Abstract
Fragrance in the grain is one of the most highly valued grain quality traits in rice, yet the origin and evolution of the betaine aldehyde dehydrogenase gene (BADH2) underlying this trait remains unclear. In this study, we identify eight putatively nonfunctional alleles of the BADH2 gene and show that these alleles have distinct geographic and genetic origins. Despite multiple origins of the fragrance trait, a single allele, badh2.1, is the predominant allele in virtually all fragrant rice varieties today, including the widely recognized Basmati and Jasmine types. Haplotype analysis allowed us to establish a single origin of the badh2.1 allele within the Japonica varietal group and demonstrate the introgression of this allele from Japonica to Indica. Basmati-like accessions were nearly identical to the ancestral Japonica haplotype across a 5.3-Mb region flanking BADH2 regardless of their fragrance phenotype, demonstrating a close evolutionary relationship between Basmati varieties and the Japonica gene pool. These results clarify the relationships among fragrant rice varieties and challenge the traditional assumption that the fragrance trait arose in the Indica varietal group.
Keywords: BADH2, Basmati, Jasmine
Recent findings suggest that although Asian cultivated rice (Oryza sativa) is comprised of several genetically distinct groups, a number of the alleles responsible for key domestication and grain quality traits are shared among these groups. Given the complex evolutionary history of rice, the origin of these genetic alterations and how they came to exist across the highly divergent subpopulations of O. sativa remain central questions in rice evolutionary biology.
Fragrance is considered one of the most important grain quality traits in rice, as it is a key factor in determining market price and is related to both local and national identity (1, 2). Investigations into the genetic basis of fragrance in rice led to the identification of a single locus on chromosome 8 (fgr) associated with fragrance (3, 4). Fine mapping (5–7) and subsequent sequence analysis identified a betaine aldehyde dehydrogenase gene, BADH2, associated with the fragrant phenotype [nomenclature follows (8)]. The functional mutation creating the recessive badh2.1 allele was first described as three single nucleotide polymorphisms (SNPs) and an 8-bp deletion in the seventh exon of the gene that resulted in a premature stop codon and putatively truncated the BADH2 protein (9). Other sequence alignments have been used to describe this complex mutation (10, 11), and so the mutation in badh2.1 will hereafter be referred to as the functional nucleotide polymorphism (FNP). Recent surveys of diverse fragrant germplasm support the association of badh2.1 with fragrance (10, 12, 13), and transformation of a fragrant variety with the dominant nonfragrant allele has been shown to abolish fragrance (14), confirming that BADH2 is the major genetic determinant of fragrance in rice.
Over 100 volatile compounds have been detected in fragrant rice varieties, but the major compound responsible for the characteristic aroma is 2-acetyl-1-pyrroline (2AP) (15, 16). This compound, which is produced in all parts of the rice plant except the roots, has a very low odor threshold, allowing humans to detect it at minute concentrations in field-grown plants or crushed leaf tissue, as well as in the grain before, during, and after cooking (17). While the biochemical pathway leading to 2AP synthesis has not been fully resolved, it is believed the BADH2 protein catalyzes the oxidation of γ-aminobutyraldehyde (AB-ald; a 2AP precursor), so that a nonfunctional allele results in the accumulation of both AB-ald and its cyclic form, Δ1pyrroline, resulting in enhanced 2AP synthesis (14, 18).
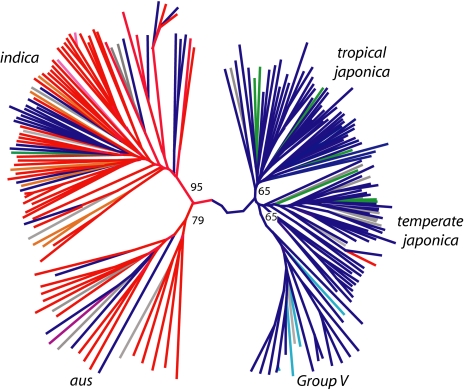
O. sativa consists of two major varietal groups, Indica (Hsien) and Japonica (Keng) (capitalized when referring to varietal groups), that have been recognized in China since ancient times (19, 20). A set of 15 isozyme markers was able to further subdivide the two major varietal groups into six genetically differentiated subpopulations corresponding to well-recognized ecotypes (21). Subsequent studies using SSRs (22) and SNPs (23) distinguished five genetically defined groups that roughly corresponded to the isozyme groups identified by Glaszmann (21): indica, aus, temperate japonica, tropical japonica, and aromatic (lowercase when referring to subpopulations) (Fig. 1). Phylogenetic analysis and FST values demonstrate a close evolutionary relationship between the aromatic, temperate japonica, and tropical japonica subpopulations, which comprise the Japonica varietal group, while the indica and aus subpopulations have a distinct ancestry and are recognized as members of the Indica varietal group (22, 23). Despite its name, the aromatic subpopulation is phenotypically diverse and includes both fragrant and nonfragrant varieties. To avoid confusion, we will hereafter refer to the aromatic subpopulation by its isozyme name, Group V (24).
Fig. 1.
