Abstract
As one of the earliest structures to degenerate in Alzheimer’s disease (AD), the hippocampus is the target of many studies of factors that influence rates of brain degeneration in the elderly. In one of the largest brain mapping studies to date, we mapped the 3D profile of hippocampal degeneration over time in 490 subjects scanned twice with brain MRI over a 1-year interval (980 scans). We examined baseline and 1-year follow-up scans of 97 AD subjects (49 males/48 females), 148 healthy control subjects (75 males/73 females), and 245 subjects with mild cognitive impairment (MCI; 160 males/85 females). We used our previously validated automated segmentation method, based on AdaBoost, to create 3D hippocampal surface models in all 980 scans. Hippocampal volume loss rates increased with worsening diagnosis (normal=0.66%/year; MCI=3.12%/year; AD=5.59%/year), and correlated with both baseline and interval changes in Mini-Mental State Examination (MMSE) scores and global and sum-of-boxes Clinical Dementia Rating scale (CDR) scores. Surface-based statistical maps visualized a selective profile of ongoing atrophy in all three diagnostic groups. Healthy controls carrying the ApoE4 gene atrophied faster than non-carriers, while more educated controls atrophied more slowly; converters from MCI to AD showed faster atrophy than non-converters. Hippocampal loss rates can be rapidly mapped, and they track cognitive decline closely enough to be used as surrogate markers of Alzheimer’s disease in drug trials. They also reveal genetically greater atrophy in cognitively intact subjects.
Introduction
There is a great interest in developing powerful methods to track the progression of Alzheimer’s disease (AD) in the living brain. Clinically, patients develop worsening symptoms over a period of several months to years, with mild memory impairments initially, progressing to debilitating decline in all cognitive domains. The medial temporal lobe structures, including the hippocampus and entorhinal cortex, are among the first to degenerate, and atrophy spreads in a characteristic sequence mirroring the spread of neurofibrillary tangles (Apostolova et al., 2006a,b, submitted for publication) – the cellular hallmarks of AD pathology. The trajectory of atrophy typically spreads from the medial temporal and limbic areas, to parietal association areas, and finally to frontal and primary cortices only late in AD (Braak and Braak, 1991; Thompson and Apostolova, 2007; Thompson et al., 2004).
The hippocampus is the target of many neuroimaging studies of AD (Barnes et al., in press), as it is one of the earliest regions to degenerate structurally. Many studies have used sequential MRI scanning to estimate rates of hippocampal volume loss, either by manually tracing the hippocampus in every scan, or with automated segmentation methods (Khan et al., 2008; Morra et al., 2008b,c, submitted for publication), or by using some combination of manual landmark identification and automated extraction (Chupin et al., 2007; Hogan et al., 2000; Mueller et al., 2007). Several computational anatomy studies have also modeled the hippocampus as a geometrical surface in 3D, plotting statistics at each surface location regarding the rate or significance of atrophy, differences in atrophic rates between groups, or factors that resist or promote degeneration (Bansal et al., 2007; Csernansky et al., 2004; Thompson et al., 2004). Such surface-based approaches can visualize 3D profiles of atrophy in AD populations. As the hippocampus is highly differentiated functionally, selective hippocampal subregions can be identified, such as the CA1 and CA2 subfields, where atrophy occurs first and correlates with early cognitive decline (Csernansky et al., 1998; Thompson et al., 2004; Wang et al., 2003). Surface-based modeling studies have even identified local atrophic patterns that predict future decline (Apostolova et al., 2006b), and have charted the dynamic trajectory of atrophy as it spreads in the hippocampus (Apostolova et al., 2006b, in press, submitted for publication).
A key goal of all AD morphometry methods is to track longitudinal brain changes with high accuracy, providing numerical measures of disease burden that correlate with, or predict, cognitive decline. Many morphometric methods employ maps to track the disease, including tensor-based morphometry (Ashburner, 2007; Hua et al., 2008a,b; Leow et al., 2006; Studholme et al., 2004), cortical thickness mapping (Dickerson et al., in press; Fischl and Dale, 2000; Lerch et al., 2008; Salat et al., 2004; Thompson et al., 2004), ventricular mapping (Carmichael et al., 2007; Chou et al., 2008), voxel-based morphometry (Alexander et al., 2006; Ashburner and Friston, 2000; Baron et al., 2001; Chetelat et al., 2005; Karas et al., 2008; Teipel et al., 2005; Whitwell and Jack, 2005), and large deformation metric mapping (Miller et al., in press). Head-to-head comparisons and meta-analysis of these methods is an objective of several initiatives (Jack et al., 2008a). Optimal MRI-based measures for clinical trials are urgently required, and each method reflects different aspects of the disease process (Barnes et al., in press).
MRI can resolve the hippocampus at millimeter resolution. Even so, extracting the hippocampus from a whole-brain MRI scan is time-consuming, and usually requires the work of an expert. This is a rate-limiting step when one is attempting to associate clinical measures with rates of hippocampal atrophy in hundreds of subjects longitudinally. Some automated hippocampal segmentation methods have already been described (Barnes et al., 2004; Crum et al., 2001; Fischl et al., 2002; Hogan et al., 2000; Powell et al., 2008; Wang et al., 2007; Yushkevich et al., 2006), but it is fair to say that none is yet in wide use.
A promising approach for automated hippocampal segmentation is pattern recognition (Duda et al., 2001). In a prior paper, we developed a pattern recognition approach for hippocampal segmentation on MRI, which was shown to produce accurate, high-quality automated segmentations in 400 subjects from the Alzheimer’s Disease Neuroimaging Initiative (Morra et al., 2008a,b,c). The approach has a high throughput and is efficient to apply, and can be distinguished conceptually from other automated labeling approaches as it uses a powerful variant of AdaBoost, a ‘machine learning’ method that learns features to guide segmentation (Schapire et al., 1998). Our AdaBoost-based methods classify voxels as belonging to the hippocampus based on thousands of features, which are learned from a training set of manually delineated data (typically 20 hand-traced scans; see Morra et al. (2008c) for details). More specifically, we used a cascaded version of the AdaBoost algorithm (Freund and Schapire, 1997) wrapped inside a different algorithm that we call the auto context model (ACM), which updates the statistical prior distributions during the segmentation. Using that approach, our recent cross-sectional study found that 40 scans were sufficient to distinguish AD patients from controls (20 per diagnosis), using hippocampal maps, and we created several spatially detailed maps of associations between diagnosis, genotype, and depression, on baseline hippocampal structure (Morra et al., submitted for publication).
Here we used the same automated segmentation approach to analyze structural changes in the hippocampus over time, using follow-up scans of 490 subjects acquired 1 year apart. These scans were not available at the time of our first study (Morra et al., in press), but now allow us to assess factors that influence rates of degeneration as it progresses over time.
In this paper, we first automatically segment the left and right hippocampus from 490 individuals at both baseline and 1-year follow-up using our automated approach. We then use a statistical mapping approach to create parametric surface maps (Bansal et al., 2007; Csernansky et al., 1998; Styner et al., 2000; Thompson et al., 2004; Wang et al., 2007) that link the 3D profile of atrophy with various covariates of interest.
We had 3 main goals: (1) to map the 3D profile of tissue loss rates (as a percentage per year) in AD, healthy comparison subjects, and in subjects with mild cognitive impairment (MCI), an at-risk group of subjects with five-fold increased risk of conversion to AD in any given year (Petersen, 2000); (2) to plot regions where progressive atrophy was statistically associated with clinical decline on standardized tests, such as the mini-mental state exam (MMSE) (Folstein et al., 1975), and clinical dementia rating (CDR) (Morris, 1993), and (3) to determine whether loss rates in our subjects were associated with factors such as blood pressure measurements, depression scores, educational level, and the ApoE genotype (2, 3, or 4). As it requires more processing to create 3D maps than analyze hippocampal volumes, we also sought to identify what added value maps might provide over simple volumetric measures. For that reason, we present results based on both volumetric analysis and statistical mapping.
Materials and methods
Subjects
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005a,b) is a large multi-site longitudinal MRI and FDG-PET (fluorodeoxyglucose positron emission tomography) study of 800 adults, ages 55 to 90, including 200 elderly controls, 400 subjects with mild cognitive impairment, and 200 patients with AD. The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California – San Francisco.
Subjects with 1-year follow-up scans
The maps in this paper are based on baseline and 1-year follow-up scans of 97 AD subjects (49 males/48 females), 245 MCI subjects (160 males/85 females), and 148 healthy control subjects (75 males/73 females). Tables 1 and 2 show the clinical scores and demographic measures for our sample. Based on the available data in the ADNI database at the time of writing (September 2008), we used the largest possible sample that we could assemble who had both baseline and 1-year follow-up scans. To avoid possible issues with combining data across field strengths, we analyzed 1.5 T scan data only. Only a quarter of the subjects enrolled in ADNI are randomized to 3 T scans, so we will compare 3 T and 1.5 T in the future when sufficiently many 3 T follow-up scans become available. We also note that 6-month follow-up scans are available in ADNI, but here we focused on the 1-year follow-ups as we knew from prior work (Thompson et al., 2004) that they would allow sufficient time for deterioration in both cognition and structure for structure–function correlations to be readily established.
Table 1.
Demographic and clinical data are shown for the subjects, broken down into the 3 diagnostic groups, at the time of their baseline scan
| Age (years) | MMSE | Global CDR | Sum-of- Boxes CDR | ApoE | |
|---|---|---|---|---|---|
| Normal (N=148) | 75.92 (4.90) | 29.14 (0.93) | 0.0 (0.0) | 0.03 (0.11) | 19/87/39 |
| MCI (N=245) | 74.99 (7.17) | 26.98 (1.78)a | 0.5 (0.0)a | 1.55 (0.87)a | 11a/103/125a |
| AD (N=97) | 75.77 (7.33) | 23.39 (1.86)a | 0.74 (0.25)a | 4.19 (1.58)a | 3a/28/64a |
Standard deviations are in parentheses, and the p-value compares each diseased group with normals. For the ApoE column, the three numbers are the number of subjects that (1) carry at least one copy of the ApoE2 genotype, which is considered protective against AD; (2) are homozygous for the ApoE3 genotype and carry no copies of E2 or E4, and (3) carry at least one copy of the ApoE4 genotype, which is considered to be a risk factor for AD. As E2 is regarded as protective and E4 as a risk for AD, the very few subjects (N=11) who carried both E2 and E4 alleles (i.e., one of each) were excluded from the ApoE analysis. Because these 2/4-carriers were excluded, the genotype group sizes do not add up to the total sample size for each group. For comparing the ApoE status of groups we used a chi-squared test. As expected, the clinical measures reflect poorer performance in the MCI and AD groups versus controls.
p<0.01.
