Abstract
SodC is one of two superoxide dismutases produced by Mycobacterium tuberculosis. This protein was previously shown to contribute to virulence and to act as a B-cell antigen. SodC is also a putative lipoprotein, and like other Sec-translocated mycobacterial proteins it was suggested to be modified with glycosyl units. To definitively define the glycosylation of SodC, we applied an approach that combined site-directed mutagenesis, lectin binding, and mass spectrometry. This resulted in identification of six O-glycosylated residues within a 13-amino-acid region near the N-terminus. Each residue was modified with one to three hexose units, and the most dominant SodC glycoform was modified with nine hexose units. In addition to O-glycosylation of threonine residues, this study provides the first evidence of serine O-glycosylation in mycobacteria. When combined with bioinformatic analyses, the clustering of O-glycosylation appeared to occur in a region of SodC with a disordered structure and not in regions important to the enzymatic activity of SodC. The use of recombinant amino acid substitutions to alter glycosylation sites provided further evidence that glycosylation influences proteolytic processing and ultimately positioning of cell wall proteins.
Keywords: Glycoprotein, lipoprotein, mycobacterium, superoxide dismutase, tuberculosis
Introduction
Several of the immunodominant antigens of Mycobacterium tuberculosis (Mtb) are reported to be glycosylated based on their ability to bind the lectin concanavalin A (ConA) (Espitia and Mancilla 1989; Fifis et al. 1991; Garbe et al. 1993; Dobos et al. 1995). Yet the presence of glycoproteins in Mtb did not gain wide acceptance until a mass spectrometry (MS)-based analysis of the 45/47-kDa MPT32/Apa protein (Rv1860) demonstrated four separate Thr residues each O-linked with a mannose, mannobiose, or mannotriose (Dobos et al. 1996). Additional glycoproteins of the Mtb complex were subsequently identified. Two adjacent Thr residues from the secreted antigen MPB83 (Mb2898) of Mycobacterium bovis were shown by MS-based methods to be modified with a total of three mannoses (Michell et al. 2003). Using alternative approaches, others have reported the glycosylation of recombinant Mtb proteins produced in Mycobacterium smegmatis. Most recently, a member of the Mtb PPE protein family (Rv3873) produced in M. smegmatis appeared to be glycosylated at its C-terminus (Daugelat et al. 2003). Herrmann et al. (1996) detected glycosylation of two Thr clusters in the recombinant 19-kDa LpqH lipoprotein (Rv3763) and showed that glycosylation of this region protected the recombinant 19-kDa lipoprotein from proteolysis. Further work in M. smegmatis identified four putative glycolipoproteins using recombinant gene fusions in combination with ConA-based analyses (Herrmann et al. 2000). One of these putative lipoglycoproteins was identified as Rv0432 or SodC. Specifically, a 40-aa N-terminal fragment of SodC was determined to be glycosylated by virtue of its ability to bind ConA.
Recently, we employed native antigen array profiling to identify and characterize serodiagnostic proteins of Mtb (Sartain et al. 2006). Through this study, four novel antigens were discovered including SodC. This protein is one of two Mtb superoxide dismutases (SODs) and is a membrane-associated lipoprotein of the Cu,Zn-dependent SOD family (D’Orazio et al. 2001). The recognition of SodC by human antibodies (Sartain et al. 2006) provides compelling evidence that this enzyme is produced in a natural infection. Furthermore, SodC is shown to contribute to the survival of Mtb in activated macrophages (Piddington et al. 2001), suggesting a role in the defense against the oxidative burst produced in vivo. Transcription of sodC is also greatly upregulated upon infection of human macrophages, providing additional evidence of an importance in early infection when the NADPH oxidase level is expected to be high (D’Orazio et al. 2001; Volpe et al. 2006). Most recently, the Mtb SodC three-dimensional structure was solved by X-ray crystallography (Spagnolo et al. 2004). Thus, a unique opportunity presents itself to characterize the glycosylation of a known Mtb enzyme with a defined 3D structure and function, as well as a putative role in virulence.
To assess glycosylation of SodC from Mtb, a recombinant form of this protein as well as specific mutants in putative glycosylated residues was produced and analyzed by lectin binding and MS. We identified six sites of glycosylation within a 13-amino-acid region of the mature N-terminus of SodC. Glycosylation at each site was with one to three α-mannose units, and for the first time in Mycobacterium spp. glycosylation of Ser residues was observed.
Results
Expression of recombinant SodC in Escherichia coli and Mtb
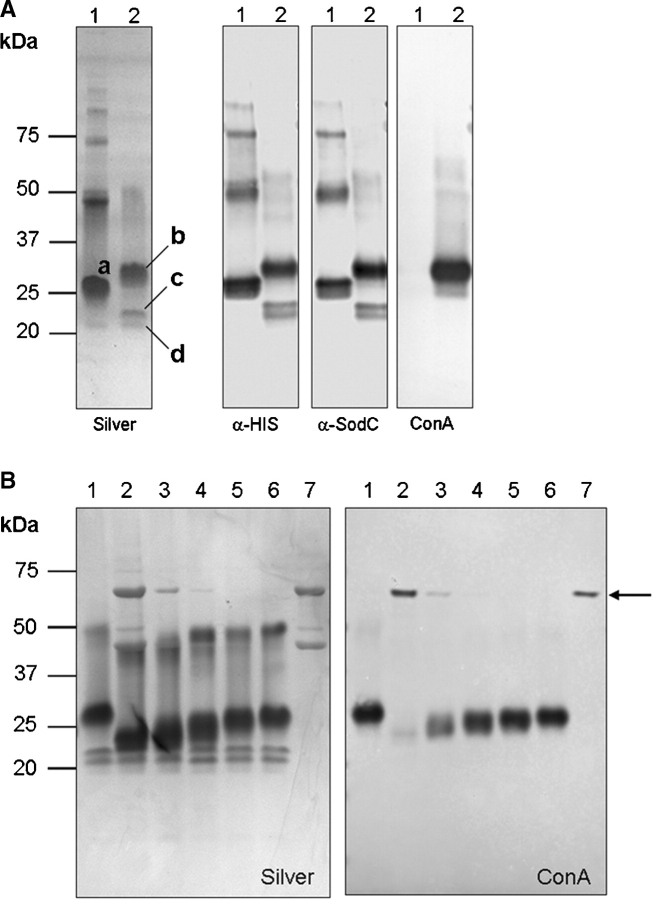
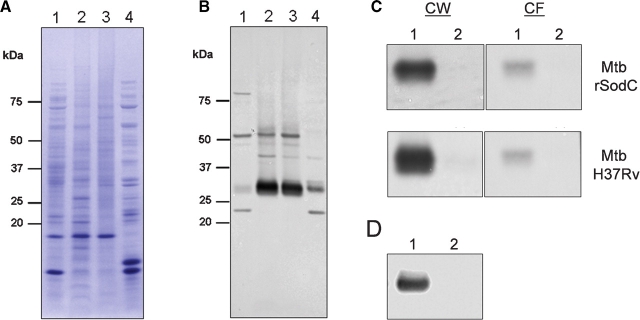
SodC was shown by others to be a lipoprotein presumably localized to the cellular envelope (D’Orazio et al. 2001), but our studies identified SodC as a potential serodiagnostic antigen present in the culture filtrate (CF) of in vitro grown Mtb (Sartain et al. 2006). To allow for isolation of SodC from the CF in quantities sufficient for biochemical analyses, a recombinant form of SodC with a signal peptide (rSodC) was produced in Mtb. For comparative studies, SodC without a signal peptide (SP-rSodC) was generated in Escherichia coli. SP-rSodC and rSodC were purified from the cell lysate of E. coli and CF of Mtb, respectively, by immobilized metal affinity chromatography (IMAC). Analysis of the recombinant proteins by SDS–PAGE revealed multiple products, regardless of source (Figure 1A). The SP-rSodC from E. coli migrated at ∼26 kDa, which is larger than the expected molecular mass of 21.8 kDa. This is a common observation for SODs in general (Battistoni and Rotilio 1995) and has been specifically described for Mtb SodC produced in E. coli (D’Orazio et al. 2001). Additionally, a putative dimer was observed at ∼47 kDa along with a series of multimers. The major rSodC product isolated from the Mtb CF migrated at ∼28 kDa, with minor products at ∼21 and 22 kDa, and a putative dimer at ∼52 kDa. All of these products reacted with the anti-His5 monoclonal antibody and SodC polyclonal antiserum. To ascertain the glycosylation status of SodC, the purified proteins were probed with the lectin ConA. The 28-kDa and 52-kDa rSodC products from Mtb displayed dominant binding to ConA, while the 21- and 22-kDa products clearly lacked ConA reactivity. As expected, the SP-rSodC from E. coli lacked ConA reactivity (Figure 1A).
Fig. 1.
