg per lane) of total leaf DNA digested with HindIII and hybridized with the probes indicated below the gels. The probe numbers refer to the nucleotides of the published Arabidopsis chloroplast genome (42). Expected fragments from restriction analysis of Col-0 ptDNA and the size of new fragments observed in variegated lines are presented below the probes. Restriction and gene maps of the reorganized regions in variegated lines are presented in Figs. S4 and S9. A lower exposition of the second panel allowing better visualization of the amplified bands in VarA is presented in Fig. S5. (B) Schematic of the possible arrangements of the reorganized ptDNA in variegated lines. A head–tail dimer and a monomeric circular molecule are represented for Var A (red) and Var B (green). Oligonucleotides used for the PCR amplification of the junctions of reorganized ptDNA are represented by small black arrows. (C) (Lower) PCR amplification of fragments containing the junctions of reorganized ptDNA in Var A and B plants. (Upper) DNA from leaves 4 (variegated) and 8 (nonvariegated) was isolated. The plastidial ycf2 gene was used as a loading control. (D) DNA gel blot analysis showing the arrangement of amplified ptDNA in the Var A line. DNA from the indicated genotypes was digested with the indicated restriction enzymes and separated on an agarose gel. The DNA was hybridized with the probe depicted in E. A 9.2-kb band corresponding to the WT DNA fragment appears in all samples digested with XhoI and PstI restriction enzymes. A band of 9.4 kb expected from digestion of the WT plastid genome with KpnI was found in all genotypes. The asterisks indicate putative circular molecules. (E) Restriction map of the reorganized regions in the Var A line. The red arrow represents the amplified region in Var A. The probe used is represented as a blue line. A portion of ptDNA is represented as a black horizontal line. The restriction sites are indicated by vertical black lines. K = KpnI; P = PstI; X = XhoI. A circular monomer is represented on the right with the expected linear digestion product of this molecule.
Reorganization of ptDNA Is Caused by Recombination Between Short Direct Repeats.
To map the extremities of the amplified DNA, we designed PCR oligonucleotides close to the ends of the amplified regions of Var A and Var B, facing opposite directions (outward-facing PCR; Fig. 2B). In a WT plastid genome, these primers would yield no product. However, in a rearranged genome where circular or head–tail concatemers are present, we predict a new fragment containing both extremities of the amplified regions. As expected, when PCR is performed with DNA from Col-0 and green KO1/3 plants, no product is observed with both Var A and Var B primers (Fig. 2C). However, when the same primers are used on Var A and Var B DNA, amplification products appear in both cases. Interestingly, more amplified products are detected in variegated versus nonvariegated leaves of the same plants, suggesting that the appearance of defective chloroplasts is related to the abundance of reorganized ptDNA molecules (Fig. 2C, compare leaves 4 and 8).
The position of the ends of amplified regions was determined by cloning and sequencing the PCR products. These positions matched perfectly with the HindIII digestion patterns of the reorganized regions (Fig. 2 A and B, Fig. S4, and Table 1). Remarkably, the regions of WT plastid genome that correspond to the amplified regions in Var A and Var B are bordered by short direct repeats of 10 and 14 bp, respectively (Table 1). In the PCR fragments, both ends of the amplified regions are joined by a single repeat, indicating that recombination occurred between these short sequences. Analysis of 3 additional variegated lines also revealed unique amplified regions with ends joined by recombination at short repeats (Table 1). Interestingly, in the Var C and Var E lines, the bordering repeats are not identical and carry a few mismatches. Altogether, our data strongly suggest that variegation is induced by an overabundance of amplified recombined ptDNA regions.
Table 1.