Table 2.
This table summarizes the change from baseline after 1 year for the same subjects whose baseline data is shown in Table 1
| Age (years) | MMSE | Global CDR | Sum-of-boxes CDR | |
|---|---|---|---|---|
| Normal | 1.078 (0.076)a | −0.027 (1.30) | 0.034 (0.161) | −0.010 (0.779) |
| MCI | 1.074 (0.064)a | −0.689 (2.57)b | 0.035 (0.197)a,b | 0.727 (1.26)a,b |
| AD | 1.072 (0.064)a | −1.88 (4.08)b | 0.201 (0.425)a,b | 1.42 (1.82)a,b |
Standard deviations are in parentheses, and the p-values compare each diseased group with normals. The The ApoE column is not reported here as it does not change with age.
p<0.01 – with respect to baseline measurements.
p<0.01 – with respect to normals.
Cognitive testing
All subjects underwent thorough clinical/cognitive assessment at the time of baseline scan acquisition, and again at follow-up 1 year later. As part of each subject’s cognitive evaluation, MMSE was administered to provide a global measure of cognitive status (Cockrell and Folstein, 1988; Folstein et al., 1975) where scores of 24 or less (out of a maximum of 30) are generally consistent with dementia. Two versions of the CDR were also used as a measure of dementia severity (Hughes et al., 1982; Morris, 1993). The global CDR represents the overall level of dementia, and a global CDR of 0, 0.5, 1, 2 and 3, respectively, indicate no dementia, questionable, mild, moderate, or severe dementia. The “sum-of-boxes” CDR score is the sum of 6 scores assessing different areas: memory, orientation, judgment and problem solving, community affairs, home and hobbies, and personal care. The sum of these scores ranges from 0 (no dementia) to 18 (very severe dementia). Memory impairment was assessed via education-adjusted scores on the Wechsler Memory Scale-Logical Memory II (WMS-R LM II; Wechsler, 1987). Subjects with MCI had objective memory loss measured by education-adjusted scores on the WMS-R LM II. AD diagnosis was made according to the National Institute of Neurologic and Communicative Disorders and Stroke and the AD and Related Disorders Association (NINCDS-ADRDA) criteria (McKhann et al., 1984). The normal subjects in our sample had MMSE scores between 25 and 30, a global CDR of 0, a sum-of-boxes CDR between 0 and 0.5 and failed to meet criteria for MCI or AD. The MCI subjects had MMSE scores ranging from 23 to 30, a global CDR of 0.5, a sum-of-boxes CDR score between 0.5 and 5, and mild memory complaints. The AD subjects in our sample had MMSE scores between 20 and 26, a global CDR between 0.5 and 1, and a sum-of-boxes CDR between 1.5 and 9.0. As such, these subjects would be considered as having mild AD. Detailed exclusion criteria, e.g., regarding concurrent use of psychoactive medications, may be found in the ADNI protocol (page 29, http://www.adni-info.org/images/stories/Documentation/adni_protocol_03.02.2005_ss.pdf). Briefly, subjects were excluded if they had any serious neurological disease other than incipient AD, any history of brain lesions or head trauma, or psychoactive medication use (including antidepressants, neuroleptics, chronic anxiolytics or sedative hypnotics, etc.).
The study was conducted according to Good Clinical Practice, the Declaration of Helsinki and U.S. 21 CFR Part 50-Protection of Human Subjects, and Part 56-Institutional Review Boards. Written informed consent for the study was obtained from all participants before protocol-specific procedures, including cognitive testing, were performed.
Training and testing sample descriptions
As previously stated, when using a pattern recognition algorithm, a set of ground truth data must be supplied in order to learn the patterns. In medical image segmentation, we typically use a small set of brains manually segmented by an expert, that represent a variety of hippocampi with features that are likely to be encountered when segmenting other brains. In prior studies of the asymptotic behavior of the segmentation accuracy versus the size of the training sample, we found that a training sample of size ~20 subjects’ scans was sufficient to give good segmentation accuracy, if scans were delineated by a trained rater following a standardized protocol with established inter-rater reliability as set out in Morra et al. (submitted for publication). Since we are performing a longitudinal study of patients of various ages, different sexes, and stages of disease, our training set should encompass as much variation as possible. Therefore, we trained our segmentation algorithm on 21 subjects using scans at both time points – baseline and 1 year (i.e., a total of 42 scans). The subjects were 7 AD, 7 MCI, and 7 healthy controls with age ranges and sex ratios that matched that of all other ADNI subjects. Using inter-rater reliability tests, we also found, in our validation studies (Morra et al., 2008c), that the effect of using different trained raters for training the algorithm is relatively small; intriguingly, the algorithm agrees with human raters (even those who were not involved in training it) as well as two independent human raters agree with each other, which is a reasonable benchmark for the target performance accuracy of a machine labeling a structure. For the “testing” subjects (i.e., scans not used for training but used as the basis for the results), we had 97 AD subjects (49 men/48 women), 245 MCI subjects (160 men/85 women) and 148 control subjects (75 men/73 women). Table 1 gives an overview of baseline statistics (including cognitive scores, genotype, and other demographics) and Table 2 gives an overview of the follow-up information 1 year later.
MRI acquisition and pre-processing
The MRI acquisition and pre-processing steps were exactly as in Morra et al., (2008b), but they are summarized here for completeness. All subjects were scanned with a standardized MRI protocol, developed after a major effort evaluating and comparing 3D T1-weighted sequences for morphometric analyses (Jack et al., 2008a; Leow et al., 2006).
High-resolution structural brain MRI scans were acquired at multiple ADNI sites using 1.5 T MRI scanners manufactured by General Electric Healthcare, Siemens Medical Solutions, and Philips Medical Systems. ADNI also collects data at 3.0 T from a subset of subjects, but, as noted earlier to avoid having to model field strength effects in this initial study, only 1.5 T images were used. All scans were collected according to the standard ADNI MRI protocol (http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml). For each subject, two T1-weighted MRI scans were collected using a sagittal 3D MP-RAGE sequence. Typical 1.5 T acquisition parameters are repetition time (TR) of 2400 ms, minimum full excitation time (TE), inversion time (TI) of 1000 ms, flip angle of 8°, 24 cm field of view, acquisition matrix was 192×192×166 in the x-, y-, and z-dimensions yielding a voxel size of 1.25×1.25×1.2 mm3 (Jack et al., 2008a). In-plane, zero-filled reconstruction (i.e., sinc interpolation) yielded a 256×256 matrix for a reconstructed voxel size of 0.9375×0.9375×1.2 mm3. The ADNI MRI quality control center at the Mayo Clinic (in Rochester, MN, USA) selected the MP-RAGE image with higher quality based on standardized criteria (Jack et al., 2008a). Additional phantom-based geometric corrections were applied to ensure spatial calibration was kept within a specific tolerance level for each scanner involved in the ADNI study (Gunter et al., 2006).
Additional image corrections were also applied, using a processing pipeline at the Mayo Clinic, consisting of: (1) a procedure termed GradWarp for correction of geometric distortion due to gradient non-linearity (Jovicich et al., 2006), (2) a “B1-correction”, to adjust for image intensity non-uniformity using B1 calibration scans (Jack et al., 2008a), (3) “N3” bias field correction, for reducing intensity inhomogeneity (Sled et al., 1998), and (4) geometrical scaling, according to a phantom scan acquired for each subject (Jack et al., 2008a), to adjust for scanner- and session-specific calibration errors. In addition to the original uncorrected image files, images with all of these corrections already applied (GradWarp, B1, phantom scaling, and N3) are available to the general scientific community, as described at http://www.loni.ucla.edu/ADNI. Ongoing studies are examining the influence of N3 parameter settings on measures obtained from ADNI scans (Boyes et al., 2008).
Image pre-processing
To adjust for global differences in brain positioning and scale across individuals, all scans were linearly registered to the stereotactic space defined by the International Consortium for Brain Mapping (ICBM-53) (Mazziotta et al., 2001) with a 9-parameter (9P) transformation (3 translations, 3 rotations, 3 scales) using the Minctracc algorithm (Collins et al., 1994). Globally aligned images were resampled in an isotropic space of 220 voxels along each axis (x, y, and z) with a final voxel size of 1 mm3. Both baseline and follow-up scans were independently registered to the same ICBM template.
Algorithm description
Throughout this paper we use an automated segmentation approach derived from AdaBoost to segment the hippocampus from a whole brain MRI. For a complete description of the algorithm including statistical motivations, please see our previous work (Morra et al., 2008c). We used the exact same feature database, along with the same method, a cascade of AdaBoosts wrapped inside of the auto context model (ACM), and the same model parameters. Fig. 1 gives an overview of AdaBoost, while Fig. 2 gives an overview of ACM.
Fig. 1.
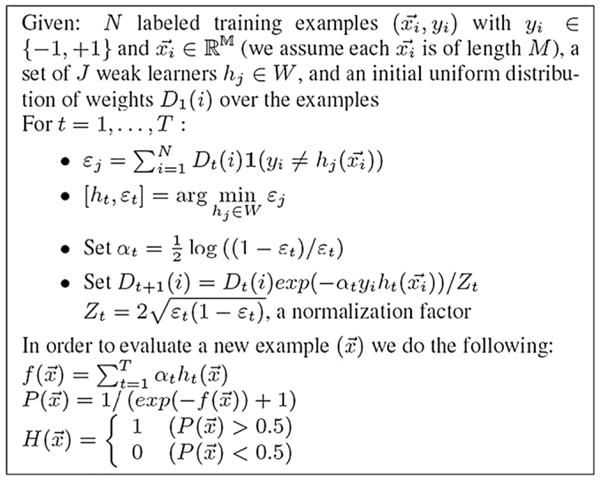
An overview of the AdaBoost algorithm. The x vectors are the feature vectors at each voxel (there are N voxels), and the y values are the ground truth classifications (+1 for hippocampus, −1 for non-hippocampus). Weak learners are defined to be classifiers that give binary outputs regarding a voxel’s class, based on one single feature and a threshold value for that feature. Weak learners are classification functions based on any feature that can help to classify a structure correctly with an accuracy slightly better than chance. The algorithm gives an update rule for the weightings given to each of the labeled examples, in training this set of weak learners, and the epsilon terms are the sum of the weights. The Dt vector represents the importance of each example, and examples misclassified at one iteration of the algorithm receive more weight on subsequent iterations, and those that are correctly classified receive less weight in the subsequent iterations. 1 is an indicator function, returning 1 if the expression is true and 0 otherwise. The function f is a function combining the outputs of all the weak learners using weights (the alpha terms). P is a probability function that gives the Bayesian maximum a posteriori (MAP) estimate of the labeling (Morra et al., 2008c). The H function thresholds the posterior distribution P at the threshold of 1/2, returning a decision as to whether each voxel x belongs to the hippocampus (1 for yes and 0 for no).