ConA reactivity demonstrates glycosylation of Mtb rSodC. (A) Purified rSP-SodC (lane 1) and purified rSodC (lane 2) analyzed by SDS–PAGE and silver staining (panel 1) and Western blot analyses with anti-His5 monoclonal antibody (panel 2), polyclonal SodC antiserum (panel 3), and ConA as probes (panel 4). The proteins displayed are (a) the major 26-kDa product from E. coli; (b) the major 28-kDa product from Mtb; and (c and d) the minor 22- and 21-kDa products from Mtb. (B) Silver-stained SDS–PAGE (panel 1) and ConA Western blot (panel 2) analyses of purified rSodC from Mtb treated with various amounts of α-mannosidase. Lane 1, untreated rSodC; lanes 2–6, rSodC (10 μg) treated with 10, 2, 0.2, 0.1, and 0.02 μg α-mannosidase; and lane 7, α-mannosidase alone. The 44- and 66-kDa subunits of α-mannosidase are observed in the silver-stained gel (panel 1), and the 66-kDa subunit modified with a high-mannose-type-glycan (Kimura et al. 1999) displays ConA reactivity (denoted by arrow).
The presence of lower molecular mass rSodC products retaining reactivity against the anti-His5 monoclonal antibody suggested truncation occurring at the N-terminus. Therefore, N-terminal amino-acid sequencing of each product was performed to determine their full intact sequences. N-Terminal sequencing of rSodC from Mtb revealed a sequence of 41TVPGXXPXI49 for the dominant 28-kDa product, while sequencing of the two smaller products was more ambiguous with a mixture of products beginning with Gly58 and Ser60. (Note: numbering of amino acids begins with fMet1 of the full sodC gene product without processing.) The 26-kDa SP-rSodC product from E. coli produced an NH2-terminus of 33CSSPQ37, directly corresponding to the cloned gene fragment with processing of the fMet encoded by the ATG start codon (Link et al. 1997; Wasinger and Humphery-Smith 1998). These N-terminal sequence data demonstrated that the Mtb CF form of rSodC was not acylated. Additionally, the major product from Mtb was ∼2 kDa greater in mass than that from E. coli, yet was eight amino acids shorter. This suggested the presence of another modification on the Mtb product, presumably glycosylation. In support of this, jack-bean-α-mannosidase treatment of rSodC from Mtb resulted in a decrease in molecular mass (as observed by SDS–PAGE) and loss of ConA reactivity for the 28- and 52-kDa products (Figure 1B). The magnitude of losses in mass and ConA-reactivity correlated with increasing concentrations of α-mannosidase. Thus, glycosylation appeared to significantly alter the mobility of SodC in SDS–PAGE. Treatment with α-mannosidase did not have any effect on the migration pattern of the lower mass (21- and 22-kDa) products (Figure 1B). This with the lack of ConA reactivity indicated that the 21- and 22-kDa products lacked glycosylation and that glycosylation of the 28-kDa product may be localized to the N-terminal region.
MS-based identification of rSodC glycopeptides
To identify SodC glycosylated regions(s), a variety of MS-based approaches were utilized. Purified rSodC from Mtb was digested with trypsin, chymotrypsin, and thermolysin and analyzed by liquid chromatography-electrospray-ionization-tandem mass spectrometry (LC-ESI-MS/MS), and the resulting MS/MS data searched using SEQUEST (Yates et al. 1995) and X! Tandem (Fenyo and Beavis 2003) software against the rSodC protein sequence that started with the experimentally determined Thr41 N-terminus. As summarized in Figure 2, interrogation of the MS/MS data for peptide identification, without inclusion of potential O-glycosylation, resulted in the recombinant protein being mapped in its entirety except for the N-terminal Thr41-Leu59 sequence and the peptide 123CEPNS127 (Table SI). The inability to identify unmodified N-terminal peptide(s) was consistent with the hypothesized glycosylation of the N-terminal region.
Fig. 2.
The deduced amino acid sequence (beginning with Thr41) of the 28-kDa rSodC protein product purified from Mtb and alignment with the peptides generated by chymotrypsin (Chy), trypsin (Try), and thermolysin (TL) digestion. The solid lines indicate the location of individual unmodified peptides identified by LC-ESI-MS/MS, and the boxes indicate unidentified sequences. The dashed lines indicate two predicted chymotrypsin-derived peptides used for the remainder of this work.
MS and MS/MS of rSodC Chy2 Glycopeptides
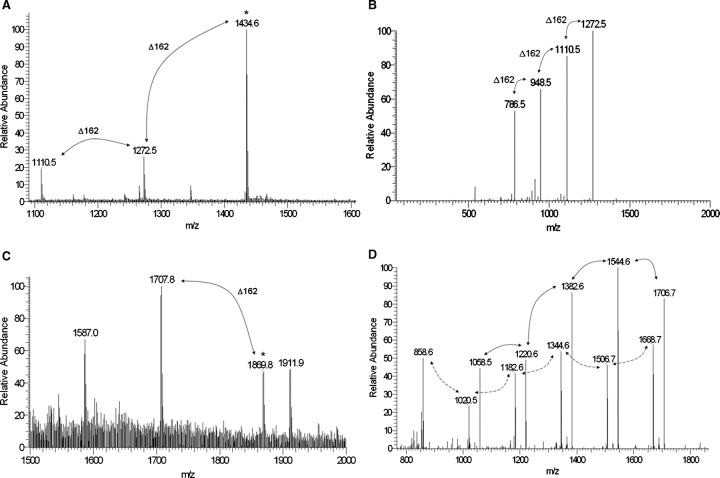
To identify glycopeptides, the MS/MS data of chymotrypsin-digested rSodC were searched for neutral losses of 162 amu, a diagnostic property of the dissociation of hexose units from a singly charged glycopeptide (Dobos et al. 1996). Three major [M+H]+1 molecular ions (m/z 1110.5, 1272.5, and 1434.6) were found that differed by 162 amu and corresponded to the peptide designated Chy2 (51TGSPAPSGL59) with two, three, and four hexose units, respectively (Figures 2 and 3A, Table I). MS/MS fragmentation of the largest molecular ion (m/z 1434.6) generated the [M+H]+1 daughter ions of m/z 1272.5, 1110.5, 948.5, and 786.5 arising from neutral losses of 162 amu (Figure 3B and Table I). The m/z 786.5 fragmentation ion corresponded to the nonglycosylated Chy2 peptide. A similar search of the MS/MS data for molecular ions and neutral losses corresponding to a Chy1 glycopeptide was unsuccessful.
Fig. 3.
MS and MS/MS of glycopeptides generated by chymotrypsin digestion of rSodC. (A) An ESI-MS spectrum averaged over 80 scans corresponding to the Chy2 (51TGSPAPSGL59) peptide demonstrates primarily glycosylated forms modified with two, three, and four hexoses. (B) The m/z 1434.6 [M+H]+1 molecular ion marked by an asterisk in (A) was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu. The m/z 786.5 molecular ion represents the fully deglycosylated peptide. (C) An ESI-MS spectrum averaged over 96 scans corresponding to the Chy1 (41TVPGTTPSIW50) peptide demonstrates primarily glycosylated forms modified with four and five hexoses. (D) The m/z 1869.8 [M+H]+1 molecular ion marked by an asterisk in (C) was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu (marked with solid arrows). The m/z 1058.5 molecular ion represents the fully deglycosylated peptide. In addition, an ion series corresponding to a glycosylated y8 ion series with differences of 162 amu was observed (marked with dashed arrows).
Table I.
Summary of rSodC glycopeptide MS data
| Peptide | Sequence | Predicted [M+H]+1 ion | Observed [M+H]+1 ion | MS/MS ionsa |
|---|---|---|---|---|
| Chy1 | TVPGTTPSIW | 1059.2 | Not observed | |
| Chy1 | TVPGTTPSIW + 1Hex | 1221.3 | Not observed | |
| Chy1 | TVPGTTPSIW + 2Hex | 1383.5 | Not observed | |
| Chy1 | TVPGTTPSIW + 3Hex | 1545.6 | Not observed | |
| Chy1 | TVPGTTPSIW + 4Hex | 1707.8 | 1707.8 | 1544.6, 1382.6, 1220.6, 1058.6, 1506.6*,1344.6*, |
| 1182.6*, 1020.5*, 858.5* | ||||
| Chy1 | TVPGTTPSIW + 5Hex | 1869.9 | 1869.8 | 1706.7, 1544.6, 1382.6, 1220.6, 1058.5, 1668.7*, |
| 1506.7*, 1344.6*, 1182.6*, 1020.5*, 858.6* | ||||
| Chy2 | TGSPAPSGL | 786.9 | Not observed | |
| Chy2 | TGSPAPSGL + 1Hex | 949.0 | Not observed | |
| Chy2 | TGSPAPSGL + 2Hex | 1111.1 | 1110.5 | 948.5, 786.5 |
| Chy2 | TGSPAPSGL + 3Hex | 1273.3 | 1272.5 | 1110.5, 948.5, 786.5 |
| Chy2 | TGSPAPSGL + 4Hex | 1435.4 | 1434.6 | 1272.5, 1110.5, 948.5, 786.5 |
aThe ions listed correspond to neutral losses of 162 amu.
Ions marked with an asterisk are part of the glycosylated y8 ion series.