Characteristics of amplified regions in variegated plants
| Plant line | Length of amplified region. bp | Direct repeats positions* | Short direct repeat sequences and junction | Direct repeat length, bp (and mismatches) |
|---|---|---|---|---|
| Var A | 6250 | 54514 | atctcattagCCTTTTTTTTcgtattttca | (0) |
| Recombinant | agcattcattCCTTTTTTTTcgtattttca | 10 bp | ||
| 60764 | agcattcattCCTTTTTTTTattttctatc | (0) | ||
| Var B | 19275 | 63165 | ttttgtagttTTCTTTTTTTTTTTttcaattttg | (0) |
| Recombinant | tagttctttaTTCTTTTTTTTTTTttcaattttg | 14 bp | ||
| 82450 | tagttctttaTTCTTTTTTTTTTTcagttgctac | (0) | ||
| Var C | 25940 | 9992 | gttctagttgCTCTTGAATACCTTCTTTcaaaaagctt | (0) |
| Recombinant | attctcttatCTCTTGAATACCTTCTTTcaaaaagctt | 18 bp | ||
| 35932 | attctcttatCTCTTGAATTCATTCGTTgcagatccaa | (3) | ||
| Var D | 19216 | 61921 | tgaagcagttGTTAAAATTCCTTATgatatgcaac | (0) |
| Recombinant | cccatgataaGTTAAAATTCCTTATgatatgcaac | 15 bp | ||
| 80507 | cccatgataaGTTAAAATTCCTTATtgttctataa | (0) | ||
| Var E† | 46310 | 124537 | tcaataatgtAAAAGTATACTGTCTCTTGgttagactaa | (3) |
| Recombinant | tatattacaaAAAAGTGTAG-GACTCTTGgttagactaa | 18 bp | ||
| 6019 | tatattacaaAAAAGTGTAG-GACTCTTGtaaaatagaa | (0) |
Additional DNA hybridization experiments were performed to determine the arrangement of the amplified DNA. DNA from Col-0, green KO1/3, and Var A plants was digested either with XhoI and PstI restriction enzymes that cut on both sides of the Var A region in WT ptDNA or with KpnI that cleaves once in this region (Fig. 2 D and E). A probe specific for the Var A-amplified region was used for detection. Intact Col-0 and green KO1/3 DNA migrated within the unresolved compression zone (above ≈10 kb). Six bands were detected in undigested Var A DNA, including a fast-migrating band of ≈4.5 kb. When DNA was digested with XhoI and PstI, the Var A-specific bands were unaffected (Fig. 2E), indicating that these DNA molecules are extrachromosomal. Finally, after KpnI digestion, all Var A-specific bands collapsed into a single 6.25-kb band, showing that these forms are concatemers of the same repeat unit. This result also suggests that the fast migrating band in undigested Var A DNA corresponds to a circular molecule, possibly a monomeric one. Similar results were obtained with the Var B line (data not shown). We thus conclude that amplified ptDNA is present as both circular and/or head–tail concatemers in variegated plants.
Illegitimate Recombination Is Increased in the Absence of Arabidopsis ptWhirlies.
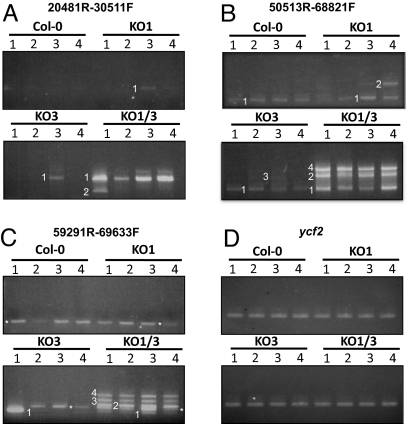
Our results suggest that the absence of ptWhirlies induces plastid genome instability through an increase in recombination between short direct repeats. One could therefore expect to detect low-level recombination events in nonvariegated Whirly mutant plants. We tested this hypothesis by using the outward-facing PCR approach described above, on each of 4 independent DNA pools from Col-0, KO1, KO3, and green KO1/3 plants. Representative results of the PCR amplification are shown in Fig. 3 A–C. Rearranged products were observed in all genotypes.
Fig. 3.
Illegitimate recombination is increased in the absence of Arabidopsis ptWhirlies. For each genotype, PCRs using outward or inward-facing PCR primers were performed on 4 pools of DNA from 4 different plants. Reactions were run on agarose gels containing ethidium bromide. (A–C) Representative PCRs are shown. The oligonucleotides used are indicated above each panel. Individual bands (white numbers) were cut, cloned, and sequenced. Each band represents a unique recombination product (Table S2). The asterisks indicate nonrecombinant products arising from nonspecific hybridization of the 69633F primer at positions 58720–58733 of the plastid genome. (D) The plastidial ycf2 gene was used as a loading control.
