Abstract
A key step in the assembly of many viruses is the packaging of DNA into preformed procapsids by an ATP-powered molecular motor. To shed light on the motor mechanism we used single-molecule optical tweezers measurements to study the effect of mutations in the large terminase subunit in bacteriophage λ on packaging motor dynamics. A mutation, K84A, in the putative ATPase domain driving DNA translocation was found to decrease motor velocity by ≈40% but did not change the force dependence or decrease processivity substantially. These findings support the hypothesis that a deviant “Walker A-like” phosphate-binding motif lies adjacent to residue 84. Another mutation, Y46F, was also found to decrease motor velocity by ≈40% but also increase slipping during DNA translocation by >10-fold. These findings support the hypothesis that viral DNA packaging motors contain an adenine-binding motif that regulates ATP hydrolysis and substrate affinity analogous to the “Q motif” recently identified in DEAD-box RNA helicases. We also find impaired force generation for the Y46F mutant, which shows that the Q motif plays an important role in determining the power and efficiency of the packaging motor.
Keywords: optical tweezers, phage, single molecule, virus
The detailed function of viral packaging motors, which package dsDNA inside viral capsids against a large internal force into near crystalline density, has generated much interest recently in part because of the advent of single-molecule techniques capable of directly measuring DNA translocation dynamics (1–4). Studies of dsDNA packaging of bacteriophages will be helpful in understanding similar medically important mammalian viruses including herpesviruses and adenoviruses.
Bacteriophage λ is an important model system because its genetics and biochemistry have been studied extensively (5). An integral part of the λ packaging motor is its translocase, often referred to as a “terminase complex.” It is composed of multiple heterotrimer units each containing 1 gpA “large subunit,” a 641-aa residue protein, and 2 gpNu1 “small subunit” proteins composed of 181 residues each. This ATP-powered complex mediates a number of different critical activities: Terminase first binds to a concatemer of dsDNA at a specific site (cos), introduces staggered nicks 12 bp apart, separates the cohesive ends, and forms a tight complex at the left chromsome end. This “complex I” docks on to the portal protein of a procapsid and sponsors the packaging of the DNA. The active packaging complex is referred to as “complex II.” Packaging terminates when sufficient filling is achieved and the motor encounters the downstream cos site, introduces single-strand nicks, separates the cohesive ends, and releases a DNA-filled head.
The details of the mechano-chemical coupling and translocation of dsDNA in this class of molecular motors remain poorly understood. Ideally one would link the structure of the motor to its functional capabilities. Such studies could, for example, help in the understanding of how the ATPase center and its various functional domains are responsible for DNA packaging in bacteriophage λ. Answers to these questions have thus far proved to be elusive. We have previously found evidence for a critical ATPase in gpA using 8-Azido-ATP photolabeling (6). Among our findings was that residues Y46 and K84 interact with ATP. Mutant terminases with changes at these sites revealed that while the endonuclease and helicase activities of terminase remained the same, virion assembly was abolished by the Y46F change in gpA and was reduced 40-fold by the K84A change. Subsequent work using bulk packaging and ATPase assays discovered that the procapsid-dependent translocation ATPase activity (DNA packaged/ATP hydrolyzed) of mutants with Y46F, Y46A, and K84A changes was similar to wild type (WT) (7). The mutant enzymes showed mild DNA packaging defects in bulk DNase protection assays: The Y46F and Y46A enzymes sponsored DNA packaging that was ≈30% the WT level, and the K84A motor's efficiency was ≈5% that of WT. Because the ATPase activity of the mutants was measured to be similar to WT, the nature of the packaging defects was left unresolved. The profound virion assembly defects of Y46F and Y46A mutants sharply contrasted with the mild packaging defects, while the virion assembly and DNA packaging defects of the K84A enzyme were consistent.
In light of these results, the definitive determination of the location and characteristics of the ATPase center that powers translocation has thus far been elusive because it is difficult to measure the motor translocation dynamics in bulk biochemical assays. In particular, the DNase protection assays measure only the total amount and minimum lengths of DNA packaged, as judged by gel electrophoresis of restriction fragments after a long reaction incubation time of typically tens of minutes to hours. The times taken for assembly of terminase on the DNA, docking of the terminase DNA complex on the procapsid, initiation of translocation, translocation (with rate changes, stalling, pausing, and slipping), and termination are convolved together and cannot easily be separated. Furthermore, the bulk assay measures the “ensemble average” behavior and cannot discern heterogeneity in dynamics and lack of synchronization of these processes occurring in different individual complexes. The single-molecule optical tweezers assay, on the other hand, directly measures initiation of packaging and DNA translocation in individual complexes in real time, affording the characterization of the heterogeneity in the packaging dynamics. Our previous optical tweezers assays with WT motor complexes revealed that packaging could be triggered to occur within seconds of bringing a target DNA molecule into contact with a procapsid–terminase complex and that packaging at a high speed averaging 580 bp/s usually proceeded within seconds (8). Similar optical tweezers assays were previously applied to measuring packaging dynamics in bacteriophage phi29 and have also recently been applied to bacteriophage T4 (9). These optical tweezers assays also revealed that all of these viral motors were very strong, exerting >50 pN of force during packaging. These initial measurements pave the way for further detailed investigations on the mechanisms of packaging for the bacteriophage λ system.