Subpopulation structure in O. sativa. Unrooted neighbor-joining tree constructed from 169 nuclear SSRs (22). Branch color corresponds to chloroplast haplotype. Bootstrap values (out of 100) are indicated at the branch points. This tree clearly illustrates the major division between the two varietal groups (Indica and Japonica), which are further subdivided into the five rice subpopulations: indica, aus, tropical japonica, temperate japonica, and Group V (aromatic). Reproduced and modified, with permission (22).
Fragrant accessions have been identified within at least three of the distinct genetic subpopulations of rice, including Group V (i.e., “Basmati” and “Sadri” varieties), indica (i.e., “Jasmine” varieties), and tropical japonica. In surveys of diverse fragrant rice accessions, nearly all have been shown to carry the badh2.1 allele, suggesting that this allele is common by descent in fragrant rice varieties (9, 10, 12). Recently, a second mutation in the BADH2 gene, badh2.2, was found to be associated with fragrance within a limited set of germplasm from China (13). Evidence that there may be additional mutations in the pathway leading to 2AP synthesis comes from a rigorous study involving a diverse panel of fragrant germplasm that identified several accessions, mostly from Southeast Asia, that had elevated levels of 2AP but did not carry the badh2.1 allele (12). Given that a single allele is largely responsible for fragrance in rice, the goal of this study was to investigate the origin of this allele and trace its ancestry among the genetically divergent subpopulations of rice. We also set out to identify additional functional mutations in the BADH2 gene that may be responsible for independent, local origins of the fragrant phenotype.
Results
Frequency of the badh2.1 Allele in Diverse Rice Germplasm.
We examined the occurrence of the badh2.1 allele in 280 accessions of wild rice (Oryza rufipogon/Oryza nivara) and found that it was absent from all wild genotypes, except for a single accession that was heterozygous for the allele (Table 1). This wild accession exhibited several traits characteristic of domesticated rice, including white pericarp, suggesting it was the result of a recent hybridization event with a fragrant accession of O. sativa. A diverse collection of 176 O. sativa accessions was also surveyed, and the subpopulation identity of these cultivars was determined using a set of genome-wide SSR and SNP markers (22, 23). Overall, the badh2.1 allele was detected in 17 (10%) of these accessions, with the fragrant allele detected at the highest frequency in Group V and at the lowest frequency in temperate japonica and aus (Table 1).
Table 1.
Frequency of badh2.1 allele in wild and cultivated rice
| No. individuals |
% badh2.1 | ||
|---|---|---|---|
| Total | badh2.1 | ||
| O. rufipogon/O. nivara | 280 | 0.5 | 0.2 |
| O. sativa | 176 | 17 | 10.0 |
| indica | 54 | 3 | 6.0 |
| aus | 23 | 0 | 0 |
| Group V (aromatic) | 10 | 6 | 60.0 |
| temperate japonica | 36 | 0 | 0 |
| tropical japonica | 53 | 8 | 15.0 |
Origin of the badh2.1 Allele.
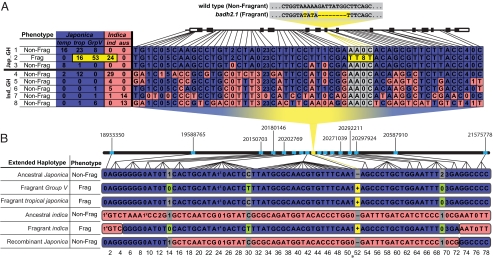
Given that an identical badh2.1 allele was detected in both the Japonica and Indica varietal groups, it was our goal to determine in which group it had originated. To address this question, we sequenced across the BADH2 gene in a panel of 242 O. sativa accessions, which included the original panel and additional accessions known to possess the badh2.1 allele (12) (Table S1). In ≈5 kb of aligned sequence, we detected 106 SNP, insertion-deletion (indel) and SSR polymorphisms, 54 of which were present at a frequency >5%. These polymorphisms were used to construct eight gene haplotypes (GH), and these haplotypes were clearly differentiated into two distinct clusters (Fig. 2A and Table S2). Within the first cluster, all accessions carrying the wild-type allele were from the Japonica varietal group (Jap_GH), while the majority of accessions from the second cluster (74%) were from the Indica varietal group (Ind_GH). Every accession carrying the badh2.1 allele fell within the Jap_GH cluster, regardless of subpopulation identity. Within the Jap_GH cluster, fragrant accessions differed from the ancestral group of nonfragrant accessions only at the FNP that defines the badh2.1 allele (highlighted in yellow in Fig. 2A). These data therefore support a single origin of the badh2.1 allele in a Japonica-like genetic background.
Fig. 2.
