Abstract
For most associations of common single nucleotide polymorphisms (SNPs) with common diseases, the genetic model of inheritance is unknown. The authors extended and applied a Bayesian meta-analysis approach to data from 19 studies on 17 replicated associations with type 2 diabetes. For 13 SNPs, the data fitted very well to an additive model of inheritance for the diabetes risk allele; for 4 SNPs, the data were consistent with either an additive model or a dominant model; and for 2 SNPs, the data were consistent with an additive or recessive model. Results were robust to the use of different priors and after exclusion of data for which index SNPs had been examined indirectly through proxy markers. The Bayesian meta-analysis model yielded point estimates for the genetic effects that were very similar to those previously reported based on fixed- or random-effects models, but uncertainty about several of the effects was substantially larger. The authors also examined the extent of between-study heterogeneity in the genetic model and found generally small between-study deviation values for the genetic model parameter. Heterosis could not be excluded for 4 SNPs. Information on the genetic model of robustly replicated association signals derived from genome-wide association studies may be useful for predictive modeling and for designing biologic and functional experiments.
Keywords: Bayes theorem; diabetes mellitus, type 2; meta-analysis; models, genetic; polymorphism, genetic; population characteristics
When the association between a genetic marker and a trait is evaluated in a population-based study, there is rarely a priori biologic evidence supporting a particular genetic model of inheritance for the risk allele. Investigators may present and analyze the results of genetic association studies in various ways. If the risk is the same for heterozygotes, carrying 1 copy of the high-risk allele a, as for homozygotes, then the underlying genetic model is dominant, and therefore the data are dichotomized into “carriers” versus “noncarriers.” If 2 copies of a are required for the risk to be different from the baseline risk, then the genetic model is recessive. The additive model assumes that on a log scale, the risk in carriers of 2 copies of a is double the risk in heterozygotes. Usually a strong preference for 1 genetic model is unjustified (1). Exceptions exist, as in the case of null genotypes of enzyme-coding genes, where extensive data on enzymatic activity may be available, but typically the model of inheritance used is suggested by convenience or even tradition in the research field. For example, it is common in the analysis of associations with rare susceptibility alleles to analyze data assuming a dominant model, perhaps because it is recognized that a recessive-model analysis would have negligible statistical power. Usually the rationale used in choosing 1 particular model is not discussed at all.
In theory, one could try to examine the fit of different models to the data. However, the ability to draw inferences from a single study's data is limited. A meta-analysis of many studies improves the power to demonstrate associations consistently and may also allow for a stronger exploration of how the available data fit different models of inheritance, along with obtaining a summary effect size. In this regard, a Bayesian model has been suggested; the genetic model in a meta-analysis is represented by an unknown parameter which, when estimated across studies, allows us to learn about the underlying model (2–4).
Until now, this method has been applied primarily to meta-analyses of genetic associations from the candidate gene era (3, 5). This has posed difficulties in exploiting its full potential, given the poor replication record of such associations (6). If an association is not confirmed, modeling of the best-fitting genetic model may produce a fit to the noise and errors in the data rather than the true underlying biology. However, the advent of genome-wide association studies (7) and large-scale consortia (8) has transformed the evidence on genetic associations. For several diseases, we now have robust evidence on a number of common genetic variants. Using data from associations with robust statistical support and large-scale evidence from many data sets, one can revisit the question of genetic model fit more efficiently. This can yield some useful insights into the underlying biology of the identified associations and may also be informative with regard to the best analysis plan for genome-wide association studies. In the setting of an agnostic approach, investigators often pick 1 genetic model for analyzing the data. Most often this is the additive (per-allele or codominant) model, because of statistical power considerations, and thus most associations derived from genome-wide association studies are usually presented as per-allele risks (9). It would be useful to examine whether other models might fit these data equally well or better.
Our aim in this paper was to explore the potential of Bayesian meta-analysis to inform us about the underlying genetic model for 17 single nucleotide polymorphisms (SNPs) that have robust statistical support for an association with type 2 diabetes. Type 2 diabetes is a prime paradigm in which a large number of common variants have already been identified with a successful application of large-scale collaborative research. We analyzed data from the DIAGRAM [Diabetes Genetics Replication and Meta-Analysis] consortium that incorporate, through meta-analysis, data from 3 genome-wide association studies and from replication data sets (10–14).
MATERIALS AND METHODS
Type 2 diabetes data
The field of type 2 diabetes genetics has witnessed rapid progress in the identification of robustly associated susceptibility loci over the last few years. The list of established disease-associated variants continues to grow. We examined genotype data from 19 case-control studies for 17 of these established type 2 diabetes loci (see Web Table 1, posted on the Journal’s Web site (http://aje.oxfordjournals.org/)). We obtained data generated principally through the efforts of the DIAGRAM consortium. Details on the contributing data sets can be found elsewhere (10–14). For each study, we used the raw genotype data in cases and controls. The data sets were derived from 3 genome-wide association studies (10–12) and additional replication teams. Results regarding the strength of association for each of these polymorphisms in each of the discovery genome-wide association studies are provided elsewhere (14). For the 3 genome-wide association studies, SNPs were excluded if the controls violated Hardy-Weinberg equilibrium at P < 0.000001 (P < 0.0001 in the Wellcome Trust Case Control Consortium study), given the large multiplicity of analyses; the threshold for Hardy-Weinberg equilibrium testing in the replication data sets was P < 0.001 (P < 0.05 in the Nurses’ Health Study).