On the basis of sequence alignments, it was hypothesized that gpA's Y46 residue is located in a putative “adenine-binding” domain or “Q motif” identified in many different viral terminases (10). This domain was recently discovered in DEAD box proteins, members of SF2 RNA helicases (11). Q motifs are located ≈17 aa upstream of the Walker A motif and have highly conserved aromatic residues ≈17 aa upstream of the Gln. The aromatic residues were proposed to aid in hydrophobic stacking interactions with adenine. Q motifs were found to be involved in the efficient binding of ssRNA and for conformational changes that are driven by nucleotide and ATP hydrolysis (11–14). It was further proposed that the Q motif functions as a molecular on–off switch for ATP hydrolysis and helicase activity (14). Mutation studies in the Q motif in RNA helicases concluded that, at minimum, it directly regulates the affinity of the protein for the substrate (RNA) through conformational changes associated with nucleotide binding (14, 15). Also, through sequence alignments of various terminases, Mitchell and Rao suggested that bacteriophage λ may have a deviant Walker A motif (76KSARVGYS83 unlike the classic Walker GXXXXGKS/T) responsible for interactions with the β and γ phosphates of ATP (10, 16, 17). Do such motifs indeed exist in viral packaging motors and other related motors? And what exactly are their roles?
In this study, single-molecule optical tweezer packaging assays are used to uncover relationships between mutations of potentially located functional domains of packaging ATPases and their effects on the function of the viral DNA packaging motor in λ bacteriophage. Among the mutants we have recently characterized are a slightly defective terminase with mild changes in velocity of packaging (K84A) and an inefficient terminase (Y46F) with impaired force generation and processivity while packaging DNA. Below, we link our single-molecule measurements to functional domains in phage packaging motors that appear highly related to those of RNA helicases. In particular, we present evidence that a “deviant” Walker A is responsible for interacting with the ATP phosphate and that a Q motif governs force generation and substrate interaction (motor–DNA interaction).
Results
Many measurements were done using a force–clamp mode [supporting information (SI) Fig. S1] in which feedback control was used to change the separation between 2 optical traps as DNA is translocated under a small constant load force (≈5 pN) (1). With these measurements, the distance of DNA translocated during packaging could be tracked with time. Additional measurements of motor velocity vs. load dependence were made using a passive mode. The optical traps were held in a fixed position at a minimum starting force of 3–7 pN in the vicinity of the expected full tether length, and packaging was detected when the DNA tension increased with time. Using the known elasticity of dsDNA, the compliance of the traps, and the separation distance of the traps, the change in tether length is calculated from the force (1). To test the functions of the putative adenine-binding (Q) and Walker A motifs, we used mutants Y46F, K76R, and K84A (see Methods) (6, 18). Sequence alignments of the viral packaging motors and the location of the mutants in their putative functional domains are shown in Fig. 1. The DNA translocation dynamics of the terminase–procapsid complexes were then measured using optical tweezers. Fig. 2 shows representative raw data traces of packaging events measured for the terminases studied.
Fig. 1.
Sequence alignments (performed by Rao and coworkers, refs. 10 and 28) comparing the various functional domains of selected SF2 helicases and dsDNA packaging phage/virus motors. In color are the Q or adenine-binding motif (green), the Walker A motif (orange), the Walker B motif (purple), catalytic carboxylate (red), and the C motif (blue). Indicated with asterisks are mutants of phage λ's gpA examined in this study. In parentheses are the numbers of residues between adjacent domains indicated in the sequence alignments. According to Sun et al., structurally based alignments of functional domains of T4's gp17 crystal structure and nucleotide-binding domains of various other ATPases are mostly consistent with amino acid sequence alignments performed by Mitchell et al. (10, 23).
Fig. 2.
