Abstract
Iron release from human serum transferrin (hTF) has been studied extensively; however, the molecular details of the mechanism(s) remain incomplete. This is in part due to the complexity of this process, which is influenced by lobe–lobe interactions, the transferrin receptor (TFR), the salt effect, the presence of a chelator, and acidification within the endosome, resulting in iron release. The present work brings together many of the concepts and assertions derived from previous studies in a methodical, uniform, and visual manner. Examination of earlier work reveals some uncertainty due to sample and technical limitations. We have used a combination of steady-state fluorescence and urea gels to evaluate the effect of conformation, pH, time, and the soluble portion of the TFR (sTFR) on iron release from each lobe of hTF. The use of authentic recombinant monoferric and locked species removes any possibility of cross-contamination by acquisition of iron. Elimination of detergent by use of the sTFR provides a further technical advantage. We find that iron release from the N-lobe is very sensitive to the conformation of the C-lobe, but is insensitive to the presence of the sTFR or to changes in pH (between 5.6 and 6.4). Specifically, when the cleft of the C-lobe is locked, the urea gels indicate that only about half of the iron is completely removed from the cleft of the N-lobe. Iron release from the C-lobe is most affected by the presence of the sTFR and changes in pH, but is unaffected by the conformation of the N-lobe. A model for iron release from diferric hTF is provided to delineate our findings.
Keywords: Cooperativity, Urea gels, Steady-state tryptophan fluorescence, Transferrin/transferrin receptor complex, Iron-release model
Introduction
Human serum transferrin (hTF) is a bilobal 80-kDa iron binding glycoprotein responsible for delivering iron to cells by clathrin dependent receptor mediated endocytosis. The single-chain polypeptide folds into two homologous lobes (N- and C-lobes) connected by a short peptide linker. Each lobe is further divided into two subdomains (NI, NII and CI, CII) that come together to form the metal binding cleft. The iron binding ligands are identical in the two lobes (a histidine, an aspartic acid, two tyrosines, as well as two oxygen atoms from the synergistic anion, carbonate). However, the mechanism and rate(s) of iron release differ between the N- and C-lobes, owing in large part to differences in the amino acids surrounding the liganding residues, termed the “second shell” [1]. In healthy individuals, hTF is present in the serum at a concentration of 25–50 µM, but is only approximately 30% saturated with iron. The distribution of the pool is 27% diferric, 23% monoferric N-lobe, 11% monoferric C-lobe, and 40% apo [2, 3]. The source of this uneven distribution is not completely understood. At pH 7.4, diferric hTF preferentially binds to the hTF receptor (TFR) with nanomolar affinity, the two monoferric species bind approximately 40-fold weaker (although each lobe contributes equally to the binding energy of the interaction with the TFR), and apo hTF does not compete for binding [4]. However, at the putative endosomal pH of 5.6, apo hTF remains bound to the TFR and is recycled back to the plasma membrane.
The precise details of the steps by which iron is released from the two lobes of hTF have been elusive in spite of much research. Although there has been a great deal of progress, some controversy exists with regard to the relative importance of various factors, which minimally include pH (protonation of various key residues in each lobe is an initial step within the endosome), ionic strength (anions or inert salt must be present for iron release to occur), the identity of a chelator (critical to extraction of the iron in the physiologically observed time frame of less than 3 min), and the TFR [5, 6]. The bilobal composition of hTF adds complexity by introducing cooperativity (negative or positive) between the lobes [7–9]. What is clear is the crucial role of the second-shell residues in the release of iron from each lobe. Thus, in the N-lobe, two lysine residues, Lys206 and Lys296 (residing on opposite sides of the binding cleft), constitute the “dilysine trigger.” These two lysine residues share a hydrogen bond at neutral pH which is protonated at low pH and triggers cleft opening [10, 11]. Likewise in the C-lobe, Lys534 and Arg632 are found in positions that are homologous to those of Lys206 and Lys296 [1, 12]. Mutation of Lys206 to glutamate in the N-lobe or of Arg632 to alanine in the C-lobe completely prevents iron release from that lobe on a relevant timescale, allowing targeted measurement of iron release from the opposite lobe.
In transitioning from a “closed” iron-bound state to an “open” iron-free state, the N- and C-lobes each undergo large conformational changes upon iron release [13, 14]. The intrinsic tryptophan fluorescence of hTF increases dramatically upon iron removal and can be used to monitor iron removal from hTF [15]. Some of the eight tryptophan residues in hTF (three in the N-lobe and five in the C-lobe) are strongly quenched by the bound Fe3+ through radiationless transfer of electronic excited-state energy [16]. This energy is transferred to an absorption band that overlaps the tryptophan fluorescence and is created by the metal–tyrosine interaction. Of course, tryptophan fluorescence is also sensitive to changes in the local environment around each residue, so it is possible to monitor conformational changes both before and after iron release [17]. As an example, we have reported the contribution of each of the three tryptophan residues in the isolated N-lobe to the change in fluorescence when iron is released [18]. Similarly, we have described the role of each of the five tryptophan residues in the C-lobe (in the absence and presence of the soluble portion of the TFR, sTFR) in the increase in fluorescence as a result of iron release at pH 5.6 [19].
Over 30 years ago, the serendipitous observation was made that 6 M urea gels could be used to distinguish the iron status of the two lobes of hTF [20]. Migration through these gels is influenced by the shape, charge, and the disulfide bond content of the two lobes. Thus diferric hTF, which is the most compact form of hTF, migrates the farthest into the gel before being denatured by the urea. The least compact, apo, form (both lobes open) is denatured soon after exposure to the 6 M urea and stays near the top of the gel; in part owing to the fact that the N-lobe has eight disulfide bonds and the C-lobe has 11, the two monoferric species migrate at differing intermediate positions. Since their discovery, many studies have used urea gels to monitor the effect of a variety of factors on the iron status of hTF [2, 9, 21–27].