Fig. 2.

An overview of the auto context model. Here, H() is a cascade of AdaBoosts. Essentially, the labeling at each iteration of AdaBoost is fed back into the learning process as a new feature along with neighborhood-based information calculated on this map, which allows neighboring voxels to influence each other probabilistically. Convergence criteria and more details are presented in our previous work (Morra et al., 2008c).
Statistical maps
After segmentation was completed, we created various maps to link different disease-related factors with the rate of hippocampal atrophy over time. This is accomplished by first creating 3D parametric surface maps of each individual’s hippocampus; a medial curve is then threaded down the center of the hippocampus. For each surface model, a medial curve was derived from the line traced out by the centroid of the boundary for each hippocampal surface model (Thompson et al., 2004). The local radial size was defined as the radial distance between each boundary point and its associated medial curve. This radial measure may be interpreted as a local measure of thickness, and has been shown to be sensitive to the volumetric atrophy at different locations along the anterior to posterior axis. Both this method (Thompson et al., 2004) and others (Liu et al., 2008; Shi et al., 2007; Terriberry et al., 2007; Vaillant and Glaunes, 2005; Wang et al., 2007) are currently under development to link radial atrophy with covariates of interest.
The current method has been proven capable of detecting disease-related differences in AD (Apostolova and Thompson, 2007; Thompson et al., 2004), MCI (Apostolova et al., 2007; Becker et al., 2006; Morra et al., 2008c), pre-MCI (Apostolova et al., in press), frontotemporal dementia (Frisoni et al., 2006), and in other neurobiological or neuropsychiatric disorders such as epilepsy, depression, bipolar illness, psychopathy, and autism (Ballmaier et al., 2008; Bearden et al., 2008; Boccardi et al., submitted for publication; Lin et al., 2005; Nicolson et al., 2006; Ogren et al., submitted for publication).
Mapping atrophic rates
To create spatially detailed maps of atrophic rates, the parametric meshes imposed on each subject’s hippocampal anatomy were compared between baseline and follow-up scans, and at each surface point, a measure of the estimated change in radial distance over time was plotted as a percentage of the baseline value. As in prior work (Thompson et al., 2004), group average maps of hippocampal loss rates were computed by using the parametric grids to associate corresponding anatomy across subjects, pooling data on atrophic rates, by averaging the percentage loss rates across subjects (other definitions of average loss are possible, but this is a reasonable operational definition). These maps were color-coded and visualized separately for each hippocampus and for each group, prior to making statistical comparisons, as detailed next.
Mapping statistical effects on rates of atrophy
Rates of surface contractions over time were statistically compared between groups at equivalent locations using Student’s t-tests (1-tailed), and were correlated with different clinical characteristics including diagnosis, ApoE genotype, cognitive scores, and declines in these scores over the 1-year interval. Because multiple covariates were assessed, we divided them into two categories – those for which prior hypotheses and some prior evidence exist regarding the association (e.g., change in MMSE should link with change in volume), and others that were more exploratory (e.g., blood pressure may be associated with rates of volume loss); for the exploratory correlations, a Bonferroni correction for multiple comparisons was enforced, to avoid inflating the probability of false positive findings.
In each statistical map, a p-value was assigned to each surface point that associates the rate of change (in percent per year) in the radial distance at that point to the covariate of interest. For visualization, the associated p-values describing the uncorrected significance of these statistics were plotted onto geometric average hippocampi created from the surface models of all subjects involved in each statistical test.
Permutation and multiple comparisons correction
Overall p-values, for the spatial pattern of effects observed in each map, were computed using a permutation testing approach. Permutation methods measure the distribution of features in statistical maps that would be observed by accident if the subjects were randomly assigned to groups (Thompson et al., 2003) and provide a p-value for the observed effects that is corrected for multiple comparisons. All our permutation tests are based on measuring the total area of the hippocampus with suprathreshold statistics, after setting the threshold at p<0.01. To correct for multiple comparisons and assign an overall p-value to each p-map (Nichols and Holmes, 2002; Thompson et al., 2003), permutation tests were used to determine how likely the observed level of significant atrophy (proportion of suprathreshold statistics, with the threshold set at p<0.01) within each p-map would occur by chance (Thompson et al., 2003, 2004). The number of permutations N was chosen to be 100,000, to control the standard error SEp of the omnibus probability p, which follows a binomial distribution B(N, p) with known standard error (Edgington and Onghena, 2007). When N=8000, the approximate margin of error (95% confidence interval) for p is around 5% of p; to improve upon this, we ran 100,000 permutations, with 0.01 chosen as the significance level. We prefer to use the overall extent of the suprathreshold region, as we know that atrophy is relatively broadly distributed over the hippocampus, and a set-level inference is more appropriate for detecting diffuse effects with moderate effect sizes at many voxels, rather than focal effects with very high effect sizes (which would generally be better detected using a test for peak height in a statistical map).
When reporting permutation test results, one-sided hypothesis testing was used, i.e., we only considered statistics in which the AD or MCI group showed more rapid atrophy than the controls, in line with prior findings. Likewise, the correlations are reported as one-sided hypotheses, i.e., statistics are shown in the map where the correlations are in the expected direction, e.g., greater atrophy associated with lower MMSE scores, and with higher CDR scores.
Throughout this paper we report whether or not a linkage is detected between the local rate of hippocampal atrophy and different covariates of interest, including diagnosis (normal, MCI, AD), MMSE, global CDR, sum-of-boxes CDR, changes (over 1 year) in these scores, the ApoE genotype, depression severity assessed using the Geriatric Depression Scale (GDS) (Yesavage et al., 1982), systolic and diastolic blood pressure, plasma homocysteine levels, and educational level.
Results
Volumetric analyses
First we performed a volumetric analysis of overall hippocampal volumes. We aimed to confirm that the volumetric loss rate for each group was (1) progressively greater, in the order: control<MCI<AD, and (2) in the expected numerical range based on a recent MRI-based meta-analysis of hippocampal loss rates (Barnes et al., in press).
Fig. 3 shows that, in line with many prior longitudinal studies of AD versus controls (Jack et al., 1998, 2004) and versus MCI subjects (Jack et al., 2000, 2004), hippocampal tissue loss is detected in all 3 diagnostic groups. As noted in Fig. 3, the mean hippocampal loss rate increases with worsening diagnosis (AD=5.59%/year [95% confidence interval, CI: +/−1.44%]; MCI=3.12%/year [95% CI: +/−0.79%]; healthy controls=0.66%/year [95% CI: +/−0.96%]).
Fig. 3.
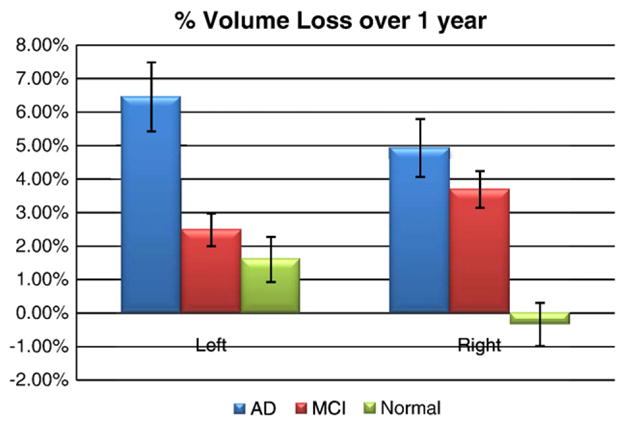
The percentage of hippocampal volume loss over a 1-year interval. As expected, the AD group had the most rapid tissue loss, followed by MCI and then normal subjects.
When comparing baseline volume to follow-up volume, all groups were losing tissue with statistical significance (p<0.01) except for the normals on the right side, where no significant change was detected. This is not entirely surprising as many prior studies report low or undetectable rates of loss in cognitively intact, healthy controls. These values are marginally higher than those reported in the meta-analysis by Barnes et al. (in press), who included nine studies from seven centers, with data from a total of 595 AD and 212 matched controls. In Barnes et al. (in press), mean (95% CIs) annualized hippocampal atrophy rates were found to be 4.66% (95% CI 3.92, 5.40) for AD subjects and 1.41% (0.52, 2.30) for controls. The difference between AD and control subjects in this rate was 3.33% (1.73, 4.94). These slight differences may depend on the anatomical definition of the hippocampus, or on the severity of dementia in the AD group. Also, the left/right asymmetry in the loss rates proved to be statistically significant (p<0.05) in the normal and MCI group, at least in this sample. Although a left/right difference in rates has been found before, what is surprising is that the asymmetry is reversed in MCI compared to controls (and compared to AD for that matter). This reversal was not hypothesized and requires independent confirmation; if true, it may suggest that the right hemisphere may lag behind the left in its rate of atrophy initially, but then may accelerate in MCI to exceed that on the left.
Clinical correlations
Next, we examined the correlations between annual hippocampal volume loss and annual change in MMSE, global CDR and sum-of-boxes CDR across the full sample and separately within each diagnostic group (Tables 3 and Table 4). All correlations were highly significant, and in the expected directions. Their absolute values were in the range |r|=0.10–0.23 for all measures. In the full sample with all diagnostic groups combined (N=490), there is a strong correlation between every clinical score and the rate of change in hippocampal volume. Even so, this is not surprising as there is a group difference in atrophic rate and the clinical data strongly correlate with group. It is more useful to know whether these correlations were maintained when the sample is broken down by diagnosis – Table 4 shows that correlations are generally maintained in the non-AD groups. This suggests that progressive hippocampal degeneration is more strongly correlated with clinical scores in individuals who have not yet lost a large proportion of their hippocampus (non-AD subjects). Also, the power to detect these correlations depends on the sample size, and the MCI sample size was almost twice as large as the number of AD subjects.
Table 3.