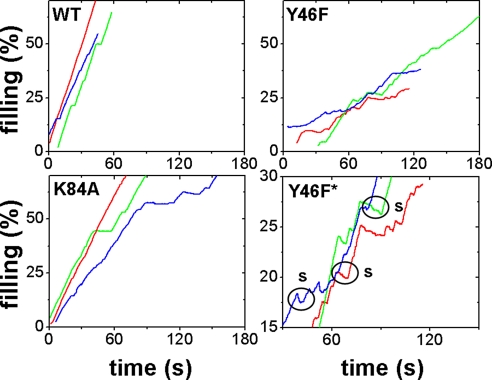
Representative raw data of individual packaging events (filling vs. time) of WT, Y46F, and K84A terminases. Percentage of filling means percentage of the wild-type genome length (48,502 bp) packaged. Data are acquired using force feedback at a small constant load (≈5 pN) while packaging is taking place. Y46F* is a zoomed-in version of the graph depicting Y46F packaging events, where large (>100 bp) slips are marked with circles.
Packaging Dynamics of K84A (Walker A Motif).
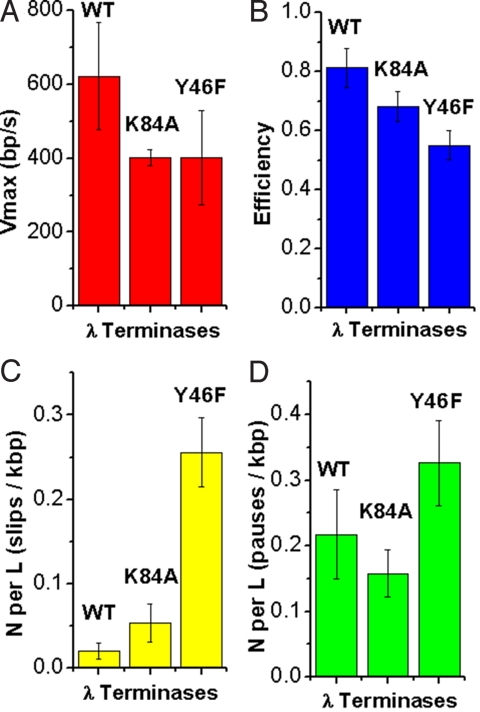
The maximum translocation rate of K84A is ≈40% less than that of WT (Fig. 3). The K84A mutant was found to slip much less frequently than theY46F mutant and only slightly more frequently than WT within error (N/L = 0.054 ± 0.023 compared to 0.020 ± 0.009). To assess the effects of slipping and pausing on the time it takes for terminase to package DNA, we define an efficiency factor:
This factor quantifies how much slower packaging occurs because of pausing and slipping and gives a value for how continuous packaging is with the various terminases (see Methods). The normalized efficiency of packaging, relative to WT measurements, is 0.839, which shows there is only a minor increase in the amount of time lost in packaging because of pausing and slipping relative to WT.
Fig. 3.
(A) Vmax of various terminases studied. Measurements were taken at saturating ATP (1 mM) and only at ≤10% filling to avoid effects from capsid expansion. Errors are SD. (B) Efficiency of various terminases as described in text using Eq. 2. Errors are SE. (C) Number of large slips (>100 bp) per kilobase pair of DNA translocated. Errors are SE. (D) Number of long pauses (>0.2 s) per kilobase pair of DNA translocated. Errors are SE.
Using a passive mode in our optical tweezers measurements, we investigated the force-generating ability of the mutant motors. These measurements yield a force–velocity (F-V) curve that shows that an opposing force against translocation increases the height of the reaction barrier and slows the reaction rate. Within a minimal Kramer's-type model of thermal activation over multiple reaction free energy barriers (19, 20), we find that a function with at least 2 rate-limiting transitions is needed to fit the F-V data,
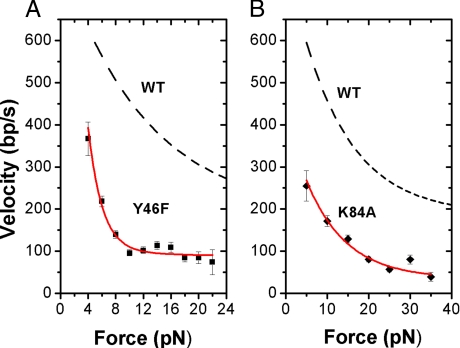
where Δx1 and Δx2 are the characteristic distances the DNA must move against the force F to reach the position of the energy barrier maximums, v1 and v2 are maximum velocities at zero load, and kT is the thermal energy. According to Fig. 4, the F-V curve measured for K84A has a similar steepness to WT: The Δx1 deduced from the force–velocity curve of K84A (0.43 ± 0.17 nm) is similar to WT (≈0.34 nm, Table 1). At low forces, the first transition is rate limiting and force dependent, indicating that this transition involves DNA translocation. At high forces, the second transition becomes rate limiting, but this transition appears to be independent of force (because Δx2 ≈0, for both K84A and WT), implying that this transition must not involve DNA translocation. The maximum power generated by K84A is less than WT: P = 3,222 pN·bp·s−1 = 1.10 × 10−18 W = (34.3% of WT). The loss of power of K84A is related to a decrease in Vmax rather than the steepness of its F-V curve.
