CONSPECTUS
The thymine-uracil exchange constitutes one of the major chemical differences between DNA and RNA. Although these two bases form the same Watson-Crick base pairs with adenine and are equivalent for both information storage and transmission, uracil incorporation in DNA is usually a mistake that needs to be excised. There are two ways for uracil to appear in DNA: thymine replacement and cytosine deamination. Most DNA polymerases readily incorporate dUMP as well as dTMP depending solely on the availability of the d(U/T)TP building block nucleotides. Cytosine deamination results in mutagenic U:G mismatches that must be excised. The repair system, however, also excises U from U:A “normal” pairs. It is therefore crucial to limit thymine-replacing uracils.
dUTP is constantly produced in the pyrimidine biosynthesis network. To prevent uracil incorporation into DNA, representatives of the dUTP nudeotidohydrolase (dUTPase) enzyme family eliminate excess dUTP. This Account describes recent studies that have provided important detailed insights into the structure and function of these essential enzymes.
dUTPases typically possess exquisite specificity and display an intriguing homotrimer active site architecture. Conserved residues from all three monomers contribute to each of the three active sites within the dUTPase. Although even dUTPases from evolutionary distant species possess similar structural and functional traits, in a few cases, a monomer dUTPase mimics the trimer structure through an unusual folding pattern. Catalysis proceeds by way of an SN2 mechanism; a water molecule initiates in-line nucleophilic attack. The dUTPase binding pocket is highly specific for uracil. Phosphate chain coordination involves Mg2+ and is analogous to that of DNA polymerases. Because of conformational changes in the enzyme during catalysis, most crystal structures have not resolved the residues in the C-terminus. However, recent high-resolution structures are beginning to provide in-depth structural information about this region of the protein.
The dUTPase family of enzymes also shows promise as novel targets for anticancer and antimicrobial therapies. dUTPase is upregulated in human tumor cells. In addition, dUTPase inhibitors could also fight infectious diseases such as malaria and tuberculosis. In these respective pathogens, Plasmodium falciparum and Mycobacterium tuberculosis, the biosynthesis of dTMP relies exclusively on dUTPase activity.
Introduction
The Reason for Negative Discrimination against Uracil in DNA
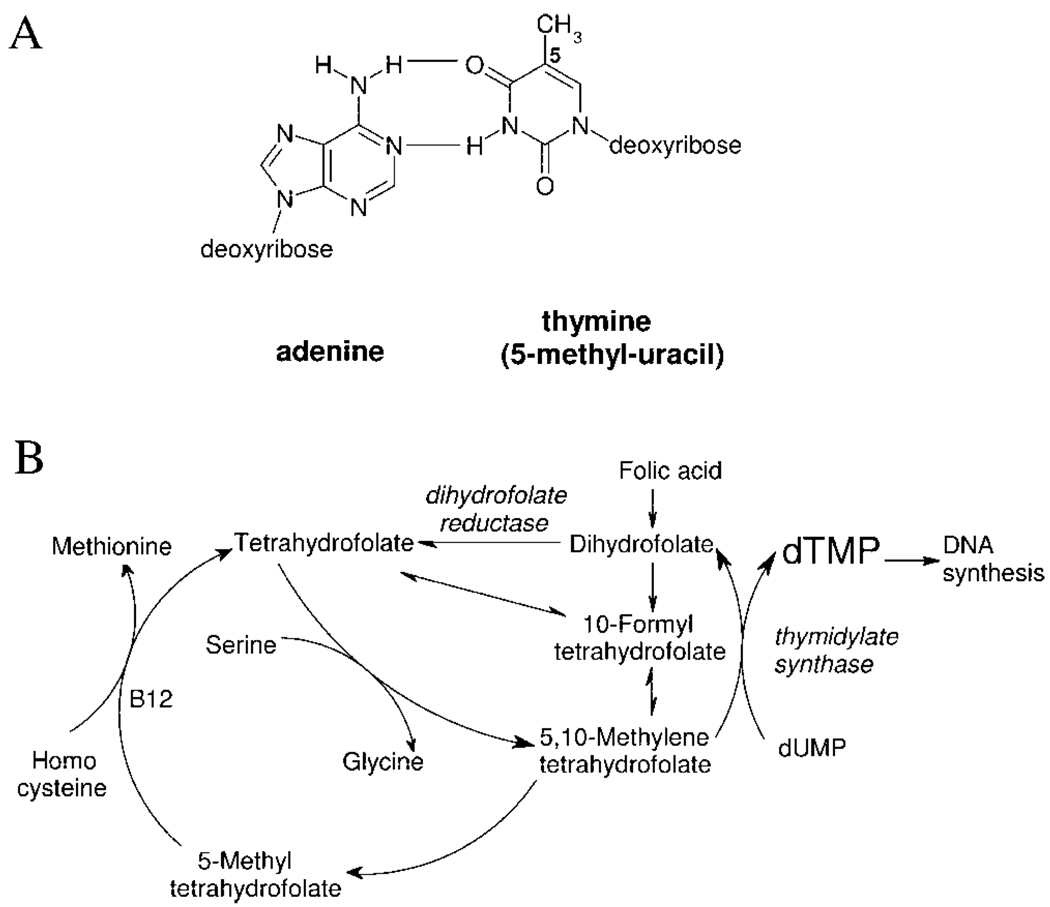
The thymine ↔ uracil exchange constitutes one of the major chemical differences between DNA and RNA. However, these two bases are equivalent for both information storage and transmission as they form the same H-bonded base pairs with adenine (Figure 1A). The 5-methyl group, that is, the difference between the two bases, has no effect on the interaction with adenine. In fact, DNA synthesized in a thymine-less environment has the same coding function,1 and bacteriophage PBS 1/2 relies completely on thymine-less DNA that contains uracil instead of thymine.2 De novo biosynthesis of thymine is an intricate and energetically expensive process that requires dUMP as the starting material and a complex array of two enzymes and cofactors (Figure 1B). It is therefore straightforward to ask: is there any specific reason that justifies this costly and seemingly equivalent replacement of uracil by thymine in DNA?
FIGURE 1.
(A) Watson-Crick base pairing between adenine and thymine (i.e., 5-methyl-uracil) and (B) de novo biosynthesis of dTMP.
It is generally accepted that negative discrimination against uracil in DNA is caused by the chemical instability of cytosine.3 Deamination of cytosine, a rather frequent process that readily occurs under physiological circumstances, gives rise to uracil (Figure 2). Unless corrected, this mutagenic transition will result in a C:G into U(T):A base-pair change, that is, a stable point mutation. To deal with this problem, a highly efficient repair process (uracil-excision repair, see ref 4, for example) has evolved that starts with uracil–DNA glycosylase (UDG) (Figure 2). The importance of this repair process is well-reflected in two observations. One, cytosine deamination is one of the most frequent spontaneous mutations in DNA.5,6 Two, UDG activity resides in at least four families of enzymes:7,8 redundancy may be required for specific circumstances.
FIGURE 2.