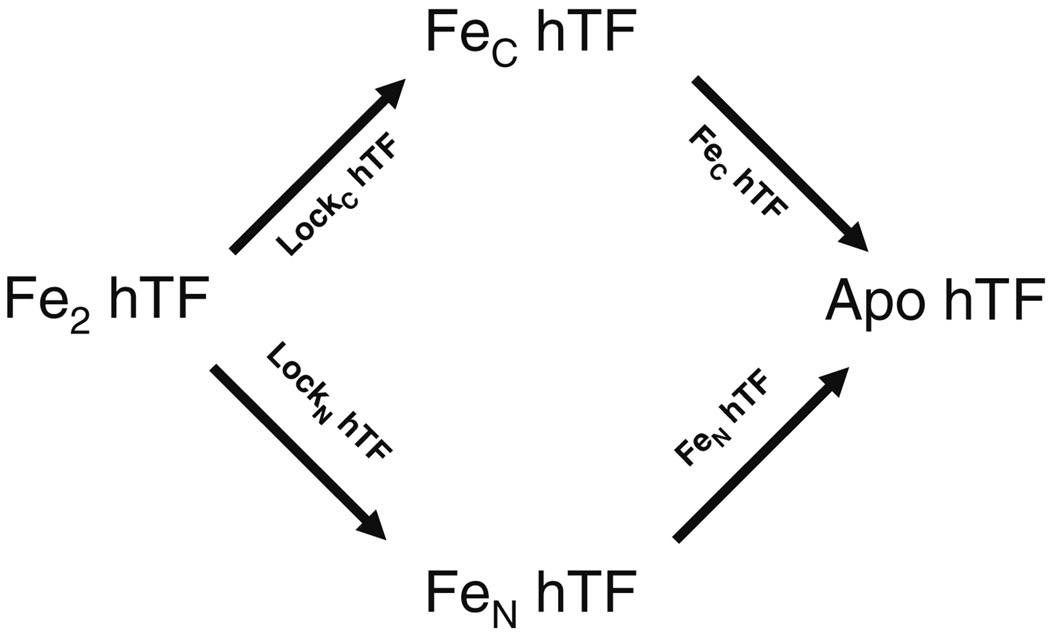
In the present study, we have substantiated much previous work in a concise and semiquantitative manner using a unique combination of urea gels and steady state fluorescence measurements to examine hTF and a complex of hTF bound to the sTFR as a function of lobe conformation, pH, and time to qualitatively examine iron release from each lobe. Deciphering the role of the TFR in iron release has previously been limited by the availability of TFR from natural sources and the need for detergent to keep the TFR in solution [28, 29]. Availability of non-glycosylated recombinant diferric hTF (Fe2 hTF), authentic monoferric TFs (designated FeN hTF and FeC hTF), and constructs with either N- and/or C- “locked” lobes (designated LockN hTF and LockC hTF), as well as the sTFR (see abbreviations for a full description of these constructs) allow a comprehensive assessment of the contributions of pH, the TFR and the conformation of each lobe to the release of iron. As shown in Fig. 1, use of these constructs allows a rational and precise assessment of pathway(s) available to transition from fully iron loaded, to monoferric, to fully iron free hTF. The quality of the results obtained from commercially available urea gels is improved by the absence of carbohydrate in the recombinant hTF samples (the asparagine linkage sites at positions 413 and 611 in the C-lobe are mutated to aspartic acid). A model of iron release from diferric hTF is provided. The current work foreshadows our more quantitative work on this system by providing concepts and end points to generate the models we are developing by analysis of kinetic data from stopped-flow fluorescence studies.
Fig. 1.
Pathways available to transition from Fe2 hTF to apo hTF. Recombinant constructs used to specifically monitor each route are indicated above/below the arrows
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium containing Ham F-12 nutrient mixture (DMEM-F12), antibiotic–antimy-cotic solution (100×), and trypsin solution were from the GIBCO-BRL Life Technologies Division of Invitrogen. Fetal bovine serum was obtained from Atlanta Biologicals (Norcross, GA, USA). Ultroser G is a serum replacement from Pall BioSepra (Cergy, France). Nickel nitrilotriacetic acid resin was purchased from QIAGEN. Corning expanded surface roller bottles and Dynatech Immunolon 4 Removawells were obtained from Fisher Scientific. The Hi-Prep 26/60 Sephacryl S-200HR and S-300HR columns were from Amersham Pharmacia. Amicon Ultra-4 (30-kDa cutoff) ultrafiltration concentrators were from Millipore. Novex 6% tris(hydroxymethyl)aminomethane (Tris)–borate–EDTA (TBE)–urea gels, 2× TBE–urea gel sample buffer, and 5× TBE–urea gel running buffer were from Invitrogen. All other chemicals and reagents were of analytical grade.
Protein production and purification
The DNA manipulations used to generate Fe2 hTF, FeN hTF, FeC hTF, LockN hTF, LockC hTF, and the sTFR have been described in detail previously [1, 27, 30, 31]. Briefly, to produce recombinant hTF and all of the mutants, baby hamster kidney (BHK) cells transfected with the pNUT plasmid containing the appropriate complementary DNA sequence were placed into two to four expanded surface roller bottles. Adherent BHK cells were grown in DMEM-F12 containing 10% fetal bovine serum. This medium was changed twice at 2-day intervals, followed by addition of DMEM-F12 containing the serum substitute Ultroser G (1%) and 1 mM butyric acid. The presence of 1 mM butyric acid has been shown to increase the production of recombinant protein from BHK cells [30]. The amount of protein produced was determined using a competitive immunoassay [32]. The His6-tagged recombinant protein from the tissue culture medium was captured by passage over a nickel nitrilotriacetic acid column, followed by final purification on a gel filtration column (S-200HR for hTF constructs and S-300HR for sTFR). Polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate was used to verify the homogeneity of the all of the recombinant proteins. Proteins were brought to a nominal concentration of 15 mg/mL using the published absorption coefficients determined by the modified Edelhoch method [33].
Complexes of hTF/sTFR were prepared by combining the sTFR with a small molar excess of hTF (Fe2 hTF, FeN hTF, FeC hTF, LockN hTF, and LockC hTF) and isolated by passage over an S-300HR column [31]. The concentrations of the complexes were adjusted to a nominal concentration of 15 mg/mL with respect to hTF.
Urea gel analysis
The iron binding status of the hTF constructs as a function of pH or time was examined by urea gel electrophoresis using Novex 6% TBE–urea minigels, run in 90 mM Tris–90 mM borate, pH 8.4, containing 2 mM EDTA. For pH titrations, samples (1 µg/µL) were incubated for 15 min in 100 mM 2-morpholinoethanesulfonic acid (MES) buffer ranging from pH 5.6 to 6.4 (in increments of 0.2 pH units), also containing 300 mM KCl and 4 mM EDTA. The reaction was stopped by addition of 2× TBE–urea gel sample buffer (final concentration of sample 0.5 µg/µL). The composition of the 2× sample buffer was 45 mM Tris and 45 mM borate, containing 1 mM EDTA, 6% Ficoll 400, 0.005% bromophenol blue, 0.025% xylene cyanol, and 3.5 M urea. For time-based experiments, sample (1 µg/µL) was added to pH 5.6 iron-removal buffer (100 mM MES, pH 5.6 containing 300 mM KCl and 4 mM EDTA) and incubated for the designated times (0, 3, 6, 12, and 15 min) at room temperature (note that time zero is the construct in sample buffer). At each time point, the iron-removal process was stopped by the addition of sample buffer as stated above and the sample was placed on ice (final concentration of sample 0.5 µg/µL). This time course was chosen because it results in complete removal of iron during stopped-flow kinetic experiments (unpublished results). Approximately 2.5 µg of sample was loaded per lane and the gels were electrophoresed for 2.25 h at 125 V. Protein bands were visualized by staining with Coomassie blue.
Steady-state fluorescence
Steady-state fluorescence spectra were obtained for each sample using a Quantamaster 6 spectrofluorimeter from Photon Technology International (South Brunswick, NJ, USA). Iron-bound protein (500 nM) was added to a cuvette (1.8 mL final volume) containing 100 mM N-(2-hydroxyethyl) piperazine-N′-ethanesulfonic acid, pH 7.4. An identical amount of sample was incubated for at least 15 min in iron-removal buffer of appropriate pH (as above) to generate apoprotein. Samples were excited at 280 nm and emission was monitored between 300 and 400 nm. Slit widths of 1 nm (excitation) and 6 nm (emission) were used with a 320-nm cut-on filter in front of the emission monochromator. All emission spectra were corrected for Raman scattering by subtraction of a buffer blank spectrum [18].
