Abstract
Staphylococcus aureus mastitis in dairy sheep ranges from subclinical mastitis to lethal gangrenous mastitis. Neither the S. aureus virulence factors nor the host-factors or the epidemiological events contributing to the different outcomes are known. In a field study in a dairy sheep farm over 21 months, 16 natural isolates of S. aureus were collected from six subclinical mastitis cases, one lethal gangrenous mastitis case, nasal carriage from eight ewes and one isolate from ambient air in the milking room. A genomic comparison of two strains, one responsible for subclinical mastitis and one for lethal gangrenous mastitis, was performed using multi-strain DNA microarrays. Multiple typing techniques (pulsed-field-gel-electrophoresis, multiple-locus variable-number, single-nucleotide polymorphisms, randomly amplified polymorphic DNA, spa typing and sas typing) were used to characterise the remaining isolates and to follow the persistence of the gangrenous isolate in ewes’ nares. Our results showed that the two strains were genetically closely related and they shared 3 615 identical predicted open reading frames. However, the gangrenous mastitis isolate carried variant versions of several genes (sdrD, clfA-B, sasA, sasB, sasD, sasI and splE) and was missing fibrinogen binding protein B (fnbB) and a prophage. The typing results showed that this gangrenous strain emerged after the initial subclinical mastitis screening, but then persisted in the flock in the nares of four ewes. Although we cannot dismiss the role of host susceptibility in the clinical events in this flock, our data support the hypothesis that S. aureus populations had evolved in the sheep flock and that S. aureus genetic variations could have contributed to enhanced virulence.
Keywords: subclinical mastitis, gangrenous mastitis, dairy sheep, Staphylococcus aureus, microarray
1. INTRODUCTION
Staphylococci are the main aetiological agents of small ruminant intramammary infections (IMI), and Staphylococcus aureus is the most frequent isolate from clinical IMI cases and coagulase-negative species are the most frequent in subclinical IMI. The annual incidence of clinical IMI in dairy sheep is generally lower than 5%, but in a small percentage of herds the incidence may exceed 30–50% of the animals, causing mortality (gangrenous mastitis) or culling of up to 70% of the herd [3]. In addition subclinical or hidden S. aureus IMI cases occur in 3% to 37% of dairy sheep, and are important for economic, hygienic (dairy products consumers) and legal reasons in Europe (EU Directives 46/92 and 71/94 defining the bacteriological quality of milk) [3]. In ewe flocks on farms with small-scale production of raw milk cheese, culling for S. aureus subclinical IMI is not the general rule but when a gangrenous case occurs, the death of the ewe due to the infection is common, even with emergency antibiotic therapy. If an animal survives the acute disease, the affected gland becomes necrotic and gradually separates from the surrounding tissue; in this case, healing can take many months [4, 39]. Why some cases should remain subclinical while others become gangrenous is not known. Bacterial features, which can be identified by genetic content, could be responsible [4], or alternatively, ewe factors might be important.
There has been some conflicting data about whether some S. aureus strains are more virulent than others. A large study of 161 human nasal carriages of S. aureus in healthy people versus isolates that caused invasive disease failed to identify genetic markers associated with infection [18]. In contrast, some human clones of methicillin-resistant S. aureus (MRSA), such as the USA300 and TW isolates, clearly cause unique types of infection [8, 33]. In rabbits, differences in virulence between S. aureus strains have clearly been demonstrated in experimental infection studies as well as in the field [20, 34].
In bovine IMI, the virulence of S. aureus differs among strains according to previous studies but no specific virulence factor or combination of factors has been strongly associated with the severity of mastitis [13].
Because most previous studies have focussed on well known virulence factors (e.g. exoenzymes, toxins and adhesions) to try to explain the difference of S. aureus virulence between subclinical and acute cases of mastitis, the aim of this study was to explore if other genetic features could be found (e.g. genes evolved in cellular processes, cell-wall synthesis, transport and intermediary metabolism) that might contribute to virulence differences. This study was made possible because genetically similar S. aureus isolates caused either subclinical mastitis or gangrenous mastitis within the same flock. Here we used microarray technologies for the first time to probe the genomes of two isolates. These strains were then compared to additional isolates over a 21 month period using a variety of typing techniques to investigate the evolution of S. aureus in this flock.
2. MATERIALS AND METHODS
2.1. Clinical examination and sample collection
The field study was carried out over a 21 month period in a dairy sheep farm with small-scale production of raw milk cheese located in the southeast of France. The sheep were a crossbreed of the Lacaune breed and the “Rouge du Péone” breed. Three visits were made. The first was in January 2002 to sample 80 ewes for S. aureus subclinical mastitis. The second was in November 2002 to sample a primipare ewe with S. aureus gangrenous mastitis. This ewe died within 24 h in spite of systemic and intramammary therapies. The last visit to the farm was made one year later in October 2003 to look for the gangrenous S. aureus strain in the flock after one dry period for the ewes (absence of milk production during five months). The isolates were recovered from the anterior nares of the ewes and the air of the milking room. The shepherd stopped the exploitation of the farm at the end of 2003.