Uracil-excision repair. The deaminated cytosine is excised by uracil-DNA glycosylase. AP endonuclease nicks the DNA phosphodiester backbone at the abasic site, creating a free 3′-OH. 5′-phosphodiesterase removes the sugar from the abasic site, and the gap is filled by DNA polymerase. Ligase completes the repair.4
UDG-initiated repair deals with cytosine instability; however, it also inherently defines all uracils as mistakes to be removed. Although mismatch-oriented (U:G/T:G) glycosylases do exist,9 the most efficient, UNG (the major uracil-DNA glycosylase; ung gene product) protein, excises all uracils, and Nature therefore had to derive yet another addition to this system: label the correct uracils with a methyl group (thymine) to distinguish them from deaminated cytosines. Once the methyl label was introduced, its van der Waals characteristics could also be exploited in interactions with DNA-binding proteins. For short-term storage of genetic information, as in modern RNAs, cytosine deamination rates do not pose a serious problem; therefore uracil does not need to be discriminated against.
The Role of dUTPase
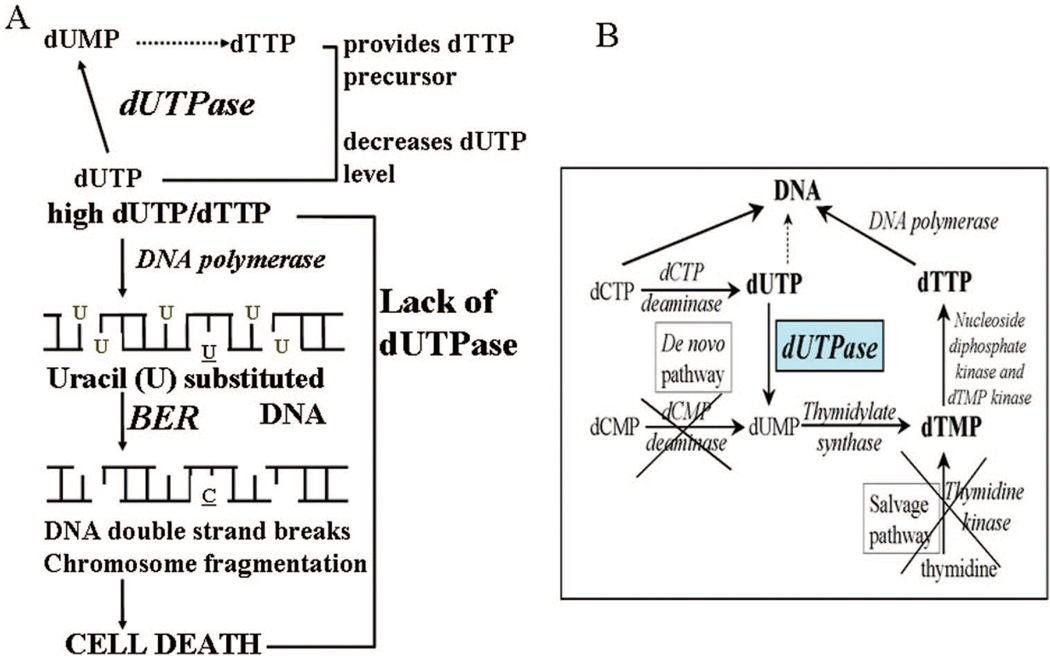
The thymine “invention” required a de novo biosynthesis route (Figure 1B). However, mere availability of dTTP is not enough to prevent uracil incorporation into DNA. Most DNA polymerases cannot distinguish between thymine and uracil. It is the relative level of the respective dNTPs (dUTP/dTTP) that defines incorporation ratios, and therefore, to keep dUMP out of DNA, dUTP levels have to be strictly regulated. The enzyme dUTPase (dUTP nucleotidohydrolase) is responsible for this task: it catalyzes the cleavage of dUTP into dUMP and inorganic pyrophosphate thereby fulfilling a dual role (Figure 3A).10 On one hand, dUTPase controls dUTP level. On the other hand, the product dUMP is the precursor for dTMP biosynthesis. This is of exceptional significance in Mycobacteria and Plasmodia where dUTPase-catalyzed reaction is the only biosynthetic route leading to dUMP (Figure 3B). The precursor for de novo pyrimidine biosynthesis is the cytosine ring that first needs to be deaminated to result in uracil upon which the methyl group can be added to form thymine (Figure 3B). Cytosine deamination can occur at different levels.11,12 In most organisms, dCMP deaminases are present that provide a direct input into the thymidylate synthase reaction. In these organisms, dUMP supply from the dUTPase-catalyzed reaction is of less importance. In enterobacteria, Mycobacteria and Plasmodia, however, cytosine deamination occurs at the dCTP level, as catalyzed by dCTP deaminases, to result in dUTP that has to be converted into dUMP by dUTPase, hence the increased importance of dUTPase in these organisms.
FIGURE 3.
A. Dual role of dUTPase. In the absence of dUTPase (i.e., when dUTP/dTTP ratio is high), thymine-replacing uracils will be reincorporated during base excision repair (BER), but cytosine deamination can be correctly repaired (underlined bases U, C). B. De novo and salvage pathways for dTTP biosynthesis. Enzymes not present in Mycobacteria and Plasmodia are crossed out.
Deficiency of dUTPase results in a high level of dUMP incorporation into DNA (Figure 3A). Uracil-substituted DNA is subjected to uracil-excision repair; however, under high dUTP/dTTP ratio, thymine-replacing uracils will be reincorporated during repair synthesis. Transformation of uracil-excision repair into a hyperactive futile cycle finally leads to cell death via double-stranded DNA breaks (thymine-less cell death) (Figure 3A). Accordingly, dUTPase knockouts are lethal in Escherichia coli and yeast alike.13,14 In human cells, siRNA silencing of dUTPase increased sensitivity to 5-fluoro-deoxyuridine, an anticancer drug that, upon phosphorylation into 5-fluoro-dUMP, perturbs dUTP/dTTP levels by inhibiting thymidylate synthase.15 This observation indicates that thymidylate metabolism may be affected at different levels and combination therapies directed against more enzymes in this pathway may be synergistic.