Fig. 4.
Force–velocity curves of (left) Y46F (2-pN bins) and (right) K84A (5-pN bins). In red are fits to Eq. 2 described in the text. Dashed lines are the fits to the F-V curve found for WT (Fuller et al., ref. 9) used for comparison to the mutant. Errors are SE. Measurements were taken under a position clamp mode described in the text.
Table 1.
Parameters extracted from fits of force–velocity curves of various terminases using Eq. 2
| Terminase | V1, bp/s | V2, bp/s | Δx1, nm |
|---|---|---|---|
| WT | 615 ± 120 | 187 ± 155 | 0.34 ± .12 |
| Y46F | 2,084 ± 1,330 | 102 ± 56 | 2.02 ± 0.62 |
| K84A | 392 ± 131 | 35 ± 21 | 0.43 ± 0.17 |
Packaging Dynamics of Y46F (Adenine-Binding/Q Motif).
Similar to K84A, the mutant Y46F displays a slower translocation rate (≈40% less than WT). In contrast to K84A, Y46F has a dramatic loss in processivity and slips >10 times more frequently than WT per length of DNA (0.237 ± 0.059 compared to 0.020 ± 0.009 slips/kbp DNA packaged, Fig. 3). No significant differences were resolved in pausing behavior between Y46F and other terminases studied (within standard errors). Combining the time lost from slipping and pausing, the efficiency of packaging (Fig. 3b) is decreased to 67% of WT. As shown by Fig. 4, velocity drops much more steeply with force for Y46F than for WT, unlike K84A, as indicated by the much larger Δx1 fit parameter (≈2 nm in Y46F vs. ≈0.34 nm in WT, Table 1). Y46F approaches minimum velocity near 22 pN (77 bp/s), while WT can still reach a velocity of 208 bp/s at higher forces (>45 pN) (8). The maximum power generated by the Y46F motor has decreased dramatically relative to WT [WT, 9,400 pN·bp·s−1 = 3.22 × 10−18 W; Y46F, 1,745 pN·bp·s−1 = 5.97 × 10−19 W (18.6% of WT)], which is related to the dramatic increase in steepness of its F-V curve. We note that the increase in the value of Δx1 obtained by fitting the simple model of Eq. 2 could possibly correspond to an increased step size but could also be due to a change in the dependence of the ATP turnover rate on force (if stepping is tightly coupled to hydrolysis and force slows stepping) or a change in the dependence of the coupling efficiency (probability that the motor makes a step per hydrolysis cycle) on force. In any case, as discussed by Wang et al. and Chemla et al., our finding of an altered force dependence for Y46F implies that the rate-limiting transition affected by the mutation is a mechanical one that involves DNA translocation (a conformational change that couples the ATP hydrolysis cycle to DNA translocation) as opposed to a purely chemical transition (2, 20).
To further investigate the biochemical kinetic parameters changed by the mutation, we measured how varying ATP concentrations affected the velocity of translocation. Fits to the Hill equation,
revealed Michaelis–Menten kinetics for both WT and Y46F (Fig. S2 and Fig. S3, Table S1) and a Hill coefficient of ≈1 (Table 2), which is consistent with recent studies on the bacteriophage phi29 motor (2). To investigate if the Y46F mutation leads to an ATP binding defect, we can compare Vmax/Km, which is proportional to kb, an effective rate of ATP binding within simple models (2, 21, 22). We find similar values of Vmax/Km (17 bp·s−1·μM−1 for WT and 18 bp·s−1·μM−1 for Y46F), suggesting that there is no apparent ATP binding defect.
Table 2.
Parameters extracted from fits to the Hill equation
| Terminase | Vmax, bp/s | Km, μM | Vmax/Km, bp·s−1·μM−1 | Hill coefficient |
|---|---|---|---|---|
| WT | 580 | 33.8 ± 8.6 | 17.1 ± 4.3 | 0.75 ± 0.15 |
| Y46F | 402 | 22.2 ± 2.1 | 18.3 ± 1.7 | 1.17 ± 0.13 |
Vmax is measured from force clamp measurements and is imposed for fits. Fits without imposing a Vmax constraint for WT and Y46F, while adding more uncertainty to Km, similarly yield Hill coefficients ≤1. Velocities at concentrations <10 μM for Y46F were not measured because of the low efficiency of initiation at low ATP concentrations.