Results
Iron release as a function of pH
To elucidate the pathway of iron release as a function of pH, each of the hTF constructs described below was incubated in 100 mM MES containing 300 mM KCl and 4 mM EDTA at pH values ranging from 5.6 to 6.4. In all cases, iron removal was evaluated by electrophoresis on 6 M urea gels that specifically show bands corresponding to apo, each monoferric, and Fe2 hTF conformations. More globally, steady state emission scans monitor the increase in the intrinsic tryptophan fluorescence signal as a function of pH. Bar graphs are used to show the end points derived from the steady state emission spectra (Fig. 2, Fig. 3, Fig. 4). (Note that the source spectra from which the bar graphs are derived are provided as electronic supplementary material.) It is important to recognize that Fe2 hTF with iron bound in both lobes at pH 7.4 should be maximally quenched and that the same sample at pH 5.6 should be maximally unquenched. Accordingly, at pH 7.4 LockN hTF and LockC hTF would also both be quenched to the same extent as Fe2 hTF, but would only reach a value that is approximately half of that of the maximally unquenched state (the monoferric species). Likewise, the two monoferric constructs (FeN hTF and FeC hTF) would start at this intermediate value of fluorescence intensity and upon iron removal would reach the unquenched apo state.
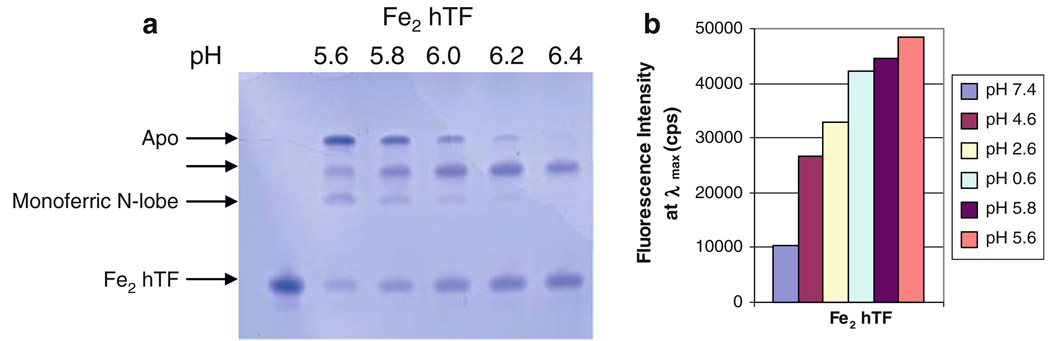
Fig. 2.
Effect of lowering pH on iron release from Fe2 hTF. a 6 M urea gel. All samples were incubated for 15 min in iron-removal buffer before being loaded on the gel (2.5 µg per lane). b Bar graph of steady-state emission intensity at λmax for each spectrum. The emission spectra from which the end points shown in the bar graph were obtained are provided as electronic supplementary material. All samples were incubated for 15 min in iron removal buffer [100 mM MES pH 5.6, 300 mM KCl, 4 mM EDTA], before the emission was monitored. Samples were excited at 280 nm and emission was monitored between 300 and 400 nm using a 320-nm cut-on filter. As an important control, we analyzed a construct with iron locked in both lobes and observed that at pH 5.6 no iron was removed and that the fluorescence intensity was equal to the intensity at pH 7.4, i.e., no iron was removed (data not shown)
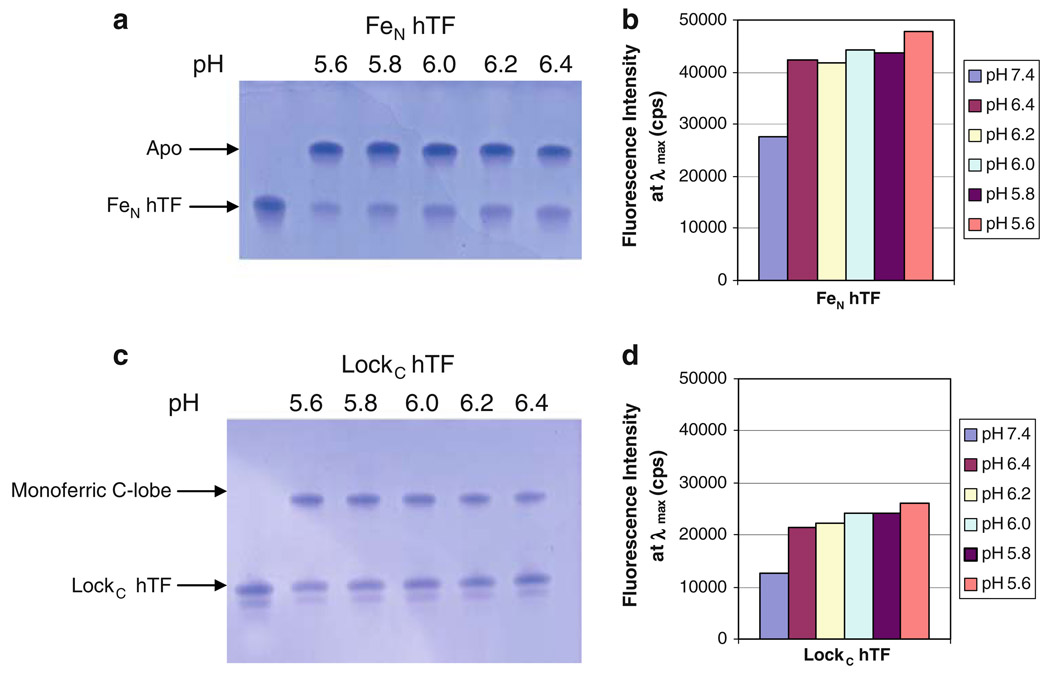
Fig. 3.