Cloning and sequencing of rearranged DNA confirmed that illegitimate recombination is strongly increased in green KO1/3 plants, where 40 different recombination products were identified out of 30 PCRs (Tables S2, S3, and S4). Recombination products were also detected in single KOs, although less frequently, indicating that the depletion of AtWhy1 or AtWhy3 is sufficient to increase spurious recombination. Surprisingly, 2 recombination events were detected in Col-0 samples. These were present in all genotypes tested (Table S2, S3, and S4), suggesting the presence of a small subpopulation of rearranged ptDNA even in WT plants.
Illegitimate Recombination Is Increased in Maize Whirly Mutants.
We verified in 3 maize ZmWhy1 mutant lines whether mutation of monocot ptWhirlies also affects plastid genome stability. The lines ZmWhy1–1, ZmWhy1–2, and the heteroallelic progeny of complementation crosses ZmWhy1–2/-1 all have a reduced level of the plastid-localized protein ZmWhy1. These lines exhibit ivory (ZmWhy1–1), pale green (ZmWhy1–2), and intermediate (ZmWhy1–2/-1) leaf phenotypes (16). Our Southern hybridizations did not reveal amplified plastid genome regions in any of the 3 lines (Fig. S6), indicating that the defect in chloroplast biogenesis in ZmWhy1 mutants is not linked to the presence of amplified ptDNA regions. Outward-facing PCR analysis of the B73 control maize line revealed a single short-repeat mediated illegitimate recombination event for the 19 primer pairs tested (Tables S5 and S6). The same primers revealed 3 events in lines ZmWhy1–2 and ZmWhy1–2/-1. Illegitimate recombination was highest in the most severe mutant line (ZmWhy1–1), with a total of 14 different events (Tables S5 and S6). These results suggest that Whirlies are also involved in stabilizing the plastid genome in maize.
Discussion
Whirlies Prevent Short Homology-Dependent Illegitimate Recombination.
Accumulation of reorganized DNA through illegitimate recombination has also been observed in bacteria. In Escherichia coli, stress can induce selective amplification of genome regions conferring tolerance to the applied stress, a phenomenon called adaptive amplification. These regions (≈10–30 kb) are first duplicated through illegitimate recombination between microhomologous repeats (5–15 bp) and are then further replicated through a mechanism requiring the homologous recombination machinery (reviewed in ref. 19). Another recombination process, called “short homology-dependent illegitimate recombination” exists in bacteria (reviewed in ref. 20). It also requires microhomology (3–20 bp) at recombination sites, and its occurrence is increased by various DNA-damaging agents. Similarly, microhomology-mediated recombination has recently been proposed to trigger segmental DNA duplications that are linked to some genetic diseases in humans and to copy number variations among individual genomes (21–23). It was also observed in yeast cells undergoing segmental duplications (24). Interestingly, in bacteria, DNA-RRR proteins, such as the subunits of the UvrAB complex, the RecQ helicase, the histone-like H-NS, and the SSB can suppress microhomology-mediated recombination (25–29). However, the genetics requirements for this suppression may vary with the system used to monitor recombination. We propose a similar role for Whirlies in plastids, where they contribute to maintain genome stability by preventing accumulation of illegitimate recombination products.
A Model for the Variegation in Arabidopsis Plants Lacking ptWhirlies.
The direct repeat sequences at Arabidopsis ptDNA junctions are too short to serve as substrates for homologous recombination (30, 31). Recently, an alternative model was proposed to explain how microhomologies may generate large DNA rearrangements in E. coli, yeast, and human (21). In this model, the collapse of a replication fork leads to the generation of a 3′ protruding ssDNA end. This 3′ tail may anneal to a microhomologous ssDNA sequence at a different location, allowing reinitiation of replication at this new site. When such template switching occurs between 2 different DNA molecules, either duplications (switch to a position behind the fork) or deletions (switch to a position ahead of the fork) will occur. Alternatively, switching on the same DNA molecule behind a stalled replication fork leads to rolling-circle replication that produces concatemers of subgenomic regions and eventually circular DNA products (Fig. 2D). In addition to duplication/circularization events that were detected by DNA hybridization and outward-facing PCR, deletions were identified by using inward-facing PCR in Arabidopsis Whirly mutants (Table S4). In summary, the above replication-based mechanism might explain the variety of microhomology-mediated rearrangements observed in the plastid genome of Whirly mutants.