Statistical methods
We extended a model initially suggested by Minelli et al. (2). Consider a biallelic locus, with A being the “low-risk” allele and a the allele associated with higher risk of type 2 diabetes. The association of the locus with the disease is then reflected in 2 odds ratios (ORs); choosing the homozygotes AA as a reference group, ORAa is the odds ratio for the heterozygotes and ORaa is the odds ratio for the homozygotes aa in comparison with the reference group. The underlying genetic model refers to the relation between these 2 odds ratios. In a general case, log(ORaa) = λlog(ORAa), with λ = 1 for a dominant model, λ = 0.5 for an additive model, and λ = 0 for a recessive model for the diabetes risk allele. However, one may argue that λ may be left unspecified and let the data inform the model. Each study, however, provides only 1 genotype-specific estimate of the effect and therefore only 1 estimate of λ.
If there is little rationale assuming that the genetic model would vary across studies, a fixed-effects summary estimate of λ could be obtained from a meta-analysis. However, if there are reasons to believe that the model of inheritance might vary across studies (if, for example, the studies refer to different ethnic groups), a hierarchical random-effects model for λ can be fitted.
The meta-analysis model is outlined below. We fit it within a Bayesian framework to take advantage of its flexibility. An important asset is its ability to incorporate full uncertainty in all model parameters (including the heterogeneity parameter τ2 and the genetic model parameter λ). Eventually, it is possible to estimate the probability of each genetic model's being the true one.
Genetic model-free meta-analysis model
The evaluated type 2 diabetes studies had a case-control design, and therefore they would be appropriately analyzed using a retrospective likelihood approach. Consider the 2 vectors of the observed distribution of the cases and controls for the 3 genotypes (AA, Aa, aa), cai = (caAAi,caAai,caaai) and coi = (coAAi,coAai,coaai), for a study i. The basic parameters to model are 2 probability vectors for the 3 genotypes, given the case or control status (15). The likelihood is multinomial for both cases and controls. In study i, the cases (denoted ca) and the controls (denoted co) relate to the parameters πica = (πAAica,πAaica,πaaica) and πico = (πAAico,πAoico,πaaico) with and through the multinomial likelihoods
As discussed above, it is not necessary to assume a particular inheritance model. We can parameterize the 2 log odds ratios as log(ORAa) = λiμi and log(ORaa) = μi.
Then the 3 genetic models can be identified as follows.
Dominant: λ = 1, so that mutant homozygotes and heterozygotes have the same disease odds.
Codominant: λ = 0.5, so that homozygotes have double the odds (on the log scale).
Recessive: λ = 0, suggesting that heterozygotes have no higher disease odds than wild-type homozygotes.
Generally it should be reasonable to assume that risks are inherited in a similar way in different populations, so we have a common effect for λ in our analyses; that is, λi = λ, and we will refer to this as model 1, which represents our main analysis. To allow for heterogeneity in the underlying inheritance model across studies, we also examined as sensitivity analyses random effects, where random λi parameters underlie each study, and these are drawn from a common distribution, where λi may be restricted to lie between 0 and 1 (i.e., λi∼N(λR,τλ2)I(0, 1) (model 2)) or could be more unrestricted and thus also take negative values and values above 1 (i.e., λi∼N(λU,τλ2) (model 3)). When λi is restricted to lie between 0 and 1, the risk conferred by heterozygosity is forced to range between no risk and the risk conferred by homozygosity. With more unrestricted values of λi, additional possibilities are allowed; for example, heterozygotes may have a protective effect while homozygotes have increased risk (negative values of λ), or heterozygotes may have more increased risk than homozygotes (λ > 1).
The case probabilities are parameterized in terms of the log odds ratios and the probabilities in the controls:
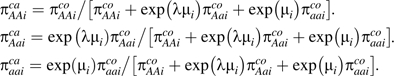 |
A fixed-effects meta-analysis may be undertaken assuming μi = μ for each i or a random-effects meta-analysis assuming μi∼N(μ,τ2), with τ measuring the extent of heterogeneity.
Prior distributions and model fit
We used minimally informative normal priors centered at 0 for the location parameters μi,μ. For the probabilities πica, we used priors that approximated the Dirichlet distribution and gave equal prior probabilities of the diabetes condition to all 3 genotypes (). For the heterogeneity standard deviation, we placed a half-normal prior τ∼N(0,1), τ > 0.