Evolution and Occurrence of dUTPases
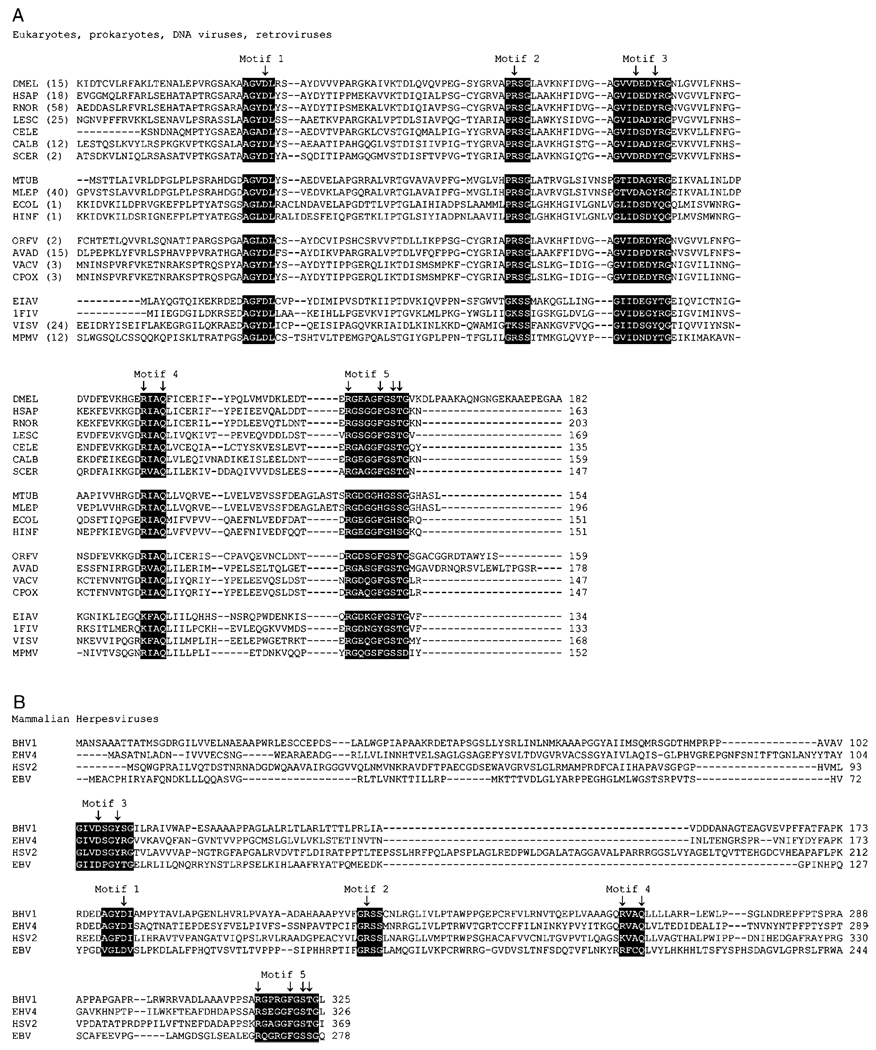
All free-living organisms and also diverse DNA and RNA (including retro-) viruses encode dUTPase.16 The dUTPase protein (product of the dut gene) contains five hallmark conserved sequence motifs (Figure 4A,B).17 Elongated additional conservation patterns can also be recognized in several distinct evolutionary branches.18 Common features of all enzymes that contain these motifs include strict specificity for cleavage of the α–β phosphate-ester linkage of dUTP, practically no cleavage of dUDP, and a β-hairpin motif to accommodate the uracil ring with high specificity.
FIGURE 4.
Sequence alignments of dUTPases. Conserved motifs appear in white lettering on black background. Arrows indicate conserved residues shown in Figure 5–Figure 7.
Some trypanosomal parasites lack the dut gene.19 Here, the dUTPase function is provided by a protein of highly distinct character without any of the hallmark dUTPase motifs. This protein is capable of equally cleaving dUDP and dUTP (hence the term dUDPases/dUTPases), and recognition of uracil is provided by a different structural solution.
In the primate lentiviruses, like in HIV, the dut gene is not present in the genome. (Interestingly, the nonprimate immunodeficiency viruses, for example, feline immunodeficiency virus, or other nonprimate lentiviruses, for example, equine infectious anemia virus, do encode the dut gene.20) In the absence of their own dUTPase, viruses probably rely on the host cell to provide dUTPase activity. Because dUTPase expression in eukaryotes is usually regulated during the cell cycle such that activity is present mostly in dividing cells, these viruses should either target nondifferentiated cells or switch on the host dUTPase in differentiated cells. Such a switch was proposed to operate during HIV infection that was suggested to turn on expression of an endogenous retroviral dut gene.21
Isoenzymes and Cellular Trafficking
In some eukaryotes, two dUTPase isoenzymes are generated by mRNA alternative splicing22 or by use of alternative promoters.23 In the latter case, human cells contain nuclear and mitochondrial isoforms possessing cognate localization signals. The nuclear isoform is under cell cycle control, while the mitochondrial isoform is constitutive,23 probably reflecting the semi-independent nature of mitochondrial DNA repair. Drosophila melanogaster cells also contain two dUTPase isoforms, generated by alternative splicing, but these are both expressed under the control of the same cell-cycle-dependent promoter, and both are therefore present only in actively dividing cells.22 One of these isoforms contains a nuclear localization signal, while the other one lacks any detectable localization signal and is usually located in the cytoplasm. The role of this “cytoplasmic” isoform needs further investigation to understand why the dUTPase function is located in a cellular compartment separate from the site of DNA synthesis and repair. The nuclear localization signal found on human and Drosophila nuclear dUTPase isoforms22,24 is rather unusual but is well preserved in other eukaryotic dUTPase sequences indicating that in most cases, the enzyme can be transported to its physiologically cognate cellular compartment, that is, the nucleus.
Post-transcriptional regulation of dUTPase expression may also operate. In Drosophila melanogaster, dUTPase protein and mRNA levels show an intriguing discrepancy: in most larval stages, the protein is under detection level, while mRNA levels do not decrease significantly.22 These data suggest either mRNA-processing or fast protein degradation.
Structural Basis of Specificity
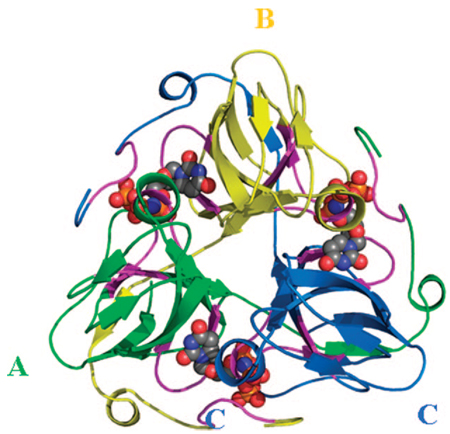
Most dUTPases are homotrimers where subunits fold into a jelly-roll β sheet25–31 (Supplementary Table 1, Supporting Information, Figure 5–Figure 7). The unique characteristic of this dUTPase fold is the intricate interaction pattern among subunits: the C-terminal β-strand of subunit A leaves its protomer and reaches out to contact the surface of subunit C. This strand becomes incorporated into the β-sheet of subunit C by forming main chain contacts with the N-terminal β-strand of subunit C. The last ten residues of subunit A (the segment that follows the terminal β-strand) contact one of the enzyme-bound substrate molecules, also accommodated by conserved motifs from subunit B. The homotrimer forms three active sites in a symmetric fashion. Substrate in each of the active sites is contacted by conserved sequence motifs from all the three subunits (Figure 5). Therefore, although each subunit contains all necessary residues for substrate binding, trimer formation is indispensable to bring these residues in proximity for the cognate binding site.
FIGURE 5.