Discussion
A Deviant Walker A Motif in Bacteriophage λ's Packaging Motor.
Our finding an abolishment of packaging with the K76R mutant of gpA is consistent with the hypothesis that the sequence 76KSARVGYS83 comprises a deviant Walker A region (18). The phosphate-binding Lys residue in gpA is located in the beginning of the Walker A loop, rather than adjacent to the Ser, as in the consensus Walker A sequence. In the consensus sequence, the Lys of GXXXXKS/T was proposed to interact with the negatively charged β and γ phosphates of ATP.
We found that optical tweezers measurements of packaging could not be initiated with the K76R mutant. In addition, in extensive genetic screening experiments, no revertants of this mutant were recovered. These results further confirm the importance of this residue in DNA packaging and support the hypothesis that K76 is a critical residue in the Walker A region that interacts with ATP phosphates. In contrast, we were able to initiate packaging in the optical tweezers assays with the K84A mutant. In accordance with the assignment of residues 76–83 of gpA as a deviant Walker A region, the residue K84 of lambda would be directly adjacent to this region. It is reasonable to propose that the K84A mutation would slightly perturb the structure of the ATP binding pocket, which could slow ATPase activity. This is supported by our finding of a modest decrease in Vmax found in the K84A mutant (60–70% of the WT Vmax, Fig. 3a). In addition, a structural model of λ's gpA, which aligns with T4's gp17 crystal structure, indicates that the K76 residue, not K84, would be equivalent to the conserved Lys that is critical for ATPase and translocation activities, giving further evidence of a deviant Walker A motif (28).
However, the large reduction in DNA packaged by K84A found in bulk packaging assays (5% compared to WT) is unexplained. Taking into account the measured modest decrease in speed, pausing, and stalling behavior (Fig. 3 c and d), our present optical tweezers measurements show that some other step in the process besides DNA translocation must account for the defect. A defect in initiation is possible, but seems unlikely because we find that the K84A mutant terminase initiates packaging in our optical tweezers assays with similar frequency to WT and no significant change in time between binding and initiation of translocation is observed (Fig. S4). Also, we do not observe any significant change in pausing frequency per length of DNA, and we do observe less severe slipping frequency changes than with Y46F. Therefore, there does not appear to be a major structural defect in this mutant that affects translocation uncovered by measurements taken at a low applied load (5 pN). However, upon further examination, the F-V data show that the K84A mutation slows down packaging appreciably at high forces, which may prevent completion of packaging (Fig. 4). It is important to note, however, this is not due to changes in the steepness of the F-V curve (Δx1 is similar to WT), which suggests no intrinsic changes in the efficiency of force generation have occurred. Interestingly, unlike Y46F, K84A has similar virion assembly efficiency to WT after taking into account packaging defects (7). Because of the similar reduced velocity of K84A to Y46F and otherwise mild translocation defects relative to WT, in this study K84A serves as a “control mutant” to contrast the different types of defects seen in the Y46F mutant that are discussed below.
Implications of the Decreased Velocity of the Y46F Mutant.
Mitchell and Rao have proposed that the residue Y46 of gpA is part of an adenine binding region (YQ) that helps bind ATP for hydrolysis (10). This hypothesis was based on sequence alignments of phage T4's gp17, comparing the amino acid sequence of ATPase domains of various phages with DEXH/D box proteins. Using these methods, they mapped several motifs: an adenine-binding motif (or YQ loop), Walker A, Walker B, and motif III (a.k.a. ATPase coupling or C motif) in the large terminase subunit (Fig. 1). It was suggested that in the case of bacteriophage T4's gp17, the Tyr Y142 of the “YQ loop” interacts with the adenine base on an ATP substrate through hydrophobic interactions. Meanwhile the glutamine residue (Q) was proposed to be involved in hydrogen binding with the N6 and N7 of the adenine nucleotide (14). It has been suggested that the putative YQ loop may be involved in opening and closing of the ATP binding pocket for product release and substrate capture, respectively (23). The crystal structure of T4's gp17 showing this loop and its interaction with ATP is provided in Fig. S4 (courtesy of K. Kondabagil and V. Rao). We have previously found that the nonconservative change Y46E of λ's gpA completely abolished DNA packaging, implicating Y46 as an important residue involved in packaging. Previous measurements of ATP affinity of Y46F using ATP titrations indicated that it was similar to WT, and the apparent ATP consumption per base pair packaged for WT and Y46F was indistinguishable (7). Here we show directly that the average motor velocity is reduced ≈40%, which could account for both binding and hydrolysis defects combined (Fig. 3a).