Haplotype analysis of the BADH2 gene region. (A) BADH2 gene haplotypes. Sequence reads across the BADH2 gene were aligned for 242 O. sativa accessions and all polymorphisms (frequency >5%) were concatenated and used to create eight gene haplotypes. Letters in each haplotype represent alternative nucleotides at a SNP site; numbers indicate the size of a deletion (0 = no deletion), with the relative position of each polymorphism indicated along the BADH2 gene model. Haplotypes are numbered 1 through 8 followed by the corresponding fragrance phenotype, and the number of accessions from each subpopulation possessing that haplotype. Two gene haplotype clusters were identified: The Japonica Gene Haplotype Cluster (Jap_GH) and Indica Gene Haplotype Cluster (Ind_GH). Blue cells represent polymorphisms characteristic of Jap_GH, while red cells represent those characteristic of Ind_GH. The badh2.1 FNP is depicted in gray/yellow, with yellow representing the fragrant allele. (B) Extended haplotypes. Out of 17 total extended haplotypes for BADH2, six “consensus” haplotypes are depicted with the phenotype of each indicated. Letter/number designations and color coding are as in Fig. 2A (with the following abbreviated deletions: 1x = 28, 1y = 12, 1z = 48). Breaks in coloration indicate positions where recombination was detected. Position 51, marked with a star, represents the MITE polymorphism described previously (10). The badh2.1 FNP is highlighted in yellow at site 52, with “+” referring to the derived (fragrant) allele and “−” referring to the wild-type (nonfragrant) allele. The polymorphisms highlighted in green at positions 14, 30, and 69 are those that were fixed within all fragrant Group V and indica accessions carrying the badh2.1 allele. The relative positions and locations (on the TIGR v.5 pseudomolecule) of the 16 markers that provided AIPs are depicted as light blue dots along a black bar, representing the stretch of chromosome 8 that was sample-sequenced.
All fragrant varieties from the Indica varietal group that carried the badh2.1 allele clustered with Jap_GH, creating an apparent contradiction between the gene and genome phylogenies of these accessions. This incongruence can be explained if the fragrant indica accessions are found to carry a defined region of Japonica-like DNA around the BADH2 gene within an Indica genetic background. We therefore broadened the scope of our haplotype analysis by sequencing 24 amplicons in a genomic region spanning 3.2 Mb upstream and 2.1 Mb downstream of BADH2 in our panel of 242 O. sativa accessions. In this 5.3-Mb region flanking BADH2, a total of 426 SNP, indel, and SSR polymorphisms were identified across ≈13 kb of aligned sequence, with 271 polymorphisms present at a frequency >5%. From these, 78 ancestrally informative polymorphisms (AIPs) were identified and used to create extended haplotypes (see Materials and Methods and Table S3). These extended haplotypes were then summarized into “consensus” extended haplotypes representing six major haplotype classes (Fig. 2B). The extended haplotypes were consistent with the gene haplotypes, with all badh2.1-containing accessions having an extended region of Japonica DNA around the BADH2 gene. In the 24 indica accessions carrying the badh2.1 allele, the Japonica region was bordered by recombination breakpoints ≈650 kb upstream and ≈330 kb downstream, with the flanking regions identical to the ancestral Indica extended haplotype (Fig. 2B). This supports the hypothesis that the badh2.1 allele was transferred via introgression into indica.
Group V accessions carrying the badh2.1 allele possessed an extended haplotype identical to ancestral Japonica, with the exception of the FNP and three unique polymorphisms at sites 14, 30, and 69 (highlighted in green; Fig. 2B). These same three signature polymorphisms flanking the FNP were detected within the Japonica-like region found in all indica accessions carrying the badh2.1 allele. The single wild accession from Myanmar that was heterozygous for the badh2.1 allele segregated 1:2:1 in the next generation, and haplotype analysis confirmed the chromosome carrying the badh2.1 allele also contained the same three Group V-specific polymorphisms described above. We were therefore able to trace the ancestry of the badh2.1 allele in the heterozygous wild accession and all fragrant indica accessions to a Group V ancestor.
There were several nonfragrant Japonica accessions that grouped with the Ind_GH cluster, which was otherwise made up of nonfragrant Indica accessions (Fig. 2A). These nonfragrant accessions from the Japonica varietal group contained an Indica-like genomic region around the BADH2 gene, and all showed recombination back to ancestral Japonica in the flanking regions (Fig. 2B; “Recombinant Japonica”). This provides an explanation for the number of Japonica accessions having a BADH2 gene haplotype that clustered with Ind_GH.
Reduced Nucleotide Diversity and Elevated Linkage Disequilibrium at the badh2.1 Allele.
To examine evidence of selection around the badh2.1 allele, we analyzed the nucleotide diversity across the BADH2 gene in our panel of 242 accessions. We compared the total nucleotide diversity (θπ) in the ≈5 kb of aligned sequence across the BADH2 gene between accessions carrying the wild-type and badh2.1 alleles. Fragrant accessions carrying the badh2.1 allele exhibited a 97.4% reduction in diversity on average compared to the nonfragrant accessions (Table 2).
Table 2.