Effect of lowering pH, and the conformation of the C-lobe, on iron release from the N-lobe of hTF. a 6 M urea gel of FeN hTF, b bar graph of steady-state emission intensity, c 6 M urea gel of LockC hTF, d bar graph of steady-state emission intensity. All samples were prepared as described in the legend to Fig. 2. The appearance of a double band in c is ascribed to the extreme sensitivity of urea gels to charge heterogeneity. We have observed that this heterogeneity seems to increase as a function of the age of the sample and most likely results from oxidation and/or deamination of amino acid side chains
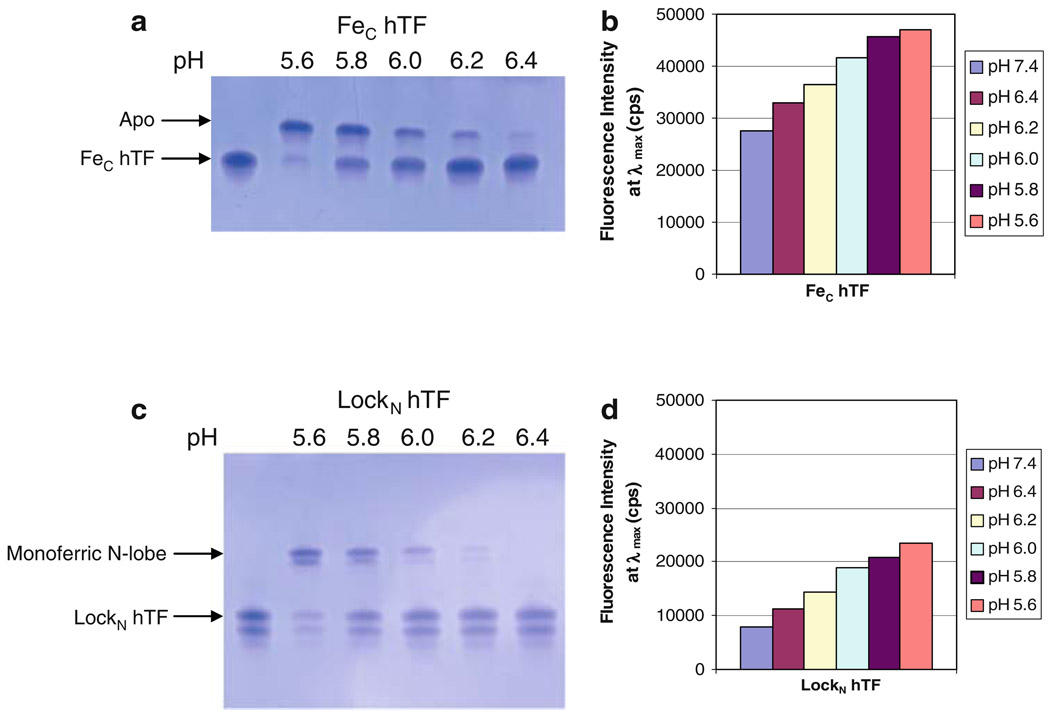
Fig. 4.
Effect of lowering pH, and the conformation of the N-lobe, on iron release from the C-lobe of hTF. a 6 M urea gel of Fec hTF, b bar graph of steady-state emission intensity, c 6 M urea gel of LockN hTF, d bar graph of steady-state emission intensity. All samples were prepared as described in the legend to Fig. 2. See the legend to Fig. 3 for explanation of the appearance of the double band in c in the bands corresponding to the monoferric N-lobe and the diferric species (LockN hTF)
Studies evaluating iron release from Fe2 hTF
As shown by the urea gel in Fig. 2a, at pH 5.6, the majority of the iron was removed from Fe2 hTF as indicated by the band corresponding to the apo form of hTF. Additionally, equal amounts of monoferric C-lobe, monoferric N-lobe, and diferric species were observed. At pH 6.4, approximately half was diferric hTF and half was monoferric C-lobe. At the intermediate pH values, there was a pH-dependent decrease in apo and monoferric N-lobe and a corresponding increase in monoferric C-lobe and diferric species, indicating that at higher pH values iron removal from the N-lobe of Fe2 hTF was favored (Fig. 2a). At pH 5.6, the steady-state data indicated a substantial increase (368%) in the fluorescence intensity relative to the diferric form at pH 7.4 (Fig. 2b). Collectively, we can conclude that iron removal from Fe2 hTF is very sensitive to pH (Fig. 2a, b).
Studies evaluating iron release from the N-lobe
To systematically evaluate iron release from the N-lobe, constructs in which the C-lobe was completely open or was locked in a closed conformation were used (Fig. 1). These constructs are designated “FeN hTF,” which is recombinant monoferric N-lobe hTF with the two tyrosine ligands disabled by mutation (Y426F and Y517F), thereby precluding iron binding in the C-lobe, and “LockC hTF,” which is recombinant Fe2 hTF with the arginine at position 632 mutated to alanine to effectively lock iron in the C-lobe.
As shown by the urea gel in Fig. 3a, following a 15-min incubation iron removal from FeN hTF was nearly complete and was equivalent regardless of the pH (5.6 up to 6.4). The steady-state data indicated a 74% increase in the fluorescence signal at pH 5.6 relative to pH 7.4 (iron bound) (Fig. 3b). As would be predicted from the urea gel results, there were minimal differences in the fluorescence intensity at the intermediate pH values (Fig. 3b), indicating nearly equivalent iron removal at each pH after 15 min.
In contrast, as shown by the urea gel in Fig. 3c, it appears that only about half of the iron was removed from the N-lobe of LockC hTF, although, again, the results did not seem to be very sensitive to the pH. At pH 5.6, the steady-state data indicated a 106% increase in the fluorescence emission intensity relative to pH 7.4 (iron bound) (Fig. 3d). In contrast to the urea gel results, the intensity at λmax at pH 5.6 after the 15-min incubation appears to be consistent with complete iron removal from the N-lobe (in this construct which retains iron in the C-lobe) (compare Fig. 3d, pH 5.6, LockC hTF, with Fig. 4b, pH 7.4, FeC hTF, both approximately 27,000 counts). The most likely explanation for the discrepancy between the results from the urea gel and the steady-state fluorescence is that in the steady-state experiment the low pH and excess chelator promote and result in irreversible iron release. In contrast, in the urea gel format, addition of the pH 8.4 sample buffer to the sample to “quench” iron release could result in rebinding of some portion of the iron. We suggest that this potential experimental anomaly is strongly and uniquely promoted by the locked C-lobe, which restricts the opening of the N-lobe, allowing a ternary complex (protein/metal/EDTA) to persist (see “Discussion”). As observed for FeN hTF, there was very little difference in the fluorescence intensity at the intermediate pH values. Collectively, we conclude that iron release from the N-lobe is relatively insensitive to pH and that considerably more iron is completely removed from the N-lobe when the C-lobe is open than when the C-lobe is locked (compare Fig. 3a, c).
Studies evaluating iron release from the C-lobe
To systematically evaluate iron release from the C-lobe, constructs in which the N-lobe was completely open or was locked in a closed conformation were used (Fig. 1). These constructs are designated “FeC hTF,” which is recombinant monoferric C-lobe hTF with the two tyrosine ligands disabled by mutation (Y95F and Y188F), thereby precluding iron binding in the N-lobe, and “LockN hTF,” which is recombinant Fe2 hTF with the lysine at position 206 mutated to a glutamate to effectively lock iron in the N-lobe.
As shown by the urea gel in Fig. 4a, after a 15-min incubation at pH 5.6 all of the iron is removed from the C-lobe when the N-lobe is open, whereas at pH 6.4 none of the iron is removed (Fig. 4a). There is a pH-dependent decrease in iron removal between pH 5.6 and 6.4. At pH 5.6, there is a 71% increase in the steady-state fluorescence relative to pH 7.4 (iron bound) (Fig. 4b). At the intermediate pH values, there is a gradual pH-dependent decrease in the fluorescence intensity with increasing pH, which more quantitatively illustrates the sensitivity to pH of iron removal from the C-lobe when the N-lobe is open (Fig. 4b).
LockN hTF is similar to FeC hTF, in that at pH 5.6 almost all of the iron is removed from the C-lobe, while at pH 6.4 none of the iron is removed (Fig. 4c). Again, there is a pH-dependent decrease in iron removal between pH 5.6 and 6.4 after a 15-min incubation. At pH 5.6 there is a 194% increase in the steady-state fluorescence relative to pH 7.4 (iron bound) (Fig. 4d). As observed for FeC hTF, there is a substantial pH-dependent decrease in the fluorescence intensity. Thus, iron release from the C-lobe is pH-sensitive (compare Fig. 4a, c), but is completely insensitive to whether the N-lobe is open or locked. The steady-state data reinforce both of these conclusions (Fig. 4b, d) as indicated by the substantial differences in end-point intensity as a function of pH.