Mechanochemistry and ATP Binding by the Y46F Terminase.
As discussed by Chemla et al., a Hill coefficient of 1, which was found for both WT and Y46F motors, could mean that either each subunit of the ring ATPase can bind and fire independently of each other, exhibiting no cooperativity or coordination, or alternatively, for every step, one or more ATPases can bind ATP successively in coordination but not cooperatively (2). Recent high-resolution studies determined that the phi29 motor behaves in a highly coordinated manner, and the authors proposed that its Michaelis–Menten ATP dependence is consistent with ATP binding being ordered in time and separated by an irreversible “commitment” transition (3). Because of the homology of the phi29 and λ phages, sequence alignments of their functional domains, and Hill coefficients ≈1, we favor a similar mechanism with the λ WT and Y46F packaging motors. High-resolution measurements, although more difficult because of the higher velocity of the λ motor, would ultimately be needed to directly show if λ has a similar coordination mechanism to phi29. Within a minimal kinetic scheme, the simplest interpretation for the similar values of Vmax/Km found for WT and Y46F (Table 2) is that the rate of ATP docking or any kinetic rates related to binding preceding the first irreversible kinetic transition have not changed dramatically (2). It is therefore likely that energy transduction and/or other kinetic events such as ATP hydrolysis or ADP release have been impaired. (Strictly speaking, an alternative explanation for the unchanged Vmax/Km is that the mutation could introduce compensating effects on the forward or reverse rate constants that may be present before the irreversible step that leads to the next ATPase cycle, making Vmax/Km an oversimplified quantity for describing ATP binding.) Nevertheless, our results challenge the previously held view that the putative adenine-binding domain is merely responsible for binding adenine of ATP. We discuss the previously unknown functions of this motif below.
The Q Motif of the Phage λ Packaging Motor Is Involved with DNA–Motor Interactions.
Although, surprisingly, Y46F does not appear to have a measureable ATP binding defect, other details revealed in its measured translocation dynamics give further support to the idea that the viral packaging motor has a Q motif analogous to the particular functional domain found in SF2 RNA helicases. Specifically, the increased slipping and loss of efficiency of Y46F, which was not detectable in bulk assays, directly show that the mutation modifies and weakens the terminase–substrate (DNA) interaction. What is the nature of this loss of DNA–motor interaction?
The decrease in processivity (increase in slipping) suggests a possible direct interaction of Y46 with the DNA substrate during packaging, because aromatic residues could be involved in base-stacking interactions/contacts with the DNA substrate, making the Q motif an important region of contact for DNA. These interactions were found for some SF1 helicases that were cocrystallized with a DNA substrate (24, 25). It has been suggested that the residues that contact DNA during translocation should be weak, sequence nonspecific, and transient (10). Also they should be regulated with the communication with the ATPase catalytic pocket. Translocation models in which the residue Y46 is involved in nonspecific hydrophobic interactions with DNA are consistent with these criteria. However, this region contacting the DNA would be different from what was suggested in a model based on structural data obtained for gp17 of T4. In gp17, which has a homologous nucleotide-binding domain to gpA, the C-terminal domain alone was proposed to make extensive contacts with DNA while packaging (26). Because the Q motif is located in the N-terminal domain of gpA, an alternative possibility is that residues in this region have a role in the communication between ATP recognition/hydrolysis and DNA contact rather than directly making contacts to DNA.
The assignment of residue Y46 in gpA as a component of a Q motif is further supported by sequence alignments and the close relationship of viral packaging motors with monomeric helicases and DEAD-box proteins (23, 27). Although Q motifs have been assumed to be specific for DEAD-box proteins, the conserved residues including an upstream Gln relative to a Walker A region and surrounding aromatic residues found for many related proteins make it probable that there are variant Q motifs found in other helicase families. In fact, sequence alignments suggest that Q motifs (adenine-binding motifs) may be generalizable to many other additional strand, conserved E (ASCE) division P-loop ATPases, which include proteins responsible for a variety of functions such as ATP synthesis, endonuclease and helicase activities, chromosome segregation, and viral DNA packaging (27, 28). The aromatic residues (Tyr or Phe) 1, 8, 9, and 23 aa upstream of the glutamine in the Q motif of terminase may aid in hydrophobic stacking interactions with adenine and give further validity to this structural assignment. The role of the Q motif in the affinity of helicases for their substrates is congruent with the observed decreased processivity found for Y46F in λ terminase. Our data suggest that Q motifs in SF2 RNA helicases and viral packaging motors play similar roles.