MS and MS/MS of rSodC Chy1 Glycopeptides
The inability to identify a Chy1 glycopeptide by ESI-MS was not unexpected given ionization suppression often occurs with glycopeptides (Cutalo et al. 2004). Therefore, a more targeted approach was designed, whereby molecular ions with m/z values corresponding to a predicted Chy1 peptide containing zero to five hexoses were selected from the full MS scan and further subjected to MS/MS analyses. Two major [M+H]+1 molecular ions (m/z 1707.8 and 1869.8) differing by 162 amu and corresponding to the Chy1 (41TVPGTTPSIW50) peptide with four and five hexose units, respectively, were found in the MS data (Figure 3C and Table I). MS/MS analysis of the larger molecular ion (m/z 1869.8) generated the [M+H]+1 daughter ions of m/z 1706.7, 1544.6, 1382.6, 1220.6, and 1058.5 arising from neutral losses of 162 amu (Figure 3D and Table I). This provides strong evidence of a Chy1 glycopeptide possessing five hexoses. The MS/MS spectra was noted as unusual in that a second ion series with neutral losses of 162 amu was also observed (m/z 1668.7, 1506.7, 1344.6, 1182.6, 1020.5, and 858.6), and the largest ion in this series (m/z 1668.7) differed from the parent ion by m/z 201.1. This series corresponds to glycosylation of the y8 ion (m/z 858.6) that resulted from fragmentation between Val42 and Pro43. Also, observed in the parent ion series (Figure 3C) were two ions (m/z 1587.0 and 1911.9) that differ by two hexose units (324.9 amu). The m/z 1911.9 ion differs from the confirmed five-hexose-Chy1 peptide ion (m/z 1869.8) by 42.1 amu. This would indicate either an N-terminal acetylated five-hexose-Chy1 peptide or a four-hexose Chy1 peptide with a single O-acetyl-, N-acetyl-, or tri-O-methyl hexose. Studies are ongoing to determine the exact structures represented by the m/z 1587.0 and 1911.9 ion series.
MS of Intact rSodC
The overall glycoprotein structure of rSodC from Mtb was supported by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS analysis of the intact glycoprotein. Specifically, we observed singly charged molecular ions of the glycosylated rSodC 28-kDa product (N-terminus of Thr41) possessing between 6 and 10 hexose units (data not shown). Additionally, the mass spectrum shows a set of peaks assigned to singly charged molecular ions of the nonglycosylated 22- and 21-kDa rSodC products (N-termini of Gly58 and Ser60, respectively) and minor peaks assigned to nonglycosylated products with N-termini of Gly61, Asp63, Glu64, Glu65, Ser66, and Gly68 (data not shown).
Expression of site-directed mutant rSodC proteins in Mtb
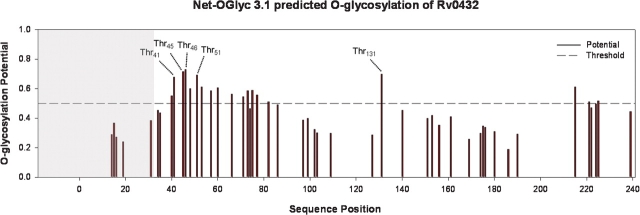
The MS-based analyses demonstrated SodC glycosylation localized to the N-terminus and were in agreement with the differential ConA reactivity profiles and N-terminal sequencing of the 21-, 22-, and 28-kDa rSodC products from Mtb. These results were further supported by analysis of the SodC sequence with the NetOglyc 3.1 program, a neural network algorithm trained on eukaryotic mucin-type O-glycosylation sites (Julenius et al. 2005) (Figure 4). In general, the highest predictive scores for glycosylation were found in the N-terminal region of the protein. Specifically, four Thr and four Ser residues predicted to be O-glycosylated were located upstream of the N-terminal amino acids of the non-ConA reactive 21- and 22-kDa products.
Fig. 4.
Prediction of O-glycosylation sites for the complete Rv0432 protein sequence using the NetOglyc 3.1 algorithm (Julenius et al. 2005). The shaded region indicates the signal peptide. The labeled Thr residues indicate the predicted glycosylation sites targeted for amino acid substitution with Ala.
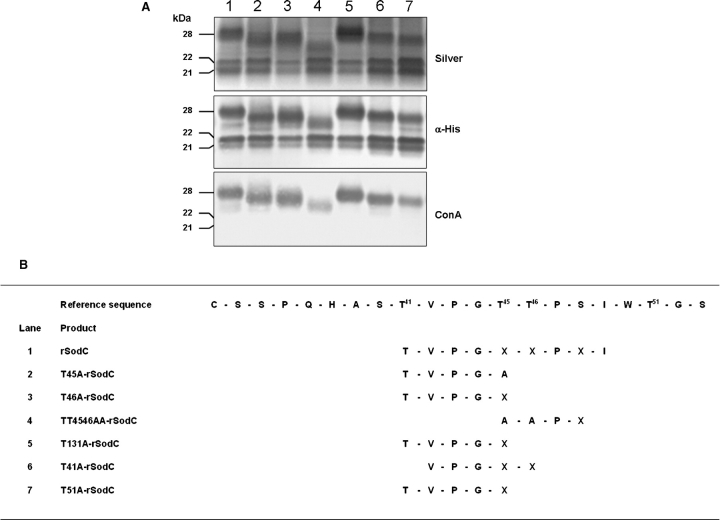
To identify specific O-glycosylated residues of SodC, those amino acids with the highest predictive NetOglyc scores (Thr41, Thr45, Thr46, and Thr51) were selected for site-directed mutagenesis to allow substitution of Thr with Ala. The more C-terminal Thr131 residue was also targeted as this residue had a high predictive score for glycosylation. Additionally, a double mutant in Thr45 Thr46 was constructed. Episomal copies of the sodC gene containing the site-directed mutations were expressed in Mtb and the proteins purified from the CF. Analysis of the recombinant proteins by SDS–PAGE and Western blot revealed multiple bands for each protein (Figure 5A). Like the nonmutated rSodC, ConA-negative products of 21- and 22-kDa were observed for all mutated rSodC proteins. The relative abundance of these two products varied among the protein constructs, but no differences in electrophoretic migration were observed. Variations in molecular mass, however, were observed by SDS–PAGE when comparing the nonmutated 28-kDa rSodC product to the corresponding products with Thr to Ala substitutions (Figure 5A). All of the altered products except T131A-rSodC displayed some reduction in mass. These mass differences were not due to truncation at the C-terminus since all of the higher molecular mass products displayed similar reactivity against the anti-His5 monoclonal antibody. To determine differences in the N-termini, each high-mass product (26–28 kDa) was subjected to N-terminal sequencing (Figure 5B). Four of the six mutated proteins (T45A-rSodC, T46A-rSodC, T51A-rSodC, and T131A-rSodC) possessed N-terminal sequences identical to the nonmutated rSodC, and T41A-rSodC differed only in the loss of the N-terminal Thr41. In contrast, a more pronounced difference was observed in the N-terminal sequence of the TT4546AA-rSodC product where a four-amino-acid truncation occurred as compared to nonmutated rSodC, and SDS–PAGE migration indicated a mass loss of approximately 2 kDa (Figure 5A, lane 4, and Figure 5B). The TT4546AA-rSodC product was the only construct that also showed a significant decrease in ConA reactivity. Therefore, the observed reduction in the mass of TT4546AA-rSodC was attributed to the absence of 41TVPG44 and decreased glycosylation. A qualitative decrease in ConA reactivity was not as apparent for the other mutated proteins (T45A-rSodC, T46A-rSodC, T51A-rSodC, and T41A-rSodC) that had modest shifts in their SDS–PAGE migration. Nevertheless, decreased glycosylation was speculated to be the cause for these molecular mass shifts since TT4546AA-rSodC retained modest levels of ConA reactivity. It was noted that amino acid assignment by Edman degradation sequencing was consistently difficult for the positions corresponding to Thr45, Thr46, and Ser48, a known characteristic of O-glycosylated residues (Abernathy et al. 1992).
Fig. 5.
Analyses of purified rSodC proteins possessing Thr to Ala substitutions. (A) Silver-stained SDS–PAGE (top panel) and Western blot analyses with anti-His5 monoclonal antibody (middle panel) and ConA (bottom panel) as probes. Lane 1, nonmutated rSodC; lane 2, T45A-rSodC; lane 3, T46A-rSodC; lane 4, TT4546AA-rSodC; lane 5, T131A-rSodC; lane 6, T41A-rSodC; and lane 7, T51A-rSodC. (B) Amino-terminal sequencing of purified rSodC products. An “X” denotes the inability to assign an amino acid. The reference sequence provided begins with the putative lipidated Cys33 residue following signal peptide cleavage. Only the high-mass (26- to 28-kDa) products were sequenced.