Bacterial examination of milk samples was performed by streaking 0.1 mL of milk on Baird-Parker rabbit plasma fibrinogen agar (BPRPFA) medium (AES, Combourg, France). Moreover, in order to increase the sensitivity of the isolation, 1 mL of milk was incubated for 24 h at 37 °C in 9 mL of Chapman selective broth (AES) prior to streaking 0.1 mL on BPRPFA medium. Ambient air was sampled by using three plates with BPRPFA medium exposed to the environment of milking room for 15 min and incubated for 24–48 h at 37 °C. For the ewes’ nares, a swab was rubbed inside each nostril and streaked directly on BPRPFA medium plates [35, 36]. One randomly chosen coagulase-positive isolate per plate was confirmed as S. aureus by PCR performed on the 23S rDNA gene [31]. On the first visit (January 2002), 6 out of 80 ewes were found to have subclinical IMI with S. aureus in their milk directly or after growth in selective broth. The six isolates were named O33, O46, O47, O54, O63 and O64. The second visit (November 2002) was for the isolation in pure culture of the S. aureus responsible of the death of the ewe with a gangrenous mastitis. This isolate was named O11. The last visit (October 2003) was to look for the S. aureus strain O11 in the nares of the ewes and in the air of the milking parlor. Eight out of 71 ewes were positive with S. aureus in their nares. These isolates were named O193-O200. The ambient air of the milking room during milking time was sampled and one isolate on the plate with BPRPFA was randomly selected and named O192.
2.2. Genomic comparison of the subclinical (O46) and gangrenous (O11) S. aureus isolates by DNA microarray studies
In the following methods, the isolate O46 (subclinical) was randomly chosen between the six isolates responsible of the subclinical mastitis in January 2002. This isolate O46 was genetically compared with O11 (gangrenous) with the DNA microarrays.
A one colour dye DNA microarray was constructed with 188 oligonucleotide probes of 65-mer, mainly designed from the putative virulence genes of S. aureus Mu50. Chromosomal DNA extraction, microarray design, hybridisation and data analysis methods have been described in detail by Vautor et al. [37]. The results can be found in the Table S1 (on line material available at www.vetres.org). In addition, the two strains were compared using the well-validated, two-colour S. aureus seven strain whole genome microarray that has been described previously [40] and contains 3 623 PCR products representing every predicted open reading frame (ORF) in the first seven genome sequencing projects: MRSA252, N315, Mu50, COL, 8325, MW2 and MSSA476. Each strain was co-hybridised with DNA from the reference strain MRSA252, and data were analysed using GeneSpring 7.2. The array design is available in BμG@Sbase (accession No. A-BUGS-171) and also ArrayExpress (accession No. A-BUGS-17). Fully annotated microarray data have been deposited in BμG@Sbase (accession No. E-BUGS-762) and in ArrayExpress (accession No E-BUGS-76).
Microarray profiles of the strains were compared to a database of previously characterised human and animal isolates, and clustered using 723 core-variable genes to identify lineages [18, 32]. The genes which were found “present” in one strain and not in the other using the DNA microarray where confirmed by PCR. Moreover, all the 16 isolates were tested by PCR for the genes found different between O11 and O46 with the DNA microarrays. The PCR was performed using primers designed with the software Primer3 [29] except for fibrinogen binding protein B (fnbB) for which the primers designed by Kuhn et al. were used [16]. Primers are listed in Table I.
Table I.
Primers used in the study to confirm the microarray data.
| Gene | Primers used in this study (5′-3′) or author’s references |
|---|---|
| SdrD (Mu50 NC_002758) | AACGATTGTACCAGCCCAAG |
| TTTGCAGTCGCAATTGTTTC | |
| FnbB | Khun et al. [16] |
| SplE (SAR1902) | CAGCCAAAGCCGAACATAAT |
| TATGTGCGCCAATTTCCATA | |
| SAS0897 | GAGAACTTGCTGAAGCTATTGGA |
| CCCTCCTTATCAAAATGAGCA | |
| SAR1558 | CAAACCAAAAACGCAACAAG |
| CAGGCGAAACGACATACTCA | |
| SAR0940 | TTTGCGGACACTGTAGGATG |
| ATTACCCGCTCTCTCACCAA | |
| SAR2100 | GCTGATGTTTTCGAGGTTGG |
| TACACCAGCAGAGACGCAAC | |
| SACOL0343 | CAAGCAATGAGGCATTCAGA |
| GTCCGATAGCATTGGTCGTT |
2.3. Typing milk, air and nasal carriage S. aureus isolates from the flock
Sixteen S. aureus isolates from mastitis milk, air and nares were compared with different discriminating typing techniques. DNA was extracted with the DNeasy®Tissue kit (Qiagen, Courtabœuf, France) according to the manufacturer’s recommendations.