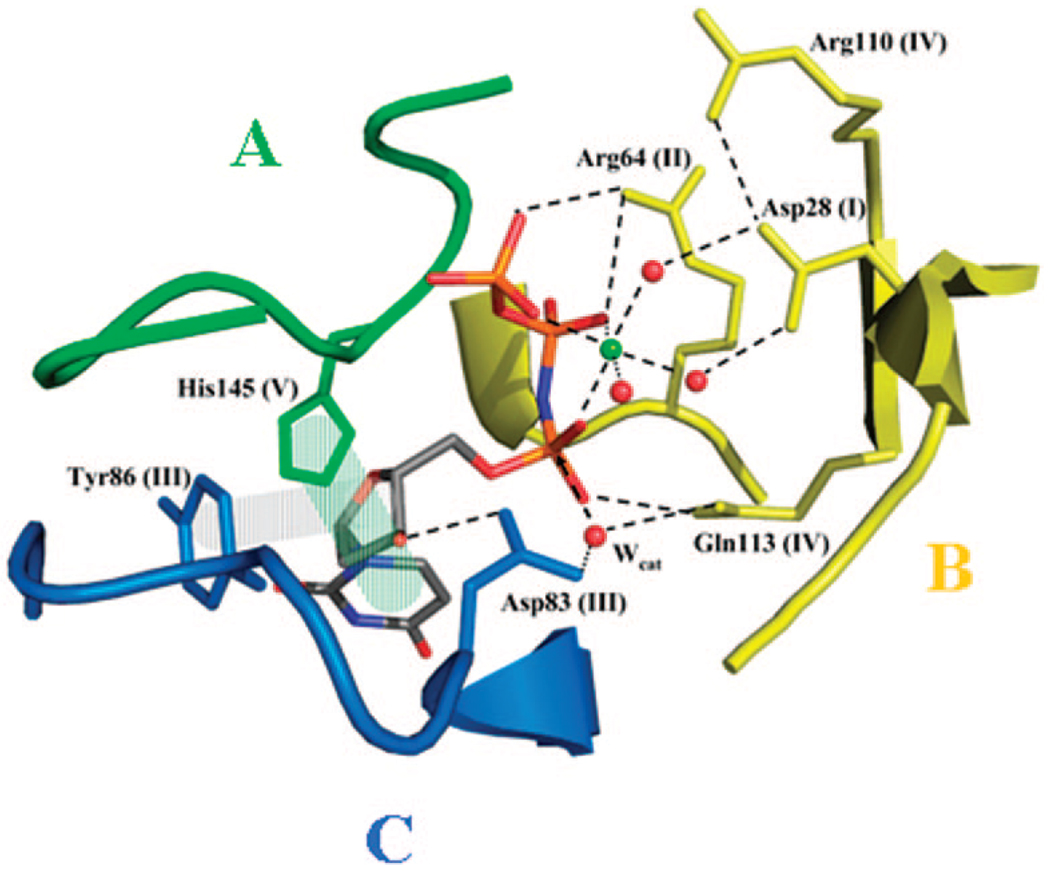
Active site close-up in M. tuberculosis dUTPase (2PY4).33 Substrate coordinating residues are labeled. Roman numerals stand for respective conserved motifs. Dashed lines indicate H-bonds; shaded rectangles indicate aromatic overlaps. In this dUTPase, the aromatic phenylalanine within motif 5 (see Figure 4) is replaced with a histidine.
FIGURE 7.
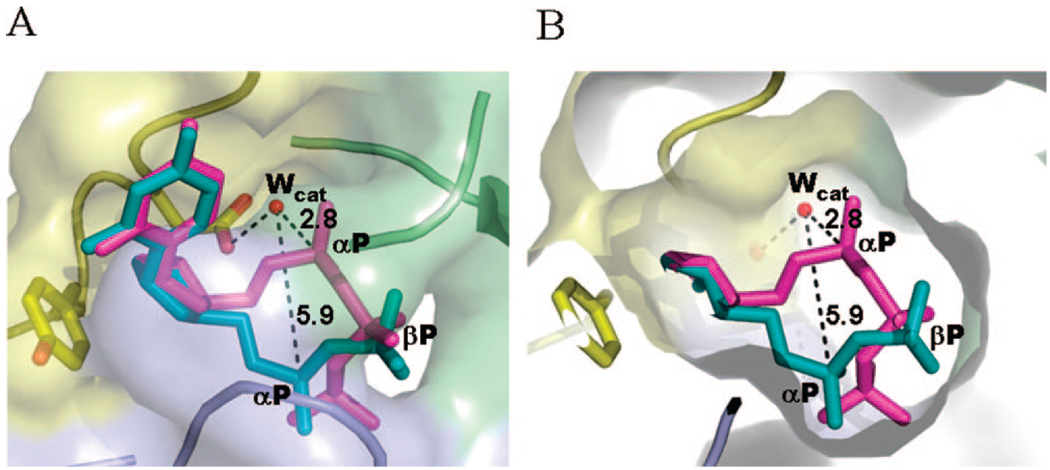
Catalytically competent and noncompetent conformation of the bound substrate. (A) dUDP (teal) and α,β-imido-dUTP (dUPNPP, magenta) complexed human dUTPase crystal structures (PDB IDs 1 Q5H and 2HQU) are superimposed to show relevant differences between the catalytically noncompetent trans (dUDP) and competent gauche (dUPNPP) binding modes within the active site. Surface and ribbon models of the protein are subunit-color-coded (A, yellow; B, green; C, blue). (B) Slab view of panel A to reveal the available inner hole that provides ample conformational space for the phosphate chain.
To prevent wasteful hydrolysis of energy-containing NTPs and dNTPs, specificity is of utmost importance for dUTPase. Two major mechanisms provide this: (i) steric exclusion of purines, thymine, and ribose and (ii) a hydrogen bonding pattern specific only for uracil. Steric hindrance is realized by residues within motif 3.25,32 This motif forms a tight β-hairpin to bind uracil and deoxyribose and to exclude purines or thymine. In addition to this main-chain steric hindrance, a tyrosine at the bottom of the β-hairpin is responsible for stacking over the deoxyribose and discriminating against ribose. Uracil-complementary hydrogen bonding is also provided by motif 3 (Figure 5). Interestingly, protein atoms that form H-bonding partners with the uracil atoms belong to the main chain (O and N from the peptide chain) and not to side chains. Therefore, main chain folding is of major importance in uracil accommodation.
Motifs 1, 2, and 4 coordinate the metal ion and the phosphate chain of the nucleotide. A conserved aspartate within motif 1 donates two coordination positions for water molecules that will coordinate the metal ion Mg(II). Motif 5 has a crucial role in flipping over the active site and creating a microenvironment rather secluded from the bulk medium. Conserved residues within motif 5 are either charged/polar (these contribute to phosphate chain binding) or apolar (a phenylalanine ring is almost invariably present (in Mycobacterium tuberculosis dUTPase, it is replaced by histidine33) and stacks over the uracil ring of the nucleotide). Active site architecture within the trimer is realized in a highly interconnected manner wherein one nucleotide molecule together with the Mg(II) ion coordinated to it receives interacting partners from motif 3 of the first monomer, motifs 1, 2, and 4 from the second monomer, and motif 5 from the third monomer.
In addition to C-terminal arm swapping, monomer interfaces also constitute interacting surfaces. Binary interactions between two neighboring monomers display both polar H-bonding and apolar contacts in a species-specific manner.18,34 Ternary interactions also occur along the inner channel within the trimer. This central channel can be either strongly apolar and tight25 or much more polar, accommodating several water molecules and also metal ions.27,28 Stability of the protein seems to depend on the character of the central channel: the E. coli enzyme is considerably more stable in either heat- or chemical-induced unfolding experiments as compared with human or Drosophila dUTPases34,35 that contain a more polar channel.
Trimer-Mimicking Monomers
The formation of the dUTPase fold may be facilitated in a very specific manner in Caenorhabditis elegans dUTPase. Here, the genome encodes three monomers of the enzyme within the same gene, with linker regions between the subunits.36 Homologous modeling of this protein suggested that the dUTPase fold can be adopted by this protein, as well.