Based on our results, we propose the following model for the emergence of variegation in KO1/3 plants (Fig. S7). In Col-0, the genome surveillance machinery repairs DNA lesions and, with the help of Whirlies, prevents accumulation of illegitimate DNA recombination products, which are rarely detected in these plants. In the single KO plants, the absence of 1 ptWhirly is enough to induce a low-level accumulation of rearranged molecules. However, when both Whirlies are eliminated, aberrant molecules accumulate to high levels. Some of these molecules, most likely circularized products, could replicate independently of the main plastid genome and eventually become overabundant. In each of the 5 variegated lines analyzed in detail (Table 1) only 1 rearranged molecule is amplified. Although a certain level of these molecules can be tolerated by the plastid, as demonstrated by their presence in green tissue of variegated plants (Fig. 2C), their accumulation eventually gives rise to plastids that are nonfunctional, thus leading to the appearance of variegated leaf sectors. No deletions resulting in variegation were detected, suggesting that their accumulation is either lethal and/or that they are more easily eliminated by gene conversion (32).
Conservation of the Role of ptWhirlies in the Maintenance of Plastid Genome Stability.
The finding that illegitimate recombination mediated by short repeats is also enhanced in a severely affected maize Whirly mutant line suggests a conserved function for Whirlies in maintaining plastid genome stability. ZmWhy1 hypomorphic mutants show a less clear trend than the ZmWhy1–1 mutant, which might be caused by the presence of a higher residual level of ZmWhy1 protein in these lines (16). This situation resembles that of the single KO1 and KO3 Arabidopsis Whirly mutants, which show less illegitimate recombination than the double KO. The low level of ZmWhy1 protein present in all ZmWhy1 mutants may also explain why no amplified ptDNA regions are found in these plants. In fact, the level of residual ptWhirly protein in the maize mutants appears similar to that present in the Arabidopsis KO1 mutant, which also does not contain amplicons. Amplified regions were observed only in KO1/3, the only line analyzed that is completely devoid of ptWhirlies, suggesting that a low level of Whirlies is sufficient to prevent amplification of recombined ptDNA molecules.
The absence of detectable amplicons and the severe first-generation phenotypes in the maize Whirly mutants indicate that a different mechanism contributes to the formation of defective chloroplasts in this species. Actually, the ZmWhy1 mutants exhibit a reduced content of plastid ribosomes, with the albino ZmWhy1–1 seedlings almost completely lacking plastid rRNA (16). This ribosome deficiency was proposed to result from a defect in the biogenesis of the large ribosomal subunit caused by an aberrant 23S rRNA metabolism, suggesting a role for Whirlies in plastid RNA metabolism (16). Although we cannot rule out such a role in Arabidopsis, it is unlikely that the more subtle variegation phenotype observed in this species is caused by aberrant rRNA metabolism as no major changes are detected in 23S and 16S rRNA levels in the Arabidopsis ptWhirly mutants (Fig. S8). Thus, Whirlies appear to form a flexible family of single-stranded nucleic acid-binding proteins that can fulfill a variety of roles, depending on the cellular context and/or plant species.
Characteristics of the Short Direct Repeats.
Some of the direct repeats reported here appear to be particularly prone to illegitimate recombination as they are used repeatedly and at multiple different positions of the plastid genome (see related events in Tables S4 and S5), which suggests that a specific sequence motif or structure promotes their use as substrates. Homopolymeric A/T stretches appear to be over-represented among the direct repeats (Tables 1, S2, S4, and S5). This type of sequence produces unusual structures called slipped-strand DNA, which can cause replication stalling (reviewed in ref. 33). Stalled replication forks can be reinitiated by template switching and can also generate double-strand ends (reviewed in ref. 34), potentially leading to an increase in illegitimate recombination. Alternatively, these poly(A)/T sequences could have a greater tendency to melt, providing easier access to invading ssDNA 3′ tails and thus favoring ptDNA rearrangements.
Whirlies Could Prevent Illegitimate Recombination by Protecting ssDNA.