The Q Motif Governs the Force-Generating Ability of a Viral Packaging Motor.
In previous work examining the Q motif of Rh1B helicase, Worrall et al. conjectured that the communication between the ATPase catalytic center and the RNA-binding surface through the Q motif governs the mechanical power and efficiency of RNA helicases and is tuned to the cellular function of the helicase (15). Also, in the crystal structure of T4's gp17 (N360-ED), it was discovered that a structural difference occurred between the apo and ADP/ATP-bound states in the adenine-interacting loop (Cys-125-Arg-140) near Q143, suggesting that T4's Q motif (shown in Fig. S5) is pivotal for translating chemical energy to mechanical energy (23).
Here, we have directly measured the effects of mutations of the Q motif on the force-generating ability of the phage λ motor that result in a tremendous loss of maximum power (>5-fold). The mild defects in packaging revealed in the packaging dynamics measured under a small load can be contrasted with the gross impairment of translocation and power generation found in packaging measurements with increasing load (the steepness of the F-V curve, as measured by Δx1, increases ≈6-fold relative to WT, Table 1). We propose that rather than simply binding adenine, the Q motif communicates ATP recognition to energy transduction driving the motor's power stroke, which leads to contact/pushing of the DNA substrate. It is an interesting possibility that Q motifs of different nucleic acid translocases have evolved to sponsor the enormous force generation needed for a motor to package dsDNA into small viral capsids.
Q Motif Mutations Are Lethal to the Phage.
We previously showed that a Y46F mutant had a severe virion assembly defect (6). It was hypothesized that the terminase enzyme failed to complete translocation and the end of the chromosome was subject to DNase digestion (7). The early stalling we observe before completion of packaging (Fig. S6) offers further support that virion assembly was abolished because the mutants failed to complete packaging. Note that we are not presently able in the optical tweezers assays to follow packaging beyond ≈90% completion because procapsids appear to rupture beyond 90% in the absence of the gpD stabilizing protein (5). Structural defects mentioned earlier that would cause terminase to lose its affinity to DNA would make processes following packaging, such as termination, less efficient. Because of the large forces necessary to package and compact the entire genome into a small viral capsid, an estimated >30–50 pN, the force-generation impairment of Y46F is lethal to the virus.
Conclusions
We have directly shown, through measurements of DNA packaging dynamics with our single-molecule optical tweezers assays, evidence that detailed functions of 2 putative functional domains in a viral packaging motor are similar to homologous domains identified in RNA helicases. In particular, mutants in the vicinity of a deviant Walker A and in a recently discovered Q motif were shown to have unique DNA packaging phenotypes, which show that these regions have roles in ATP recognition/hydrolysis and/or communication of ATP recognition to DNA interaction. Moreover, we show that the mechanical force and power generated by viral packaging motors may be controlled by their degree of recognition of ATP and its proper orientation in the catalytic pocket and that these factors are largely determined by the Q and Walker A motifs. Our findings may ultimately be generalizable to other ASCE division P-loop NTPases, challenging some of the previously held views held on their functional domains.
Methods
cos Cleavage.
A 34.7-kbp biotinylated λ DNA construct was prepared as described by Fuller et al. (8). Biotinylated DNA molecules (2 μL DNA, 30–40 μg/mL) were attached to streptavidin-coated microspheres (2.1 μm diameter) by incubation for 30 min. These DNA-coated microsphere complexes were incubated with integration host factor (33 nM final concentration) and terminase (267 nM final concentration) for 20 min to 1 h in a buffer consisting of 20 mM Tris-HCl (pH 8.0), 10 mM magnesium, 7 mM β-mercaptoethanol. This reaction afforded the maturation of λ DNA and the binding of the terminase motor complex to a cos site located near one end of the DNA. Protein G microspheres (2.1 μm) were incubated with IgG λ antibodies to create IgG λ microspheres. To make procapsid-bound microspheres, procapsids (12 nM final concentration) were incubated with IgG λ microspheres for 30 min.
Initiation of Packaging in an Optical Tweezers Assay.
Two microspheres are held in 2 laser traps, one of which was moved relative to the other using an acousto-optic deflector (29). Viral DNA packaging was initiated by bringing prohead microspheres into near contact with DNA/terminase-coated microspheres for several seconds (Fig. S1). The viral DNA packaging assay used purified (WT, Y46F, K84A) or crude extract (WT, K76R) terminase and proheads. This assay induced the assembly of the terminase complex on λ DNA and the maturation of this DNA through its endonuclease and helicase activities. Packaging was measured with saturating ATP (1 mM) in a buffer consisting of 20 mM Tris-HCl and 10 mM MgCl2 at pH 7.5.