Another peculiar case for trimer-mimicking monomer is found in mammalian herpes viruses:17,37,38 the conserved motifs are still preserved but are arranged in a different fashion (Figure 4B), possibly due to gene duplication followed by point mutations. Interestingly, the active site architecture and three-dimensional fold closely resembles the usual dUTPase trimeric fold in such a way that the N-terminal segment acts as the first monomer, most of the C-terminal segments play the role of the second monomer and the last C-terminal residues mimic the third monomer in closing the active site by the fifth conserved motif.38
Nucleocapsid dUTPases
In contrast to other retroviruses, β-retroviruses encode the dut gene at the junction of gag and pro reading frames.30,39,40 Ribosomal frame shifting results in the translation of the GAG–PRO polyprotein, which is cleaved into the mature viral proteins by the retroviral protease. A bifunctional protein is created: the retroviral nucleocapsid protein is fused to the N-terminus of the dUTPase. The trimeric organization dictated by the dUTPase core is preserved, and correct functions of both domains are displayed. Coupling of nucleic acid binding and dUTPase activity within one single polypeptide may facilitate dUTPase anchoring to reverse transcription sites where DNA synthesis occurs. The trimeric fold of the dUTPase is modulated in this case in a specific manner to allow for the presence of the N-terminal additional polypeptide segment: the C-terminal β-strand that usually reaches over to the active site of another subunit has an altered orientation and interacts with the active site of the same monomer.30
The dCTP Deaminase/dUTPase Bifunctional Enzymes
Coupling of dCTP deaminase and dUTPase activities within the same active site excludes the possibility of dUTP escape into the cellular milieu for DNA synthesis. Such bifunctional enzymes exist in thermophilic Archaea,41 where removal of dUTP is of increased significance since archeal DNA polymerases are strongly inhibited by the presence of uracil in DNA.42 Recently, the dCTP deaminase from M. tuberculosis was also shown to possess dUTPase activity.43 M. tuberculosis also encodes a bona fide dUTPase,31 the catalytic power of which exceeds that of the bifunctional dCTP deaminase/dUTPase by several orders of magnitude.33 The bifunctional enzyme represents a direct channeling pathway from dCTP into dUMP for thymidylate synthase. However, for efficient removal of dUTP, generated by alternative pathways, from the DNA polymerase pathway, the bona fide dUTPase with its excessively high affinity for dUTP33 is indispensable. The relative significance of these two enzymes was assessed in knockout studies where dUTPase was shown to be essential while the bifunctional enzyme was dispensable for viability of M. tuberculosis.44
Catalytic Mechanism of Homotrimeric dUTPases
dUTPase is an efficient catalyst: kcat/KM is (1–4) × 107 M−1 s−1 (compare diffusion-controlled limit (1–2) × 108 M−1 s−1), while turnover rates vary between 0.5 and 25 s.1,26,30,37,40,45–50 This time range can be best studied using high-end rapid kinetic tools and allows investigation of the fundamental enzymatic steps.47
All homotrimeric dUTPases characterized so far seem to share a similar enzymatic mechanism. In several crystal structures, a water molecule is positioned for in-line nucleophilic attack on the αP29,31 coordinated by a strictly conserved aspartate side chain of motif 3 (Figure 6B). Aspartate to asparagine mutation abolishes the ability to accommodate the catalytic water and leads to practically complete loss of hydrolytic activity while retaining wild-type substrate binding affinity.29 It was also shown that the 18O-label appears in the product dUMP when catalysis occurs in 18O-labeled water.51 dUTP hydrolysis on the enzyme therefore occurs through an associative SN2 mechanism, and catalysis is brought about by relative stabilization of a pentacovalent reaction intermediate. Computational studies are in agreement with the proposed stabilization of this intermediate within the dUTPase active site.52 A series of crystallographic snapshots along the catalytic cycle of dUTPase was collected to conclude this issue. These data are now being analyzed and show the presence of a transient product conformation reflecting SN2 mechanism with in-line attack (Barabas, O.; Vertessy, B. G., manuscript in preparation).
FIGURE 6.
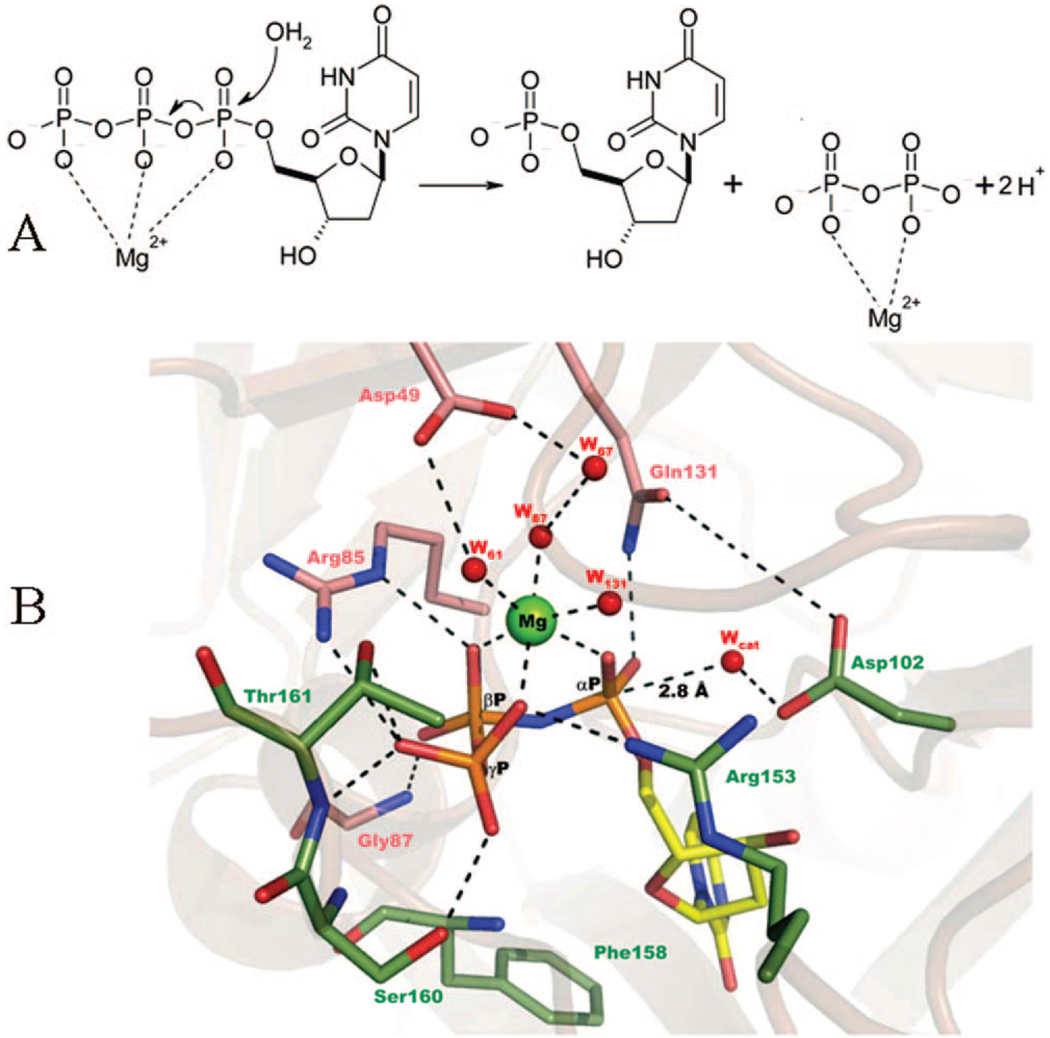
Mechanism of dUTPase-catalyzed dUTP hydrolysis. (A) Reaction scheme of dUTP hydrolysis showing Mg(II) coordination to the triphosphate chain of the substrate. Mg(II) probably dissociates from the enzyme with the product PPi and not with dUMP because no Mg(II) could be observed in dUTPase–dUMP crystal structures. (B) Active site of human dUTPase (2HQU).53 Conserved residues responsible for coordination of the relevant water molecules and the phosphate chain are shown as sticks connected to the ribbon model of the entire protein. Non-carbon atoms are color-coded as oxygen, red; nitrogen, blue; phosphorus, orange; and Mg, light green. Residues of monomers B and C are shown in pink and green, respectively. Substrate carbons are yellow. H-bonds are shown as dashed lines. The catalytic water molecule (Wcat) is positioned for an in-line attack on the αP.