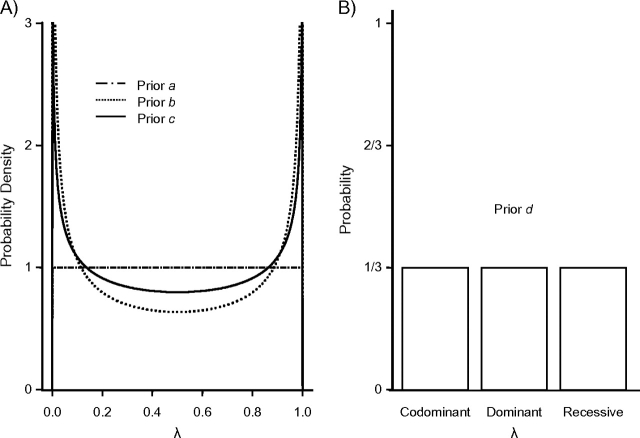
For the genetic model parameter λ, which is the focus of our research, we implemented 4 prior probabilities. The first 3 use the flexible Beta distribution (previously used by Minelli et al. (2)) that returns λ values between 0 and 1:
a) λ∼Beta(1, 1);
b) λ∼Beta(0.5, 0.5);
c) λ∼Beta(0.7, 0.7).
The first prior has a flat uniform shape between 0 and 1. However, when the model is recessive or dominant (i.e., λ is at the edges of the distribution), the first prior (prior a) tends to shift the parameter values toward the mean of the distribution (0.5). Therefore, the second prior, Beta(0.5, 0.5), gives higher probabilities at the upper and lower ends of the interval. This prior has the drawback that when the true model is additive, it tends to shift the estimate towards the edges of the distribution. The third prior represents a compromise between the above 2 situations; we used the third prior in the main analysis and the other 2 in sensitivity analyses.
We further introduce another discrete distribution approach. This fourth prior reflects situations in which λ is allowed to take discrete values only, those corresponding to the 3 genetic models. Therefore,
d) λ∼cat(0, 0.5, 1), with corresponding probabilities pR,pC,pD, where we set all models to be equally probable and thus pR = pC = pD = 1/3.
Figure 1 presents the 4 priors.
Figure 1.
Prior probabilities for λ, the genetic model parameter. The first 3 priors are based on a Beta distribution (part A), and the fourth prior is a categorical prior (part B).
For the 2 models in which it is assumed that there are random small differences in λi, the prior on the mean of the random-effects distribution λR for the restricted case is a Beta prior λR∼Beta(0.7, 0.7). For the unrestricted case, the prior is reflecting the ability to incorporate heterosis and negative λ values but it is truncated to the interval −1 to 2 (λU∼N(0, 1,000)I( − 1, 2)), since values outside of this very wide range are not very plausible. For the genetic model heterogeneity standard deviation τλ, we placed a half-normal prior: τλ∼N(0, 1), τλ ≥ 0.
We then estimated the posterior distribution of λ. For the first 3 priors, we obtained the median value and its 95% credibility interval and also evaluated the impact on the estimated odds ratios and heterogeneity parameter (τ). For prior d, the posterior distribution shows directly the probability of each model, as the probability that λ takes each 1 of the 3 alternative values.
Table 1 presents schematically all of the alternative models and their combinations with the priors.
Table 1.
Alternative Models for the Genetic Model Parameter
| Model | Effects | Restriction on Study-Specific λi | Prior on the Fixed/Average λ |
| 1 | Fixed | Not applicable | λ∼Beta(1, 1) |
| λ∼Beta(0.5, 0.5) | |||
| λ∼Beta(0.7, 0.7) | |||
| λ∼cat(0, 0.5, 1) | |||
| 2 | Random | λi lies within (0, 1) | λR∼Beta(0.7, 0.7) |
| 3 | Random | No restriction | λU∼N(0, 1,000)I( − 1, 2) |
Eight SNPs have been approximated in some studies by genotyping a nearby SNP in high linkage disequilibrium (Web Table 1). Since the suggested model might be affected by the use of proxies, we performed sensitivity analysis by including only studies in which the investigators had genotyped the main SNP of interest.
We fitted the model with WinBUGS software (16), using 100,000 Markov chain Monte Carlo cycles after 10,000 burn-in iterations. The WinBUGS code can be found at http://www.dhe.med.uoi.gr/software.htm.
RESULTS
Posterior probabilities for λ—main analysis
In Table 2, we present the odds ratios for heterozygotes and homozygotes and the median values and 95% credibility intervals for λ using model 1 and prior c (i.e., λ ∼ Beta(0.7, 0.7)); we also show the discrete probability of each of the 3 models of inheritance based on prior d. All odds ratios refer to the high-risk allele for each SNP, and each meta-analysis comprises 19 studies, unless stated otherwise. Of the 312 data sets of controls, 30 had P values less than 0.05 in Hardy-Weinberg equilibrium testing; only 4 had P < 0.001 in such testing.
Table 2.