Average Nucleotide Diversity across BADH2 gene (θπ per kb)
| indica | aus | Group V (aromatic) | temperate japonica | tropical japonica | All | |
|---|---|---|---|---|---|---|
| Badh2 (non-fragrant) (n = 151) | 3.79 | 5.25 | 6.75 | 2.13 | 4.50 | 6.64 |
| badh2.1 (fragrant) (n = 93) | 0.03 | None | 0.06 | None | 0.29 | 0.17 |
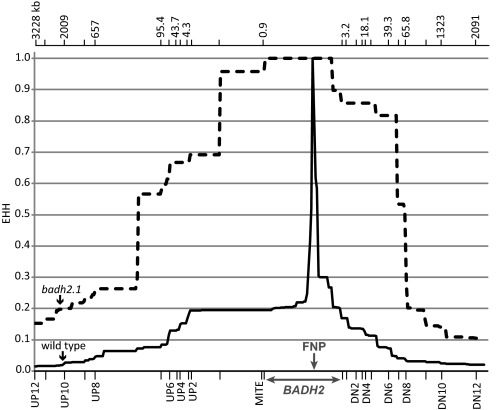
Extended haplotype homozygosity (EHH) estimates the probability that two randomly chosen genomic regions are identical by descent, and allows the measurement of linkage disequilibrium (LD) decay with increasing distance from a functional mutation (25). We calculated the EHH for the BADH2 gene in our germplasm panel by comparing the extended haplotypes of the fragrant (badh2.1) and nonfragrant accessions. The fragrant accessions carrying the badh2.1 allele exhibited a large block of extended LD around the FNP, while LD around the wild-type allele declined rapidly (Fig. 3). This same pattern was observed when the extent of LD was compared between accessions containing the wild-type or the badh2.1 allele within individual subpopulations (Fig. S1). The dramatically reduced nucleotide diversity at badh2.1, coupled with the extended region of linkage disequilibrium surrounding this allele, provide evidence for strong positive selection for the badh2.1 FNP.
Fig. 3.
EHH across the BADH2 genomic region. EHH values were calculated for the 242 O. sativa accessions examined in this study based on haplotype data across a 5.3-Mb genomic region surrounding the BADH2 gene. Solid and dashed lines indicate the combined EHH values of accessions having the wild-type and badh2.1 alleles, respectively. The position of the BADH2 gene is indicated, and badh2.1 FNP is indicated with an arrow. Dashes along the x-axis represent the locations of each amplicon used to obtain haplotype data (Table S5). The physical distances of selected amplicons from the BADH2 gene (in kb) are depicted across the top.
Additional Mutations in the BADH2 Gene.
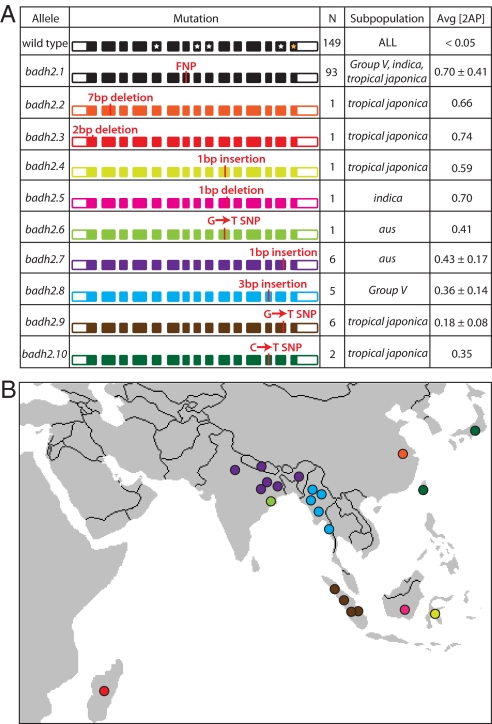
To detect additional mutations in BADH2 that may be responsible for enhanced 2AP synthesis, the BADH2 gene was sequenced in 26 fragrant accessions that lacked either of the previously described derived alleles conferring fragrance (12). Sequence alignments from these 26 accessions with the 242 previously sequenced accessions revealed eight nonsynonymous polymorphisms, four of which were frameshift-inducing indels and one of which was a SNP creating a premature stop codon, all putatively resulting in a truncated BADH2 protein (Fig. 4A and Table S4). The other three potentially functional polymorphisms included a 3-bp insertion and two different SNPs in the coding region. While several of these polymorphisms were found and confirmed in only a single accession, four were found in multiple fragrant accessions, and these polymorphisms appear to have strong geographic associations (Fig. 4B). There were two accessions for which we do not, as yet, have candidate functional mutations that could explain the presence of elevated 2AP levels.
Fig. 4.
BADH2 allelic diversity. (A) Coding Mutations in the BADH2 gene. Sequencing across 26 fragrant accessions that lacked either known mutation in BADH2 identified eight mutations in the coding region of the gene that are putatively responsible for the fragrance phenotype. The gene model for BADH2, with 15 exons and 14 introns, is depicted for each allele along with the location and identity of the coding mutation. Four white stars in the gene model of the wild-type allele illustrate the locations of critical catalytic or substrate-binding domains, while the orange star indicates the location of an oligimerization domain (14). For each allele, the number (N) and genetic subpopulation(s) of accessions having that allele are shown, along with the average 2AP concentration of the accessions possessing that allele (with standard deviations). We obtained accurate 2AP measurements for only one of the accessions carrying the badh2.10 allele, so no standard deviation is indicated. (B) Geographic locations of accessions possessing mutations in BADH2. Each accession having a mutation in BADH2 that is predicted to cause fragrance was placed on a map of Asia using all available collection/passport data for the accessions. The clustering of accessions with a given mutation within the same geographical region suggests that the fragrance trait was selected on multiple occasions, but these alleles remained isolated within local rice gene pools.
Discussion
A Japonica Origin of Fragrance in Rice.