The key resolvable enzymatic steps include (i) rapid substrate binding followed by (ii) a relatively slow substrate-induced isomerization to the catalytically competent active site conformation, (iii) the rate-limiting hydrolysis step, and (iv) rapid release of the products.47 Dissociation rate constants suggested nonordered release of the products from the enzyme-products complex.47 Both products (dUMP and PPi) are competitive inhibitors of the dUTPase reaction, which again implies nonsequential release. Nevertheless, the hypothesis of PPi leaving first and dUMP leaving second has been commonly accepted on the basis of mostly intuitive structural considerations.
The specificity of dUTP binding to dUTPase is provided by the uridine moiety, which precisely fits into the active site (Figure 5). The phosphate chain, however, may adopt multiple conformations (Figure 7). The trans geometry of the αP is catalytically noncompetent.31,53,54 In the so-called gauche conformation, αP is in-line with the catalytic water thus allowing formation of the pentacovalent hydrolysis intermediate (Figure 7). In the presence of Mg(II), the gauche conformation is exclusive and the catalytic efficiency is maximal. In the absence of the cation, the phosphate chain may adopt both gauche and trans conformations in most dUTPases investigated,54 while the catalysis rate is reduced to ∼500%.33,40,46,53,55 These observations imply that the equilibrium of trans to gauche transition will define the frequency of catalytic events in the absence of cation.
In the catalytically competent gauche conformation of the αP, basic residues within motifs 2 and 4 participate in polar interactions with the phosphate chain oxygens in such a way that the phosphate chain is bent in a semicircle with the metal ion in the middle (Figure 6B). Such tridentate Mg(II) coordination pattern is rather exceptional but also observed in DNA polymerases that catalyze the same type of reaction: dNTP cleavage between the α–β phosphate groups.56
Mg(II) also plays a role in increased substrate binding affinity through an extended H-bonding network formed between the substrate and the enzyme in its presence (Figure 6). Another important role of the metal cofactor is leaving group stabilization (Figure 6A). The original coordination symmetry to the triphosphate chain is broken during catalysis and Mg(II) stays coordinated to the pyrophosphate product within the enzyme–products complex. Replacement of Mg(II) with VO(II) results in a 2–3-fold increase in turnover rate probably due to the increased affinity (1000×) of the pyrophosphate leaving group to VO(II)46
The highly conserved sequence (RGxxGF/YGS/TT/SG) in the C-terminal arm (motif 5) is similar to that of P-loops found in other nucleotide hydrolyzing enzymes.57 Glycine positions are of utmost importance in these sequences to adopt the correct tertiary structure for the accommodation of the γP. Both structural and solution dynamics data indicate that the C-terminal motif 5 is the single largest protein fragment that undergoes relatively large conformational changes during the enzymatic cycle, while only subtle changes can be detected in the protein core.55,58,59
Although much studied, the exact role of motif 5 is still not clear. Nucleophilic attack on the α-phosphorus was suggested to be facilitated by closure of the arm upon the active site.55 Conformational studies of this segment during the catalytic cycle of the human enzyme showed that although flexibility is altered (depending on the bound reaction intermediate), overall structure and proximity to the protein core are not.47 Another possible role was attributed to the stacking interaction between the uracil ring of the substrate and the conserved phenylalanine/tyrosine in motif 5. This so-called phenylalanine/tyrosine lid would facilitate dUMP dissociation after hydrolysis by pulling it out of the nucleotide pocket.27 These suggestions may be experimentally tested using dUTPase mutants and transient kinetic methods. Such experiments will hopefully also explain why dUDP is not hydrolyzed by dUTPase despite the fact that the α–β phosphate bond of dUDP and dUTP is positioned similarly in dUTPase crystal structures in the presence of Mg(II).
Concluding Remarks
dUTPases satisfy the cellular requirement for low dUTP/dTTP levels. The catalytic mechanism involves in-line attack by an activated nucleophilic water and a specific configuration of the substrate α-phosphorus. Due to its essential role, the enzyme is a promising target for several strategies. First, since dUTPase is up-regulated in tumor cells,60 the human enzyme is a straightforward target for anticancer therapies. dUTPase inhibitors have also been proposed to fight infectious diseases of worldwide concern, such as tuberculosis and malaria.31,33,61 Here, dUTPase targeting offers special promise since thymidylate biosynthesis in both Mycobacterium tuberculosis and Plasmodium falciparum relies exclusively on the dUTPase-catalyzed pathway.
Supplementary Material
A comprehensive list of dUTPase 3D structures deposited in the PDB. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by grants from Hungarian Scientific Research Fund (No. K68229), Howard Hughes Medical Institute (Nos. 55005628 and 55000342), Alexander von Humboldt Foundation, Grants GVOP-3.2.1.-2004-05-0412/3.0 and JÁP-TSZ-071128-TB-INTER from the National Office for Research and Technology Hungary, Grants FP6 STREP 012127 and FP6 SPINE2c LSHG-CT-2006-0-31220 from the EU to B.G.V, and EMBO Long Term Postdoctoral Fellowship to J.T.
Biographies
Beáta G. Vértessy was born in Budapest, Hungary. She obtained her M.Sc. from the University of Chicago in 1987, Ph.D./C.Ss. from the Eötvös Loránd University and the Hungarian Academy of Sciences in 1991, and D.Sc. from the Hungarian Academy of Sciences in 2001. Her laboratory focusing on Genome Metabolism and Repair at the Institute of Enzymology, Budapest, Hungary, was started in 2000 and is funded by Howard Hughes Medical Institute, U.S.A., Wellcome Trust, U.K., Alexander von Humboldt-Stiftung, Germany, and Hungarian Scientific Research Fund. The current research in her group aims to understand prevention, recognition, and repair of uracil in DNA from perspectives of structural and cell biology.
Judit Tóth was born in Oroszlány, Hungary. She obtained her Ph.D. degree in Structural Biochemistry (2006) in a joint graduate program at Eötvös Loránd University, Hungary, and at the National Institutes of Health, USA. She currently holds an EMBO Postdoctoral Fellowship. Her current research focuses on enzyme kinetics of various nucleotide-hydrolyzing enzymes including dUTPase.