Iron release as a function of time and the sTFR
To elucidate the pathway of iron release as a function of time and to determine the effect of the sTFR on each lobe, the various hTF constructs alone and in complex with the sTFR were incubated in 100 mM MES containing 300 mM KCl and 4 mM EDTA at pH 5.6. In all cases, iron removal was specifically evaluated by electrophoresis on 6 M urea gels (Fig. 5, Fig. 6, Fig. 7).
Fig. 5.
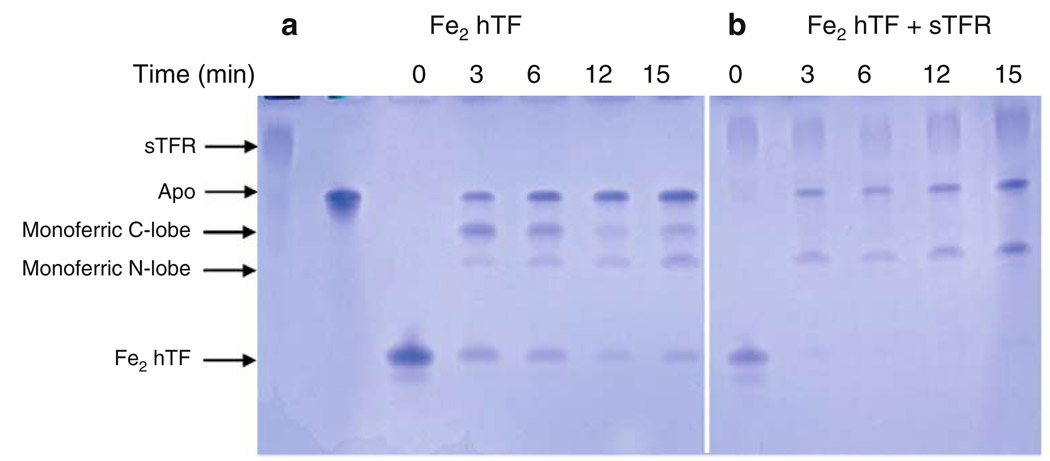
Influence of the soluble portion of the transferrin receptor (sTFR) on time-based iron release from Fe2 hTF at pH 5.6. a Fe2 hTF alone, b Fe2 hTF/sTFR complex. All samples were incubated for the designated time courses in 100 mM MES pH 5.6, 300 mM KCl, 4 mM EDTA. Iron release was quenched by the addition of sample buffer
Fig. 6.
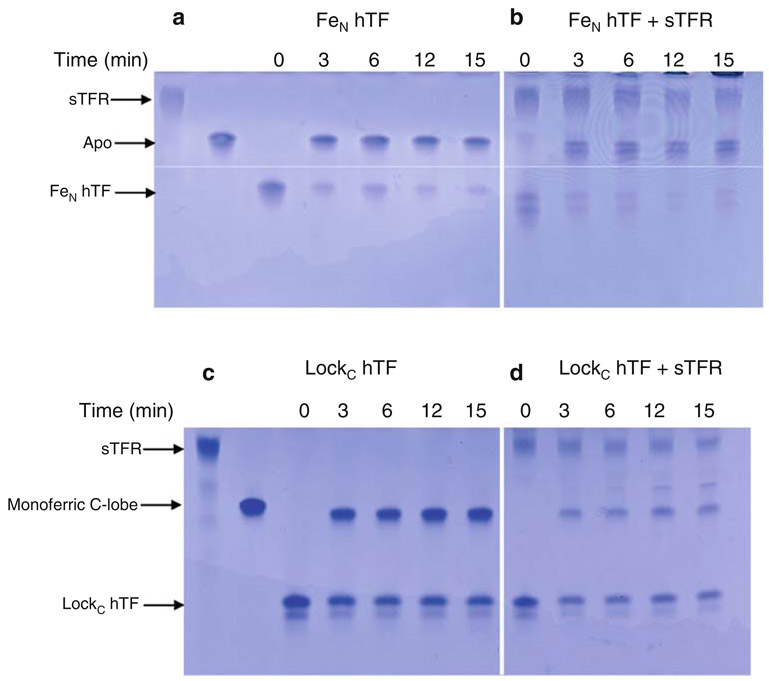
Influence of the sTFR on time-based iron release from the N-lobe at pH 5.6. a FeN hTF, b FeN hTF/sTFR complex, c LockC hTF, d LockC hTF/sTFR complex. All samples were incubated for the designated time courses in iron-removal buffer (100 mM MES pH 5.6, 300 mM KCl, 4 mM EDTA). Iron release was quenched by the addition of sample buffer. See the legend to Fig. 3 for explanation of the appearance of the double band in b and c
Fig. 7.
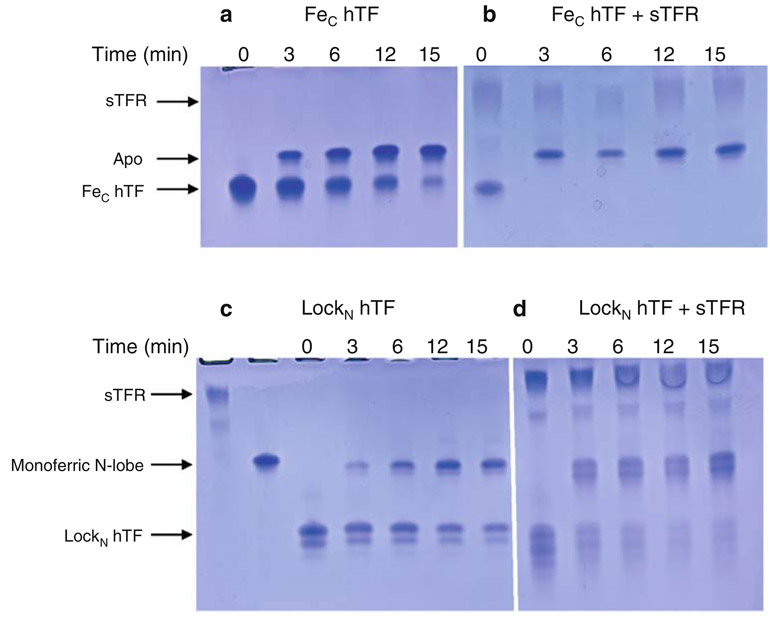
Influence of the sTFR on time-based iron release from the C-lobe at pH 5.6. a FeC hTF, b FeC hTF/sTFR complex, c LockN hTF, d LockN hTF/sTFR complex. All samples were incubated for the designated time courses in 100 mM MES pH 5.6, 300 mM KCl, 4 mM EDTA. Iron release was quenched by the addition of sample buffer. See the legend to Fig. 3 for explanation of the appearance of the double band in c and d
Studies evaluating iron release from Fe2 hTF
As shown by the urea gel in Fig. 5a, iron removal from Fe2 hTF was time-dependent, with all four species visible at the 3-min time point. Additionally, there is a time-dependent increase in apo hTF such that at the 15-min time point the predominant species was apo with an approximately equal distribution of the other three species (Fig. 5a). In the presence of the sTFR, apo and monoferric N-lobe were the only species present (Fig. 5b). In the presence of the sTFR, iron removal from the C-lobe was complete within the first 3 min. Thus, the sTFR reverses the order of iron release, strongly favoring removal from the C-lobe.