This study presents an in-depth survey of the genetic diversity of the BADH2 gene in a large panel of genetically and geographically diverse rice germplasm from across Asia. Fragrant accessions carrying the badh2.1 allele exhibited a dramatic reduction in nucleotide diversity (97%) and elevated linkage disequilibrium around the gene compared to nonfragrant accessions, consistent with strong positive selection for the badh2.1 allele. The intensity of selection on the badh2.1 allele was similar to reports from other rice genes controlling grain morphology traits (i.e., wx, rc, gs3) (26–28).
Haplotype analysis allowed us to demonstrate that the badh2.1 allele arose in the genetic background of the Japonica varietal group. Extended haplotype analysis revealed a clear introgression of a Japonica genomic region encompassing the badh2.1 allele in all fragrant indica accessions, including Jasmine varieties. All fragrant indica also possessed three polymorphisms flanking the badh2.1 allele that were diagnostic of the Group V subpopulation, demonstrating that this important grain quality allele originated in the Group V subpopulation (i.e., Basmati) and was introgressed into indica (i.e., Jasmine).
The derived polymorphism at site 69 that was ancestrally informative and fixed within fragrant Group V accessions was also fixed within all tropical japonica accessions carrying the badh2.1 allele. This suggests that: (i) The badh2.1 allele may have arisen in an ancestor common to both the Group V and tropical japonica lineages, and the two Group V-specific polymorphisms (sites 14 and 30; Fig. 2B) arose in the Group V lineage following the divergence of these groups or (ii) that a genomic region containing the badh2.1 allele and one downstream polymorphism (site 69) may have been introgressed from Group V to tropical japonica. The extended region of identical sequence flanking the BADH2 gene between fragrant Group V and tropical japonica accessions precludes the detection of recombination breakpoints that would define an introgression from Group V to tropical japonica (or vice versa).
Evidence from this study suggests that the badh2.1 allele was selected as a de novo mutation in O. sativa after domestication from its wild progenitor, and presumably after the divergence of the Japonica subpopulations, given the high frequency of the allele in Group V and vanishingly low frequency in temperate japonica. In our collection of 280 diverse accessions of O. rufipogon/O. nivara from across Asia, we found only a single accession from Myanmar that was heterozygous for the badh2.1 allele. Sequence analysis of this accession demonstrated that the chromosome carrying the badh2.1 allele contained the three signature polymorphisms fixed in all fragrant Group V accessions bordered by DNA sequence characteristic of Indica. While badh2.1 has been identified in other wild rice germplasm from Southeast Asia (29–31), it is likely that these represent introgressions of the derived allele into wild and weedy rice populations growing in the vicinity of fragrant cultivars (10). Gene flow of alleles from O. sativa into wild rice populations has been reported on multiple occasions and is now thought to be ubiquitous (32, 33).
This study improves our understanding of the genetic relationships between O. sativa subpopulations and further clarifies the evolutionary history of Asian cultivated rice. Basmati varieties from the Group V subpopulation are often erroneously referred to as members of the Indica varietal group (34–36). While varieties from the indica and Group V subpopulations are widely grown in South and Central Asia and both may exhibit long, slender grain morphology, researchers have long recognized the high levels of hybrid sterility in crosses between these two groups (37–40). Glaszmann postulated an explanation for these hybrid incompatibilities by demonstrating with isozyme markers that Group V was clearly distinct from Indica and that it grouped more closely with Japonica (21, 24). Additional examinations of the genetic diversity in O. sativa using chloroplast markers, nuclear SSRs, and SNPs independently determined that the Group V subpopulation is a unique genetic entity closely related to the Japonica varietal group (22, 23, 41). Our haplotype analysis in this study demonstrated that the Group V accessions, both fragrant and nonfragrant, cluster with the ancestral Japonica accessions both across the BADH2 gene and across the entire 5.3-Mb region of chromosome 8 surveyed. This provides further evidence that Group V, despite its morphological similarities and geographical distribution that overlaps with Indica, is genetically a member of the Japonica varietal group. Interestingly, the overlapping distributions of Group V and Indica varieties in South Asia may have provided a corridor for the transfer of the major fragrance allele to Indica.
Independent Origins of Fragrance in Rice.
The presence of rice varieties exhibiting elevated 2AP levels, but lacking any known nonfunctional allele of BADH2, raised the possibility that there might be additional fragrance-causing alleles of BADH2 (12). Analysis of 26 fragrant accessions that lacked any known fragrance allele identified eight polymorphisms in the coding region of the gene that are predicted to alter the BADH2 protein. It had been previously shown that only the full-length BADH2 transcript, resulting in an intact 503-aa protein, was capable of inhibiting 2AP production (14). Four of the eight coding polymorphisms identified in this study (creating alleles badh2.3-2.6) are predicted to cause premature transcript termination, which would putatively abolish protein function and result in fragrance. These mutations all result in truncation of the BADH2 protein before critical residues that form the catalytic and/or substrate binding domains (14). Additionally, the badh2.7 allele, which was shared by six aus accessions, is also predicted to result in a shortened transcript that would eliminate an oligomerization domain of the protein. The other three alleles (badh2.8–2.10) either result in an additional in-frame amino acid (badh2.8) or amino acid substitutions (badh2.9–2.10). Despite the association of fragrance with these coding mutations in the BADH2 gene, future work is needed to confirm the effect of these mutations on the degree of 2AP accumulation.