Estimated Odds Ratios for Type 2 Diabetes (With Associated 95% Credibility Intervals) According to the Underlying Genetic Model (Parameter λ), Assuming a Beta(0.7, 0.7) Priora
| SNP | Risk Allele | No. of Studies | Chromosome | Gene/Region | ORAa | ORaa |
λ |
Probabilityb, % |
||||||||
| 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | Recessive | Additive | Dominant | |||||
| rs10923931 | T | 19 | 1 | NOTCH2 | 0.99 | 1.07 | 1.15 | 0.99 | 1.11 | 1.28 | 0.32 | 0.65 | 0.99 | 0 | 75 | 25 |
| rs7578597 | T | 19 | 2 | THADA | 1.00 | 1.07 | 1.27 | 1.15 | 1.26 | 1.49 | 0.01 | 0.29 | 0.62 | 39 | 61 | 0 |
| rs1801282 | C | 19 | 3 | PPARG | 1.01 | 1.13 | 1.34 | 1.05 | 1.26 | 1.56 | 0.20 | 0.55 | 0.71 | 1 | 98 | 1 |
| rs4607103 | C | 19 | 3 | ADAMTS9 | 1.01 | 1.08 | 1.17 | 1.09 | 1.17 | 1.27 | 0.09 | 0.47 | 0.70 | 4 | 96 | 0 |
| rs4402960 | T | 19 | 3 | IGF2BP2 | 1.05 | 1.11 | 1.17 | 1.10 | 1.20 | 1.32 | 0.39 | 0.55 | 0.77 | 0 | 100 | 0 |
| rs10010131 | G | 18 | 4 | WFS1 | 1.00 | 1.05 | 1.11 | 1.09 | 1.17 | 1.26 | 0.04 | 0.31 | 0.53 | 17 | 83 | 0 |
| rs10946398 | C | 17 | 6 | CDKAL1 | 1.09 | 1.13 | 1.18 | 1.21 | 1.29 | 1.40 | 0.34 | 0.48 | 0.65 | 0 | 100 | 0 |
| rs864745 | T | 19 | 7 | JAZF1 | 1.05 | 1.09 | 1.15 | 1.12 | 1.18 | 1.25 | 0.31 | 0.53 | 0.76 | 0 | 100 | 0 |
| rs13266634 | C | 19 | 8 | SLC30A8 | 1.05 | 1.12 | 1.19 | 1.14 | 1.21 | 1.29 | 0.35 | 0.58 | 0.76 | 0 | 100 | 0 |
| rs10811661 | T | 19 | 9 | CDKN2A/B | 1.09 | 1.22 | 1.37 | 1.31 | 1.46 | 1.64 | 0.32 | 0.53 | 0.66 | 0 | 100 | 0 |
| rs12779790 | G | 19 | 10 | CDC123/CAMK1D | 1.05 | 1.10 | 1.14 | 1.08 | 1.16 | 1.27 | 0.34 | 0.62 | 0.98 | 0 | 85 | 15 |
| rs5015480 | C | 19 | 10 | HHEX/IDE | 1.03 | 1.09 | 1.14 | 1.12 | 1.20 | 1.28 | 0.24 | 0.45 | 0.63 | 0 | 100 | 0 |
| rs7901695 | C | 17 | 10 | TCF7L2 | 1.19 | 1.31 | 1.45 | 1.36 | 1.61 | 1.91 | 0.50 | 0.58 | 0.66 | 0 | 100 | 0 |
| rs5219 | T | 19 | 11 | KCNJ11 | 1.07 | 1.11 | 1.16 | 1.18 | 1.26 | 1.35 | 0.31 | 0.44 | 0.59 | 0 | 100 | 0 |
| rs7961581 | C | 18 | 12 | TSPAN8/LGR5 | 1.02 | 1.07 | 1.13 | 1.04 | 1.12 | 1.23 | 0.31 | 0.59 | 0.97 | 0 | 89 | 11 |
| rs757210 | A | 14 | 17 | TCF2 | 1.01 | 1.06 | 1.14 | 1.01 | 1.10 | 1.20 | 0.32 | 0.70 | 0.99 | 1 | 68 | 31 |
| rs8050136 | A | 19 | 16 | FTO | 1.12 | 1.16 | 1.21 | 1.25 | 1.33 | 1.41 | 0.42 | 0.53 | 0.66 | 0 | 100 | 0 |
Abbreviations: OR, odds ratio; rs, reference SNP; SNP, single nucleotide polymorphism.
All results pertain to 19 studies (14), unless stated otherwise.
Probabilities of λ being 0, 0.5, and 1 (representing the recessive, additive, and dominant models, respectively) according to a categorical prior (prior d).
In the majority of cases, the suggested most likely model was the additive model. For 4 SNPs, the underlying model seemed to lie between the additive model and the dominant model for the risk allele (at the NOTCH2, CDC123/CAMK1D, TSPAN8/LGR5, and TCF2 loci), with corresponding probabilities supporting the dominant model being 25%, 15%, 11%, and 31%. These are the 4 SNPs for which the estimated ORaa’s for homozygotes were the weakest, ranging from 1.10 to 1.16. For 2 SNPs (at the THADA and WFS1 loci), the model seemed to lie between the additive and recessive models for the risk allele, with probabilities supporting the recessive model being 39% and 17%, respectively. In both cases, the ORAa for heterozygotes was weak (1.07 and 1.05, respectively).
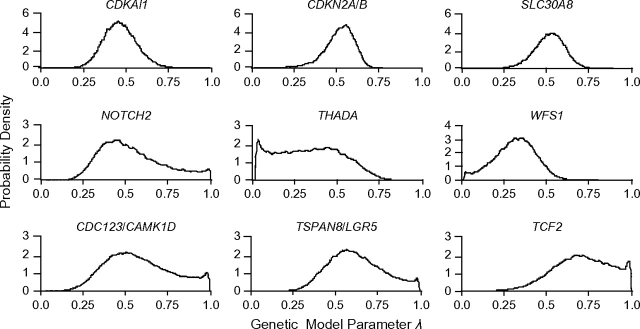
For all 17 SNPs, at least 1 of either the dominant or recessive models could be excluded. Figure 2 shows representative posterior distributions according to prior c.