Correlations (Pearson’s r values) associating clinical covariates (and changes in the same clinical covariates over 1 year) and the rates of hippocampal atrophy in percent per year
| Left | Right | N | |
|---|---|---|---|
| MMSE | −0.191a | −0.168a | 490 |
| Change in MMSE | 0.117a | 0.136a | 490 |
| gCDR | 0.134a | 0.225a | 490 |
| Change in gCDR | −0.171a | −0.100b | 490 |
| sobCDR | 0.173a | 0.181a | 490 |
| Change in sobCDR | −0.174a | −0.171a | 489 |
Since higher MMSE scores tend to indicate a more cognitively intact individual, the observed negative correlation, in the case of MMSE, is to be expected. The opposite is true for both of the CDR measurements (global and sum-of-boxes CDR scores), where higher scores indicate greater cognitive impairment. For the change in a score, one would expect an opposite correlation also (i.e., more change in MMSE would be expected to be associated with a greater rate of loss in the hippocampus), and this is borne out by all the data; all of these correlations are in the expected directions. One subject did not have a sum-of-boxes CDR score at follow-up, so we included 489 subjects for that one measure.
p<0.01.
p<0.05.
Table 4.
Correlations (Pearson’s r values) between clinical covariates (and change over 1 year) and the rates of hippocampal atrophy in percent per year
| AD |
N | MCI |
N | Normal |
N | ||||
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | Left | Right | Left | Right | ||||
| MMSE | −0.149 | 0.021 | 97 | 0.017 | 0.023 | 245 | −0.144a | −0.149a | 148 |
| Change in MMSE | 0.039 | 0.098 | 97 | 0.195b | 0.133a | 245 | −0.184a | −0.022 | 148 |
| gCDR | −0.077 | 0.015 | 97 | N.A. | N.A. | 245 | N.A. | N.A. | 148 |
| Change in gCDR | −0.193a | −0.104 | 97 | −0.099 | −0.072 | 245 | −0.059 | −0.040 | 148 |
| sobCDR | 0.031 | 0.019 | 97 | −0.006 | 0.011 | 245 | 0.013 | 0.062 | 148 |
| Change in sobCDR | −0.093 | −0.150 | 97 | −0.131a | −0.112a | 245 | −0.144a | 0.023 | 147 |
N.A. (not applicable) is reported for the global CDR in the MCI and AD groups because all MCI and AD patients had the same global CDR values (0.5 and 0 respectively). One normal subject did not have a sum-of-boxes CDR follow-up score, hence the 147 subjects for that one measure.
p<0.05.
p<0.01.
Genetic influences on volumetric loss rates
An active area of AD research focuses on whether the ApoE4 genotype predicts future mental decline, whether the ApoE2 genotype is protective against dementia, and whether the characteristic morphometric signatures of AD (such as profiles of loss rates) differ by genotype. Answering this question has practical value, because a drug may be more effective in patients with a certain genotype, or, it may be easier to detect a drug effect in genetic subgroups for which the annual rate of decline is greater. For all experiments involving ApoE, individuals with the 2/4 genotype (one protective and one high-risk allele) were excluded because it is not clear how to group these subjects. First, Table 5 shows that there is a highly significant correlation between the ApoE4 genotype and the rate of decrease in hippocampal volume, when all individuals are pooled together. This is somewhat expected, as individuals with ApoE4 were more likely to have AD (Table 1). As such, this correlation is not as interesting, as it simply reflects the over-representation of E4-carriers in the AD group. What is more interesting is that this same correlation (on the left side; Table 5, top row) was still detected in the healthy normal group. This suggests that even cognitively intact individuals carrying ApoE4 have faster hippocampal loss. In groups split by diagnosis, E4-carriers lost hippocampal tissue faster than non-carriers, except in the case of the right hippocampus in MCI (Table 4).
Table 5.
Associations (Pearson’s r) ApoE genotype and hippocampal atrophy rates
| Left | Right | N | |
|---|---|---|---|
| ApoE 3v4 NC | 0.164a | 0.120 | 126 |
| ApoE 3v4 MCI | 0.131a | −0.001 | 228 |
| ApoE 3v4 NC & MCI | 0.144b | 0.085 | 354 |
| ApoE 3v4 All | 0.187b | 0.111b | 446 |
ApoE4 carriers had faster atrophic rates in the left hippocampus in all groups, and effect sizes were generally greater for the left hippocampus. Even healthy ApoE4 carriers showed faster decline in hippocampal volume (at least on the left).
p<0.05.
p<0.01.
It has also been hypothesized that ApoE2 is a protective factor against the development of AD; in prior studies with a related morphometric technique, called tensor-based morphometry, we found that ApoE2 gene carriers – 1/6 of the normal group – showed reduced ventricular expansion, suggesting a protective effect (Hua et al., 2008b). Here we found no significant effects of ApoE2, perhaps due to the low number of individuals with the ApoE2 genotype (Table 6). The association between ApoE2 and left hippocampal loss rates in normals was just outside of trend level (p=0.109), but may be detectable in an even larger sample (Fig. 4).
Table 6.
Correlations (Pearson’s r) between atrophic rates and having the ApoE2 allele (2/3, 3/2, 2/2) versus the ApoE3 allele (3/3)
| Left | Right | N | |
|---|---|---|---|
| ApoE 2v3 NC | 0.121 | 0.054 | 106 |
| ApoE 2v3 MCI | −0.065 | 0.000 | 114 |
| ApoE 2v3 NC and MCI | 0.024 | 0.057 | 220 |
| ApoE 2v3 All | −0.022 | 0.052 | 251 |
None of these values is significant, and the correlation with atrophic rates in the left hippocampus in normals is just outside trend level (p=0.109). In a future studies with more ApoE2 subjects, correlations may be detectable (Hua et al., 2008b).
Fig. 4.
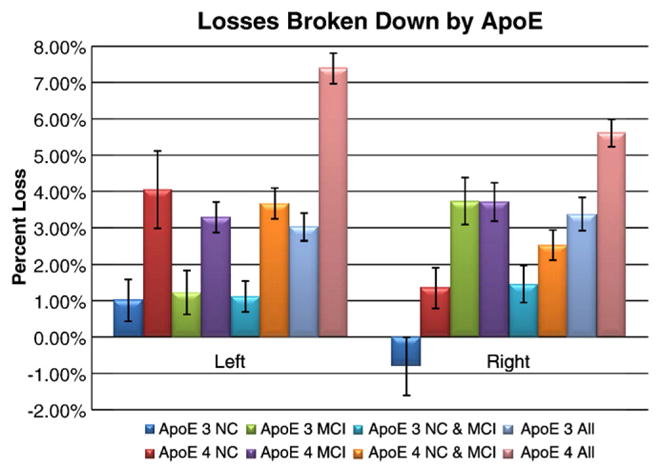
Hippocampal loss rates broken down by ApoE genotype. Values here correspond to Table 5; for all cases except MCI on the right side, ApoE4 carriers lost hippocampal volume more rapidly than non-carriers. One surprising aspect here is the left/right difference in the effect of ApoE4 in MCI: as seen in Fig. 3, the MCIs have right faster than left atrophy, overall. Within this MCI group, the E3 carriers (green bars) have right faster than left atrophy, but the E4 carriers (purple bars) have atrophy on the left that is almost as fast as on the right. Most investigators would be surprised to see a lateralized effect of ApoE, but it remains possible that the left hippocampus, which atrophies significantly faster than the right hippocampus in controls, maintains its rate of atrophy in MCI E3 carriers, but accelerates its rate of atrophy in MCI E4 carriers. If this is true, then the accelerative effect of the ApoE4 risk genotype may not be simply superimposed on the rate of atrophy in all groups uniformly in both hemispheres, but may accelerate the pregression of left hippocampal atrophy more than the right in MCI. As these asymmetries in the ApoE4 effect were not expected, they require independent replication.
Other factors associated with hippocampal volume loss rates
In addition to severity of disease measures and ApoE genotype, we investigated associations between other clinical factors and hippocampal loss rates. We chose to examine conversion from MCI to AD (treated as a binary variable over the 1-year follow-up interval), the Geriatric Depression Rating scale (GDS) (Yesavage et al., 1982), homocysteine levels, baseline systolic and diastolic blood pressure, and years of education. As multiple measures were tested, these correlations should be regarded as exploratory and were subjected to a Bonferroni-type correction for multiple comparisons. Table 7 shows the correlation between these covariates and atrophic rates in all subjects tested; Table 8 shows them broken down by diagnosis. First, those MCI subjects who converted to AD showed a faster rate of reduction in hippocampal volume on the right side. This effect, only significant on the right side, is consistent with an independent study (Apostolova et al., in press), in which we found that the right CA1 radial distance is predictive (increases the hazards) of conversion to AD in MCI. None of the other correlations were significant in the whole sample; some correlations were detected in the subgroups broken down by diagnosis. Although failure to detect a correlation does not mean that there is no correlation in the overall population, the fact that correlations were not detectable in a sample of N=490 subjects scanned twice suggests that effects of these covariates on hippocampal loss rates is likely to be relatively small, or that a longer follow-up interval, more than 1 year, or a more accurate measurement method, may be required to detect their influence on hippocampal degeneration.
Table 7.
Pearson’s r values associating a decline in volume (hippocampal loss rates in percent per year) with conversion from MCI to AD, geriatric depression (GD) scores, change in GD scores, homocysteine level (Homocys), systolic blood pressure, diastolic blood pressure, and years of education
| Left | Right | N | |
|---|---|---|---|
| MCI to AD | 0.093 | 0.130a | 245 |
| GDS | −0.020 | 0.063 | 490 |
| GDS change | −0.114 | −0.070 | 490 |
| Homocysteine | −0.044 | 0.002 | 490 |
| Systolic BP | 0.012 | −0.033 | 490 |
| Diastolic BP | 0.058 | −0.007 | 490 |
| Education | −0.060 | −0.064 | 490 |
The MCI to AD converters were the only type of conversion tested because there were too few subjects with any other types of change in diagnosis. Over our 1-year interval, 47 subjects converted from MCI to AD, which is marginally higher than the annual conversion rate typically reported for MCI. The significant association with conversion is in line with prior hypotheses.
p<0.05.
Table 8.