The geographic association between accessions carrying the same mutant alleles at BADH2 suggests fragrance was selected independently on multiple occasions in different geographic regions. Interestingly, while we now know the predominant allele responsible for enhanced 2AP production in rice originated within the Japonica varietal group, it appears that other fragrant alleles of BADH2 were identified within the Indica varietal group, exemplified by the badh2.7 allele, which was found in several aus accessions. These and other findings suggest that the Group V and aus lineages harbor useful alleles that have yet to be fully exploited in rice improvement (22, 27, 47, 48).
Out of the 26 accessions exhibiting fragrance but lacking any of the previously identified fragrance alleles, there were two accessions for which we could find no mutation in the coding or promoter regions that would be predicted to alter the BADH2 protein or its expression. It is therefore possible that these accessions contain genetic lesions elsewhere in the metabolic pathway controlling 2AP synthesis; mapping experiments are underway to test this hypothesis. Identifying additional genes responsible for fragrance would provide opportunities to tailor rice grain quality to suit diverse cultural preferences (1).
BADH2 in the Broader Context of Rice Domestication and Varietal Differentiation.
A variety of functional alleles at genes associated with rice domestication and grain quality enhancement have been described in rice. Alleles at some of these loci appear to have been selected from standing variation in the wild, while others have been selected de novo from within a cultivated gene pool (42, 43). Regardless of their origin, these alleles either remained isolated or became widely disseminated among the subpopulations of O. sativa. Many of the derived alleles characterized thus far in rice show evidence of a single origin within one subpopulation, but are shared by both the Indica and Japonica varietal groups, consistent with the process of introgression. The rc allele, which is responsible for white pericarp in 98% of modern O. sativa varieties, offers an example of a domestication-related gene that arose in a Japonica genetic background followed by dissemination into other, genetically divergent subpopulations (27). The average size of the rc-containing introgression in the Indica varietal group was around 1 Mb, similar to the size of the badh2.1 introgression in indica as reported in this study. Similarly, a mutation in the GS3 gene conferring long grain originated in a Japonica ancestor, with subsequent introgression into indica cultivars (26). In this study, we demonstrate that the evolutionary history of the badh2.1 allele is also consistent with this pattern. These alleles would have been transferred between the rice subpopulations through the process of introgressive hybridization, facilitated by the higher outcrossing rates among early rice cultivars and the physical proximity of divergent (Indica and Japonica) cultivars as a result of population expansion and human migration throughout Asia (42, 44–46). It is noteworthy that despite the apparent importance of hybridization and gene flow during rice evolution, opposing forces have maintained the genetic divergence among the subpopulations of O. sativa. Solving this paradox will require future research to identify the key factors that contribute to subpopulation isolation, as well as to provide insight into the dynamics of genetic exchange among these groups.
Materials and Methods
Plant Materials.
Our germplasm panel consisted of a total of 280 O. rufipogon and 242 O. sativa accessions collected from 38 countries across Asia. We also obtained 26 fragrant accessions lacking the badh2.1 allele from a previous study (12). A complete list of the accessions used in this study can be found in Table S1. All O. sativa accessions not previously reported were analyzed with diagnostic SSR markers to determine their subpopulation identities.
DNA Extraction, PCR, and Sequencing.
DNA extraction was performed using a potassium acetate-SDS protocol for leaf tissue (49) and a modified protocol for milled seeds (50). The functional marker for badh2.1 (51) was used to genotype our germplasm panel. For gene haplotype analysis, eight ≈700-bp amplicons were sequenced across the coding region of the BADH2 gene, resulting in ≈5 kb of aligned sequence. For extended haplotype analysis, 24 regions were sequenced, spanning 3.2 Mb upstream and 2.1 Mb downstream of BADH2, and resulting in over 13 kb of aligned sequence. A previously described MITE polymorphism (10) was also genotyped in the O. sativa panel. A complete list of the primers for the 32 sequenced regions can be found in Table S5. PCR products were purified and sequencing was performed on ABI Prism 3700/3100 DNA analyzers (Applied Biosystems) at the Cornell Life Sciences Core Laboratories Center. Sequences were aligned using the CodonCode Aligner program (CodonCode), and the ends of amplicons were trimmed to remove low quality sequences. Singletons and ambiguous sites were resequenced as necessary. The nonsynonymous polymorphisms found in the fragrant accessions lacking any known nonfunctional allele at BADH2 were resequenced several times for confirmation.
2AP Phenotyping.
Extraction of 2AP with dichloromethane was performed using a modified method (52). Chemically synthesized 2AP was provided by Dr. T. Yoshihashi (Japan International Research Centre for Agricultural Sciences, Ibaraki, Japan) and was used to quantify 2AP in the samples. Each sample was extracted and analyzed on six different occasions and from at least three biological replicates.
Haplotype and Genetic Diversity Analysis Across the BADH2 Region.