Many ssDNA-binding proteins bind nonspecifically to DNA and play important roles in recombination processes. Bacterial SSBs and their eukaryotic nuclear counterparts such as RPA play essential protective roles in genome biology by protecting ssDNA from damage and preventing spurious DNA annealing. They also serve to recruit a large number of genome maintenance proteins (reviewed in refs. 35 and 36). In E. coli, SSB also suppresses short homology-dependent illegitimate recombination (27, 28). It is thus tempting to speculate that in plastids, Whirlies play a role similar to that of SSB in E. coli. This idea is supported by our data (Fig. S3 and ref. 15) and those of Prikryl et al. (16), indicating that Whirlies bind DNA in a nonsequence specific manner. In addition, similar to bacterial SSBs, AtWhy1 protects ssDNA against degradation by nucleases in vitro (L. Cappadocia, A.M., É. Lepage, J. Sygusch, and N.B., unpublished results). We therefore propose that in plastids the absence of Whirlies enhances the availability of free ssDNA, which might lead to increased DNA damage caused by the vulnerability of the exposed DNA with a subsequent increase in recombination–repair mechanisms. More importantly, the damaged ssDNA might increase the rate of illegitimate recombination by increasing the frequency of collapsed replication forks (reviewed in ref. 37). In any case, stresses able to induce DNA damage would also increase ssDNA availability, as happens after processing of double-strand breaks, leading to increased recombination (28).
Our experiments define Whirlies as important components of the plastid genome maintenance machinery. As a follow-up, it will be interesting to determine whether stresses that alter the integrity of ptDNA also lead to increased illegitimate recombination in both Whirly mutants and plants deficient for putative ptDNA-RRR proteins. This will help determine the function of Whirlies in DNA protection/repair processes and permit identification of new regulators of plastid genome stability.
Materials and Methods
Mutant Characterization.
The Salk Institute Genomic Analysis Laboratory (La Jolla, CA) provided the sequence-indexed T-DNA insertion line (38). The Seattle TILLING Project (39) provided plants with mutations in the AtWhy3 gene. The maize mutant lines ZmWhy1–1, ZmWhy1–2, and ZmWhy1–2/-1 were obtained from Alice Barkan (University of Oregon, Eugene) (16).
Antibody Production.
Recombinant AtWhy1 was purified as described (40). Rabbits were immunized and antiserum was collected. For protein gel blot analysis, the antiserum was used at a concentration of 1:4,000.
DNA Gel Blots.
DNA was isolated from plants by using a Cetyl trimethylammonium bromide DNA extraction protocol (41). Running of the samples and blotting of the gels were performed as described (15).
Detection of DNA Rearrangements by PCR in Arabidopsis.
PCRs were conducted by using the Taq polymerase from Genscript and a series of outward-facing oligonucleotides spaced by ≈10–20 kb. Inward-facing primers spaced by 10–20 kb were used to detect deletion events. Thirty reactions were performed on each of 4 independent DNA samples from Col-0, KO1, KO3, and green KO1/3 plants and analyzed by gel electrophoresis. All visible DNA bands were isolated, cloned, and sequenced. See SI Appendix.
Supplementary Material
Acknowledgments.
We thank Dr. C. S. Hardtke for help with the Arabidopsis crosses; Dr. R. M. Gaafar for help with genotyping; S. Grondin, P. Tan, and A. Moreau for technical assistance; L. Forget for the sequencing of ptDNA; and Dr. A. Barkan for the ZmWhy1 mutant lines. A.M. was supported by scholarships from the Natural Science and Engineering Research Council of Canada and the Fonds Québécois de la Recherche sur la Nature et les Technologies. This work was supported by a grant from the Natural Science and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901710106/DCSupplemental.