Pulsed-field-gel-electrophoresis (PFGE) of the chromosomal DNA was performed with the restriction enzyme SmaI and subsequent analysis as described by Vautor et al. [36].
Randomly amplified polymorphic DNA (RAPD) typing was performed three times for each isolates [27]. The RAPD pattern with at least one band of difference was considered as one type and named (R, R1, R2 or R3).
For spa typing, the polymorphic X region of the protein A gene (spa) was amplified using the primers spa-1113f (5′ TAA AGA CGA TCC TTC GGT GAG C 3′) and spa-1514r (5′ CAG CAG TAG TGC CGT TTG CTT 3′) and sequenced3. Applying the recently developed algorithm BURP (Based Upon Repeat Patterns), spa types (spa-t) were clustered into different groups with calculated cost between members of a group less than or equal to five. BURP spa clonal complexes (spa-CC) were automatically assigned by Ridom Staph Type software using the code system described on the Ridom SpaServer website.
Multiple-locus variable-number (MLVA) tandem repeats analysis for sdrD, sdrC, fnb, clfA, clfB and SAV1078 genes was performed according to Gilbert et al. [10] and for coa according to Callon et al. [6]. The MLVA profiles for the S. aureus strains were determined by the combination of types of allele found for each gene analysed.
A genotyping method was also used, based on single-nucleotide polymorphisms (SNP) of exotoxin genes (ssl) [1]. The sequences of PCR-amplified internal fragments of three different ssl genes (ssl2, ssl4, ssl9) were compared. These genes encode S. aureus superantigen-like proteins (ssl), belong to a family of exotoxins called staphylococcal exotoxins. For coherence with the literature, these genes, originally named set2, set5 and set7 by Aguiar-Alves et al. [1], have been renamed ssl4 (432 bp), ssl9 (467 bp) and ssl2 (496 bp), respectively [17]. Sequences were compared for SNP using the BioEdit Software4. Finally, a multiplex PCR assay was used for the detection of prophages in the genomes of lysogenic S. aureus strains. These PCR results allow the prophages to be classified into serogroups A, B, Fa, Fb, L or D [23].
S. aureus surface protein typing (sas typing) was implemented using the method described by Robinson and Enright [26]. Briefly, gene fragments from seven putative or proven surface protein-encoding loci (sasA, sasB, sasD, sasE, sasF, sasH, sasI) were PCR-amplified and sequenced. Unique nucleotide sequences defined sas alleles, and unique series of alleles defined sas sequence types.
3. RESULTS
3.1. Genomic comparison of the subclinical (O46) and gangrenous (O11) S. aureus isolates by DNA microarray studies
Genomic differences between the two strains O11 and O46 are summarized in Table II along with PCR screening results for the other strains. The core genes (except SAR0940), the core-variable genes (except sdrD, fnbB, splE), and the mobile genetic elements (plasmids, staphylococcal cassette chromosome, transposons, S. aureus pathogenicity islands, but excepting some bacteriophage genes) hybridize with similar intensity to the genomic DNA of both strains. PCR confirmed the genetic difference between the strains listed in Table II, including the presence of SAS0897, SAR1558, SAR2100, SACOL0343 in strain O46 but not in O11; these genes are typical of S. aureus lysogenic bacteriophage (hydrolase, lipoprotein, repressor, helicase dnaB). There were different weak comparative signals in the DNA array for MW0387 (exotoxin), SAR2036 (CHIPS) and SACOL0886 (enterotoxin K) but these targets were not confirmed by PCR.
Table II.
Typing study of sixteen S. aureus isolates in a dairy sheep flock over 21 months.