Aligned sequences for the eight amplicons across the coding region of the BADH2 gene in the 242 O. sativa accessions were imported into the TASSEL program to extract all polymorphisms present at a frequency above 5% in the sample for constructing gene haplotypes (Table S1). The highly polymorphic SSR at position 20249280 was either long (TA11–14) or short (TA6–8), which corresponded to the Japonica and Indica varietal groups, respectively, and so was coded as “1” if the allele had greater than eight repeats and “0” if the allele had eight or fewer repeats. A total of eight gene haplotypes were inferred using 238 accessions, with four recombinant haplotypes not included in Fig. 2A. The Bayesian clustering program STRUCTURE was used to analyze the gene haplotypes and the highest likelihood was obtained at K = 2 clusters, which were then labeled Japonica Gene Haplotype Cluster (Jap_GH) and Indica Gene Haplotype Cluster (Ind_GH).
For the extended haplotype analysis, we sequenced 24 regions flanking BADH2 in the same 242 O. sativa accessions as described above (Table S5). TASSEL was used to extract the 216 polymorphisms present at a frequency above 5% in the sample. To reduce the number of haplotypes, we applied the following criteria to select AIPs: A polymorphism must (i) have a significant frequency difference between the Japonica varietal group and the indica subpopulation (P < 0.00003) and (ii) be present at a frequency above 20% in the population. A total of 78 polymorphisms from 16 of the sequenced amplicons, the FNP, and the MITE polymorphism met these criteria, and were used in Fig. 2B and Table S3.
Polymorphism data from the 24 sequenced regions flanking BADH2 was used to calculate EHH across the region in the 242 O. sativa accessions, following the method described previously (25).
Supplementary Material
Acknowledgments.
We thank Dr. T. Yoshihashi for providing 2AP standards and Keyan Zhao for computational assistance. We are grateful to Martha Hamblin, Rebecca Nelson, and Gurdev Khush for valuable critical analysis of this manuscript and Lois Swales for administrative assistance. This work was supported by the Plant Genome Program of the National Science Foundation Award 0606461 and the European Union Project META-PHOR FOOD-CT-2006-036220.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the EMBL/GenBank database (accession nos. FJ697177–FJ705056).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904077106/DCSupplemental.
References
- 1.Fitzgerald MA, McCouch SR, Hall RD. Not just a grain of rice: The quest for quality. Trends Plants Sci. 2009;14:133–139. doi: 10.1016/j.tplants.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee P, Singhal RS, Kulkarni PR. Basmati rice: A review. Intl J Food Sci Tech. 2002;37:1–12. [Google Scholar]
- 3.Kadam BS, Patankar VK. Inheritance of aroma in rice. Chronica Bot. 1938;4:32. [Google Scholar]
- 4.Jodon NE. The inheritance of flower fragrance and other characters in rice. Amer Soc Agron. 1944;36:844–848. [Google Scholar]
- 5.Ahn SH, Bollich CN, Tanksley SD. RFLP tagging of a gene for aroma in rice. Theor Appl Genet. 1992;84:825–828. doi: 10.1007/BF00227391. [DOI] [PubMed] [Google Scholar]
- 6.Lorieux M, Petrov M, Huang N, Guiderdoni E, Ghesquière A. Aroma in rice: Genetic analysis of a quantitative trait. Theor Appl Genet. 1996;93:1145–1151. doi: 10.1007/BF00230138. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Wu J, Yang Y, Shi W, Xu M. The fgr gene responsible for rice fragrance was restricted within 69 kb. Plant Sci. 2006;171:505–514. doi: 10.1016/j.plantsci.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.McCouch S. Gene nomenclature system for rice. Rice J. 2008;1:72–84. CGSNL. [Google Scholar]
- 9.Bradbury LMT, Fitzgerald TL, Henry RJ, Jin Q, Waters DLE. The gene for fragrance in rice. Plant Biotech J. 2005;3:363–370. doi: 10.1111/j.1467-7652.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- 10.Bourgis F, et al. Characterization of the major fragance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice. Theor Appl Genet. 2008;117:353–368. doi: 10.1007/s00122-008-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amarawathi Y, et al. Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.) Mol Breed. 2008;21:49–65. [Google Scholar]
- 12.Fitzgerald MA, Sackville Hamilton NR, Calingacion MN, Verhoeven HA, Butardo VM. Is there a second fragrance gene in rice? Plant Biotech J. 2008;6:416–423. doi: 10.1111/j.1467-7652.2008.00327.x. [DOI] [PubMed] [Google Scholar]
- 13.Shi W, Yang Y, Chen S, Xu M. Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol Breed. 2008;22:185–192. [Google Scholar]
- 14.Chen S, et al. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell. 2008;20:1850–1861. doi: 10.1105/tpc.108.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paule CM, Powers JJ. Sensory and chemical examination of aromatic and nonaromatic rices. J Food Sci. 1989;54:343–346. [Google Scholar]
- 16.Buttery RG, Lilng LC, Juliano BO. 2-Acety-1-pyrroline: An important aroma component of cooked rice. Chem Ind. 1982:958–959. [Google Scholar]
- 17.Buttery RG, Ling LC, Juliano BO, Turnbaugh JG. Cooked rice aroma and 2-acetyl-1-pyrroline. J Ag Food Chem. 1983;31:823–826. [Google Scholar]