Studies evaluating iron release from the N-lobe
As shown by the urea gel in Fig. 6a and b, almost all of the iron was removed by the 3-min time point from the N-lobe of FeN hTF with an open C-lobe. In addition, there is little time dependence following the initial iron release and the sTFR had no effect.
As shown by the urea gel in Fig. 6c, by the 3-min time point only about half of the iron was removed from the N-lobe of LockC hTF, with no further time-dependent increase in the absence or presence of the sTFR (Fig. 6c, d). We note that there was a small amount of apo species present at the 6-min time point (Fig. 6d) which was not observed in the absence of the sTFR. In conclusion, it appears that more iron was removed from the N-lobe when the C-lobe was open than when it was locked (compare Fig. 6a–c), and there was no time-dependent increase. Iron removal from either FeN hTF or LockC hTF in the presence of the sTFR is equivalent to iron removal in the absence of the sTFR (compare Fig. 6a–d).
Studies evaluating iron release from the C-lobe
As shown by the urea gel in Fig. 7a, iron removal from the C-lobe of FeC hTF (with an open N-lobe) was time-dependent as indicated by an increase in the amount of apo hTF; however, complete iron removal was not observed at the 15-min time point. In contrast, in the presence of the sTFR, all of the iron was removed by the 3-min time point (Fig. 7b).
Iron removal from the C-lobe of LockN hTF was time-dependent as indicated by an increase in the monoferric N-lobe band on the urea gel in Fig. 7c, although complete iron removal was not observed at the 15-min time point (Fig. 7c). In the presence of the sTFR, iron removal was nearly complete by the 3-min time point, with a further increase as a function of time (Fig. 7d). Thus, a similar amount of iron is removed from the C-lobe whether the N-lobe is open or locked in the absence of the sTFR (compare Fig. 7a, c). The presence of the sTFR drives iron release from the C-lobe of FeC hTF to completion within the first 3 min (compare Fig. 7a, b). Similarly, almost all of the iron is removed from the C-lobe of LockN hTF in the presence of the sTFR (compare Fig. 7c, d), although complete iron removal is not achieved by the 3-min time point.
Discussion
In the current study, we have analyzed iron release from a variety of recombinant hTF constructs at different pH values, including the putative endosomal pH of approximately 5.6. The use of authentic monoferric constructs unable to bind iron in one lobe, diferric constructs with iron locked in one lobe, and the soluble portion of the specific TFR has allowed us to more thoroughly dissect the system. This work provides the most comprehensive and unambiguous assessment of the effect of pH, the sTFR, and lobe–lobe interactions on iron release that has been carried out to date. The use of urea gels combined with the data for the increase in the fluorescence intensity (as a result iron removal) allows direct visualization of the global effects of iron release. As detailed below, it solidifies many of the prevailing “truths” gathered in a less systematic fashion.
The studies described herein definitively demonstrate that iron release from the N-lobe is highly dependent on the conformation of the C-lobe. We clearly showed that there is a difference in iron removal from the N-lobe as a function of the conformation of the C-lobe [34]. Thus, when the cleft of the C-lobe is locked, we observe by urea gels that the N-lobe appears to be unable to undergo the conformational changes needed to ensure irreversible iron removal (either in the absence or in the presence of the sTFR) at pH 5.6 (Fig. 6c, d). This apparent incomplete removal of iron from the N-lobe of LockC hTF is independent of pH (Fig. 3c), time (Fig. 6c), and salt concentration (data not shown, see below). In agreement with previous work [26], when the C-lobe is in an open conformation, nearly all of the iron is removed from the N-lobe (Fig. 3a, Fig. 6a, b).
In contrast to iron release from the N-lobe, iron release from the C-lobe is independent of the conformation of the N-lobe (open or locked), but is highly dependent on pH (Fig. 4), time (Fig. 7a, c), and salt concentration (data not shown). As reported previously [34], the presence of the sTFR greatly accelerates iron release from the C-lobe (Fig. 7).
In the case of Fe2 hTF, pH, time, and the sTFR affect iron removal (Fig. 2, Fig. 5). As in numerous studies, we showed that iron is first released from the N-lobe and then the C-lobe of Fe2 hTF (Fig. 5a) [26, 35]. In agreement with previous work, at low pH the presence of the sTFR switches the order of iron release from the N-lobe then the C-lobe, to the C-lobe then the N-lobe (Fig. 5b) [26, 34]. These results are not in accord with chemical relaxation studies monitoring iron release which suggest that iron is released from the N-lobe before the C-lobe of diferric hTF whether alone or in a complex [36]. In this work more drastic changes in the pH were used and the TFR was isolated from placenta, requiring detergent to maintain solubility. Whether micelles could interfere with the iron release process is unknown.