| O54, O33 (sub.) | O47 (sub.) | O63 (sub.) | O64 (sub.) | O46 (sub.) | O11 (gangrenous) | O193, O194, O195, O196 (nares) | O197 (nares) | O198 (nares) | O200 (nares) | O192 (air) | O199 (nares) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SdrD | − | − | − | − | − | + | + | − | − | + | + | + |
| FnbBa | + | + | + | + | + | − | − | + | + | − | − | − |
| SplE | − | − | − | − | − | + | + | − | − | + | + | + |
| SAS0897 | + | + | + | + | + | − | − | + | + | + | − | + |
| SAR1558 | + | + | + | + | + | − | − | + | + | − | − | − |
| SAR0940 | + | + | + | + | + | − | − | + | + | + | − | + |
| SAR2100 | + | + | + | + | + | − | − | + | + | − | − | − |
| SACOL0343 | + | + | + | + | + | − | − | + | + | + | + | − |
| Spa types | 3568 | 3568 | 3568 | 3568 | 3568 | 524 | 524 | 3568 | 3568 | 524 | 524 | 524 |
| Spa clonal complex | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 | 1773 |
| RAPD typesb | R | R2 | R | R | R | R1 | R1 | R3 | R2 | R1 | R1 | R1 |
| PFGE typesc | OV | OV | OV | OV | OV | OV’ | OV’ | OV’’’ | OV | OV’’ | OV’ | OV’ |
| MLVA types (coa, sdrD, sdrC, fnb, clfA, clfB, SAV 1078)d | B | F | E | D | B | A | A | C | B | A | A | A |
| Prophagese | A, B, Fb | A, B, Fb | A, B, Fb | A, B, Fb | A, B, Fb | A, Fb | A, Fb | A, B, Fb | A, B, Fb | A, B, Fb | A, Fb | A, Fb |
| Ssl9 SNPf | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ssl4 SNP | 1 | N.D. | N.D. | N.D. | 1 | 1 | 1 | N.D. | 1 | N.D. | 1 | 1 |
| Ssl2 SNP | 1 | N.D. | N.D. | N.D. | 1 | 1 | 1 | N.D. | 1 | N.D. | 1 | 1 |
| Sas typesg | N.D. | N.D. | N.D. | N.D. | I | II | II | N.D. | N.D. | N.D. | N.D. | N.D. |
The absence/presence of the genes, sdrD, splE, SAS0897, SAR0940, SAR1558, SAR2100 and SACOL0343 were evaluated with primers designed in this study.
[16].
[27]: at least one band of difference for each RAPD types.
[35]: the PFGE profiles were named, OV, OV’, OV’’and OV’’’.
[6, 10]: the letters correspond to a unique pattern made by the combination of the variable number-number tandem repeats of each gene.
[23]: bacteriophages serogroups. + positive with PCR, − negative with PCR; sub.: isolates recovered in a S. aureus subclinical mastitis case (January 2002); O192-O200 are the isolates recovered in the last visit to look for the gangrenous strain O11 in the ewes’nares and in the air of the milking room (October 2003).
[1]: single nucleotide polymorphism (SNP). The same exotoxin sequence type had an identical numerical number, N.D.: not done.
[26]: unique sequences defined alleles and unique series of alleles defined a sequence type (named I or II).
The two strains O11 and O46 clustered with isolates of the CC130 lineage from cows (data not shown). The two strains had in common 3 615 identical predicted ORF as designed from the first seven genome sequencing project, including redundant ORF printed from multiples strains on the arrays. Moreover, both strains O11 and O46 had no evidence of free plasmids (data not shown).
3.2. Typing S. aureus isolates from milk, air and nasal carriage in the flock
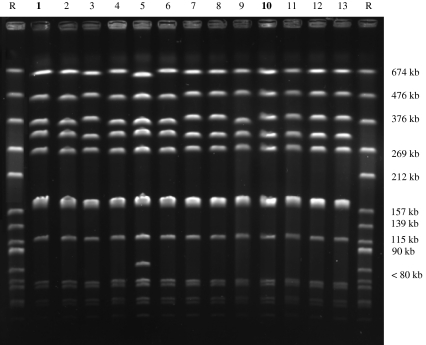
The PFGE results comparing the 16 S. aureus isolates recovered over 21 months are illustrated in Figure 1. Four patterns with more than one band difference were identified and named OV, OV’, OV’’ and OV’’’. The strain O46 (OV) and the strain O11 (OV’) had a PFGE pattern within three-band difference of each other. The strain O11 and the strains O193, O194, O195 and O196 had an identical PFGE pattern (OV’).
Figure 1.
Representative example of S. aureus PFGE pattern of DNA digested by SmaI. Lane R: reference strain of S. aureus (strain CIP57.10); 1: O46 (subclinical strain); 2: O64 (sub.); 3: O200 (nares); 4: O54 (sub.); 5: O197 (nares); 6: O198 (nares); 7: O192 (ambient air); 8: O199 (nares); 9: O47 (sub); 10: O11 (gangrenous strain); 11: O194 (nares); 12: O195 (nares); 13: O196 (nares). The subclinical isolates (O46, O47, O64, O54) had been recovered in January 2002, the gangrenous isolates (O11) had been recovered in November 2002, the nares isolates (O200, O197, O198, O199, O194, O195, O196) and the ambient air isolate (O192) had been recovered in October 2003 in a dairy sheep farm.
Two spa-t were found: t3568 with the repeats 04-39-17 and t524 with the repeats 04-17. These two spa-t, by clustering with BURP, belonged to the spa-CC 1773. The other genes found by DNA microarray results in O46 or in O11 (Tab. II), distinguishing these two isolates, were used to track by PCR the gangrenous strains in the ewes’ nares.
The MLVA tandem repeats showed six types named A–F. With the MLVA, the patterns between O11 and O46 were found different for the genes sdrD, fnb, clfA and clfB (data not shown).
The RAPD typing technique showed four types named R-R3. Between the strain O11 (R1) and the strain O46 (R) three bands of difference were found. The strain O11 had the same RAPD pattern than O193, O194, O195 and O196.