MS studies of mutant rSodC proteins
MS of T51A-rSodC
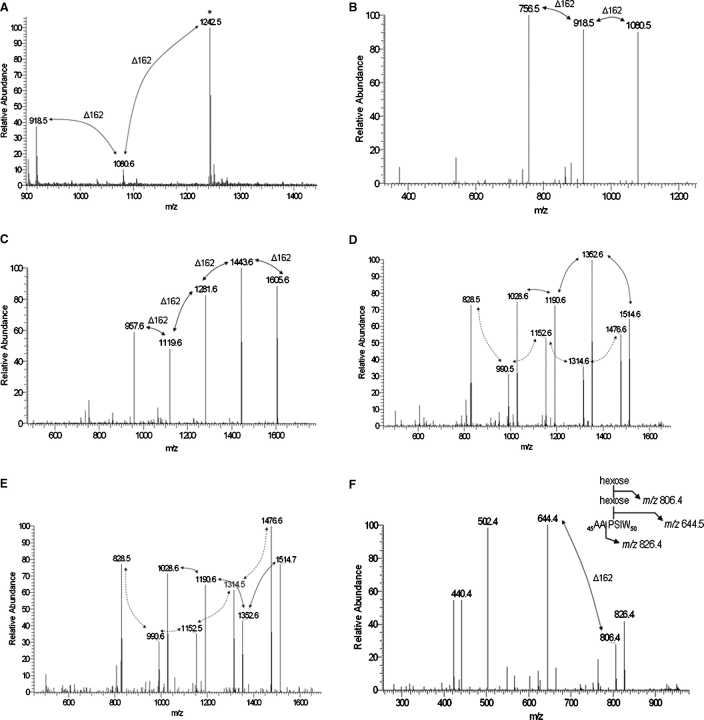
To provide a more definitive measure of the level of glycosylation as well as to identify specific O-glycosylated residues, chymotrypsin digestions of the mutant rSodC proteins were analyzed by LC-ESI-MS/MS and the data analyzed in the same manner as that for the nonmutated rSodC. Searching the T51A-rSodC MS/MS data for the spectra arising from neutral losses of 162 amu revealed three major [M+H]+1 molecular ions (m/z 918.5, 1080.6, and 1242.5) that differed by 162 amu and corresponded to the T51A-Chy2 peptide (51AGSPAPSGL59) with modifications of one, two, and three hexoses, respectively (Figure 6A and Table II). MS/MS fragmentation of the largest molecular ion (m/z 1242.5) generated the [M+H]+1 daughter ions of m/z 1080.5, 918.5, and 756.5 arising from neutral losses of 162 amu (Figure 6B). Therefore, the Thr51 to Ala51 substitution resulted in the loss of only one hexose compared to nonmutated rSodC, indicating that Thr51 possessed a single hexose. Interestingly, the absence of a Thr residue on the T51A-Chy2 peptide indicated glycosylation of either or both Ser53 and Ser57 with one to three hexoses. This unexpected finding was further supported by the MS/MS data of the nonmutated rSodC Chy2 peptide where the y4, y6, and y7 fragmentation ions corresponding to glycosylated forms of 56PSGL59, 54PAPSGL59, and 53SPAPSGL59, respectively, were observed (data not shown).
Fig. 6.
MS and MS/MS of glycopeptides generated by chymotrypsin digestion of rSodC products with Thr to Ala substitutions. (A) An MS-ESI spectrum averaged over 89 scans corresponding to Chy2 (51AGSPAPSGL59) peptide of T51A-rSodC demonstrates primarily glycosylated forms modified with one to three hexoses. (B) The m/z 1242.5 [M+H]+1 molecular ion marked by an asterisk in (A) was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu. The m/z 756.5 molecular ion represents the fully deglycosylated peptide. (C) The m/z 1768.8 [M+H]+1 molecular ion corresponding to the Chy1 (42VPGTTPSIW50) peptide of T41A-rSodC with five hexoses was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu. The m/z 957.6 molecular ion represents the fully deglycosylated peptide. (D) The m/z 1677.7 [M+H]+1 molecular ion corresponding to the Chy1 (41TVPGATPSIW50) peptide of T45A-rSodC with four hexoses was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu (marked with solid arrows). The m/z 1028.6 molecular ion represents the fully deglycosylated peptide. In addition, an ion series corresponding to a glycosylated y8 ion series with differences of 162 amu was observed (marked with dashed arrows). (E) The m/z 1677.7 [M+H]+1 molecular ion corresponding to the Chy1 (41TVPGTAPSIW50) peptide of T46A-rSodC with four hexoses was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu (marked with solid arrows). The m/z 1028.6 molecular ion represents the fully deglycosylated peptide. In addition, an ion series corresponding to a glycosylated y8 ion series with differences of 162 amu was observed (marked with dashed arrows). (F) The m/z 969.0 [M+H]+1 molecular ion corresponding to the Chy1 (45AAPSIW50) peptide of TT4546AA-rSodC with two hexoses was selected for ESI-MS/MS resulting in a fragmentation pattern containing neutral losses of 162 amu. The m/z 644.4 molecular ion represents the fully deglycosylated peptide. The inset shows the fragmentation of this glycopeptide.
Table II.
The rSodC Thr to Ala substitutions resulted in reduced levels of glycosylation
| Peptide | Expected peptide | Observed peptide sequence | Observed [M+H]+1 ions | Peptide glycosylation statusa |
|---|---|---|---|---|
| sequence/structure | (no. of hexose units) | |||
| Nonmutated Chy1 | TVPGTTPSIW | TVPGTTPSIW | 1707.8, 1869.8 | 4–5 |
| T41A-Chy1 | AVPGTTPSIW | VPGTTPSIW | 1606.7, 1768.8 | 4–5 |
| T45A-Chy1 | TVPGATPSIW | TVPGATPSIW | 1353.5, 1515.6, 1677.7 | 2–4 |
| T46A-Chy1 | TVPGTAPSIW | TVPGTAPSIW | 1353.5, 1515.6, 1677.7 | 2–4 |
| TT4546AA-Chy1 | TVPGAAPSIW | AAPSIW | 806.9, 969.0 | 1–2 |
| Nonmutated Chy2 | TGSPAPSGL | TGSPAPSGL | 1110.5, 1272.5, 1434.6 | 2–4 |
| T51A-Chy2 | AGSPAPSGL | AGSPAPSGL | 918.5, 1080.6, 1242.5 | 1–3 |
aThe number of hexose units is based on the observed [M+H]+1 ions and was confirmed by MS/MS (Figure 6).
MS of T41A-rSodC
Similar to nonmutated rSodC, the mutant rSodC Chy1 peptides were consistently found in low abundance, and an approach was taken to selectively scan for [M+H]+1 molecular ions corresponding to the predicted Chy1 peptides modified with zero to five hexoses taking into account the experimentally determined N-termini. For T41A-rSodC, two major [M+H]+1 molecular ions (m/z 1606.7 and 1768.8) were found that corresponded to the 42VPGTTPSIW50 peptide with four or five hexose units, respectively (Table II). Fragmentation and MS/MS of the m/z 1768.8 ion generated the [M+H]+1 daughter ions of m/z 1605.6, 1443.6, 1281.6, 1119.6, and 957.6, confirming the presence of five hexose residues (Figure 6C). Thus, the N-terminal peptides of T41A-rSodC and nonmutated rSodC possessed identical levels of glycosylation demonstrating that Thr41 is not glycosylated. In support of this, N-terminal sequencing was consistently successful in assigning a Thr residue to this position in the nonmutated 28-kDa rSodC, an assignment otherwise blocked if this residue was glycosylated.
MS of T45A-rSodC, T46A-rSodC, and TT4546AA-rSodC
For both T45A-rSodC and T46A-rSodC, three major [M+H]+1 molecular ions (m/z 1353.5, 1515.6, and 1677.7) were found that differed by 162 amu and corresponded to the 41TVPGATPSIW50 and 41TVPGTAPSIW50 peptides with two, three, and four hexoses, respectively (Table II). Fragmentation and MS/MS of the m/z 1677.7 ion generated the [M+H]+1 daughter ions of m/z 1514.6–1514.7, 1352.6, 1190.6, and 1028.6 arising from neutral losses of 162 amu (Figure 6D and E). This demonstrated that an amino acid substitution at Thr45 or Thr46 resulted in the loss of one hexose unit. Similar to the nonmutated rSodC Chy1 peptide, these MS/MS spectra contained a second ion series with neutral losses of 162 amu (m/z 1476.6, 1314.5–1314.6, 1152.6–1152.6, 990.5–990.6, and 828.5), and this ion series corresponded to glycosylation of the y8 ion (m/z 828.5) that resulted from fragmentation between Val42 and Pro43. For TT4546AA-rSodC, two major [M+H]+1 molecular ions (m/z 806.9 and 969.0) were found that differed by 162 amu and corresponded to the 45AAPSIW50 peptide with one and two hexoses, respectively. Fragmentation and MS/MS of the m/z 969.0 ion generated [M+H]+1 daughter ions of m/z 806.4 and 644.4 demonstrating the presence of two hexoses on this peptide (Figure 6F). In summary, the data generated for all mutants in the Thr45 Thr46 cluster were in agreement and revealed that each Thr residue is modified with one to two hexoses and collectively the Thr45 Thr46 doublet is modified with a total of three hexoses. The remaining glycosylation on the 45AAPSIW50 peptide is only explained if Ser48 is modified with one to two hexoses.