Correlations (Pearson’s r) between the loss rate in hippocampal volume and various biological and environmental covariates broken down by diagnosis
| AD |
N | MCI |
N | Normal |
N | ||||
|---|---|---|---|---|---|---|---|---|---|
| Left | Right | Left | Right | Left | Right | ||||
| GD baseline | 0.071 | 0.030 | 97 | −0.069 | 0.030 | 245 | −0.135 | −0.055 | 148 |
| GD change | −0.190a | −0.122 | 97 | −0.127a | −0.068 | 245 | −0.018 | 0.017 | 148 |
| Homocys | 0.073 | −0.018 | 97 | −0.114a | −0.066 | 245 | −0.011 | 0.105 | 148 |
| Systole | −0.003 | −0.219a | 97 | 0.052 | −0.007 | 245 | −0.051 | 0.030 | 148 |
| Diastole | 0.010 | −0.093 | 97 | 0.060 | 0.022 | 245 | 0.091 | −0.017 | 148 |
| Education | −0.131 | 0.036 | 97 | −0.021 | −0.038 | 245 | 0.039 | −0.138a | 148 |
None of these associations proved significant when all 3 diagnostic groups were combined (Table 7), but some were significant in groups split by diagnosis. Given that 18 statistical tests were performed, a Bonferroni correction should be applied (0.05/18), so these linkages should be regarded as exploratory due to the large number of covariates tested, and require independent confirmation. No association exceeded p=0.01.
p<0.05.
3D hippocampal maps
Linking volumetric measurements to clinical covariates is interesting, but fails to give a picture of how the hippocampus is degenerating, and which specific regional changes link with worsening cognition. Therefore in this section we present a complementary analysis to the volumetric measures using 3D statistical maps (p-maps). First, percent change maps are presented, broken down by diagnosis, to show how the hippocampus is changing. Next, we have investigated, in 3D, the correlations between disease severity and other clinical covariates and atrophic rates. The global significance levels for all p-maps, adjusted for multiple comparisons, were established by permutation testing (Table 9).
Table 9.
Factors associated with hippocampal loss rates
| Left | Right | |
|---|---|---|
| AD baseline vs. follow up | 0.0091 | 0.0006 |
| MCI baseline vs. follow up | 0.0018 | 0.0001 |
| Normal baseline vs. follow up | 0.0001 | 0.0003 |
| AD vs. normal | 0.0022 | 0.0161 |
| AD vs. MCI | 0.0741 | 0.4712 |
| MCI vs. Normal | 0.1788 | 0.1943 |
| MMSE | 0.0002 | 0.0231 |
| Global CDR | 0.0024 | 0.0421 |
| Sum-of-boxes CDR | 0.0003 | 0.0646 |
| Change in sum-of-boxes CDR | 0.0484 | 0.1027 |
| Years of education | 0.3335 | 0.0051 |
| ApoE3 vs. 4 (Normals) | 0.3532 | 0.2411 |
| ApoE3 vs. 4 (MCI) | 0.3042 | 0.7694 |
| ApoE3 vs. 4 (Normals and MCI) | 0.1292 | 0.8491 |
| ApoE3 vs. 4 (All) | 0.0071 | 0.4886 |
Overall significance levels for all the p-maps shown, after correction for multiple comparisons by permutation testing. Values below 0.05 are shown in bold font, and below 0.01 are shown in bold italics. All changes over time are significant, and the rate of change is statistically faster in AD than in controls. Lower baseline MMSE and CDR scores were associated with faster loss rates over the subsequent year. The ApoE4 gene was associated with faster atrophic rates on the left, and more years of education were associated with slower atrophic rates on the right.
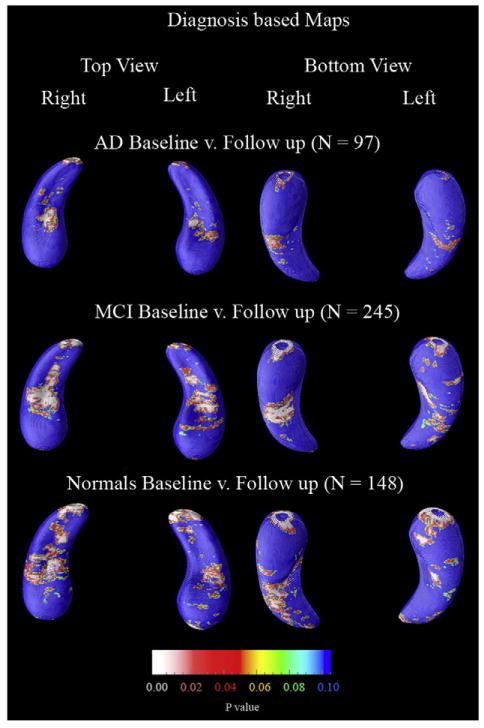
The first statistical maps to look at are the correlations between baseline and follow up. Fig. 5 shows that for all three diagnostic groups baseline hippocampi were significantly different from follow up, which is to be expected due to the volumetric results of Fig. 3.
Fig. 5.
Statistical maps (p-maps) showing the significance of progressive atrophy over 1 year broken down by diagnosis. All maps showed significant changes overall by permutation testing (Table 9).
Diagnostic and clinical maps
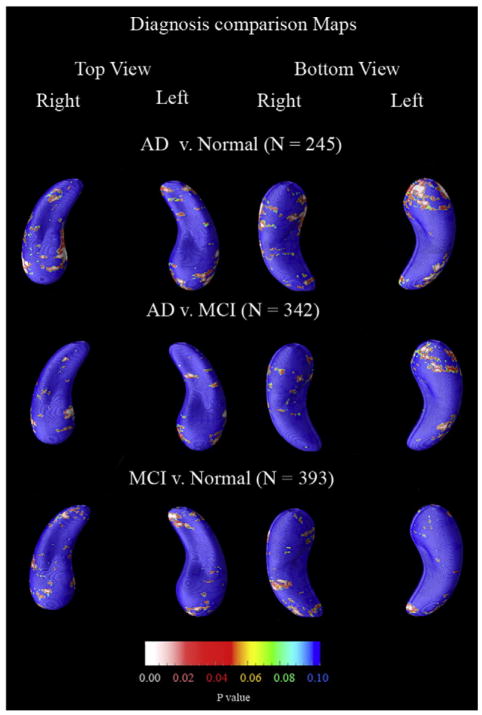
Between-group comparisons of hippocampal loss rates and correlations with various other clinical covariates are shown in Fig. 6 and Fig. 7 respectively.
Fig. 6.
P-maps showing between-group differences in atrophic rates. Overall only the two most diagnostically different groups proved to be significantly different (AD vs. Normals) as shown in Table 9.
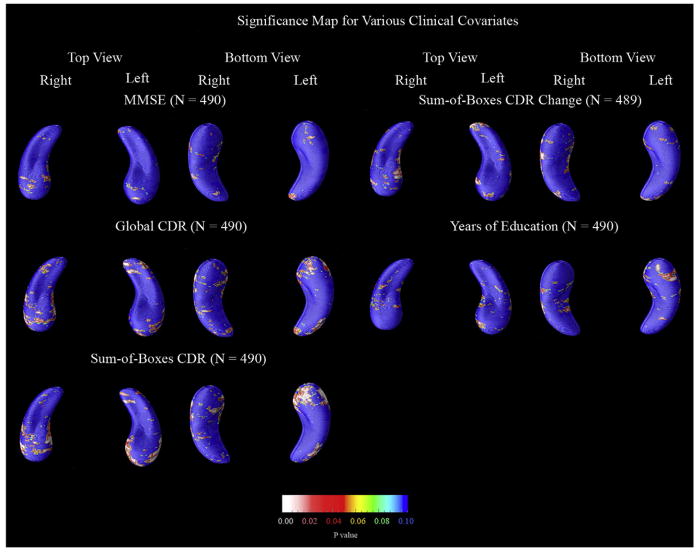
Fig. 7.
Maps of clinical and demographic factors that could be associated with hippocampal atrophy rates. These maps show the significance of correlations between a change in radial distance at each hippocampal surface point and various clinical covariates. Each map presented here was statistically significant, on at least one side, after multiple comparisons correction by permutation testing (Table 9). Maps are shown for all clinical covariates that proved significant, among those examined in this study. The maps for the sum-of-boxes CDR change were based on only 489 subjects (one less than the other maps) because one subject’s sum-of-boxes CDR score was not obtained at follow-up.
First, Fig. 6 shows differences in atrophic rates between the two most diagnostically dissimilar groups (AD and normals); AD patients lost hippocampal tissue more rapidly than normals, in line with our volumetric results (Fig. 3). No other comparisons proved significant by permutation testing, although there was a trend on the left for faster loss rates in AD v. MCI groups (p=0.07; Table 9), suggesting an acceleration in the loss rate between these stages. Consistent with this, AD and MCI groups showed a greater difference in hippocampal volume on the left side than the right side (Fig. 3).
Next, Fig. 7 shows the statistically significant correlations between atrophic rates and various clinical covariates. The four clinical tests (MMSE, global CDR, sum-of-boxes CDR, and change in sum-of-boxes CDR) showed correlations with numerically greater effect sizes on the left side than the right suggesting either that the left hippocampus is changing more as the disease progresses, or that such changes have a greater impact on cognition. When educational level was used as the covariate, years of education were linked with atrophic rates for the right hippocampus only. The correlation between education and hippocampal loss rates was only detected in the p-maps, and not in the overall volumetric analysis, underscoring the fact that volumetric comparisons and p-maps may reveal different linkages, but requiring future confirmation in independent samples.
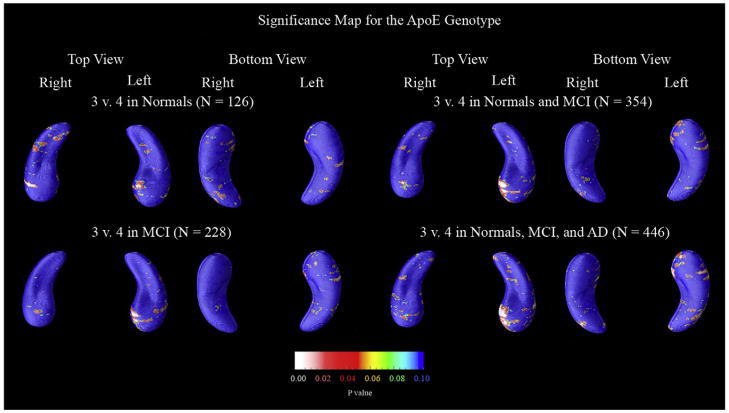
ApoE maps
Our last set of p-maps, in Fig. 8, show hippocampal regions where atrophic rates differed according to the presence of the ApoE4 gene. Only when all subjects are pooled was the map significant (and only on the left side). However, as noted before, this is not an especially meaningful association as ApoE4 carriers are present in higher proportions among those with AD than among controls, so that over-representation in the disease group alone would explain the gene effect on rates of atrophy. What is more interesting is whether ApoE4 carriers show faster atrophic rates if we split up the groups diagnostically – and none of the split maps proved significant. Nevertheless the same area showed genotype effects at the voxel level in each diagnostic split (see the top of the left hippocampal head of both the control group and MCI group). Because this same area appears to be correlated with hippocampal decline in all diagnostic groups, it may serve as a region of interest for independent confirmation in future studies.