References
- 1.Bendich AJ. Circular chloroplast chromosomes: The grand illusion. Plant Cell. 2004;16:1661–1666. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Day A, Madesis P. DNA replication, recombination, and repair in plastids. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol 19. Berlin: Springer; 2007. pp. 65–119. [Google Scholar]
- 3.Palmer JD. Chloroplast DNA exists in 2 orientations. Nature. 1983;301:92–93. [Google Scholar]
- 4.Staub JM, Maliga P. Long regions of homologous DNA are incorporated into the tobacco plastid genome by transformation. Plant Cell. 1992;4:39–45. doi: 10.1105/tpc.4.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao J, Combs C, Jagendorf AT. The chloroplast-located homolog of bacterial DNA recombinase. Plant Cell Physiol. 1997;38:1319–1325. doi: 10.1093/oxfordjournals.pcp.a029124. [DOI] [PubMed] [Google Scholar]
- 6.Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saotome A, et al. Characterization of four RecQ homologues from rice (Oryza sativa L. cv. Nipponbare) Biochem Biophys Res Commun. 2006;345:1283–1291. doi: 10.1016/j.bbrc.2006.04.134. [DOI] [PubMed] [Google Scholar]
- 8.Wall MK, Mitchenall LA, Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc Natl Acad Sci USA. 2004;101:7821–7826. doi: 10.1073/pnas.0400836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaegel V, et al. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006;18:3548–3563. doi: 10.1105/tpc.106.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishibashi T, Kimura S, Sakaguchi K. A higher plant has three different types of RPA heterotrimeric complex. J Biochem. 2006;139:99–104. doi: 10.1093/jb/mvj014. [DOI] [PubMed] [Google Scholar]
- 11.Desveaux D, Maréchal A, Brisson N. Whirly transcription factors: defense gene regulation and beyond. Trends Plants Sci. 2005;10:95–102. doi: 10.1016/j.tplants.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell. 2000;12:1477–1489. doi: 10.1105/tpc.12.8.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo HH, Kwon C, Lee MM, Chung IK. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 2007;49:442–451. doi: 10.1111/j.1365-313X.2006.02974.x. [DOI] [PubMed] [Google Scholar]
- 14.Krause K, et al. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005;579:3707–3712. doi: 10.1016/j.febslet.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 15.Maréchal A, et al. Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function. BMC Plant Biol. 2008;8:42. doi: 10.1186/1471-2229-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang TL, et al. Characterization of primary lesions caused by the plastome mutator of Oenothera. Curr Genet. 1996;30:522–530. doi: 10.1007/s002940050165. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Zapater JM, Gil P, Capel J, Somerville CR. Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell. 1992;4:889–899. doi: 10.1105/tpc.4.8.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastings PJ. Adaptive amplification. Crit Rev Biochem Mol Biol. 2007;42:271–283. doi: 10.1080/10409230701507757. [DOI] [PubMed] [Google Scholar]
- 20.Ehrlich SD. Illegitimate recombination in bacteria. In: Berg DE, Howe MM, editors. Mobile DNA. Washington D.C.: American Society for Microbiology; 1989. pp. 799–832. [Google Scholar]
- 21.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 24.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanada K, Iwasaki M, Ihashi S, Ikeda H. UvrA and UvrB suppress illegitimate recombination: Synergistic action with RecQ helicase. Proc Natl Acad Sci USA. 2000;97:5989–5994. doi: 10.1073/pnas.100101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanada K, et al. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukaihara T, Enomoto M. Deletion formation between the two Salmonella typhimurium flagellin genes encoded on the mini F plasmid: Escherichia coli ssb alleles enhance deletion rates and change hot-spot preference for deletion endpoints. Genetics. 1997;145:563–572. doi: 10.1093/genetics/145.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy M, Gowrishankar J. Identification and characterization of ssb and uup mutants with increased frequency of precise excision of transposon Tn10 derivatives: Nucleotide sequence of uup in Escherichia coli. J Bacteriol. 1997;179:2892–2899. doi: 10.1128/jb.179.9.2892-2899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shanado Y, Hanada K, Ikeda H. Suppression of γ ray-induced illegitimate recombination in Escherichia coli by the DNA-binding protein H-NS. Mol Genet Genomics. 2001;265:242–248. doi: 10.1007/s004380000399. [DOI] [PubMed] [Google Scholar]
- 30.Lovett ST, Hurley RL, Sutera VA, Jr, Aubuchon RH, Lebedeva MA. Crossing over between regions of limited homology in Escherichia coli RecA-dependent and RecA-independent pathways. Genetics. 2002;160:851–859. doi: 10.1093/genetics/160.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liskay RM, Letsou A, Stachelek JL. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987;115:161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- 33.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Persky NS, Lovett ST. Mechanisms of recombination: Lessons from E. coli. Crit Rev Biochem Mol Biol. 2008;43:347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- 38.Alonso JM, et al. Genomewide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 39.Till BJ, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desveaux D, et al. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 41.Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2002. [Google Scholar]
- 42.Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- 43.Kunnimalaiyaan M, Nielsen BL. Fine mapping of replication origins (ori A and ori B) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res. 1997;25:3681–3686. doi: 10.1093/nar/25.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.