Subcellular localization
The SodC of Mtb is annotated as a lipoprotein and would be expected to be associated with the cell wall or membrane of Mtb. However, we initially identified this antigenic protein in the CF of Mtb (Sartain et al. 2006). To assess the subcellular location of SodC, Mtb H37Rv cytosol (Cyt), membrane (Mem), cell wall (CW), and CF fractions were isolated (Hirschfield et al. 1990), separated by SDS–PAGE, electroblotted, and probed with polyclonal antiserum raised against SodC (Figure 7A and B). Native SodC was dominant in the Mem and CW fractions, with a 27-kDa product being the most abundant form. Native SodC was also equally distributed between the Mem and CW fractions. This is likely due to mixing of the Mem and CW subcellular fractions during the mechanical breakage of cells. The protein was also present in the CF, but in much smaller quantities (Figure 7B). In the CF, a 21-kDa product was also observed. The 1.1-kDa C-terminal 241KLHHHHHH248 sequence present only in recombinant forms likely accounts for the difference in size between native and recombinant products. The rSodC showed a subcellular distribution pattern similar to the native protein (data not shown), and the 28-kDa rSodC product isolated from the whole-cell lysate (WCL) had an N-terminal sequence of 41TVPG44. Additionally, molecular masses of the recombinant products did not differ between WCL and CF forms, indicating that the WCL and CF forms of rSodC were truncated in the same manner (data not shown). The native SodC was believed to be acylated in part based on previous detergent-phase partitioning experiments (D’Orazio et al. 2001). We found that both the native 27-kDa SodC and 28-kDa rSodC products from both WCL and CF partitioned to the detergent phase, even though the recombinant proteins lacked the putative acylated Cys33 residue (Figure 7C). To confirm this finding, the rv0432 gene without the signal sequence was expressed in M. smegmatis and analysis of the 26-kDa SP-rSodC product in the WCL demonstrated that SodC partitions to the detergent phase even in the absence of an acylated Cys residue (Figure 7D). Collectively, these data bring into question whether this protein is truly acylated.
Fig. 7.
SodC subcellular localization. Subcellular fractions of Mtb H37Rv (lane 1, Cyt; lane 2, Mem; lane 3, CW; and lane 4, CF) were separated by SDS–PAGE and stained with Coomassie brilliant blue (A) or electroblotted and probed with polyclonal SodC antiserum (B). (C) Western blot with polyclonal SodC antiserum of TX-114 detergent phase partitioning (lane 1, detergent phase; lane 2, aqueous phase) performed on CW (left panels) and CF (right panels) fractions of Mtb expressing rSodC (top panels) and wild type Mtb H37Rv (bottom panels). (D) Western blot with polyclonal SodC antiserum of TX-114 detergent phase partitioning (lane 1, detergent phase; lane 2, aqueous phase) performed on WCL of M. smegmatis SP-rSodC.
Discussion
The detailed evaluation and description of glycosylation patterns for the MPT32 and MPB83 lipoprotein were accomplished with native products from filtrates of in vitro grown cells (Dobos et al. 1995, 1996; Michell et al. 2003). In contrast, SodC is not a dominant protein of the CF, had previously escaped detection in proteome-wide evaluation of Mtb CF proteins (Sonnenberg and Belisle 1997; Jungblut et al. 1999; Rosenkrands et al. 2000), and was only found in this extracellular preparation after extensive enrichment via multidimensional chromatography (Sartain et al. 2006). Thus, to obtain sufficient quantities for analyses, the sodC gene was overexpressed in its natural host (Mtb) and recombinant protein purified from the CF. N-Terminal sequencing and analysis of the resulting three products with the lectin ConA suggested that glycosylation was localized to the 19-amino-acid N-terminus, and the NetOglyc algorithm (Julenius et al. 2005) predicted specific Ser and Thr residues to be O-glycosylated in this region. Taking these observations into account and considering the previous discovery of only O-glycosylated Thr residues in mycobacteria (Dobos et al. 1995, 1996; Michell et al. 2003), five Thr residues of SodC with the highest predictive scores were substituted with Ala. None of the Thr to Ala substitutions resulted in a complete loss of glycosylation as assessed by ConA reactivity and MS. Nevertheless, the substitutions allowed for the assignment of glycosylated residues and determination of the extent of glycosylation at each residue. Furthermore, the full pattern of glycosylation could only be adequately explained with O-glycosylation of Ser residues, a finding that is in contrast to earlier studies where only Thr residues of mycobacterial proteins were shown to be glycosylated (Dobos et al. 1995, 1996; Herrmann et al. 1996; Michell et al. 2003). Arguably the Thr to Ala substitutions might have altered the natural glycosylation state of surrounding sequences and in effect forced glycosylation onto neighboring Ser residues. However, the MS/MS data of the nonmutated rSodC Chy2 peptide showed that this phenomenon did not occur. Our data located three Thr and two to three Ser residues possessing modifications with one to three hexose units, with the total number of hexose units ranging from 6 to 10. Figure 8 summarizes these results.
Fig. 8.
Working model for the posttranslational modification of Mtb SodC. The region underlined with a solid line denotes the Type II signal peptide preceding the putative N-terminal acylated Cys (C33) described in D’Orazio et al. (2001). The region underlined with the dashed line denotes the experimentally determined truncation that resulted in an N-terminal Thr (T41). Glycosylation sites modified with mannose (Man) are indicated. Parentheses denote variable levels of glycosylation. The shaded region indicates the folded enzymatic structure, for which a more detailed annotation can be found in Figure S1 and Spagnolo et al. (2004).
The glycosylation pattern of SodC showed several similarities to previously described mycobacterial glycoproteins. Consistent with oligosaccharide lengths of MPT32 and MPB83, each site of O-glycosylation on SodC was populated with a mono-, di- or tri-hexose. Based on reactivity to ConA and the specificity of the jack-bean-α-mannosidase used to deglycosylate the protein, mannose was likely the major glycosylating entity of SodC. Similar to the 19-kDa lipoprotein, MPT32, and MPB83, a Thr doublet near the N-terminus was glycosylated (Dobos et al. 1995; Herrmann et al. 1996; Michell et al. 2003). Furthermore, the glycosylation pattern of the SodC Thr doublet showed great similarity to the MPB83 Thr doublet, where each Thr is variably modified with a mannose or a mannobiose, and the Thr doublet is collectively modified with a total of three mannose units (Michell et al. 2003). Indeed, the overall glycosylation pattern of the five SodC sites closely mirrored the generalized structural features of glycosylated mycobacterial residues described by Belisle et al. (2005). Specifically, three of the five glycosylation sites (treating the Thr doublet as one site) were present one or two amino acids downstream of a Pro residue, and all five sites were localized to Pro-, Ala-, Thr-, and Ser-rich domains. Finally, similar to other mycobacterial glycoproteins, SodC possesses multiple O-glycosylated residues within a short region (five to six residues in a 13-amino-acid sequence).
An unresolved question from our current studies is the nature of the carbohydrate linkages in SodC. The Mtb-secreted MPT32 was found to contain α(1→2)-linked mannose units (Dobos et al. 1996) while the mannobiose of the M. bovis lipoprotein MPB83 was found to possess an α(1→3) linkage (Michell et al. 2003). Future studies of SodC sugar linkages may help to address whether differences occur in glycoprotein biosynthesis between mycobacterial species, or whether glycosylation of lipidated versus nonlipidated secreted proteins differs.
SodC is considered one of the few well-characterized lipoproteins of Mtb (Sutcliffe and Harrington 2004) and evidence for its acylation comes from radiolabeling experiments on recombinant SodC produced in E. coli and a subsequent comparison with Mtb SodC by native PAGE (D’Orazio et al. 2001). We found SodC primarily associated with the cell envelope. However, the cell-associated rSodC possessed the same N-terminal sequence, two-dimensional SDS–PAGE pattern, and MS-based glycosylation pattern as the nonlipidated rSodC purified from the CF (data not shown). These data (in particular, that from N-terminal sequencing) demonstrated that cell-associated rSodC was not lipidated. The association of SodC with the cell wall in the absence of acylation may have resulted from interactions with extended hydrophobic stretches of the protein. This possibility is supported by our observations that the nonacylated rSodC was localized to the detergent fraction of a TX-114 biphasic partitioning. Alternatively, the cell wall association could have resulted from specific interactions of the glycosylated N-terminus with cell wall constituents. Such a phenomenon would also explain the recent observation that the nonlipidated glycoprotein MPT32/Apa associates with the cell surface (Ragas et al. 2007).
The lack of acylated rSodC could be the result of a process termed “proteolytic shaving” (Tjalsma and van Dijl 2005). This has been reported for Bacillus subtilis where amino acids downstream of the acylated N-terminal Cys direct the release of lipoproteins from membranes through proteolysis. Although protein glycosylation was not reported to be involved in proteolytic shaving, it is proposed that glycosylation of Mtb proteins may serve as a signal for proteolytic cleavage, or that glycosylation prevents further amino-terminal degradation following proteolytic cleavage from the acylated anchor (Herrmann et al. 1996; Michell et al. 2003). Interestingly, substitution of the rSodC glycosylated Thr doublet (Thr45 Thr46) with Ala residues resulted in a new, further truncated N-terminus and was similar to the reported truncation of the 19-kDa lipoprotein induced by substitution of two glycosylated Thr clusters with Val residues (Herrmann et al. 1996). Therefore, the data presented here for the rSodC protein produced in Mtb provide further evidence for the hypothesis that amino acids in the N-terminal region and downstream of the acylated Cys influence proteolytic shaving and that glycosylation impacts this process. Alternatively, the truncated forms of rSodC could be explained through differential signal peptidase processing. Signal peptidase II may recognize other cleavage sites or act independently of acylation as recently described for Listeria monocytogenes (Baumgartner et al. 2007). However, this phenomenon has not been demonstrated in Mtb.