For the ssl SNP typing, only ssl9 showed a difference, which was detected by the nucleotide replacement of T for G. The detection of prophages by the multiplex PCR showed that the isolates present on the farm over 21 months had prophages of serotypes A, B and Fb. The gangrenous isolate O11 was missing prophage B compared to the subclinical mastitis isolate O46.
The last typing technique used was the highly variable sas genes, which was done only for the strains found identical with all the previous typing techniques (i.e. O11, O193-O196) and O46. The strain O46 had sas type I (sasA26, sasB28, sasD20, sasE26, sasF33, sasH36 and sasI29), whereas the strains O11 and O193-O196 had sas type II (sasA34, sasB27, sasD19, sasE26, sasF33, sasH36 and sasI28). The sasA and sasB alleles differed at two and one nucleotide sites, respectively, whereas the sasD and sasI alleles differed by insertion-deletion events.
In summary, with all the techniques used to discriminate the S. aureus isolates, the gangrenous isolate O11 was found identical to the isolates O193, O194, O195 and O196 recovered in the nares of the ewes during the last visit. It was also very closely related to O192, O199 and O200 isolated in the last visit, varying only in bacteriophage profiles. No isolate in the first visit was identical to O11. In contrast, the subclinical mastitis isolate O46 was found to be indistinguishable from subclinical mastitis isolates O33, O54 (recovered during the first visit) and was not found in ewes’ nares (the last visit). Some variations in MLVA and ssl5 SNP were seen in the remaining isolates from the first and final visits, although they looked closely related to all of the isolates.
4. DISCUSSION
In ewe mastitis it is suspected that S. aureus strains have different virulence potential5 [2], but the genetic features associated with the different outcomes were unknown. This is the first comprehensive genomic comparison of an ovine S. aureus strain responsible for subclinical mastitis versus a related strain responsible for a case of acute gangrenous mastitis. We found genomic differences between these genetically closely related strains but we did not find differences in genes evolved in cellular processes, cell-wall synthesis, transport, or intermediary metabolism.
As defined by Lindsay et al. by microarray [18], the two strains O11 and O46 were closely related and clustered into the same lineages as two bovine isolates of MLST Clonal Complex 130 [32]. The PFGE and the spa typing results confirmed that the two isolates were genetically closely related (Fig. 1 and Tab. II) [33, 37] although there are allelic variants between the strains as detected by MLVA, sslSNP and sas genes. The microarray results found that the gangrenous strain O11 was positive for the sdrD gene, which belongs to the Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMM) family. The human protein that SdrD binds to is not known, but a study on human strains [30] showed that the strains carrying sdrD and sdrE had an increased association with bone infection. In fact, O46 had a variant copy of the sdrD gene as determined by the MLVA (data not shown). One limitation of the microarray studies was that these microarrays were designed using only genes from seven human S. aureus strains [32] so we were unable to detect animal S. aureus-specific genes potentially present in O11 and O46. Moreover these microarrays do not provide information about the position of genes in the chromosome.
Serine protease-like E (splE) is one of several highly homologous spl genes found on the S. aureus genomic island beta (GIβ), and is predicted to be a substrate-specific serine protease [25]. splE was only found in the gangrenous strain O11. The product of this splE gene might participate in the difference in disease manifestation between O11 and O46, as some Spl proteases possess restricted substrate specificity similar to that of the V8 protease and epidermolytic toxins [25].
fnbB was missing in the gangrenous O11 compared to O46. fnbB also belongs to the MSCRAMM family, and can bind to host proteins fibrinogen, fibronectin and elastin, and also play a role in biofilm production [22]. In rabbit flocks with S. aureus infections (e.g. pododermatis, subcutaneous abscesses and mastitis), there are “low virulence” strains where the infection remains limited to a small number of animal, and “high virulence” strains which spread throughout the rabbitry [20, 34]. Vancraeynest et al. [34] showed that fnbB was less common in high virulence isolates, and is associated with reduced spread of infection. It is interesting to notice that the O11 gangrenous strain did not have the fnbB compared to O46. Sung et al. [32] have also shown that animal strains of S. aureus generally encode variant types of fnbB compared to human S. aureus strains.