REFERENCES
- 1.el-Hajj HH, Wang L, Weiss B. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 1992;174:4450–4456. doi: 10.1128/jb.174.13.4450-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan BK, Warner HR. Metabolism of uracil-containing DNA: degradation of bacteriophage PBS2 DNA in Bacillus subtilis. J. Virol. 1977;22:835–838. doi: 10.1128/jvi.22.3.835-838.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearl LH, Savva R. The problem with pyrimidines [news] Nat. Struct. Biol. 1996;3:485–487. doi: 10.1038/nsb0696-485. [DOI] [PubMed] [Google Scholar]
- 4.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T. Instability and decay of the primary structure of DNA [see comments] Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 6.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 7.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325(part 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L, Koonin EV. The alpha/beta fold uracil DNA glycosylases: A common origin with diverse fates. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-4-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: A comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vertessy BG, editor. dUTPases. Special Issue. Vol. 2. Bentham Science Publishers; 2001. pp. 277–397. [Google Scholar]
- 11.Bianchi V, Pontis E, Reichard P. Regulation of pyrimidine deoxyribonucleotide metabolism by substrate cycles in dCMP deaminase-deficient V79 hamster cells. Mol. Cell. Biol. 1987;7:4218–4224. doi: 10.1128/mcb.7.12.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donovan GA, Edlin G, Fuchs JA, Neuhard J, Thomassen E. Deoxycytidine triphosphate deaminase: characterization of an Escherichia coli mutant deficient in the enzyme. J. Bacteriol. 1971;105:666–672. doi: 10.1128/jb.105.2.666-672.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.el-Hajj HH, Zhang H, Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadsden MH, Mcintosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehler SE, Ladner RD. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol. Pharmacol. 2004;66:620–626. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 16.Baldo AM, McClure MA. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J. Virol. 1999;73:7710–7721. doi: 10.1128/jvi.73.9.7710-7721.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGeoch DJ. Protein sequence comparisons show that the pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiser A, Vertessy BG. Altered subunit communication in subfamilies of trimeric dUTPases. Biochem. Biophys. Res. Commun. 2000;279:534–542. doi: 10.1006/bbrc.2000.3994. [DOI] [PubMed] [Google Scholar]
- 19.Camacho A, Arrebola R, Pena-Diaz J, Ruiz-Perez LM, Gonzalez-Pacanowska D. Description of a novel eukaryotic deoxyuridine 5′-triphosphate nucleotidohydrolase in Leishmania major. Biochem. J. 1997;325:441–447. doi: 10.1042/bj3250441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elder JH, Lerner DL, Hasselkus-Light CS, Fontenot DJ, Hunter E, Luciw PA, Montelaro RC, Phillips TR. Distinct subsets of retroviruses encode dUTPase. J. Virol. 1992;66:1791–174. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mcintosh EM, Haynes RH. HIV and human endogenous retroviruses: An hypothesis with therapeutic implications. Acta Biochim. Pol. 1996;43:583–592. [PubMed] [Google Scholar]
- 22.Bekesi A, Zagyva I, Hunyadi-Gulyas E, Pongracz V, Kovari J, Nagy AO, Erdei A, Medzihradszky KF, Vertessy BG. Developmental regulation of dUTPase in Drosophila melanogaster. J. Biol. Chem. 2004;279:22362–22370. doi: 10.1074/jbc.M313647200. [DOI] [PubMed] [Google Scholar]
- 23.Ladner RD, Caradonna SJ. The human dUTPase gene encodes both nuclear and mitochondrial isoforms. Differential expression of the isoforms and characterization of a cDNA encoding the mitochondrial species. J. Biol. Chem. 1997;272:19072–19080. doi: 10.1074/jbc.272.30.19072. [DOI] [PubMed] [Google Scholar]
- 24.Tinkelenberg BA, Fazzone W, Lynch FJ, Ladner RD. Identification of sequence determinants of human nuclear dUTPase isoform localization. Exp. Cell Res. 2003;287:39–46. doi: 10.1016/s0014-4827(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 25.Larsson G, Svensson LA, Nyman PO. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP) Nat. Struct. Biol. 1996;3:532–538. doi: 10.1038/nsb0696-532. [DOI] [PubMed] [Google Scholar]
- 26.Dauter Z, Persson R, Rosengren AM, Nyman PO, Wilson KS, Cedergren-Zeppezauer ES. Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex. J. Mol. Biol. 1999;285:655–673. doi: 10.1006/jmbi.1998.2332. [DOI] [PubMed] [Google Scholar]
- 27.Mol CD, Harris JM, Mcintosh EM, Tainer JA. Human dUTP pyrophosphatase: Uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 28.Prasad GS, Stura EA, Elder JH, Stout CD. Structures of feline immunodeficiency virus dUTP pyrophosphatase and its nucleotide complexes in three crystal forms. Acta Crystallogr. D: Biol. Crystallogr. 2000;56:1100–1109. doi: 10.1107/s0907444900009197. [DOI] [PubMed] [Google Scholar]
- 29.Barabas O, Pongracz V, Kovari J, Wilmanns M, Vertessy BG. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. J. Biol. Chem. 2004;279:42907–42915. doi: 10.1074/jbc.M406135200. [DOI] [PubMed] [Google Scholar]
- 30.Nemeth-Pongracz V, Barabas O, Fuxreiter M, Simon I, Pichova I, Rumlova M, Zabranska H, Svergun D, Petoukhov M, Harmat V, Klement E, Hunyadi-Gulyas E, Medzihradszky KF, Konya E, Vertessy BG. Flexible segments modulate co-folding of dUTPase and nucleocapsid proteins. Nucleic Acids Res. 2007;35:495–505. doi: 10.1093/nar/gkl1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan S, Segelke B, Lekin T, Krupka H, Cho US, Kim MY, So M, Kim CY, Naranjo CM, Rogers YC, Park MS, Waldo GS, Pashkov I, Cascio D, Perry JL, Sawaya MR. Crystal structure of the Mycobacterium tuberculosis dUTPase: Insights into the catalytic mechanism. J. Mol. Biol. 2004;341:503–517. doi: 10.1016/j.jmb.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Vertessy BG, Persson R, Rosengren AM, Zeppezauer M, Nyman PO. Specific derivatization of the active site tyrosine in dUTPase perturbs ligand binding to the active site. Biochem. Biophys. Res. Commun. 1996;219:294–300. doi: 10.1006/bbrc.1996.0226. [DOI] [PubMed] [Google Scholar]
- 33.Varga B, Barabas O, Takacs E, Nagy N, Nagy P, Vertessy BG. Active site of mycobacterial dUTPase: structural characteristics and a built-in sensor. Biochem. Biophys. Res. Commun. 2008;373:8–13. doi: 10.1016/j.bbrc.2008.05.130. [DOI] [PubMed] [Google Scholar]
- 34.Takacs E, Grolmusz VK, Vertessy BG. A tradeoff between protein stability and conformational mobility in homotrimeric dUTPases. FEBS Lett. 2004;566:48–54. doi: 10.1016/j.febslet.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 35.Kovari J, Barabas O, Takacs E, Bekesi A, Dubrovay Z, Pongracz V, Zagyva I, Imre T, Szabo P, Vertessy BG. Altered active site flexibility and a structural metal-binding site in eukaryotic dUTPase: kinetic characterization, folding, and crystallographic studies of the homotrimeric Drosophila enzyme. J. Biol. Chem. 2004;279:17932–17944. doi: 10.1074/jbc.M313643200. [DOI] [PubMed] [Google Scholar]