A large number of SodC homologs from multiple prokaryotic and eukaryotic species have been studied, and the amino-acid sequence from Cys33 to Gln70 of Mtb SodC is conserved among SodC homologs of Mycobacterium spp., but not with Cu,Zn SODs of other genera (Spagnolo et al. 2004). Further, the Mtb SodC is the only member of the Cu,Zn SOD family shown to be glycosylated. This suggests that the N-terminal portion of the mycobacterial SodC proteins serves a unique function. Recent elucidation of the crystal structure of a secreted but nonacylated form of Mtb SodC produced in E. coli (Spagnolo et al. 2004) failed to provide a defined structure for the N-terminal region (Cys33 through Gln70) of the mature protein. This was attributed to the unordered and flexible characteristics of this region. Prediction of intrinsically folded sequences within the SodC mature protein sequence using the FoldIndex© tool (Prilusky et al. 2005) agreed with this observation (Figure S1). A physical separation between the unordered glycosylated N-terminus of SodC and its folded enzymatic domain would suggest that glycosylation does not directly influence the catalytic activity of SodC. In fact, a recombinant form of this protein produced in E. coli where glycosylation is not possible was shown to be fully functional (Spagnolo et al. 2004). Thus, the predicted structure of the O-glycosylated region of SodC lends support to the hypothesis that glycosylation of this enzyme modulates proteolytic cleavage within the unstructured SodC N-terminus and influences SodC localization and stability.
Material and methods
Bacterial growth and subcellular fractionation
Recombinant clones of M. smegmatis mc2155 were selected on Luria-Bertani (LB) agar containing kanamycin (25 μg mL−1). For isolation of WCL of M. smegmatis, cells were grown in 2 L of glycerol-alanine-salts medium (GAS) (Takayama et al. 1975) containing kanamycin (25 μg mL−1) at 37°C for 3 days with gentle shaking, and bacterial pellets were suspended in PBS and passed through a French press four times at 2000 psi.
Mtb strain H37Rv cells used for the generation of electrocompetent stocks were grown in Middlebrook 7H9 broth (Difco, Detroit, MI) supplemented with oleic acid-dextrose catalase (OADC, Difco) and 0.05% Tween80. Recombinant clones of Mtb H37Rv were selected on a Middlebrook 7H11 medium (Difco) supplemented with OADC, hygromycin (100 μg mL−1), and kanamycin (25 μg mL−1). Growth of Mtb for recombinant protein purification was achieved by propagation in 2 L of GAS containing kanamycin (25 μg mL−1) at 37°C with gentle shaking for 14 days. Growth of Mtb for isolation of subcellular fractions was achieved by propagation in 14 L of GAS at 37°C with gentle shaking. Cells were harvested at 14 days of growth, and individual subcellular fractions of Cyt, Mem, CW, and CF were isolated as previously described (Hirschfield et al. 1990; Sonnenberg and Belisle 1997). Final protein concentrations were determined using the bicinchoninic (BCA) protein assay (Smith et al. 1985).
E. coli strains TOP10 (Invitrogen) and XL10-Gold Ultracompetent (Stratagene, La Jolla, CA) containing various recombinant plasmids were selected on LB agar containing kanamycin (25 μg mL−1). For recombinant protein purification, E. coli strain BL21(DE3) (Invitrogen, Carlsbad, CA) was grown in LB broth containing ampicillin (100 μg mL−1).
Bacterial transformation
For preparation of electrocompetent Mtb H37Rv, a 1 L culture was grown to an OD600 of 0.6 and the cells were harvested by centrifugation. After washing three times in a 10% ice-cold glycerol solution, the bacterial pellet was suspended in 5 mL of cold 10% glycerol. An aliquot (90 μL) of cells was mixed with 0.5 μg plasmid DNA, incubated in a 0.1 cm gap electroporation cuvette (Invitrogen) for 10 min at room temperature, and electroporated (1.25 kV, 25 μF, 1000 Ω) in a Gene Pulser (Bio-Rad, Hercules, CA). Cells were transferred to a 5 mL 7H9 medium, allowed to recover overnight at 37°C with gentle shaking, and plated onto a solid medium containing antibiotics.
Electroporation of M. smegmatis mc2155 was achieved by the method of Snapper et al. (1990).
Construction of recombinant plasmids
Recombinant plasmid constructs were created according to standard protocols (Sambrook and Maniatis 1989). PCR amplifications of rv0432 or rv0432 gene fragments were performed with PfuTurbo DNA polymerase (Stratagene) using Mtb H37Rv genomic DNA as the template. Table SII lists PCR primers used in these studies. All PCR products were first cloned into pCR4Blunt-TOPO according to the manufacturer's protocols (Invitrogen) and subsequently recovered by restriction enzyme digestions that targeted the restriction site linker sequences designed into the primers (Table SII). To generate the E. coli expression construct for recombinant production of SP-rSodC, the rv0432 gene minus the region encoding the N-terminal signal peptide was amplified using primer pair SodC(-SP)F and SodCR1. The 627 bp fragment isolated from the pCR4Blunt intermediate plasmid construct by digestion with NdeI/XhoI was ligated into the NdeI/XhoI sites of pET23b (Novagen, San Diego, CA) to generate pMRLB60. To generate a mycobacterial-expression construct that encoded mature and fully modified rSodC, the full-length rv0432 gene was amplified using primer pair SodC(+SP)F and SodCR2. The 720 bp fragment isolated from the pCR4Blunt intermediate plasmid construct by digestion with NdeI/HindIII was ligated into the NdeI/HindIII sites of the mycobacterial expression vector pVV16 (Schulbach et al. 2001) to generate pMRLB61. To generate a mycobacterial-expression construct for production of SP-rSodC, the rv0432 gene minus the region encoding the N-terminal signal peptide was amplified using the primer pair SodC(-SP)F and SodCR2. The 627 bp fragment isolated from the pCR4Blunt intermediate plasmid construct by digestion with NdeI/HindIII was ligated into the NdeI/HindIII sites of pVV16 to generate pMRLB62.
Mycobacterial expression constructs that allowed for production of rSodC with altered sites of glycosylation were based on pMRLB61 with point mutations in rv0432. Point mutations were generated using the QuikChangeTM II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations. To generate the construct for T41A-rSodC production, the primer pair SodCT41AF and SodCT41AR was used resulting in pMRLB61.1. To generate the construct for T45A-rSodC production, the primer pair SodCT45AF and SodCT45AR was used resulting in pMRLB61.2. To generate the construct for T46A-rSodC production, the primer pair SodCT46AF and SodCT46AR was used resulting in pMRLB61.3. To generate the construct for TT4546AA-rSodC production, the primer pair SodCTTAAF and SodCTTAAR was used resulting in pMRLB61.4. To generate the construct for T51A-rSodC production, the primer pair SodCT51AF and SodCT51AR was used resulting in pMRLB61.5. To generate the construct for T131A-rSodC production, the primer pair SodCT131AF and SodCT131AR was used resulting in pMRLB61.6. All plasmid constructs were confirmed by nucleotide sequencing through Macromolecular Resources Facility, Colorado State University.
Recombinant protein purification
For production of SP-rSodC in E. coli, plasmid pMRLB60 was transformed into E. coli BL21(DE3) (Studier et al. 1990). A 2 L culture was grown at 37°C to an OD600 of 0.5, and the recombinant gene was expressed via the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h. The cells were harvested, lysed with lysozyme and probe sonication in the presence of RNase, DNAse, and protease inhibitors, and centrifuged at 16,000 × g for 30 min. Inclusion bodies were solubilized with a denaturing binding buffer (5.0 mM imidazole, 0.5 M NaCl2, 6.0 M urea, 20 mM Tris–Cl, pH 7.9), and the protein applied to a 1 mL column packed with His-bind resin (Novagen) equilibrated in the denaturing binding buffer. The recombinant protein was purified by washing with 20 column volume (CV) of denaturing binding buffer, 25 CV of denaturing wash buffer (60 mM imidazole, 0.5 M NaCl2, 6.0 M urea, 20 mM Tris–Cl pH 7.9), 10 CV of Tris buffer (pH 8.0), 10 CV of 0.5% ASB-14 (Calbiochem, San Diego, CA) in the Tris buffer (pH 8.0), and again in 10 CV of Tris buffer (pH 8.0) to remove detergent. SP-rSodC was eluted from the column by the addition of 10 CV of denaturing elution buffer (1.0 M imidazole, 0.5 M NaCl2, 6.0 M urea, 20 mM Tris–Cl, pH 7.9). Each fraction was concentrated 10-fold and exchanged with 10 mM ammonium bicarbonate by ultrafiltration.