Packaging Dynamics of Purified Wild-Type Terminase.
In this study, we also verified that there were no noticeable differences observed in our optical tweezers measurements in translocation behavior of DNA between purified and crude extracts of λ terminase. Purified WT λ terminase was measured to have a Vmax of 623 ± 148 bp/s (n = 37 complexes), which is consistent with the Vmax = 580 ± 120 bp/s (n = 97 complexes) measured for assays using crude extracts of terminase (8). In addition, the pausing behavior and high processivity in packaging previously observed in the crude system were similar to what was observed for purified terminases.
Data Analysis.
Large slips (>100 bp) and long pauses (>0.2 s) were marked and compiled using custom-made software in Matlab. The tpauses and tslips marked in Fig. 3b and used in efficiency calculations (Eq. 1) were the durations of pausing that end upon resuming of packaging (tpauses) and durations for which the motor slips, resumes packaging, and reaches its previous maximum length translocated before slipping occurs (tslips).
Mutant Purification.
Terminase was purified from 1 L of Escherichia coli strain OR1265 containing pQH101 or mutant derivatives of pQH101 (30, 31). Mutations that result in the Y46F, K76R, and K84A changes were introduced by standard recombinant DNA techniques. Purification of the Y46F and K84A terminases has been described previously (6). WT terminase was purified in a similar manner, except 50 mM sodium phosphate buffer was used instead of 10 mM Tris-HCl in the steps before purification on a HisTrap HP column (GE Healthcare, 1 mL bed volume). Crude extracts of WT and K76R terminases were prepared as described in Fuller et al. (8).
Supplementary Material
Acknowledgments.
We thank Derek Fuller, Arielle Yablonovitch, Al Schweitzer, and Dorian Raymer for technical assistance and Jeff Moffitt, Venigalla Rao, and Carlos Catalano for helpful discussions. We also thank Shelley Grimes and Helene Gaussier for providing initial reagents used in this study. J.M.T. was supported by a National Institutes of Health postdoctoral fellowship award (F32GM083440). J.S. and M.F. were supported by National Institutes of Health Grant GM-51611 and National Science Foundation Grant MCB-0717620.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904364106/DCSupplemental.
References
- 1.Smith DE, et al. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 2.Chemla YR, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 3.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 5.Feiss M, Catalano C. Bacteriophage lambda terminase and the mechanism of viral DNA packaging. In: Catalano C, editor. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Georgetown TX: Landes Bioscience; 2005. pp. 5–33. [Google Scholar]
- 6.Hang JQ, Tack BF, Feiss M. ATPase center of bacteriophage lambda terminase involved in post-cleavage stages of DNA packaging: Identification of ATP-interactive amino acids. J Mol Biol. 2000;302:777–795. doi: 10.1006/jmbi.2000.4086. [DOI] [PubMed] [Google Scholar]
- 7.Dhar A, Feiss M. Bacteriophage lambda terminase: Alterations of the high-affinity ATPase affect viral DNA packaging (vol 347, pp 71–80; 2005) J Mol Biol. 2005;354:738. doi: 10.1016/j.jmb.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Fuller DN, et al. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit barge force generation, high velocity, and dynamic variability. Proc Natl Acad Sci USA. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30:4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 12.Benz J, Trachsel H, Baumann U. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae - the prototype of the DEAD box protein family. Structure. 1999;7:671–679. doi: 10.1016/s0969-2126(99)80088-4. [DOI] [PubMed] [Google Scholar]
- 13.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worrall JAR, Howe FS, McKay AR, Robinson CV, Luisi BF. Allosteric activation of the ATPase activity of the Escherichia coli RhIB RNA helicase. J Biol Chem. 2008;283:5567–5576. doi: 10.1074/jbc.M708620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell MS, Rao VB. Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology. 2004;321:217–221. doi: 10.1016/j.virol.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy C, Feiss M. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 19.Keller D, Bustamante C. The mechanochemistry of molecular motors. Biophys J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang MD, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 21.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 22.Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 23.Sun SY, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage t4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E-coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 25.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 26.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J Mol Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Fuller DN, et al. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc Natl Acad Sci USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow S, Daub E, Murialdo H. The overproduction of DNA terminase of coliphage lambda. Gene. 1987;60:277–289. doi: 10.1016/0378-1119(87)90236-8. [DOI] [PubMed] [Google Scholar]
- 31.Hang Q, Woods L, Feiss M, Catalano C. Cloning, expression, and biochemical characterization of hexahistidine-tagged terminase proteins. J Biol Chem. 1999;274:15305–15314. doi: 10.1074/jbc.274.22.15305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.