Fig. 8.
P-maps of the effect of ApoE4 status on hippocampal loss rates. For these maps, subjects that had a genotype of 3/3 were categorized as ApoE3, and those that had 3/4, 4/3, or 4/4 were considered as ApoE4. Only when all subjects are pooled is the gene effect significant – on the left side. Even so, the same area of the hippocampus has low p-values in all groups (even when normal and MCI patients are split), suggesting that this area may be a region of interest for gene effects in future samples.
We also attempted to show that ApoE2 was a protective factor, but no maps proved significant, perhaps due to the small number of ApoE2 individuals in this study.
To better understand whether the relatively faster atrophic rates in the healthy ApoE4 risk gene carriers was simply because the ApoE4 carriers in the group were more cognitively impaired than the non-carriers, we ran a logistic regression to determine what percentage of the variation in baseline MMSE scores was explainable based on the subjects’ genotype. Intriguingly, in the healthy controls, genotype did not explain a significant proportion of variance in baseline MMSE scores (r=0.049; p=0.58; N=126), nor was there any association between genotype and baseline MMSE scores in the MCI group (r=0.08; p=0.23; N=228) or in the AD group (r=0.015; p=0.88; N=92).
This means that the ApoE4-carrying controls had significantly faster atrophic rates than non-carriers even without any detectable cognitive differences on the MMSE test at the time of the baseline scan. This should underscore the potential relevance of genetic testing in identifying those with genetically accelerated but latent neurode-generation especially at a time when no sign of accelerated decline can be detected on a global cognitive test such as the MMSE.
Discussion
Here we presented the largest hippocampal mapping study to date (using 980 scans from 490 subjects). There were four main findings. First, our results with an automated analysis technique agreed well with prior studies in smaller samples, performed using manual or other relatively labor intensive methods. We found that the mean hippocampal volume loss rates increased with worsening diagnosis (AD: 5.59%/year; MCI: 3.12%/year; normal: 0.66%/year). Our estimated hippocampal loss rates in AD are a little high compared to some other studies (Du et al., 2003; Jack et al., 2000, 1998), but are nonetheless close to the average rates reported in a recent meta-analysis. Barnes et al. (in press) included nine studies from seven centers, pooling data from a total of 595 AD and 212 matched controls. They found that mean (95% CIs) annualized hippocampal atrophy rates were 4.66% (95% CI 3.92, 5.40) for AD subjects and 1.41% (0.52, 2.30) for controls. Factors that might affect these rates include differences in anatomic delineation criteria — for example, some protocols include the hippocampal tail and others do not. Also, if hippocampal loss rates truly accelerate as the disease progresses, it is plausible that any study of AD patients who are more severely affected may find a greater mean rate of atrophy. Even so, this is not a compelling argument in our case, as our AD patients are relatively mildly impaired and yet show atrophic rates slightly higher than the average rate in the Barnes et al. meta-analysis. Second, all of our change measures were correlated with MMSE, global and sum-of-boxes CDR, and with changes in these measures. All such correlations were in the anticipated directions. The automated measures satisfied a further criterion expected of a reliable measure of disease progression — reliable correlation with cognitive decline. Third, we found that ApoE4 carriers had faster rates of volume loss, both in the full sample of 490, and when only cognitively intact controls were examined. This finding may have a practical benefit for treatment trials, because samples pre-selected to over-represent E4 carriers may show greater interval changes on MRI and may therefore be better powered to detect a statistical reduction in loss rates (Jack et al., 2008b). In addition, one might argue that those carrying the risk gene are better candidates for early intervention as an accelerated disease process is already detectable on MRI, at least at the group level. Even so, the case for using ApoE4 as an enrichment criterion in addition to using information on baseline cognitive deficits also depends on whether the knowledge of a person’s genotype provides added information for predicting atrophic rates relative to what can be predicted from cognition alone; this requires further study. Fourth, of the 245 MCI subjects examined, those who converted to AD during the 1-year evaluation period had greater loss rates than non-converters.
We presented several analyses using both hippocampal volumes and 3D significance maps, to determine whether maps provide more information than simple volumetric summaries. For the main effects, both maps and volumes gave similar results, for example regarding the changes over time in all 3 diagnostic groups, and the correlations with MMSE, CDR scores and interval changes in the scores. The map data also showed similarly localized regions of ongoing hippocampal loss in controls, MCI and AD groups (Fig. 5).
The cases where maps and volumes gave different answers were typically when detecting effects of subtle factors that were associated with loss rates with borderline significance. In the full sample, a higher educational level was associated with slower right hippocampal loss rates (p=0.0051), an effect detected in the maps but not in the volumetric analysis. This significance level is borderline when corrected for multiple comparisons (0.0051×10=0.051), so it requires independent replication.
ApoE4 gene carriers showed faster loss rates in the volumetric analysis, even in controls, but these effects did not reach the corrected significance threshold in the maps (after correction for multiple comparisons by permutation testing). Even so, local effects of genotype were consistently found in the same regions in controls, MCI and AD groups, providing a possible search region for future studies of gene effects.
A major surprisewas that the maps comparing atrophy rates between diagnostic groups did not differentiate MCI from controls. The volumetric analyses showed that atrophy rates increased in the order CTL, MCI, AD, and all groups showed progressive loss (except for normals on the right side). While both the volumetric and the 3D analyses showed that AD subjects have significantly greater atrophy rates vs. controls, only the volumetric data showed a difference between MCI and controls. While maps tend to outperform volumetric summaries when detecting effects that are relatively concentrated or localized, as the most affected subregions may show higher effect sizes than an overall volumetric measure, maps may not outperform overall numerical summary measures when the effects are relatively diffuse. Numeric summaries such as hippocampal volume often involve an implicit averaging that may counteract any highly localized boundary segmentation errors, which can deplete the power of maps in the same regions. In addition, the inability to distinguish MCI from controls based on the maps alone may reflect the clinical heterogeneity of the MCI group, as a combination of various conditions in a sample likely yields a regionally diffuse mean atrophy pattern for the group as a whole. This highlights the need for both volumetric measures and map-based statistical analysis.
To obtain the most powerful statistics from map-based statistical analysis, it may be beneficial in the future to use these thresholded maps as search regions of interest for factors that influence the rates of atrophy. By focusing on the regions that are changing the most, it may be possible to statistically define surface subregions of interest in training samples to develop more powerful or precise measures of hippocampal atrophy in future independent samples. In this sense, this is the same logic as using AdaBoost to label the hippocampus, by combining classifiers with the greatest capacity for error reduction.
Some comparison is warranted between this approach and other methods for hippocampal mapping. Other hippocampal mapping studies by our group have contrasted the atrophy patterns in Lewy Body dementia, vascular dementia (Scher et al., in press), amnestic MCI (Becker et al., 2006), fronto-temporal dementia (Frisoni et al., 2006), and in those at genetic risk for AD (Boccardi et al., 2004) revealing morphometric signatures characteristic of each condition. These prior reports used the same surface parameterization methods and radial distance measures as were used in this report (statistics of radial atrophy), but the studies relied on manual tracing rather than automated segmentation, which greatly accelerates the rate at which scans can be analyzed.
Our work is related to that of Styner, Gerig, and Yushkevich on M-reps (medial representations) (Styner et al., 2003, 2005; Yushkevich et al., 2005), in which a medial curve is defined through a structure, and distances of boundary points to the centerline are examined. The radial atrophy mapping technique is robust to small rotational or translational errors in registering the images across time, as the radial distances are always measured with respect to a central line threading down the center of the structure. Other methods, such as voxel-based morphometry, for example, may incorrectly pick up global shifts of the hippocampus as compressions or expansions inside the hippocampus, as the nonlinear deformations that register structures are spatially regularized (smooth) and may not be precise enough to register the hippocampal boundaries exactly. As radial atrophy is measured with respect to the medial curve, the distance measure reflects the thickness of the hippocampus in a given section, and it is not exactly the same as a volume difference. For example, any anterior-to-posterior shortening of the hippocampus would be detected by a volume measure, but the radial distance measure is not sensitive to this type of change — it only measures the radial thickness of the structure relative to a centerline. This may in general be considered a benefit rather than a limitation, as there are some variations across normal subjects in the anterior–posterior extent of the hippocampus, and these variations will be discounted by the radial mapping approach and will not be a source of confounding variance. Even so, discounting information on the anterior–posterior extent of the hippocampus may not be beneficial when measuring within-subject rates of change, unless the benefit of the information is outweighed by variations in segmentation accuracy across time that tend to cause errors in reproducibly defining the posterior limit of the hippocampus.
A related approach using large-deformation diffeomorphic metric mapping (Joshi and Miller, 2000) has been used to deform labeled anatomical templates of the hippocampus onto new images, using a combination of manual landmarking of points on the hippocampus and 3D fluid image registration (Csernansky et al., 2000; Haller et al., 1996; Wang et al., 2007). The surface of the hippocampus was parcellated a priori on a neuroanatomical template into three zones approximating the locations of underlying subfields, and Large Diffeomorphic Metric Mapping (LDDMM) was used to generate the hippocampal surfaces of all subjects and to register the surface zones across subjects. In a cross-sectional study, Wang et al. (2006) found that inward deformities of the hippocampal surface in proximity to the CA1 subfield and subiculum may be used to distinguish subjects with questionable AD from nondemented subjects. In a longitudinal study similar to ours, Wang et al. (2003) used LDDMM to analyze hippocampal change over time in 18 subjects with questionable AD (CDR 0.5) and 26 age-matched nondemented controls (CDR 0) scanned 2 years apart. In CDR 0 subjects, they observed shape changes between baseline and follow-up largely confined to the head of the hippocampus and subiculum, while in the CDR 0.5 subjects, shape changes involved the lateral body of the hippocampus as well as the head region and subiculum. In a subanalysis of 9 subjects from the same sample, who converted from the nondemented (CDR 0) to the questionable AD (CDR 0.5) state, Qiu et al. (2008) found that compared to the non-converters, the lateral aspect of the left hippocampal tail showed inward surface deformation in the converters. With a similar method, Csernansky et al. (2005) found that inward deformation of the left hippocampal surface in a zone corresponding to the CA1 subfield is an early predictor of the onset of DAT in nondemented elderly subjects. This is consistent with our finding of more rapid hippocampal loss rates in MCI converters than non-converters. Surface-based maps may be used in the future to define hippocampal subregions where changes predict imminent cognitive decline or the onset of dementia. Using the radial distance method we have likewise demonstrated that CA1 and subicular atrophy associates with future conversion from MCI to AD (Apostolova et al., 2006b) and from normal cognition to MCI (Apostolova et al., in press).