A major genomic difference between the O11 and O46 isolates was the missing prophage B in the gangrenous isolate. Because only a few genes on the microarray correlated with this phage, we presume it is relatively unrelated to those in the sequenced genomes. Bacteriophages are among the most abundant inhabitants of the biosphere, considering that an environmental sample contains nearly 10-fold more phage particles than prokaryotes [5]. All sequenced strains of S. aureus carry between one and four bacteriophage in their genomes [19]. The contribution of these prophages to pathogenesis is probably multifactorial. Many prophage carry putative virulence genes on them. For example, S. aureus phages of the phi3 family carrying immune evasion genes (coding for staphylokinase [11], enterotoxins [15], chemotaxis inhibitory protein CHIPS [7], antiphagocytic protein SCIN [28]), yet integrate specifically into the beta-haemolysin gene, potentially offsetting the virulence afforded by carriage of the immune evasion genes. However, these phage are widely distributed in human isolates, but less common in animal isolates [18, 33], and not found in the ewe isolates of this study. The integration site of the prophage B is currently unknown as are the rest of the genes on this putative phage. Bacteriophages are also involved in the horizontal transfer of virulence genes between isolates, perhaps allowing for adaptation to new environmental conditions [38], and there is evidence that bacteriophage move during the course of human infections [12, 21]. A case of phage conversion of exfoliative toxin A in S. aureus isolated from cows with mastitis was documented [9]. The exfoliative toxin is the causative agent of staphylococcal scalded-skin syndrome in young children. The study suggested the possibility of horizontal transmission of eta gene by temperate bacteriophages among bovine isolates of S. aureus [9]. Under stressful conditions such as the use of antibiotics or UV light, prophage can be induced and cause lysis and death of bacterial populations, potentially decreasing pathogenic potential of a phage carrying strain. More recent data has suggested that phage could encode small RNA molecules that control gene regulation in S. aureus, and affecting virulence potential [24]. Therefore the role of bacteriophage in S. aureus pathogenesis is complex, and some phage may actually be a burden to the bacteria and reduce pathogenic potential.
The DNA array technologies used in this study were constructed from human S. aureus strains so it is possible there are differences between O11 and O46 that are not detected by these microarrays. However, recent studies suggest that animal isolates may only have a limited number of unique genes [14, 32] so one future direction for this study will be to implement massively parallel sequencing. The various typing methods used showed that the isolates O11 and O46 were genetically closely related. The major differences were in the presence/absence of alleles for sdrD, splE, fnbB, clfA-B and the presence of prophage genes (SAS0897, SAR1558, SAR0940, SAR2100 and SACOL0343). Furthermore, the sas typing between O11 and O46 (variations in sasA, sasB, sasD, and sasI) indicates minor variation in additional putative surface products, beside sdrD, fnbB and clfA-B, not revealed by the DNA microarray technologies. All surface expressed proteins may play a role in binding to specific host proteins and/or in immune recognition.
The sequence-based typing methods, spa and sas typing, were the easiest methods for comparing and could be used for short-term epidemiologic studies as well as long-term epidemiologic or phylogenic studies. The difference in the spa repeat between O11 and O46 shows that O11 had a deletion of only one repeat (r39) compared with O46, indicating that they are closely related and they belong to the spa-CC 1773 (Ridom SpaServer website) [37]. The 16 strains belong to the same spa-CC 1773 underscoring that the S. aureus presence in this flock is probably to link to a common ancestor. The MLVA typing provided the greatest level of discrimination of the isolates because 6 MLVA different patterns were found, which confirms its utility in epidemiological study in a given herd [10]. PFGE as a band-based, rather than sequence-based, typing method gave subtle differences between the isolates (Fig. 1) so was not easy to compare in all cases. The S. aureus exotoxin-like protein genes (ssl) did not exhibit significant allelic variability. Variation between individual isolates indicated that individual genes or the bacteriophage were capable of changes in a relatively stable background, suggesting local evolution of S. aureus from a common ancestor over an unknown period of time.
Altogether, with all the typing techniques used in this study (PFGE, MLVA, SNPssl, RAPD, spa typing, sas typing and by PCR for all the genes found different between O11 and O46) the strain O11 was found in the nares of four ewes (O193, O194, O195 and O196) 11 months after the gangrenous mastitis case. The ewes’s nares are an ecological niche for S. aureus and are important in the epidemiology of mastitis in dairy sheep farms [36]. So with this study we suggest that this body site could serve for transmission of potentially mastitis gangrenous strains in dairy sheep flock. The subclinical mastitis genotype was common in the flock, until a gangrenous genotype emerged and caused a gangrenous disease. Subsequently, the gangrenous genotype was found in the nares of ewes in the flock. But, we could not assert that the mastitis gangrenous case was a primary case with a new emerged virulent strain or if it appear after the nasal carriage had participated to the dissemination of the gangrenous strains in the flock. Maybe S. aureus in this flock has evolved to become more virulent. Evolution of S. aureus is driven by survival of the fittest in different environments, which include host interaction, antibiotic use, and environmental reservoirs, conditions which may fluctuate over time [19].
Although we cannot dismiss the role of host susceptibility in the clinical events in this flock, this study could support the hypothesis that different strains of S. aureus may have different virulence potential in ewe mastitis. Surprisingly, absence of a prophage and particular combinations of putative surface products occurred in the isolate with enhanced virulence. It will be useful to determine whether the evolution of S. aureus in this flock may be typical of S. aureus populations in general.
ONLINE MATERIAL
Supplementary on line material only Excel file provided by the author.