Figure 2.
Posterior distributions for λ for a common (fixed) genetic model using prior c (see text). In the top 3 panels, the additive model is the most probable model and the dominant and recessive models are ruled out. For THADA and WFS1, only the dominant model is ruled out, and for NOTCH2, CDC123/CAMK1D, TSPAN8/LGR5, and TCF2, only the recessive model is ruled out.
Sensitivity analyses using different priors for λ
The 3 different priors had some impact on the median λ estimate for model 1 but not on the overall conclusions. In cases where there was high confidence about the underlying model (more than 95% probability for a specific model according to prior d)—for example, the additive models suggested for PPARG, ADAMTS9, IGF2BP2, CDKAL1, JAZF1, SLC30A8, CDKN2A/B, HHEX/IDE, TCF7L2, KCNJ11, and FTO—the absolute differences for the 3 Beta distribution priors were no more than 0.01 in the median posterior λ, no more than 0.09 in the 2.5% credibility bound, and no more than 2% in the 97.5% credibility bound. For the other 6 SNPs, the absolute differences in the median λ went up to 0.08, and the respective figures for the upper and lower 95% credibility bounds were 0.05 and 0.05 (see Web Table 2 (http://aje.oxfordjournals.org/)).
Effect estimates and their uncertainty
There was also no material variation in the odds ratio point estimates or the heterogeneity standard deviation τ with the different priors (Web Table 2). Note that Bayesian estimation of the effects incorporates full uncertainty in the estimates, including the uncertainty in the heterogeneity variance τ2, and therefore gives wider intervals than the random-effects model fitted with frequentist approaches. In the main analysis (using prior c), for 9 genetic variants (at the IGF2BP2, CDKAL1, JAZF1, SLC30A8, CDKN2A/B, CDC123/CAMK1D, TCF7L2, KCNJ11, and FTO loci), the lower bound of the 95% credibility interval for the effect of heterozygosity was higher than 1.05, and this also applied to the effect for homozygotes. The 95% credibility intervals for the effect of NOTCH2 included the null value for both heterozygotes and homozygotes. For the remaining SNPs (in/near THADA, PPARG, ADAMTS9, HHEX/IDE, TSPAN8/LGR5, and TCF2), the lower bound of the 95% credibility interval for the effect of heterozygosity was rather low (lower than 1.05), while the effect of homozygosity was giving lower bounds up to 1.15.
The variation in these results based on the alternative priors was not substantial (Web Table 2).
Sensitivity analyses excluding data on proxy SNPs
Table 3 shows sensitivity analysis results for the 8 index SNPs for which proxies have been used in some studies. In 6 cases (the CDKAL1, JAZF1, HHEX/IDE, TCF7L2, TCF2, and FTO loci), exclusion of proxies did not seem to result in material changes regarding the genetic model parameter or the probability for each genetic model, although the uncertainty in all parameters increased in some cases because of the reduced total sample size. For NOTCH2, the probability supporting the dominant model dropped from 25% to 11%. Confidence for the additive model for IGF2BP2 was slightly challenged, giving 11% probability for a dominant model.
Table 3.
Estimated Genetic Model Parameter λ (and 95% Credibility Interval) Assuming a Beta(0.7, 0.7) Priora
| No. of Studies | Gene/Region | λ |
Probabilityb, % |
||||
| 2.5% | Median | 2.5% | Recessive | Additive | Dominant | ||
| 15 | NOTCH2 | 0.24 | 0.50 | 0.97 | 0 | 89 | 11 |
| 15 | IGF2BP2 | 0.43 | 0.65 | 0.96 | 0 | 89 | 11 |
| 12 | CDKAL1 | 0.30 | 0.44 | 0.61 | 0 | 100 | 0 |
| 18 | JAZF1 | 0.31 | 0.54 | 0.78 | 0 | 100 | 0 |
| 5 | HHEX/IDE | 0.14 | 0.52 | 0.82 | 0 | 99 | 1 |
| 7 | TCF7L2 | 0.36 | 0.46 | 0.59 | 0 | 100 | 0 |
| 8 | TCF2 | 0.20 | 0.70 | 0.99 | 2 | 56 | 42 |
| 18 | FTO | 0.41 | 0.52 | 0.65 | 0 | 100 | 0 |
Studies in which proxy single nucleotide polymorphisms were used were excluded.
Probabilities of λ being 0, 0.5, and 1 (representing the recessive, additive, and dominant models, respectively) according to a categorical prior (prior d).
Heterogeneity in λi estimates across studies
We fitted the model allowing the study-specific genetic model parameters λi to vary randomly with values restricted between 0 and 1. Table 4 gives results for the median λR of the distribution and the parameter τλ, which shows the magnitude of the genetic-model heterogeneity fitted using prior c. The median heterogeneity standard deviation was no higher than 0.14. For NOTCH2, the credibility interval was shifted downwards (0.32, 0.99 became 0.23, 0.78), and the recessive model could be excluded for ADAMTS9. The CDC123/CAMK1D and TSPAN8/LGR5 upper limits for median λ were lower (0.98 and 0.97 became 0.86 and 0.84, respectively). For all other SNPs, no material changes in the median λ value were observed, and the credibility intervals were comparable to those from the fixed-effects model. Consequently, no major changes regarding the underlying model or the estimated odds ratios were observed.