Related shape modeling studies have involved modeling the hippocampal surface using spherical harmonic functions (SPHARM) (Styner et al., 2004; Thompson and Toga, 1996), and using the coefficients of the harmonic expansion to infer shape differences between dementia patients and controls. In Gutman et al. (2008), we used a support vector machine classifier based on spherical harmonics to classify 49 AD patients and 63 controls with 75.5% sensitivity and 87.3% specificity, with 82.1% correct overall. This approaches the 89–96% classification accuracy of the best diagnostic predictors (Vemuri et al., 2008). Given the proliferation of new MRI-based measures of hippocampal degeneration based on automated surface matching methods (Wang et al., 2005), automated partitions of surfaces (Shi et al., 2007), random field theory on surfaces (Bansal et al., 2007) and machine learning methods (Li et al., 2007), future studies will likely focus on defining which MR-based measures provide optimal statistical power for detecting factors that slow the progression of AD, to optimize the power of future interventional trials.
Acknowledgments
Data used in preparing this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative database (www.loni.ucla.edu/ADNI). Many ADNI investigators therefore contributed to the design and implementation of ADNI or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators is available at www.loni.ucla.edu/ADNI/Collaboration/ADNI_Citation.shtml. This work was primarily funded by the ADNI (Principal Investigator: Michael Weiner; NIH grant number U01 AG024904). ADNI is funded by the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the Foundation for the National Institutes of Health, through generous contributions from the following companies and organizations: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. Algorithm development for this study was also funded by the NIA, NIBIB, the National Library of Medicine, and the National Center for Research Resources (AG016570, EB01651, LM05639, RR019771 to PT). Author contributions were as follows: JM, ZT, LA, AG, CA, SM, NP, AT, and PT performed the image analyses; CJ, NS, and MW contributed substantially to the image acquisition, study design, quality control, calibration and pre-processing, databasing and image analysis. We thank the members of the ADNI Imaging Core for their contributions to the image pre-processing and the ADNI project.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Alexander GE, Chen K, Merkley TL, Reiman EM, Caselli RJ, Aschenbrenner M, Santerre-Lemmon L, Lewis DJ, Pietrini P, Teipel SJ, Hampel H, Rapoport SI, Moeller JR. Regional network of magnetic resonance imaging gray matter volume in healthy aging. NeuroReport. 2006;17:951–956. doi: 10.1097/01.wnr.0000220135.16844.b6. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM. Brain mapping as a tool to study neurodegeneration. Neurotherapeutics. 2007;4:387–400. doi: 10.1016/j.nurt.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer’s disease. Brain. 2006a;129:2867–2873. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006b;63:693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Akopyan GG, Partiali N, Steiner CA, Dutton RA, Hayashi KM, Dinov ID, Toga AW, Cummings JL, Thompson PM. Structural correlates of apathy in Alzheimer’s disease. Dement Geriatr Cogn Disord. 2007;24:91–97. doi: 10.1159/000103914. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Mosconi L, Thompson PM, Green AE, Hwang KS, Ramirez A, Mistur R, Tsui WH, de Leon MJ. Subregional hippocampal atrophy predicts Alzheimer’s dementia in the cognitively normal. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2008.08.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Thompson P, Green A, Hwang K, Zoumalan C, Jack C, Harvey D, Petersen RM, Thal L, Aisen P, Toga A, Cummings JL, DeCarli C. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. ADCS Group. doi: 10.1002/hbm.20905. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ballmaier M, Narr KL, Toga AW, Elderkin-Thompson V, Thompson PM, Hamilton L, Haroon E, Pham D, Heinz A, Kumar A. Hippocampal morphology and distinguishing late-onset from early-onset elderly depression. Am J Psychiatry. 2008;165:229–237. doi: 10.1176/appi.ajp.2007.07030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R, Staib LH, Xu D, Zhu H, Peterson BS. Statistical analyses of brain surfaces using Gaussian random fields on 2-D manifolds. IEEE Trans Med Imag. 2007;26:46–57. doi: 10.1109/TMI.2006.884187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Boyes RG, Frost C, Lewis EB, Rossor CL, Rossor MN, Fox NC. Differentiating AD from aging using semiautomated measurement of hippocampal atrophy rates. NeuroImage. 2004;23:574–581. doi: 10.1016/j.neuroimage.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, Thompson P, Fox NC. A meta-analysis of hippocampal atrophy rates in Alzheimer’s disease. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2008.01.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron JC, Chetelat G, Desgranges B, Perchey G, Landeau B, de la Sayette V, Eustache F. In vivo mapping of gray matter loss with voxel-based morphometry in mild Alzheimer’s disease. NeuroImage. 2001;14:298–309. doi: 10.1006/nimg.2001.0848. [DOI] [PubMed] [Google Scholar]
- Bearden CE, Thompson PM, Dutton RA, Frey BN, Peluso MA, Nicoletti M, Dierschke N, Hayashi KM, Klunder AD, Glahn DC, Brambilla P, Sassi RB, Mallinger AG, Soares JC. Three-dimensional mapping of hippocampal anatomy in unmedicated and lithium-treated patients with bipolar disorder. Neuropsychopharmacology. 2008;33:1229–1238. doi: 10.1038/sj.npp.1301507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JT, Davis SW, Hayashi KM, Meltzer CC, Toga AW, Lopez OL, Thompson PM. Three-dimensional patterns of hippocampal atrophy in mild cognitive impairment. Arch Neurol. 2006;63:97–101. doi: 10.1001/archneur.63.1.97. [DOI] [PubMed] [Google Scholar]
- Boccardi M, Sabattoli F, Testa C, Beltramello A, Soininen H, Frisoni GB. APOE and modulation of Alzheimer’s and frontotemporal dementia. Neurosci Lett. 2004;356:167–170. doi: 10.1016/j.neulet.2003.11.042. [DOI] [PubMed] [Google Scholar]
- Boccardi M, Ganzola R, Sabattoli F, Laakso MP, Repo-Tiihonen EOV, Aronen HJ, Thompson P, Frisoni GB, Tiihonen J. Abnormal hippocampal shape in offenders with psychopathy. Arch Gen Psychiatry. doi: 10.1002/hbm.20877. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DL, Bernstein MA, Thompson PM, Weiner MW, Schuff N, Alexander GE, Killiany RJ, DeCarli C, Jack CR, Fox NC. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. NeuroImage. 2008;39:1752–1762. doi: 10.1016/j.neuroimage.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Cerebral ventricular changes associated with transitions between normal cognitive function, mild cognitive impairment, and dementia. Alzheimer Dis Assoc Disord. 2007;21:14–24. doi: 10.1097/WAD.0b013e318032d2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat G, Landeau B, Eustache F, Mezenge F, Viader F, de la Sayette V, Desgranges B, Baron JC. Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. NeuroImage. 2005;27:934–946. doi: 10.1016/j.neuroimage.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Chou YY, Lepore N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. NeuroImage. 2008;40:615–630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M, Mukuna-Bantumbakulu AR, Hasboun D, Bardinet E, Baillet S, Kinkingnehun S, Lemieux L, Dubois B, Garnero L. Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: method and validation on controls and patients with Alzheimer’s disease. NeuroImage. 2007;34:996–1019. doi: 10.1016/j.neuroimage.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Cockrell JR, Folstein MF. Mini-Mental State Examination (MMSE) Psychopharmacol Bull. 1988;24:689–692. [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Crum WR, Scahill RI, Fox NC. Automated hippocampal segmentation by regional fluid registration of serial MRI: validation and application in Alzheimer’s disease. NeuroImage. 2001;13:847–855. doi: 10.1006/nimg.2001.0744. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, Grenander U, Miller MI. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci U S A. 1998;95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi S, Miller JP, Gado M, Kido D, McKeel D, Morris JC, Miller MI. Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Dementia of the Alzheimer type Neurology. 2000;55:1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi SC, Ratnanather JT, Miller MI. Computational anatomy and neuropsychiatric disease: probabilistic assessment of variation and statistical inference of group difference, hemispheric asymmetry, and time-dependent change. NeuroImage. 2004;23 (Suppl 1):S56–68. doi: 10.1016/j.neuroimage.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC. Preclinical detection of Alzheimer’s disease: hippocampal shape and volume predict dementia onset in the elderly. NeuroImage. 2005;25:783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Feczko E, Augustinack JC, Pacheco J, Morris JC, Fischl B, Buckner RL. Differential effects of aging and Alzheimer’s disease on medial temporal lobe cortical thickness and surface area. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2007.07.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Schuff N, Zhu XP, Jagust WJ, Miller BL, Reed BR, Kramer JH, Mungas D, Yaffe K, Chui HC, Weiner MW. Atrophy rates of entorhinal cortex in AD and normal aging. Neurology. 2003;60:481–486. doi: 10.1212/01.wnl.0000044400.11317.ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda D, Hart P, Stork D. Pattern Classification. 2. Wiley-Interscience; 2001. [Google Scholar]
- Edgington ES, Onghena P. Randomization tests. 4. Chapman & Hall/CRC; Boca Raton, FL: 2007. [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Freund Y, Schapire RE. A decision-theoretic generalization of online learning and an application to boosting. J Comput Sys Sci. 1997;55:119–139. [Google Scholar]
- Frisoni GB, Sabattoli F, Lee AD, Dutton RA, Toga AW, Thompson PM. In vivo neuropathology of the hippocampal formation in AD: a radial mapping MR-based study. NeuroImage. 2006;32:104–110. doi: 10.1016/j.neuroimage.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Gunter J, Bernstein M, Borowski B, Felmlee J, Blezek D, Mallozzi R. Validation testing of the MRI calibration phantom for the Alzheimer’s Disease Neuroimaging Initiative Study. ISMRM 14th Scientific Meeting and Exhibition.2006. [Google Scholar]
- Gutman B, Wang YL, Morra J, Tu Z, Jack C, Weiner M, Toga A, Thompson P. Disease Classification with Hippocampal Surface Invariants. MICCAI Workshop on Hippocampal Mapping (Chair: Paul Yushkevich and Lei Wang)..2008. [Google Scholar]
- Haller JW, Christensen GE, Joshi SC, Newcomer JW, Miller MI, Csernansky JG, Vannier MW. Hippocampal MR imaging morphometry by means of general pattern matching. Radiology. 1996;199:787–791. doi: 10.1148/radiology.199.3.8638006. [DOI] [PubMed] [Google Scholar]