For production of rSodC and rSodC site-directed mutants in Mtb, the plasmids pMRLB61, pMRLB61.1, pMRLB61.2, pMRLB61.3, pMRLB61.4, pMRLB61.5, and pMRLB61.6 were electroporated into Mtb H37Rv. Culture filtrates from 14-day (late-log) cultures of recombinant Mtb were concentrated, dialyzed against 10 mM ammonium bicarbonate, and dried completely by lyophilization (Sonnenberg and Belisle 1997). The samples were suspended in an 8 mL Ni-NTA denaturing binding buffer (0.1 M sodium phosphate buffer, 8.0 M urea, 10 mM Tris–Cl, pH 8.0) and incubated for 1 h with 1 mL of Ni-NTA His-bind resin (Novagen) equilibrated in a Ni-NTA denaturing binding buffer. The recombinant proteins were purified by washing with 18 CV of Ni-NTA denaturing binding buffer, 18 CV of Ni-NTA denaturing wash buffer (0.1 M sodium phosphate buffer, 8.0 M urea, 10 mM Tris–Cl, pH 6.3), 10 CV of Tris buffer (pH 8.0), 10 CV of 0.5% ASB-14 in the Tris buffer (pH 8.0), and again 10 CV of Tris buffer (pH 8.0) to remove detergent. Proteins were eluted from the column by the addition of 10 CV of Ni-NTA denaturing elution buffer (0.1 M sodium phosphate buffer, 8.0 M urea, 10 mM Tris–Cl, pH 4.0). Each fraction was concentrated 10-fold and exchanged with 10 mM ammonium bicarbonate by ultrafiltration.
SDS–PAGE and Western blot analyses
Rabbit polyclonal serum to SodC was generated by Strategic Biosolutions (Ramona, CA) in a standard rabbit protocol. Purified SP-rSodC from E. coli was used as the antigen.
Aliquots of protein were subjected to SDS–PAGE using 10–20% Tricine gels (Invitrogen). Gels were stained with Coomassie brilliant blue R250 (Coligan 2001) or silver nitrate (Morrissey 1981), or electroblotted to nitrocellulose membranes (Bio-Rad, Hercules, CA) as previously described (Sonnenberg and Belisle 1997). The nitrocellulose membranes were blocked with 3% nonfat milk in PBS (pH 7.2) for 2 h and exposed overnight to an anti-His5 monoclonal antibody (Qiagen, Valencia, CA) or SodC antiserum diluted 1:1000 or 1:2000, respectively, in 1% nonfat milk in PBS (pH 7.2). The blots were washed with PBS (pH 7.2) containing 0.1% Tween 20, probed for 1.5 h with alkaline phosphatase-conjugated anti-mouse IgG whole molecule (Sigma, St. Louis, MO) or anti-rabbit IgG Fc fragment (Calbiochem) both diluted 1:2000 in 1% nonfat milk in PBS (pH 7.2), and washed extensively. Antigen-antibody complexes were visualized by color development with a 5-bromo-4-chloro-indoyl-phosphatase-nitroblue tetrazolium substrate (Kirkegaard & Perry Laboratories, Gaithersburg, MD). For ConA analysis, membranes were blocked with 1% BSA in PBS (pH 7.2) for 2 h and incubated with one unit of peroxidase-conjugated ConA (Sigma) in PBS (pH 7.2) overnight at 4°C. Development was achieved with 5-bromo-4-chloronaphthol (Sigma) and H2O2.
Analytical protein methods
Proteolytic digestions were performed with 10 μg aliquots of the various rSodC products purified from Mtb. Digestions with chymotrypsin (Roche Applied Science, Mannheim, Germany) were carried out in 20 μL of 0.1 M ammonium bicarbonate (pH 7.9) at 25°C for 16 h with an enzyme to substrate (E:S) ratio of 1:20 (wt:wt). Digestions with thermolysin (Calbiochem) were identical to those with chymotrypsin except that the temperature was increased to 37°C. Digestions with modified trypsin (Roche) were carried out in 20 μL 5% acetonitrile in 0.1 M ammonium bicarbonate (pH 7.9) at 37°C for 16 h with an E:S ratio of 1:20 (wt:wt). All proteolytic digestions were terminated by the addition of 2 μL of 10% TFA. The digests were dried under vacuum, suspended in 20 μL of 5% acetonitrile in 0.1% acetic acid, and aliquots (5 μL) were applied to a capillary (0.75 × 50 mm) C18 reversed phase column (Agilent Technologies, Santa Clara, CA). The peptides were eluted with an increasing linear gradient (5–64%) of acetonitrile in 0.1% acetic acid over 70 min using an Agilent 1200 series capillary HPLC system with a flow rate of 5 μL per min and introduced directly into a ThermoFinnigan LTQ electrospray mass spectrometer (San Jose, CA) operated using Xcalibur software version 2.0 SR2. Ionization of peptides was achieved with an electrospray needle setting of 4 kV with a N2 sheath gas flow of 15 and a capillary temperature of 200°C.
Multiple MS/MS methods were employed to analyze SodC peptides and their modifications. In all cases, data-dependent scanning was used to generate fragment ions and MS/MS spectra. Standard MS/MS analyses were performed at 35% normalized collision energy on the top five most intense parent ions of the previous MS scan. Peptide identification was achieved using the SEQUEST (ThermoFinnigan, ver BioWorks 3.1) and X! Tandem (www.thegpm.org; version 2006.04.01.2) software with a SodC protein sequence (accession NP_214946) extracted from the Mtb genome database (NC_000962) and modified to begin with the experimentally determined N-terminal Thr41 and to include the additional C-terminal sequence of 241KLHHHHHH248 (vector LysLeu + hexa-His tag). The SEQUEST and X! Tandem softwares were set to evaluate peptides obtained by chymotrypsin (FWYL), trypsin (KRLNH), or thermolysin (ILMV) digestion. Oxidation of Met (+16.0 Da) was set as the only possible modification. The Scaffold software, version Scaffold-01_05_04 (Proteome Software, Portland, OR), was used to validate peptide identifications. Peptide identities were accepted only when a probability of 95.0% or greater was obtained as specified by the Peptide Prophet algorithm (Keller et al. 2002). The MS/MS data were also searched for neutral losses of m/z 162 to identify glycopeptides. Neutral loss analyses of the MS/MS spectra were performed with the Xcalibur software version 2.0 SR2.
To assess glycosylation of the Chy1 peptide (41TVPGTTPSIW50) of rSodC, the predicted [M+H]+1 molecular ions of m/z 1059.2, 1221.3, 1383.5, 1545.6, 1707.8, and 1869.9 were placed in a parent mass list, and when detected in a full MS scan, fragmentation for MS/MS was achieved using 21% normalized collision energy. The same approach was used for the Chy1 peptides of T45A-rSodC and T46A-rSodC except that the predicted [M+H]+1 molecular ions of m/z 1029.2, 1191.3, 1353.5, 1515.6, 1677.7, 1839.9 were placed in the parent mass list. Glycosylation of the Chy1 peptide of T41A-rSodC (42VPGTTPSIW50) was assessed by placing the predicted [M+H]+1 molecular ions of m/z 958.1, 1120.2, 1282.4, 1444.5, 1606.7, and 1768.8 in the parent mass list and performing MS/MS fragmentation with 21% normalized collision energy. The glycosylation analyses of the Chy1 peptide of TT4546AA-rSodC (45AAPSIW50) utilized the predicted [M+H]+1 molecular ions of m/z 644.7, 806.9, 969.0, 1131.2, 1293.3, and 1455.4 in the parent mass list.
MALDI-TOF MS was performed in the Macromolecular Resources Facility, Colorado State University, with an Ultraflex MALDI/TOF/TOF (Bruker Daltonics, Billerica, MA) with sinapinic acid as a matrix.
N-Terminal sequencing of purified proteins was performed by Edman degradation (Shively et al. 1987) with a pulsed liquid-phase sequencer from Applied Biosystems by Macromolecular Resources Facility, Colorado State University.
Mannosidase treatment and Triton X-114 phase partitioning
Digestion of the protein with α-mannosidase was performed on aliquots (10 μg) of purified rSodC solubilized in 20 μL of 0.05 M sodium acetate buffer (pH 4.5) and incubated with 10, 2, 0.5, 0.1, and 0.02 μg of jack-bean-α-mannosidase from Canavalia ensiformis (Sigma) at 37°C for 18 h. Digestions were terminated by incubating with a Laemmli sample buffer at 100°C for 10 min.
The detergent-phase partitioning properties of SodC were assessed by biphasic partitioning with Triton X-114 (Sigma). Specifically, subcellular fractions (CF, CW, and WCL) were incubated with 4% Triton X-114 in PBS (pH 7.4) for 16 h at 4°C with gentle agitation. The suspensions were incubated at 37°C for 1 h and the biphase was formed by centrifugation at 27,000 × g at 37°C for 1 h. The aqueous and detergent layers were collected, adjusted to 4% Triton X-114 in PBS (pH 7.4), and the biphasic partitioning repeated. Macromolecules in the final aqueous and detergent phases were precipitated by the addition of 4 volume of ice-cold acetone and incubation at −20°C for 16 h. Precipitates were collected by centrifugation at 27,000 × g 4°C for 10 min and washed once with ice-cold acetone. The precipitates were air-dried and suspended in PBS (pH 7.4), and protein concentrations were determined using the BCA protein assay (Smith et al. 1985).
Bioinformatics
The complete Mtb SodC protein sequence was analyzed with the NetOglyc 3.1 neural network for the prediction of O-linked glycosylation sites (http://www.cbs.dtu.dk/services/NetOGlyc/) (Julenius et al. 2005). To predict sequences which are intrinsi- cally folded, the Mtb SodC sequence beginning with Cys33 was analyzed with the FoldIndex© tool (http://bip.weizmann. ac.il/fldbin/findex) (Prilusky et al. 2005) with a window size of 25.