Acknowledgments
We acknowledge G. Rossi for excellent technical assistance. We acknowledge H. Meugnier (INSERM U851, Centre National de Référence des Staphylocoques, Lyon, France) for the spa typing and clustering. We also acknowledge BμG@S (the Bacterial Microarray Group at St. George’s, University of London) for supply of the microarray and advice and The Wellcome Trust for funding BμG@S under its Functional Genomics Resources Initiative. The support of the Réseau National GénopoleTM and the CNRS (Centre National de la Recherche Scientifique – National Center for Scientific Research) are greatly appreciated and we thank Dr P. Barbry. Caroline Le Marechal was recipient of a Ph.D. fellowship from the French National Institute for Agricultural Research (INRA) and the French Food Safety Agency (AFSSA). Part of this work was supported by INRA-AFSSA transversality project (IMISa project).
Footnotes
Bergonier D., personal communication.
References
- 1.Aguiar-Alves F., Medeiros F., Fernandes O., Gudziki Pereira R.M., Perdreau-Remington F., Riley L.W.. New Staphylococcus aureus genotyping method based on exotoxin (set) genes. J. Clin. Microbiol. 2006;44:2728–2732. doi: 10.1128/JCM.00376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amorena B., García de Jalón J.A., Baselga R., Ducha J., Latre M.V., Ferrer L.M.. et al. Infection of rabbit mammary glands with ovine mastitis bacterial strains. J. Comp. Pathol. 1991;104:289–302. doi: 10.1016/s0021-9975(08)80041-2. [DOI] [PubMed] [Google Scholar]
- 3.Bergonier D., de Cremoux R., Rupp R., Lagriffoul G., Berthelot X.. Mastitis of dairy small ruminants. Vet. Res. 2003;34:689–716. doi: 10.1051/vetres:2003030. [DOI] [PubMed] [Google Scholar]
- 4.Bor A., Winkler M., Gootwine E.. Non-clinical intramammary infection in lactating ewes and its association with clinical mastitis. Br. Vet. J. 1989;145:178–184. doi: 10.1016/0007-1935(89)90102-4. [DOI] [PubMed] [Google Scholar]
- 5.Brussow H., Hendrix R.W.. Phage genomics: small is beautiful. Cell. 2002;108:13–16. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 6.Callon C., Gilbert F.B., De Cremoux R., Montel M.C.. Application of variable number of tandem repeat analysis to determine the origin of S. aureus contamination from milk to cheese in goat cheese farms. Food Control. 2008;19:143–150. [Google Scholar]
- 7.De Haas C.J., Veldkamp K.E., Peschel A., Weerkamp F., Van Wamel W.J., Heezius E.C.. et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial anti-inflammatory agent. J. Exp. Med. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgeworth J.D., Yadegarfar G., Pathak S., Batra R., Cockfield J., Wyncoll D.. et al. An outbreak of methicillin-resistant Staphylococcus aureus (MRSA)-ST 239 associated with a high rate of bacteremia. Clin. Infect. Dis. 2007;44:493–501. doi: 10.1086/511034. [DOI] [PubMed] [Google Scholar]
- 9.Endo Y., Yamada T., Matsunaga K., Hayakawa Y., Kaidoh T., Takeuchi S.. Phage conversion of exfoliative toxin A in Staphylococcus aureus isolated from cows with mastitis. Vet. Microbiol. 2003;96:81–90. doi: 10.1016/s0378-1135(03)00205-0. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert F.B., Fromageau A., Gelineau L., Poutrel B.. Differentiation of bovine Staphylococcus aureus isolates by use of polymorphic tandem repeat typing. Vet. Microbiol. 2006;117:297–303. doi: 10.1016/j.vetmic.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Goerke C., Papenberg S., Dasbach S., Dietz K., Ziebach R., Kahl B.C., Wolz C.. Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J. Infect. Dis. 2004;189:724–734. doi: 10.1086/381502. [DOI] [PubMed] [Google Scholar]
- 12.Goerke C., Wolz C.. Regulatory and genomic plasticity of Staphylococcus aureus during persistent colonization and infection. Int. J. Med. Microbiol. 2004;294:195–202. doi: 10.1016/j.ijmm.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Haveri M., Taponen S., Vuopio-Varkila J., Salmenlinna S., Pyörälä S.. Bacterial genotype affects the manifestation and percistence of bovine Staphylococcus aureus intrammmary infection. J. Clin. Microbiol. 2005;43:959–961. doi: 10.1128/JCM.43.2.959-961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herron-Olson L., Fitzgerald J.R., Musser J.M., Kapur V.. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iandolo J.J., Worrell V., Groicher K.H., Qian Y., Tian R., Kenton S.. et al. Comparative analysis of the genomes of the temperate bacteriophages phi 11 phi 12 and phi 13 of Staphylococcus aureus 8325. Gene. 2002;289:109–118. doi: 10.1016/s0378-1119(02)00481-x. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn G., Francioli P., Blanc D.S.. Evidence for clonal evolution among highly polymorphic genes in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2006;188:169–178. doi: 10.1128/JB.188.1.169-178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lina G., Bohach G.A., Nair S.P., Hiramatsu K., Jouvin-Marche E., Mariuzza R.. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004;189:2334–2336. doi: 10.1086/420852. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay J.A., Moore C.E., Day N.P., Peacock S.J., Witney A.A., Stabler R.A.. et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 2006;188:669–676. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay J.A. Lindsay J.A. Staphylococcus: Molecular genetics. Caister Academic Press; Norfolk UK: 2008. S. aureus evolution: lineages and mobile genetic elements (MGE) pp. 45–69. [Google Scholar]