Table 4.
Median Values (and 95% Credibility Intervals) for λR and the Genetic Model Heterogeneity Parameter τλ, Assuming Random Effects for λ and Using the Restricted Model (Model 2)
| Gene/Region | λR |
τλ |
||||
| 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | |
| NOTCH2 | 0.23 | 0.45 | 0.78 | 0 | 0.12 | 0.29 |
| THADA | 0.08 | 0.36 | 0.61 | 0 | 0.07 | 0.24 |
| PPARG | 0.31 | 0.55 | 0.73 | 0.01 | 0.14 | 0.30 |
| ADAMTS9 | 0.26 | 0.49 | 0.71 | 0 | 0.10 | 0.27 |
| IGF2BP2 | 0.38 | 0.56 | 0.75 | 0.01 | 0.09 | 0.25 |
| WFS1 | 0.13 | 0.34 | 0.54 | 0 | 0.08 | 0.25 |
| CDKAL1 | 0.32 | 0.47 | 0.68 | 0 | 0.10 | 0.28 |
| JAZF1 | 0.32 | 0.52 | 0.71 | 0 | 0.13 | 0.30 |
| SLC30A8 | 0.36 | 0.56 | 0.74 | 0.01 | 0.11 | 0.28 |
| CDKN2A/B | 0.31 | 0.51 | 0.65 | 0.01 | 0.05 | 0.18 |
| CDC123/CAMK1D | 0.33 | 0.57 | 0.86 | 0.01 | 0.09 | 0.26 |
| HHEX/IDE | 0.27 | 0.46 | 0.64 | 0 | 0.12 | 0.28 |
| TCF7L2 | 0.45 | 0.56 | 0.68 | 0.03 | 0.13 | 0.25 |
| KCNJ11 | 0.30 | 0.44 | 0.60 | 0.01 | 0.08 | 0.24 |
| TSPAN8/LGR5 | 0.30 | 0.53 | 0.84 | 0 | 0.10 | 0.28 |
| TCF2 | 0.33 | 0.63 | 0.89 | 0 | 0.08 | 0.29 |
| FTO | 0.41 | 0.53 | 0.66 | 0 | 0.06 | 0.22 |
Table 5 shows results from a random-effects model in which we also allowed λ to vary randomly. Important changes were observed for the NOTCH2 locus, which now covered possible values for the median λ between recessive, additive, and dominant models and included the heterosis possibility (see Web Figure 1 (http://aje.oxfordjournals.org/)). THADA gave a 95% credibility interval that covered the majority of the allowed negative values, with a considerable shift of the median from 0.36 to 0.10. ADAMTS9 included the recessive model, whereas with an unrestricted model the lowest 95% credibility bound was at 0.26 (Web Figure 1). For 3 further loci (CDC123/CAMK1D, TSPAN8/LGR5, and TCF2), the scenario of heterosis could not be excluded. However, the unrestricted analysis with model 3 seemed to give results consistent with those from the restricted model (model 2) and the fixed-effects model (model 1) for the IGF2BP2, CDKAL1, JAZF1, SLC30A8, CDKN2A/B, KCNJ11, and FTO loci.
Table 5.
Median Values (and 95% Credibility Intervals) for λU and the Genetic Model Heterogeneity Parameter τλ, Assuming Random Effects for λ and Using the Unrestricted Model (Model 3)
| Gene/Region | λU |
τλ |
||||
| 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | |
| NOTCH2 | 0.08 | 0.51 | 1.60 | 0.02 | 0.35 | 1.27 |
| THADA | −0.84 | 0.10 | 0.58 | 0.01 | 0.16 | 0.69 |
| PPARG | 0.13 | 0.53 | 0.77 | 0.01 | 0.23 | 0.64 |
| ADAMTS9 | −0.06 | 0.44 | 0.72 | 0.01 | 0.20 | 0.72 |
| IGF2BP2 | 0.38 | 0.59 | 0.87 | 0.01 | 0.14 | 0.51 |
| WFS1 | −0.05 | 0.29 | 0.56 | 0.01 | 0.20 | 0.66 |
| CDKAL1 | 0.29 | 0.47 | 0.67 | 0.01 | 0.14 | 0.43 |
| JAZF1 | 0.25 | 0.52 | 0.76 | 0.01 | 0.22 | 0.57 |
| SLC30A8 | 0.28 | 0.56 | 0.76 | 0.01 | 0.18 | 0.52 |
| CDKN2A/B | 0.31 | 0.52 | 0.65 | 0 | 0.05 | 0.20 |
| CDC123/CAMK1D | 0.35 | 0.68 | 1.65 | 0.01 | 0.16 | 0.69 |
| HHEX/IDE | 0.17 | 0.44 | 0.66 | 0.015 | 0.17 | 0.52 |
| TCF7L2 | 0.12 | 0.55 | 0.72 | 0.05 | 0.16 | 1.12 |
| KCNJ11 | 0.28 | 0.44 | 0.61 | 0.01 | 0.10 | 0.35 |
| TSPAN8/LGR5 | 0.15 | 0.56 | 1.22 | 0.02 | 0.35 | 1.07 |
| TCF2 | 0.18 | 0.71 | 1.45 | 0.02 | 0.28 | 1.15 |
| FTO | 0.41 | 0.53 | 0.69 | 0 | 0.07 | 0.24 |
DISCUSSION
We applied and extended a genetic model-free Bayesian approach to investigate the fit of type 2 diabetes associations to various genetic models of inheritance. Regardless of the prior distribution used, our analyses found that most of the common genetic variants that show robust associations with type 2 diabetes risk fitted best to an additive model. However, several exceptions existed, where either recessive or dominant models for the risk allele also had substantial support as alternative options, besides the additive model. At least 1, if not 2, of the 3 main genetic models could be excluded with considerable certainty for all 17 associations.