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The National Institutes of Health, NIAID (grant RO1 AI-056257 and contracts NO1 AI-75320 and HHSN26620040091c).
Acknowledgments
We thank Drs. Karen Dobos and SungGu Lee for assistance with mass spectrometry and Darragh Heaslip for review of the manuscript.
Conflict of interest statement
None declared.
Abbreviations
- BCA
bicinchoninic acid
- CF
culture filtrate
- ConA
concanavalin A
- CV
column volume
- CW
cell wall
- Cyt
cytosol
- E:S
enzyme to substrate ratio
- GAS
glycerol-alanine-salts medium
- IMAC
immobilized metal affinity chromatography
- LC-ESI-MS/MS
liquid chromatography-electrospray-ionization-tandem mass spectrometry
- MALDI
matrix-assisted laser desorption ionization-time of flight
- Mem
membrane
- MS/MS
tandem mass spectrometry
- MS
mass spectrometry
- Mtb
Mycobacterium tuberculosis
- OADC
oleic acid-dextrose catalase
- rSodC
recombinant SodC purified from Mtb
- SOD
superoxide dismutase
- SodC
Cu,Zn superoxide dismutase
- SPase
signal peptidase
- SP-rSodC
recombinant SodC lacking a signal peptide purified from E. coli
- TB
tuberculosis
- WCL
whole-cell lysate
References
- Abernathy JL, Wang Y, Eckhardt AE, Hill RL. Identification of O-glycosylation sites with a gas phase sequencer. In: Abernathy JL, editor. Techniques in Protein Chemistry. New York: Academic Press; 1992. pp. 277–286. [Google Scholar]
- Battistoni A, Rotilio G. Isolation of an active and heat-stable monomeric form of Cu, Zn superoxide dismutase from the periplasmic space of Escherichia coli. FEBS Lett. 1995;374:199–202. doi: 10.1016/0014-5793(95)01106-o. [DOI] [PubMed] [Google Scholar]
- Baumgartner M, Karst U, Gerstel B, Loessner M, Wehland J, Jansch L. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J Bacteriol. 2007;189:313–324. doi: 10.1128/JB.00976-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle JT, Braunstein M, Rosenkrands I, Andersen P. The proteome of Mycobacterium tuberculosis. In: Cole ST, editor. Tuberculosis and the Tubercle Bacillus. Washington, DC: ASM Press; 2005. pp. 235–260. [Google Scholar]
- Coligan JE. Current Protocols in Protein Science. 2001. p. 10.15.11. [DOI] [PubMed] [Google Scholar]
- Cutalo JM, Deterding LJ, Tomer KB. Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1545–1555. doi: 10.1016/j.jasms.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugelat S, Kowall J, Mattow J, Bumann D, Winter R, Hurwitz R, Kaufmann SH. he RD1 proteins of Mycobacterium tuberculosis: Expression in Mycobacterium smegmatis and biochemical characterization. Microbes Infect. 2003;5:1082–1095. doi: 10.1016/s1286-4579(03)00205-3. [DOI] [PubMed] [Google Scholar]
- Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J Bacteriol. 1996;178:2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobos KM, Swiderek K, Khoo KH, Brennan PJ, Belisle JT. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect Immun. 1995;63:2846–2853. doi: 10.1128/iai.63.8.2846-2853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio M, Folcarelli S, Mariani F, Colizzi V, Rotilio G, Battistoni A. Lipid modification of the Cu, Zn superoxide dismutase from Mycobacterium tuberculosis. Biochem J. 2001;359:17–22. doi: 10.1042/0264-6021:3590017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espitia C, Mancilla R. Identification, isolation and partial characterization of Mycobacterium tuberculosis glycoprotein antigens. Clin Exp Immunol. 1989;77:378–383. [PMC free article] [PubMed] [Google Scholar]
- Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- Fifis T, Costopoulos C, Radford AJ, Bacic A, Wood PR. Purification and characterization of major antigens from a Mycobacterium bovis culture filtrate. Infect Immun. 1991;59:800–807. doi: 10.1128/iai.59.3.800-807.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe T, Harris D, Vordermeier M, Lathigra R, Ivanyi J, Young D. Expression of the Mycobacterium tuberculosis 19-kilodalton antigen in Mycobacterium smegmatis: Immunological analysis and evidence of glycosylation. Infect Immun. 1993;61:260–267. doi: 10.1128/iai.61.1.260-267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JL, Delahay R, Gallagher A, Robertson B, Young D. Analysis of post-translational modification of mycobacterial proteins using a cassette expression system. FEBS Lett. 2000;473:358–362. doi: 10.1016/s0014-5793(00)01553-2. [DOI] [PubMed] [Google Scholar]
- Herrmann JL, O’Gaora P, Gallagher A, Thole JE, Young DB. Bacterial glycoproteins: A link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. Embo J. 1996;15:3547–3554. [PMC free article] [PubMed] [Google Scholar]
- Hirschfield GR, McNeil M, Brennan PJ. Peptidoglycan-associated polypeptides of Mycobacterium tuberculosis. J Bacteriol. 1990;172:1005–1013. doi: 10.1128/jb.172.2.1005-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Jungblut PR, Schaible UE, Mollenkopf HJ, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann SH. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: Towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hess D, Sturm A. The N-glycans of jack bean alpha-mannosidase. Structure, topology and function. Eur J Biochem. 1999;264:168–175. doi: 10.1046/j.1432-1327.1999.00598.x. [DOI] [PubMed] [Google Scholar]
- Link AJ, Robison K, Church GM. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- Michell SL, Whelan AO, Wheeler PR, Panico M, Easton RL, Etienne AT, Haslam SM, Dell A, Morris HR, Reason AJ, et al. The MPB83 antigen from Mycobacterium bovis contains O-linked mannose and (1→3)-mannobiose moieties. J Biol Chem. 2003;278:16423–16432. doi: 10.1074/jbc.M207959200. [DOI] [PubMed] [Google Scholar]
- Morrissey JH. Silver stain for proteins in polyacrylamide gels: A modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect Immun. 2001;69:4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Ragas A, Roussel L, Puzo G, Riviere M. The Mycobacterium tuberculosis cell-surface glycoprotein apa as a potential adhesin to colonize target cells via the innate immune system pulmonary C-type lectin surfactant protein A. J Biol Chem. 2007;282:5133–5142. doi: 10.1074/jbc.M610183200. [DOI] [PubMed] [Google Scholar]
- Rosenkrands I, King A, Weldingh K, Moniatte M, Moertz E, Andersen P. Towards the proteome of Mycobacterium tuberculosis. Electrophoresis. 2000;21:3740–3756. doi: 10.1002/1522-2683(200011)21:17<3740::AID-ELPS3740>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Sambrook JE, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sartain MJ, Slayden RA, Singh KK, Laal S, Belisle JT. Disease state differentiation and identification of tuberculosis biomarkers via native antigen array profiling. Mol Cell Proteomics. 2006;5:2102–2113. doi: 10.1074/mcp.M600089-MCP200. [DOI] [PubMed] [Google Scholar]
- Schulbach MC, Mahapatra S, Macchia M, Barontini S, Papi C, Minutolo F, Bertini S, Brennan PJ, Crick DC. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J Biol Chem. 2001;276:11624–11630. doi: 10.1074/jbc.M007168200. [DOI] [PubMed] [Google Scholar]
- Shively JE, Miller P, Ronk M. Microsequence analysis of peptides and proteins: VI. A continuous flow reactor for sample concentration and sequence analysis. Anal Biochem. 1987;163:517–529. doi: 10.1016/0003-2697(87)90257-0. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Sonnenberg MG, Belisle JT. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect Immun. 1997;65:4515–4524. doi: 10.1128/iai.65.11.4515-4524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Toro I, D’Orazio M, O’Neill P, Pedersen JZ, Carugo O, Rotilio G, Battistoni A, Djinovic-Carugo K. Unique features of the SodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J Biol Chem. 2004;279:33447–33455. doi: 10.1074/jbc.M404699200. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: An abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28:645–659. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Takayama K, Schnoes HK, Armstrong EL, Boyle RW. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J Lipid Res. 1975;16:308–317. [PubMed] [Google Scholar]
- Tjalsma H, van Dijl JM. Proteomics-based consensus prediction of protein retention in a bacterial membrane. Proteomics. 2005;5:4472–4482. doi: 10.1002/pmic.200402080. [DOI] [PubMed] [Google Scholar]
- Volpe E, Cappelli G, Grassi M, Martino A, Serafino A, Colizzi V, Sanarico N, Mariani F. Gene expression profiling of human macrophages at late time of infection with Mycobacterium tuberculosis. Immunology. 2006;118:449–460. doi: 10.1111/j.1365-2567.2006.02378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasinger VC, Humphery-Smith I. Small genes/gene-products in Escherichia coli K-12. FEMS Microbiol Lett. 1998;169:375–382. doi: 10.1111/j.1574-6968.1998.tb13343.x. [DOI] [PubMed] [Google Scholar]
- Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.