- 20.Meulemans L., Hermans K., Duchateau L., Haesebrouck F.. High and low virulence Staphylococcus aureus strains in a rabbit skin infection model. Vet. Microbiol. 2007;125:333–340. doi: 10.1016/j.vetmic.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Moore P.C.L., Lindsay J.A.. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 2001;39:2760–2767. doi: 10.1128/JCM.39.8.2760-2767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill E., Pozzi C., Houston P., Humphreys H., Robinson D.A., Loughman A.. et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantucek R., Doskar J., Ruzickova V., Kasparek P., Oracova E., Kvardova V., Rosypal S.. Identification of bacteriophage types and their carriage in Staphylococcus aureus . Arch. Virol. 2004;149:1689–1703. doi: 10.1007/s00705-004-0335-6. [DOI] [PubMed] [Google Scholar]
- 24.Pichon C., Felden B.. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popowicz G.M., Dubin G., Stec-Niemczyk J., Czarny A., Dubin A., Potempa J., Holak T.A.. Functional and structural characterization of Spl proteases from Staphylococcus aureus . J. Mol. Biol. 2006;358:270–279. doi: 10.1016/j.jmb.2006.01.098. [DOI] [PubMed] [Google Scholar]
- 26.Robinson D.A., Enright M.C.. Evolutionary models of emergence of methicillin-resistant Staphylococcus aureus . Antimicrob. Agents Chemother. 2003;47:3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Calleja J.M., Garcia-Lopez I., Santos J.A., Otero A., Garcia-Lopez M.. Molecular and phenotypic typing of Staphylococcus aureus isolates from rabbit meat. Res. Microbiol. 2006;157:496–502. doi: 10.1016/j.resmic.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Rooijakkers S.H., Ruyken M., Roos A., Daha M.R., Presanis J.S., Sim R.B.. Immun invasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 29.Rozen S., Skaletsky H.. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 30.Sabat A., Melles D.C., Martirosian G., Grundmann H., van Belkum A., Hryniewicz W.. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J. Clin. Microbiol. 2006;44:1135–1138. doi: 10.1128/JCM.44.3.1135-1138.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straub J.A., Hertel C., Hammes W.P.. A 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J. Food Prot. 1999;62:1150–1156. doi: 10.4315/0362-028x-62.10.1150. [DOI] [PubMed] [Google Scholar]
- 32.Sung J.M., Lloyd D.H., Lindsay J.A.. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008;154:1949–1959. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 33.Tenover F.C., McDougal L.K., Goering R.V., Killgore G., Projan S.J., Patel J.B., Dunman P.M.. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 2006;44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vancraeynest D., Hermans K., Haesebrouck F.. Genotypic and phenotypic screening of high and low virulence Staphylococcus aureus isolates from rabbits for biofilm formation and MSCRAMMs. Vet. Microbiol. 2004;103:241–247. doi: 10.1016/j.vetmic.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Vautor E., Abadie G., Guibert J.M., Huard C., Pepin M.. Genotyping of Staphylococcus aureus isolated from various sites on farms with dairy sheep using pulsed-field gel electrophoresis. Vet. Microbiol. 2003;96:69–79. doi: 10.1016/s0378-1135(03)00207-4. [DOI] [PubMed] [Google Scholar]
- 36.Vautor E., Abadie G., Guibert J.M., Chevalier N., Pepin M.. Nasal carriage of Staphylococcus aureus in dairy sheep. Vet. Microbiol. 2005;106:235–239. doi: 10.1016/j.vetmic.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Vautor E., Magnone V., Rios G., Le Brigand K., Bergonier D., Lina G.. et al. Genetic differences among Staphylococcus aureus isolates from dairy ruminant species: a single-dye DNA microaaray approach. Vet. Microbiol. 2008;133:105–114. doi: 10.1016/j.vetmic.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Wagner P.L., Waldor M.K.. Bacteriophage control of bacterial virulence. Infect. Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter A.. Mastitis in ewes. In Pract. 2001;23:160–163. [Google Scholar]
- 40.Witney A.A., Marsden G.L., Holden M.T., Stabler R.A., Husain S.E., Vass J.K.. et al. Design validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl. Environ. Microbiol. 2005;71:7504–7514. doi: 10.1128/AEM.71.11.7504-7514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary on line material only Excel file provided by the author.