- Hogan RE, Mark KE, Wang L, Joshi S, Miller MI, Bucholz RD. Mesial temporal sclerosis and temporal lobe epilepsy: MR imaging deformation-based segmentation of the hippocampus in five patients. Radiology. 2000;216:291–297. doi: 10.1148/radiology.216.1.r00jl41291. [DOI] [PubMed] [Google Scholar]
- Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou YY, Brun C, Chiang MC, Barysheva M, Jack CR, Jr, Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes RG, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. NeuroImage. 2008a;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008b;43:458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer’s disease. Neurology. 1998;51:993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Tangalos EG, Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008a;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Grundman M, Jin S, Gamst A, Ward CP, Sencakova D, Doody RS, Thal LJ. Longitudinal MRI findings from the vitamin E and donepezil treatment study for MCI. Neurobiol Aging. 2008b;29:1285–1295. doi: 10.1016/j.neurobiolaging.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SC, Miller MI. Landmark matching via large deformation diffeomorphisms. IEEE Trans Image Process. 2000;9:1357–1370. doi: 10.1109/83.855431. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Karas G, Sluimer J, Goekoop R, van der Flier W, Rombouts SA, Vrenken H, Scheltens P, Fox N, Barkhof F. Amnestic mild cognitive impairment: structural MR imaging findings predictive of conversion to Alzheimer disease. AJNR Am J Neuroradiol. 2008;29:944–949. doi: 10.3174/ajnr.A0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Wang L, Beg MF. FreeSurfer-initiated fully-automated subcortical brain segmentation in MRI using large deformation diffeomorphic metric mapping. NeuroImage. 2008;41:735–746. doi: 10.1016/j.neuroimage.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. NeuroImage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Pruessner J, Zijdenbos AP, Collins DL, Teipel SJ, Hampel H, Evans AC. Automated cortical thickness measurements from MRI can accurately separate Alzheimer’s patients from normal elderly controls. Neurobiol Aging. 2008;29:23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Li S, Shi F, Pu F, Li X, Jiang T, Xie S, Wang Y. Hippocampal shape analysis of Alzheimer disease based on machine learning methods. AJNR Am J Neuroradiol. 2007;28:1339–1345. doi: 10.3174/ajnr.A0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Salamon N, Dutton RA, Lee AD, Geaga JA, Hayashi KM, Toga AW, Engel J, Jr, Thompson PM. Three-dimensional preoperative maps of hippocampal atrophy predict surgical outcomes in temporal lobe epilepsy. Neurology. 2005;65:1094–1097. doi: 10.1212/01.wnl.0000179003.95838.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mio W, Shi Y, Dinov I, Liu X, Lepore N, Lepore F, Fortin M, Voss P, Lassonde M, Thompson P. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2008. Springer; New York, NY: 2008. Models of normal variation and local differences in hippocampal anatomy; pp. 407–415. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond, B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Miller MI, Priebe CE, Qiu A, Fischl B, Kolasny A, Brown T, Park Y, Ratnanather JT, Busa E, Jovicich J, Yu P, Dickerson BC, Buckner RL. Collaborative computational anatomy: an MRI morphometry study of the human brain via diffeomorphic metric mapping. Hum Brain Mapp. doi: 10.1002/hbm.20655. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J, Tu Z, Apostolova LG, Green A, Toga A, Thompson P. Comparison of AdaBoost and support vector machines for detecting Alzheimer’s disease through automated hippocampal segmentation. IEEE Trans Med Imaging. doi: 10.1109/TMI.2009.2021941. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J, Tu Z, Apostolova LG, Green A, Avedissian C, Madsen S, Parikshak N, Hua X, Toga A, Jack C, Schuff N, Weiner M, Thompson P. Automated 3D Mapping of Hippocampal Atrophy and its Clinical Correlates in 400 Subjects with Alzheimer’s Disease, Mild Cognitive Impairment, and Elderly Controls. Hum Brain Mapp. doi: 10.1002/hbm.20708. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J, Tu Z, Apostolova LG, Green A, Avedissian C, Madsen S, Parikshak N, Hua X, Toga A, Jack C, Schuff N, Weiner MW, Thompson P. Mapping Hippocampal Degeneration in 400 Subjects with a Novel Automated Segmentation Approach. International Symposium on Biomedical Imaging; IEEE, Paris, France. 2008a. [Google Scholar]
- Morra J, Tu Z, Apostolova LG, Green A, Toga A, Thompson P. Automatic subcortical segmentation using a contextual model. In: Metaxas D, Axel L, Fichtinger G, Szekely G, editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2008. Springer; New York, New York: 2008b. pp. 194–201. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. NeuroImage. 2008c;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005a;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005b;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Stables L, Du AT, Schuff N, Truran D, Cashdollar N, Weiner MW. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4 T. Neurobiol Aging. 2007;28:719–726. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson R, DeVito TJ, Vidal CN, Sui Y, Hayashi KM, Drost DJ, Williamson PC, Rajakumar N, Toga AW, Thompson PM. Detection and mapping of hippocampal abnormalities in autism. Psychiatry Res. 2006;148:11–21. doi: 10.1016/j.pscychresns.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Ogren JA, Bragin A, Wilson CL, Hoftman GD, Lin JJ, Dutton RA, Fields TA, Toga A, Thompson P, Engel JP, Staba RJ. Bilateral 3D Hippocampal Atrophy Maps Distinguish Patients with Hypersynchronous Seizure Onsets from those with Low-Voltage Fast Seizure Onsets. Annals of Neurology submitted for publication. [Google Scholar]
- Petersen RC. Aging, mild cognitive impairment, and Alzheimer’s disease. Neurol Clin. 2000;18:789–805. doi: 10.1016/s0733-8619(05)70226-7. [DOI] [PubMed] [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. NeuroImage. 2008;39:238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Younes L, Miller MI, Csernansky JG. Parallel transport in diffeomorphisms distinguishes the time-dependent pattern of hippocampal surface deformation due to healthy aging and the dementia of the Alzheimer’s type. NeuroImage. 2008;40:68–76. doi: 10.1016/j.neuroimage.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RS, Busa E, Morris JC, Dale AM, Fischl B. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14:721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- Schapire RE, Freund Y, Bartlett JW. Boosting the margin: a new explanation for the effectiveness of voting methods. Ann Stat. 1998;26:1651–1686. [Google Scholar]
- Scher A, Xu Y, Korf E, White L, Scheltens P, Toga A, Thompson P, Hartley S, Witter M, Valentino D, Launer L. Hippocampal morphometry in Alzheimer’s vs. Vascular Dementia: A Population-Based Study. Neuroimage in press. [Google Scholar]
- Shi Y, Thompson PM, de Zubicaray GI, Rose SE, Tu Z, Dinov I, Toga AW. Direct mapping of hippocampal surfaces with intrinsic shape context. NeuroImage. 2007;37:792–807. doi: 10.1016/j.neuroimage.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Studholme C, Cardenas V, Blumenfeld R, Schuff N, Rosen HJ, Miller B, Weiner M. Deformation tensor morphometry of semantic dementia with quantitative validation. NeuroImage. 2004;21:1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Styner M, Brechbuhler C, Szekely G, Gerig G. Parametric estimate of intensity inhomogeneities applied to MRI. IEEE Trans Med Imag. 2000;19:153–165. doi: 10.1109/42.845174. [DOI] [PubMed] [Google Scholar]
- Styner M, Gerig G, Lieberman J, Jones D, Weinberger D. Statistical shape analysis of neuroanatomical structures based on medial models. Med Image Anal. 2003;7:207–220. doi: 10.1016/s1361-8415(02)00110-x. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, Pantazis D, Gerig G. Boundary and medial shape analysis of the hippocampus in schizophrenia. Med Image Anal. 2004;8:197–203. doi: 10.1016/j.media.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc Natl Acad Sci U S A. 2005;102:4872–4877. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel SJ, Flatz WH, Heinsen H, Bokde AL, Schoenberg SO, Stockel S, Dietrich O, Reiser MF, Moller HJ, Hampel H. Measurement of basal forebrain atrophy in Alzheimer’s disease using MRI. Brain. 2005;128:2626–2644. doi: 10.1093/brain/awh589. [DOI] [PubMed] [Google Scholar]
- Terriberry TB, Damon JN, Pizer SM, Joshi SC, Gerig G. Population-based fitting of medial shape models with correspondence optimization. Inf Process Med Imag. 2007;20:700–712. doi: 10.1007/978-3-540-73273-0_58. [DOI] [PubMed] [Google Scholar]
- Thompson P, Toga AW. A surface-based technique for warping three-dimensional images of the brain. IEEE Trans Med Imag. 1996;15:402–417. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Apostolova LG. Computational anatomical methods as applied to ageing and dementia. Br J Radiol. 2007;80 (Spec No 2):S78–91. doi: 10.1259/BJR/20005470. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer’s disease. J Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Vaillant M, Glaunes J. Surface matching via currents. Inf Process Med Imag. 2005;19:381–392. doi: 10.1007/11505730_32. [DOI] [PubMed] [Google Scholar]
- Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. NeuroImage. 2008;39:1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. NeuroImage. 2003;20:667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chiang MC, Thompson PM. Automated surface matching using mutual information applied to Riemann surface structures. Med Image Comput Comput Assist Interv. 2005;8:666–674. doi: 10.1007/11566489_82. [DOI] [PubMed] [Google Scholar]
- Wang L, Miller JP, Gado MH, McKeel DW, Rothermich M, Miller MI, Morris JC, Csernansky JG. Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. NeuroImage. 2006;30:52–60. doi: 10.1016/j.neuroimage.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris JC, Csernansky JG, Miller MI. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans Med Imag. 2007;26:462–470. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Memory Scale. Psychological Corp/Harcourt Brace Jovanovich; New York: 1987. [Google Scholar]
- Whitwell JL, Jack CR., Jr Comparisons between Alzheimer disease, frontotemporal lobar degeneration, and normal aging with brain mapping. Top Magn Reson Imaging. 2005;16:409–425. doi: 10.1097/01.rmr.0000245457.98029.e1. [DOI] [PubMed] [Google Scholar]
- Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, Leirer VO. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 1982;17:37–49. doi: 10.1016/0022-3956(82)90033-4. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Zhang H, Gee JC. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2008. Springer; 2005. Statistical modeling of shape and appearance using the continuous medial representation; pp. 725–732. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]