The 17 SNPs that we analyzed all had considerable statistical support, with P values less than 2 × 10−7 in joint analyses (by fixed-effects models and, for most, also by random-effects models) (13, 14). They also passed several quality checks, including Hardy-Weinberg equilibrium testing, although modest deviations from such equilibrium were still possible. Overall, the credibility of these associations was rather high.
The choice of genetic model in genome-wide association studies remains open and arbitrary, but most investigators seem to adopt an additive (per-allele) analysis. Exclusively recessive-fit and exclusively dominant-fit associations may be discovered if a more comprehensive analytical approach is followed. Studies examining variants in linkage disequilibrium with the causal variant (and not the causal variant itself) may have higher power to detect an association under the additive model. Therefore, the established type 2 diabetes variants we are investigating are likely to have a higher relative representation of such loci.
We also evaluated the possibility of between-study heterogeneity in the genetic model. For most SNPs, the median between-study standard deviation was small or modest, and its consideration did not much change the overall inferences about the most likely genetic model. An unrestricted analysis showed that heterosis is not common but still remains a plausible scenario, since it could not be excluded for 4 of the 17 SNPs. Given that genome-wide association studies use target SNPs that are unlikely to be the true culprits, different linkage disequilibrium may introduce such heterogeneity in the genetic model across different populations. This is not very likely in the examined data, since all study populations were of Caucasian descent. However, this might become a more serious issue if populations of different ancestry were to be examined.
A limitation of a genetic model-free Bayesian model is that it is driven by the data at hand in identifying the most likely genetic model. The ability to extrapolate to other data and populations is an open challenge. Moreover, the proposed modeling should be used primarily for associations that are already supported by a substantial body of evidence, based on several studies and conventional meta-analysis thereof. Application of these methods to data from associations that are likely to represent false-positives may result in overfitting to noise signals. In the absence of robust support for the presence of an association, these analyses should be recognized as exploratory.
Knowledge of the best-fitting genetic model may be important in optimizing the use of these markers for predictive purposes. At the current stage, genetic markers in type 2 diabetes explain only about 2.5% of the risk variance and would result in a predictive area under the receiver operating characteristic curve (AUC) of only 0.60, while traditional predictors (body mass index, sex, and age) already result in an AUC of 0.78 (17). With many markers accrued, proper modeling may be potentially useful to increase the predictive ability. However, the Bayesian model that we used further highlights the challenges and difficulties of using this information for predictive purposes. When we considered the full scale of uncertainty in parameters, the 95% credibility intervals of the odds ratios were considerably large. For NOTCH2, these intervals even crossed the null. This means that the effects of these genetic markers in some populations may be very small or even nonexistent. This adds an extra note of caution to the possibility of predictive testing in the general population based on this information (18).
Although some of the established type 2 diabetes susceptibility loci (like PPARG and KCNJ11) have been known for several years, the field has not progressed to unequivocal identification of the truly causal variants. Consequently, statistical inference regarding the true genetic model under which these loci act has been difficult. In addition, there is a paucity of biologic data that might help address the genetic model question. Identification of the best-fitting model through Bayesian meta-analysis may be helpful in suggesting how biologic and functional experiments should be set up and what model should be used in them.