- 36.McClure MA. Evolution of the DUT gene: Horizontal transfer between host and pathogen in all three domains of life. Curr Protein Pept. Sci. 2001;2:313–324. doi: 10.2174/1389203013381062. [DOI] [PubMed] [Google Scholar]
- 37.Bergman AC, Nyman PO, Larsson G. Kinetic properties and stereospecificity of the monomeric dUTPase from herpes simplex virus type 1. FEBS Lett. 1998;441:327–330. doi: 10.1016/s0014-5793(98)01575-0. [DOI] [PubMed] [Google Scholar]
- 38.Tarbouriech N, Buisson M, Seigneurin JM, Cusack S, Burmeister WP. The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure. 2005;73:1299–1310. doi: 10.1016/j.str.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Bergman AC, Bjornberg O, Nord J, Nyman PO, Rosengren AM. The protein p30, encoded at the gag-pro junction of mouse mammary tumor virus, is a dUTPase fused with a nucleocapsid protein. Virology. 1994;204:420–424. doi: 10.1006/viro.1994.1547. [DOI] [PubMed] [Google Scholar]
- 40.Barabas O, Rumlova M, Erdei A, Pongracz V, Pichova I, Vertessy BG. dUTPase and nucleocapsid polypeptides of the Mason-Pfizer monkey virus form a fusion protein in the virion with homotrimeric organization and low catalytic efficiency. J. Biol. Chem. 2003;278:38803–38812. doi: 10.1074/jbc.M306967200. [DOI] [PubMed] [Google Scholar]
- 41.Johansson E, Bjornberg O, Nyman PO, Larsen S. Structure of the bifunctional dCTP deaminase-dUTPase from Methanocaldococcus jannaschii and its relation to other homotrimeric dUTPases. J. Biol. Chem. 2003;278:27916–27922. doi: 10.1074/jbc.M304361200. [DOI] [PubMed] [Google Scholar]
- 42.Hogrefe HH, Hansen CJ, Scott BR, Nielson KB. Archaeal dUTPase enhances PCR amplifications with archaeal DNA polymerases by preventing dUTP incorporation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:596–601. doi: 10.1073/pnas.012372799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helt SS, Thymark M, Harris P, Aagaard C, Dietrich J, Larsen S, Willemoes M. Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis. J. Mol. Biol. 2008;376:554–569. doi: 10.1016/j.jmb.2007.11.099. [DOI] [PubMed] [Google Scholar]
- 44.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 45.Vertessy BG, Zeppezauer M. Identification of tyrosine as an active site residue involved in the catalytic mechanism of Escherichia coli dUTPase. Biochem. Soc. Trans. 1994;22:233S. doi: 10.1042/bst022233s. [DOI] [PubMed] [Google Scholar]
- 46.Mustafi D, Bekesi A, Vertessy BG, Makinen MW. Catalytic and structural role of the metal ion in dUTP pyrophosphatase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5670–5675. doi: 10.1073/pnas.1031504100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth J, Varga B, Kovacs M, Malnasi-Csizmadia A, Vertessy BG. Kinetic mechanism of human dUTPase, an essential nucleotide pyrophosphatase enzyme. J. Biol. Chem. 2007;282:33572–33582. doi: 10.1074/jbc.M706230200. [DOI] [PubMed] [Google Scholar]
- 48.Harris JM, Mcintosh EM, Muscat GE. Structure/function analysis of a dUTPase: Catalytic mechanism of a potential chemotherapeutic target. J. Mol. Biol. 1999;288:275–287. doi: 10.1006/jmbi.1999.2680. [DOI] [PubMed] [Google Scholar]
- 49.Nord J, Larsson G, Kvassman JO, Rosengren AM, Nyman PO. dUTPase from the retrovirus equine infectious anemia virus: Specificity, turnover and inhibition. FEBS Lett. 1997;414:271–274. doi: 10.1016/s0014-5793(97)00935-6. [DOI] [PubMed] [Google Scholar]
- 50.Shao H, Robek MD, Threadgill DS, Mankowski LS, Cameron CE, Fuller FJ, Payne SL. Characterization and mutational studies of equine infectious anemia virus dUTPase. Biochim. Biophys. Acta. 1997;1339:181–191. doi: 10.1016/s0167-4838(96)00229-4. [DOI] [PubMed] [Google Scholar]
- 51.Larsson G, Nyman PO, Kvassman JO. Kinetic characterization of dUTPase from Escherichia coli. J. Biol. Chem. 1996;271:24010–24016. doi: 10.1074/jbc.271.39.24010. [DOI] [PubMed] [Google Scholar]
- 52.Berente I, Beke T, Náray-Szabó G. Quantum mechanical studies on the existence of a trigonal bipyramidal phosphorane intermediate in enzymatic phosphate ester hydrolysis. Theor. Chem. Acc. 2007;118:129–134. [Google Scholar]
- 53.Varga B, Barabas O, Kovari J, Toth J, Hunyadi-Gulyas E, Klement E, Medzihradszky KF, Tolgyesi F, Fidy J, Vertessy BG. Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase. FEBS Lett. 2007;581:4783–4788. doi: 10.1016/j.febslet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 54.Kovari J, Barabas O, Varga B, Bekesi A, Tolgyesi F, Fidy J, Nagy J, Vertessy BG. Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site. Proteins. 2008;71:308–319. doi: 10.1002/prot.21757. [DOI] [PubMed] [Google Scholar]
- 55.Vertessy BG, Larsson G, Persson T, Bergman AC, Persson R, Nyman PO. The complete triphosphate moiety of non-hydrolyzable substrate analogues is required for a conformational shift of the flexible C-terminus in E. coli dUTP pyrophosphatase. FEBS Lett. 1998;421:83–88. doi: 10.1016/s0014-5793(97)01545-7. [DOI] [PubMed] [Google Scholar]
- 56.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 57.Prasad GS. Glycine rich P-loop motif in deoxyuridine pyrophosphatase. Curr. Protein Pept. Sci. 2001;2:301–311. doi: 10.2174/1389203013381017. [DOI] [PubMed] [Google Scholar]
- 58.Nord J, Nyman P, Larsson G, Drakenberg T. The C-terminus of dUTPase: Observation on flexibility using NMR. FEBS Lett. 2001;492:228–232. doi: 10.1016/s0014-5793(01)02257-8. [DOI] [PubMed] [Google Scholar]
- 59.Dubrovay Z, Gaspari Z, Hunyadi-Gulyas E, Medzihradszky KF, Perczel A, Vertessy BG. Multidimensional NMR identifies the conformational shift essential for catalytic competence in the 60-kDa Drosophila melanogaster dUTPase trimer. J. Biol. Chem. 2004;279:17945–17950. doi: 10.1074/jbc.M313644200. [DOI] [PubMed] [Google Scholar]
- 60.Chano T, Mori K, Scotlandi K, Benini S, Lapucci C, Manara MC, Serra M, Picci P, Okabe H, Baldini N. Differentially expressed genes in multidrug resistant variants of U-2 OS human osteosarcoma cells. Oncol. Rep. 2004;11:1257–1263. [PubMed] [Google Scholar]
- 61.Whittingham JL, Leal I, Nguyen C, Kasinathan G, Bell E, Jones AF, Berry C, Benito A, Turkenburg JP, Dodson EJ, Ruiz Perez LM, Wilkinson AJ, Johansson NG, Brun R, Gilbert IH, Gonzalez Pacanowska D, Wilson KS. dUTPase as a platform for antimalarial drug design: Structural basis for the selectivity of a class of nucleoside inhibitors. Structure. 2005;13:329–338. doi: 10.1016/j.str.2004.11.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A comprehensive list of dUTPase 3D structures deposited in the PDB. This material is available free of charge via the Internet at http://pubs.acs.org.