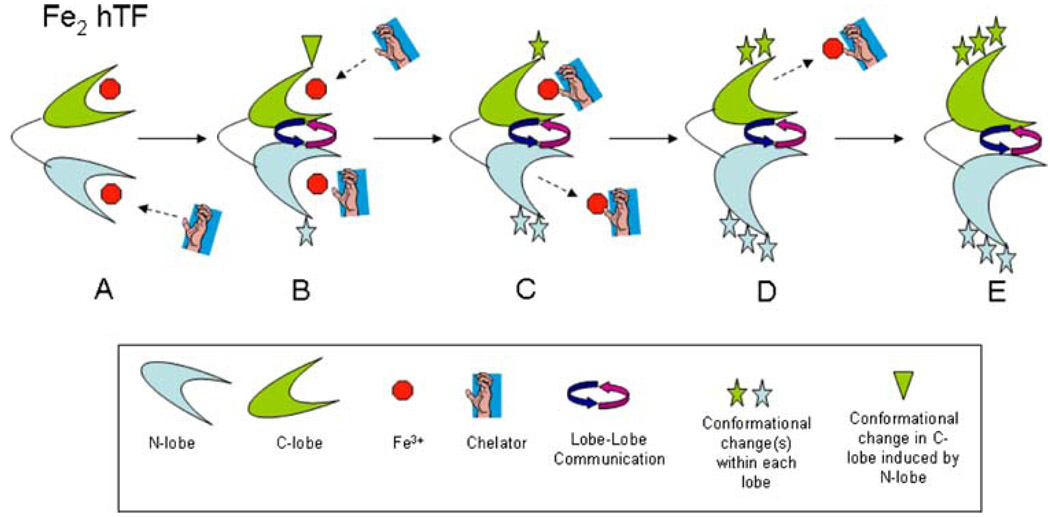
We suggest that the apparent restriction of iron release from the N-lobe of LockC hTF is due to a conformational effect of the C-lobe that disrupts the cooperativity between the lobes. Iron release is a highly dynamic process which requires that both lobes are capable of undergoing the necessary conformational changes that ultimately result in the release of iron to a chelator. It is well established that the transition from pH 7.4 to 5.6 involves a series of protonation events which allow each cleft to open and iron to be removed [5, 35]. Overlay of the apo hTF structure with the diferric pig transferrin structure suggests that most of the movement is restricted to the NII and CII subdomains, since the NI and CI subdomains closely align in the two structures. The N- and C-lobes undergo rotations of 59.4° and 49.5°, respectively, upon cleft opening [14]. X-ray scattering studies of the N-lobe indicate that a two-step process leads to cleft closure; a 20° rigid-body twist of the NII subdomain followed by a 50° hinge-bend [37]. Assuming that each lobe of hTF undergoes a similar process (in reverse) to open the cleft at pH 5.6, we suggest a possible model to explain our results. Fe2 hTF (Fig. 8, A) is shown in a fully iron bound conformation with a chelator approaching to remove the iron from the N-lobe as the pH is lowered to 5.6. Protonation events as well as the interaction of anions and the chelator with the N-lobe (Fig. 8, B) result in a conformational change, possibly the 50° hinge-bend (Fig. 8, B, blue star), which is communicated to the C-lobe, thereby changing its conformation (Fig. 8, B, green triangle). The chelator enters the iron binding cleft of the N-lobe and extracts the iron, inducing the second conformational change in the N-lobe, the 20° hinge-twist (Fig. 8, C, two blue stars). Simultaneously, anions and the chelator attack the more rigid C-lobe, triggering its equivalent hinge-bend motion (Fig. 8, C, green star) and allowing entry of the chelator. Following iron removal from the N-lobe and complete elimination of the iron bound to the chelator, the N-lobe is able to adopt its final apo conformation (Fig. 8, D, three blue stars). Subsequently, the iron is extracted from the C-lobe by the chelator as a result of an equivalent hinge-twisting motion (Fig. 8, D, two green stars). Lastly (Fig. 8, E), the iron bound to the chelator is completely removed and both lobes are in an apo conformation (Fig. 8, E, three stars).
Fig. 8.
Model of iron release from Fe2 hTF. See “Discussion” for full details
When iron is locked in the C-lobe, the chelator interacts with the N-lobe and induces the first conformational change, the hinge-bend. This change is communicated to the C-lobe, but because it is locked, it cannot undergo the necessary conformational change to promote complete iron release (including departure of the iron bound to the chelator) from the N-lobe. The C-lobe communicates back to the N-lobe that it is unable to undergo this change, thereby preventing full opening and complete iron/chelator release from the N-lobe (possibly by restricting the hinge-twist motion). Thus, the locked C-lobe restricts the movement of both lobes and partially disrupts their communication with each other. In the steady-state format the dilute sample, excess chelator, and invariable pH promote complete iron removal from the N-lobe, in spite of the restrictions imposed by the locked C-lobe. In the urea gel format we suggest that the ternary complex is not fully resolved and some of the iron rebinds to the N-lobe when the pH 8.4 sample buffer is added. As described in “Results,” this is uniquely promoted by the locked C-lobe. It is curious and not obvious why this occurs in approximately half of the sample.
A previous study, designed to identify a chelator that would be effective at pH 7.4, showed that iron release from the N-lobe critically depended on a closed C-lobe [9]. In this work, it is suggested that at pH 7.4, an open C-lobe blocks an anion binding site in the N-lobe necessary for iron release. As described above, at pH 5.6 iron release from the N-lobe appears to be partially inhibited by locking the C-lobe closed with no dependence on salt, strongly indicating that lobe-lobe communication and cooperativity change as a function of pH.
We provide further unequivocal evidence for the requirement of the sTFR for complete iron release from the C-lobe regardless of whether the N-lobe can release iron (Fe2 hTF), has no iron (FeC hTF), or has iron locked in (LockN hTF) (Fig. 5b, Fig. 7b, d). Iron release from the N-lobe of FeN hTF and LockC hTF is unaffected by the presence of the sTFR. The time-based patterns are identical to those observed for these samples in the absence of the sTFR, i.e., iron removal from the N-lobe of FeN hTF is complete and iron removal from the N-lobe of LockC hTF is restricted (Fig. 6). However, iron release from the N-lobe of Fe2 hTF bound to the sTFR is partially inhibited as indicated by the presence of the monoferric N-lobe species on the urea gel (Fig. 5b).
Although the C-lobe binds iron with higher affinity, considerably more monoferric N-lobe than monoferric C-lobe is present in the serum (23 and 11%, respectively) [2]. Our studies provide support for the suggestion [26] that this monoferric N-lobe comes from Fe2 hTF that has been taken into the cell through receptor-mediated endocytosis. Preferential release of iron from the C-lobe could result in the return of the monoferric N-lobe/TFR complex to the serum.
In summary, iron release from the N-lobe is highly dependent on the conformation of the C-lobe, but is independent of changes in pH and time. In contrast, iron release from the C-lobe is dependent on pH and time and independent of the conformation of the N-lobe. Importantly, iron release from Fe2 hTF is not a simple combination of iron release from FeC hTF and FeN hTF. Fe2 hTF can undergo all of the dynamic conformational changes required for iron release to a chelator at low pH allowing complete communication and cooperativity between the lobes. The significance and complexity of the bilobal structure of hTF is indicated by the different iron-release properties observed when the lobes are altered (to prevent iron binding or to lock iron in a lobe). Although the two lobes are homologous, their differences are further evidenced by their responses to conformation and changes in pH.
Supplementary Material
The online version of this article (doi:10.1007/s00775-009-0491-y) contains supplementary material, which is available to authorized users.
Acknowledgment
This work was supported by USPHS Grant R01 (DK 21739) to A.B.M.. Support for S.L.B. came from Hemostasis and Thrombosis Training Grant (5T32HL007594), issued to K. G. Mann at the University of Vermont by the National Heart, Lung, and Blood Institute.