Supplementary Material
Acknowledgments
Author affiliations: Clinical and Molecular Epidemiology Unit and Clinical Trials and Evidence-Based Medicine Unit, Department of Hygiene and Epidemiology, School of Medicine, University of Ioannina, Ioannina, Greece (Georgia Salanti, John P. A. Ioannidis); Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom (Lorraine Southam, Eleftheria Zeggini, Mark I. McCarthy, Andrew Morris); Institute of Musculoskeletal Sciences, Botnar Research Centre, Nuffield Orthopaedic Centre, University of Oxford, Oxford, United Kingdom (Lorraine Southam); Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, Massachusetts (David Altshuler, Kristin Ardlie, Benjamin F. Voight); Center for Human Genetic Research, Massachusetts General Hospital, Boston, Massachusetts (David Altshuler, Benjamin F. Voight); Diabetes Unit, Massachusetts General Hospital, Boston, Massachusetts (David Altshuler); Department of Molecular Biology, Massachusetts General Hospital, Boston, Massachusetts (David Altshuler); Department of Medicine, Harvard Medical School, Boston, Massachusetts (David Altshuler, Benjamin F. Voight); Department of Genetics, Harvard Medical School, Boston, Massachusetts (David Altshuler); Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, United Kingdom (Inês Barroso, Felicity Payne, Eleftheria Zeggini); Department of Biostatistics and Center for Statistical Genetics, School of Public Health, University of Michigan, Ann Arbor, Michigan (Michael Boehnke, Laura J. Scott); Department of Nutrition, Harvard School of Public Health, Boston, Massachusetts (Marilyn C. Cornelis, Frank B. Hu); Genetics of Complex Traits, Peninsula Medical School, Exeter, United Kingdom (Timothy M. Frayling); Institute of Epidemiology, German Research Center for Environmental Health, Neuherberg, Germany (Harald Grallert, Thomas Illig); Steno Diabetes Center, Gentofte, Denmark (Niels Grarup, Torben Hansen); Diabetes and Endocrinology Research Unit, Department of Clinical Sciences, Lund University, Malmö, Sweden (Leif Groop, Valeriya Lyssenko); Faculty of Health Sciences, University of Southern Denmark, Odense, Denmark (Torben Hansen); Molecular Genetics Research Group, Peninsula Medical School, Exeter, United Kingdom (Andrew T. Hattersley); HUNT Research Center, Faculty of Medicine, Norwegian University of Science and Technology, Trondheim, Norway (Kristian Hveem, Carl G. P. Platou); Department of Medicine, Levanger Hospital, The Nord-Trøndelag Health Trust, Levanger, Norway (Kristian Hveem, Carl G. P. Platou); Department of Medicine, University of Kuopio and Kuopio University Hospital, Kuopio, Finland (Johanna Kuusisto); MRC Epidemiology Unit, Institute of Metabolic Sciences, Addenbrooke's Hospital, Cambridge, United Kingdom (Markku Laakso, Nicholas J. Wareham, Claudia Langenberg); Oxford Centre for Diabetes, Endocrinology and Metabolism, Churchill Hospital, Oxford, United Kingdom (Mark I. McCarthy); Diabetes Research Centre, Biomedical Research Institute, University of Dundee, Dundee, United Kingdom (Andrew D. Morris); Pharmacogenetics Research Centre, Biomedical Research Institute, University of Dundee, Dundee, United Kingdom (Colin N. A. Palmer); and Center for Genetic Epidemiology and Modeling, Institute for Clinical Research and Health Policy Studies, Department of Medicine, School of Medicine, Tufts University, Boston, Massachusetts (John P. A. Ioannidis).
Support for this project was provided through the Tufts Clinical and Translational Science Institute under funding from the National Institutes of Health/National Center for Research Resources (grant UL1 RR025752). Dr. Eleftheria Zeggini was supported by the Wellcome Trust (grant WT088885/Z/09/Z).
Drs. Eleftheria Zeggini and John P. A. Ioannidis contributed equally to this article.
Points of view or opinions presented in this paper are those of the authors and do not necessarily represent the official position or policies of the Tufts Clinical and Translational Science Institute.
Conflict of interest: none declared.
Glossary
Abbreviations
- AUC
area under the receiver operating characteristic curve
- OR
odds ratio
- SNP
single nucleotide polymorphism
References
- 1.Attia J, Thakkinstian A, D'Este C. Meta-analysis of molecular association studies: methodological lessons for genetic epidemiology. J Clin Epidemiol. 2003;56(4):297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 2.Minelli C, Thompson JR, Abrams KR, et al. Bayesian implementation of a genetic model-free approach to the meta-analysis of genetic association studies. Stat Med. 2005;24(24):3845–3861. doi: 10.1002/sim.2393. [DOI] [PubMed] [Google Scholar]
- 3.Thakkinstian A, McEvoy M, Minelli C, et al. Systematic review and meta-analysis of the association between β2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–211. doi: 10.1093/aje/kwi184. [DOI] [PubMed] [Google Scholar]
- 4.Minelli C, Thompson JR, Abrams KR, et al. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34(6):1319–1328. doi: 10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- 5.Sagoo GS, Tatt I, Salanti G, et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am J Epidemiol. 2008;168(11):1233–1246. doi: 10.1093/aje/kwn235. [DOI] [PubMed] [Google Scholar]
- 6.Ioannidis JP, Ntzani EE, Trikalinos TA, et al. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 8.Seminara D, Khoury MJ, O'Brien TR, et al. The emergence of networks in human genome epidemiology: challenges and opportunities. Epidemiology. 2007;18(1):1–8. doi: 10.1097/01.ede.0000249540.17855.b7. [DOI] [PubMed] [Google Scholar]
- 9.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 11.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seaman SR, Richardson S. Bayesian analysis of case-control studies with categorical covariates. Biometrika. 2001;88(4):1073–1088. [Google Scholar]
- 16.Spiegelhalter D, Thomas A, Best N, et al. WinBUGS User Manual. Version 1.4, January 2003. (Upgraded to Version 1.4.3) Cambridge, United Kingdom: MRC Biostatistics Unit; 2007. ( http://www.mrc-bsu.cam.ac.uk/bugs/winbugs/manual14.pdf). (Accessed May 1, 2009) [Google Scholar]
- 17.Lango H, Palmer CN, Morris AD, et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57(11):3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle—will we get our wish? N Engl J Med. 2008;358(2):105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.