- 18.Bradbury L, Gillies S, Brushett D, Waters D, Henry R. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol Biol. 2008;68:439–449. doi: 10.1007/s11103-008-9381-x. [DOI] [PubMed] [Google Scholar]
- 19.Chou SL. China is the place of origin of rice (in Chinese) J Rice Soc China. 1948;7:53–54. [Google Scholar]
- 20.Ting Y. The origin and evolution of cultivated rice in China (in Chinese with English abstract) Acta Agron Sinica. 1957;8:243–260. [Google Scholar]
- 21.Glaszmann JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74:21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- 22.Garris A, Tai T, Coburn J, Kresovich S, McCouch SR. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:e163. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaszmann JC. In: Rice Genetics I. Banta SJ, editor. Manila, Philippines: International Rice Research Institute; 1985. pp. 83–90. [Google Scholar]
- 25.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 26.Takano-Kai N, et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics. 2009 doi: 10.1534/genetics.109.103002. Jun 9 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen KM, Purugganan MD. Molecular evidence on the origin and evolution of glutinous rice. Genetics. 2002;162:941–950. doi: 10.1093/genetics/162.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanavichit A, et al. Proceedings of the 1st International Conference on Rice for the Future; Bangkok, Thailand. 2004. [Google Scholar]
- 30.Vanavichit A. International Training Workshop, The Conservation and Utilization of Tropical/Subtropical Plant Genetic Resources. Taiwan: Taiwan Agricultural Research Institute, Council of Agriculture (TARIC); 2007. pp. 131–134. Pub No. 128. [Google Scholar]
- 31.Prathepha P. The fragrance (fgr) gene in natural populations of wild rice (Oryza rufipogon Griff.) Genet Res Crop Evol. 2008 [Google Scholar]
- 32.Song ZP, Lu B-R, Zhu YG, Chen JK. Gene flow from cultivated rice to the wild species Oryza rufipogon under experimental field conditions. New Phytol. 2003;157:657–665. doi: 10.1046/j.1469-8137.2003.00699.x. [DOI] [PubMed] [Google Scholar]
- 33.Chen LJ, Lee DS, Song ZP, Suh HS, Lu B-R. Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Annals Bot. 2004;93:67–73. doi: 10.1093/aob/mch006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maqbool SB, Husnain T, Riazuddin S, Masson L, Christou P. Effective control of yellow stem borer and rice leaf folder in transgenic rice indica varieties Basmati 370 and M 7 using the novel δ-endotoxin cry2A Bacillus thuringiensis gene. Mol Breed. 1998;4:501–507. [Google Scholar]
- 35.Garg AK, et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA. 2002;99:15898–15903. doi: 10.1073/pnas.252637799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bashir K, et al. Field evaluation and risk assessment of transgenic indica basmati rice. Mol Breed. 2004;13:301–312. [Google Scholar]
- 37.Engle LM, Chang TT, Ramirez DA. The cytogenetics of sterility in F1 hybrids of Indica/Indica and Indica/Javanica varieties of rice Oryza sativa L. Philipp Agric. 1969;53:289–307. [Google Scholar]
- 38.Shobha RN. Efforts to improve indigenous basmati rice. Rice India. 1992;1:9–15. [Google Scholar]
- 39.Singh RK, Singh US, Khush GS. Aromatic Rices. Enfield, NH: Science Publishers; 2000. [Google Scholar]
- 40.Khush GS, Dela Cruz N. In: Speciality Rices of the World: Breeding, Production and Marketing. Duffy R, editor. Enfield, NH: FAO and Science Publishers; 2001. pp. 15–18. [Google Scholar]
- 41.Jain S, Jain R, McCouch S. Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labeled microsatellite markers. Theor Appl Genet. 2004;109:965–977. doi: 10.1007/s00122-004-1700-2. [DOI] [PubMed] [Google Scholar]
- 42.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 43.Izawa T. The process of rice domestication: A new model based on recent data. Rice. 2008;1:127–134. [Google Scholar]
- 44.Izawa T, Konishi S, Shomura A, Yano M. DNA changes tell us about rice domestication. Curr Op Plant Biol. 2009;12:185–192. doi: 10.1016/j.pbi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Vaughan DA, Lu B-R, Tomooka N. The evolving story of rice evolution. Plant Sci. 2008;174:394–408. [Google Scholar]
- 46.Sang T, Ge S. The puzzle of rice domestication. J Integr Plant Biol. 2007;49:760–768. [Google Scholar]
- 47.Xu K, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 48.Garris AJ, McCouch SR, Kresovich S. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.) Genetics. 2003;165:759–769. doi: 10.1093/genetics/165.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dellaporta S, Wood J, Hicks J. A plant DNA minipreparation: Version II. Plant Mol Biol Reporter. 1983;1:19–21. [Google Scholar]
- 50.Kang HW, Cho YG, Yoon UH, Eun MY. A rapid DNA extraction method for RFLP and PCR analysis from a single dry seed. Plant Mol Biol Rptr. 1998;16:1–9. [Google Scholar]
- 51.Bradbury L, Henry R, Jin Q, Reinke RF, Waters DLE. A perfect marker for fragrance genotyping in rice. Mol Breed. 2005;16:279–283. [Google Scholar]
- 52.Bergman CJ, et al. Rapid gas chromatographic technique for quantifying 2-acetyl-1-pyrroline and hexanal in rice (Oryza sativa, L.) Cereal Chem. 2000;77:454–458. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.