Abbreviations
- BHK
Baby hamster kidney cells
- DMEM-F12
Dulbecco’s modified Eagle’s medium containing Ham F-12 nutrient mixture
- FeC hTF
Nonglycosylated recombinant monoferric C-lobe human serum transferrin (mutations Y95F and Y188F preclude binding in the N-lobe) that contains an N-terminal His6 tag
- Fe2 hTF
Nonglycosylated recombinant diferric human serum transferrin that contains an N-terminal His6 tag
- FeN hTF
Nonglycosylated recombinant monoferric N-lobe human serum transferrin (mutations Y426F and Y517F preclude binding in the C-lobe) that contains an N-terminal His6 tag
- hTF
Human serum transferrin
- LockC hTF
Nonglycosylated recombinant diferric hTF that contains an N-terminal His6 tag and where mutation R632A locks iron in the C-lobe
- LockN hTF
Nonglycosylated recombinant diferric hTF that contains an N-terminal His6 tag and where mutation K206E locks iron in the N-lobe
- MES
2-Morpholinoethanesulfonic acid
- sTFR
Soluble portion of the transferrin receptor (residues 121–760) expressed as a recombinant entity that contains an N-terminal His6 tag
- TBE
Tris(hydroxymethyl)aminomethane–borate–EDTA
- TFR
Human serum transferrin receptor
- Tris
Tris(hydroxymethyl)aminomethane
References
- 1.Halbrooks PJ, He QY, Briggs SK, Everse SJ, Smith VC, Mac-Gillivray RT, Mason AB. Biochemistry. 2003;42:3701–3707. doi: 10.1021/bi027071q. [DOI] [PubMed] [Google Scholar]
- 2.Williams J, Moreton K. Biochem J. 1980;185:483–488. doi: 10.1042/bj1850483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huebers H, Josephson B, Huebers E, Csiba E, Finch C. Proc Natl Acad Sci USA. 1981;78:2572–2576. doi: 10.1073/pnas.78.4.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason AB, Halbrooks PJ, James NG, Connolly SA, Larouche JR, Smith VC, MacGillivray RT, Chasteen ND. Biochemistry. 2005;44:8013–8021. doi: 10.1021/bi050015f. [DOI] [PubMed] [Google Scholar]
- 5.He QY, Mason AB. In: Molecular and cellular iron transport. Templeton DM, editor. Toronto: Dekker; 2002. [Google Scholar]
- 6.Dautry-Varsat A, Ciechanover A, Lodish HF. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bali PK, Harris WR. J Am Chem Soc. 1989;111:4457–4461. [Google Scholar]
- 8.Chasteen ND, Grady JK, Woodworth RC, Mason AB. Adv Exp Med Biol. 1994;357:45–52. doi: 10.1007/978-1-4615-2548-6_5. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton DH, Turcot I, Stintzi A, Raymond KN. J Biol Inorg Chem. 2004;9:936–944. doi: 10.1007/s00775-004-0592-6. [DOI] [PubMed] [Google Scholar]
- 10.Dewan JC, Mikami B, Hirose M, Sacchettini JC. Bio-chemistry. 1993;32:11963–11968. doi: 10.1021/bi00096a004. [DOI] [PubMed] [Google Scholar]
- 11.He QY, Mason AB, Tam BM, MacGillivray RT, Woodworth RC. Biochemistry. 1999;38:9704–9711. doi: 10.1021/bi990134t. [DOI] [PubMed] [Google Scholar]
- 12.Halbrooks PJ, Giannetti AM, Klein JS, Bjorkman PJ, Larouche JR, Smith VC, MacGillivray RT, Everse SJ, Mason AB. Biochemistry. 2005;44:15451–15460. doi: 10.1021/bi0518693. [DOI] [PubMed] [Google Scholar]
- 13.Hall DR, Hadden JM, Leonard GA, Bailey S, Neu M, Winn M, Lindley PF. Acta Crystallogr D Biol Crystallogr. 2002;58:70–80. doi: 10.1107/s0907444901017309. [DOI] [PubMed] [Google Scholar]
- 14.Wally J, Halbrooks PJ, Vonrhein C, Rould MA, Everse SJ, Mason AB, Buchanan SK. J Biol Chem. 2006;281:24934–24944. doi: 10.1074/jbc.M604592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan TJ, Zak O, Aisen P. Biochemistry. 1993;32:8162–8167. doi: 10.1021/bi00083a016. [DOI] [PubMed] [Google Scholar]
- 16.Lehrer SS. J Biol Chem. 1969;244:3613–3617. [PubMed] [Google Scholar]
- 17.Ross JA, Jameson DM. Photochem Photobiol Sci. 2008;7:1301–1312. doi: 10.1039/b804450n. [DOI] [PubMed] [Google Scholar]
- 18.James NG, Berger CL, Byrne SL, Smith VC, Macgillivray RT, Mason AB. Biochemistry. 2007;46:10603–10611. doi: 10.1021/bi602425c. [DOI] [PubMed] [Google Scholar]
- 19.James NG, Byrne SL, Steere AN, Smith VC, MacGillivray RTA, Mason AB. Biochemistry. 2009 doi: 10.1021/bi8022834. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makey DG, Seal US. Biochim Biophys Acta. 1976;453:250–256. doi: 10.1016/0005-2795(76)90270-1. [DOI] [PubMed] [Google Scholar]
- 21.Aisen P, Leibman A, Zweier J. J Biol Chem. 1978;253:1930–1937. [PubMed] [Google Scholar]
- 22.Evans RW, Williams J. Biochem J. 1978;173:543–552. doi: 10.1042/bj1730543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leibman A, Aisen P. Blood. 1979;53:1058–1065. [PubMed] [Google Scholar]
- 24.Chasteen ND, Williams J. Biochem J. 1981;193:717–727. doi: 10.1042/bj1930717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams J, Chasteen ND, Moreton K. Biochem J. 1982;201:527–532. doi: 10.1042/bj2010527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bali PK, Aisen P. Biochemistry. 1991;30:9947–9952. doi: 10.1021/bi00105a019. [DOI] [PubMed] [Google Scholar]
- 27.Mason AB, Miller MK, Funk WD, Banfield DK, Savage KJ, Oliver RW, Green BN, MacGillivray RT, Woodworth RC. Biochemistry. 1993;32:5472–5479. doi: 10.1021/bi00071a025. [DOI] [PubMed] [Google Scholar]
- 28.Turkewitz AP, Amatruda JF, Borhani D, Harrison SC, Schwartz AL. J Biol Chem. 1988;263:8318–8325. [PubMed] [Google Scholar]
- 29.Bali PK, Zak O, Aisen P. Biochemistry. 1991;30:324–328. doi: 10.1021/bi00216a003. [DOI] [PubMed] [Google Scholar]
- 30.Mason AB, Halbrooks PJ, Larouche JR, Briggs SK, Moffett ML, Ramsey JE, Connolly SA, Smith VC, MacGillivray RT. Protein Expr Purif. 2004;36:318–326. doi: 10.1016/j.pep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Byrne SL, Leverence R, Klein JS, Giannetti AM, Smith VC, MacGillivray RT, Kaltashov IA, Mason AB. Biochemistry. 2006;45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 32.Mason AB, He QY, Adams TE, Gumerov DR, Kaltashov IA, Nguyen V, MacGillivray RT. Protein Expr Purif. 2001;23:142–150. doi: 10.1006/prep.2001.1480. [DOI] [PubMed] [Google Scholar]
- 33.James NG, Mason AB. Anal Biochem. 2008;378:202–207. doi: 10.1016/j.ab.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bali PK, Aisen P. Biochemistry. 1992;31:3963–3967. doi: 10.1021/bi00131a011. [DOI] [PubMed] [Google Scholar]
- 35.El Hage Chahine JM, Pakdaman R. Eur J Biochem. 1995;230:1102–1110. doi: 10.1111/j.1432-1033.1995.tb20661.x. [DOI] [PubMed] [Google Scholar]
- 36.Hemadi M, Ha-Duong NT, El Hage Chahine JM. J Mol Biol. 2006;358:1125–1136. doi: 10.1016/j.jmb.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 37.Grossmann JG, Crawley JB, Strange RW, Patel KJ, Murphy LM, Neu M, Evans RW, Hasnain SS. J Mol Biol. 1998;279:461–472. doi: 10.1006/jmbi.1998.1787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version of this article (doi:10.1007/s00775-009-0491-y) contains supplementary material, which is available to authorized users.