Abstract
With genome sequencing nearing completion for the model organisms used in biomedical research, there is a rapidly growing appreciation that proteomics, the study of covalent modification to proteins, and transcriptional regulation will likely dominate the research headlines in the next decade. Protein methylation plays a central role in both of these fields, as several different residues (Arg, Lys, Gln) are methylated in cells and methylation plays a central role in the “histone code” that regulates chromatin structure and impacts transcription. In some cases, a single lysine can be mono-, di-, or trimethylated, with different functional consequences for each of the three forms. This review describes structural aspects of methylation of histone lysine residues by two enzyme families with entirely different structural scaffolding (the SET proteins and Dot1p) and methylation of protein arginine residues by PRMTs.
Keywords: protein arginine methyltransferases, protein lysine methyltransferases, SET domain proteins, S-adenosyl-L-methionine (AdoMet)
INTRODUCTION
Histones can be modified in many ways that affect gene expression, including acetylation, phosphorylation, ubiquitination, methylation (42, 115), and sumoylation (82). Evidence accumulated over the past few years suggests that such modifications constitute a “histone code” that directs a variety of processes involving chromatin (20, 31, 87, 98, 99). There are currently many known sites of lysine and arginine methylation on histones, and additional sites of modification are still being uncovered. Methylation at these sites, in combination with other modifications (or demodifications) at nearby residues, generates “modification cassettes” (22) that yield distinct patterns on chromatin for signaling downstream events (23). The best-characterized sites of histone methylation are located on the N-terminal tails of histones (such as at Lys-4 and Lys-9 of histone H3 and Arg-3 of histone H4) that protrude from the nucleosome. In contrast, Lys-79 of histone H3 is located in the core of the histone, exposed on the nucleosome disk surface.
There are many recent reviews in this fast-moving field of histone methylation (35, 53, 78, 107). In this review we summarize the progress that has been made in structural studies of protein (particularly the histone) methylation enzymes and their related sequence conservations, and we discuss, somewhat speculatively, their mechanisms.
PROTEIN LYSINE METHYLATION
SET Domain Proteins
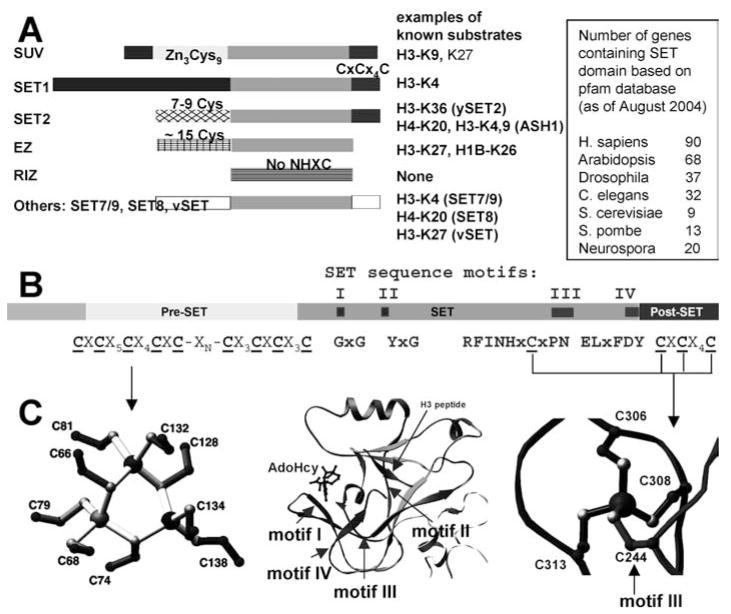
With one exception (Dot1p, see below), histone lysine (K) methyltransferases (HKMTs) contain a SET domain of approximately 130 amino acids. The SET domain was originally identified in three Drosophila genes involved in epigenetic processes, the suppressor of position-effect variegation 3–9, Su(var)3–9; an enhancer of the eye color mutant zeste, En(zeste); and the homeotic gene regulator Trithorax (32). Mammalian homologues of Drosophila Su(var) 3–9 were the first HKMTs identified, and they specifically methylate H3 at Lys-9 (73). So far, SET-containing HKMTs that methylate Lys-4, -9, -27, or -36 of histone H3 and Lys-20 of histone H4 have been identified. The SET domain is found in a large number of eukaryotic proteins (see box in Figure 1) as well as in a few bacterial proteins (112) and is not limited to HKMTs. HKMTs can be classified according to the presence or absence and the nature of sequences surrounding the SET domain that are conserved within families (4, 39). Representatives of the major families include SUV, SET1, SET2, EZ, and RIZ (Figure 1a). The SET7/9 and SET8 proteins do not fit into these families. The SUV family includes the greatest number of HKMTs.
Figure 1.
SET domain HKMTs. (a) Domain structure of SET HKMT families. (b) DIM-5 protein (one of the smallest members of the SUV family) contains four segments: a weakly conserved N-terminal region, a pre-SET domain containing nine invariant cysteines, the SET region containing four signature motifs, and the post-SET domain containing three invariant cysteines. (c) Illustration of pre-SET Zn3Cys9 triangular zinc cluster (left panel); ribbon diagram of DIM-5 SET domain, with arrows indicating locations of conserved motifs, the cofactor binding and substrate histone H3 peptide, and the pseudo knot formed by motifs III and IV (middle panel); and post-SET zinc center (right panel).
Sequence alignment of SET proteins revealed four strongly conserved sequence motifs. We have termed them SET motif I (GxG), SET motif II (YxG), SET motif III (RFINHxCxPN), and SET motif IV (ELxFDY) (Figure 1b). The tertiary structure of SET proteins shows that these conserved residues are clustered together (Figure 1c, middle panel) and involved in one of the three steps in the methylation reaction: AdoMet binding (motif I: GxG; the first half of motif III: RFINH; and the last Tyr of motif IV), catalysis of methyl transfer (the Tyr of motif II), and formation of the hydrophobic target lysine-binding channel (the second half of motif III: CxPN, and motif IV: ELxFDY).
Structures of SET Domain Proteins
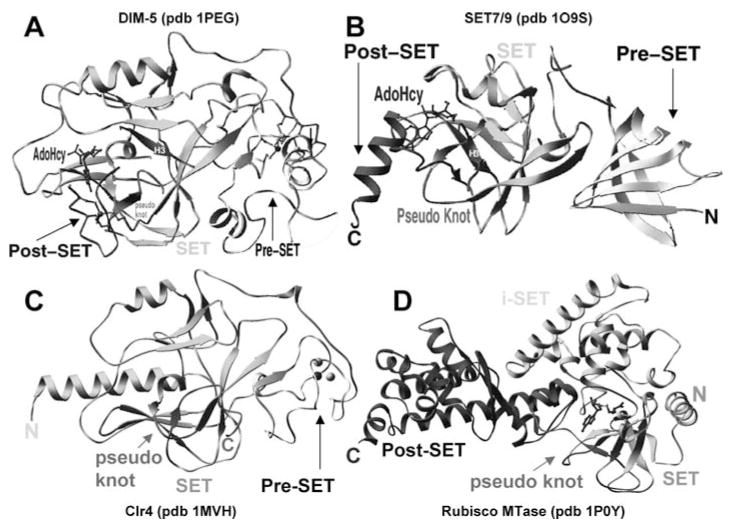
Currently known structures of SET proteins include the crystal structures of two SUV family proteins, Neurospora crassa DIM-5 (112, 113) and Schizosaccha-romyces pombe Clr4 (57); four human SET7/9 structures in various configurations (30, 41, 105, 106); an NMR structure of a viral protein that contains only the SET domain (vSET) (52); and a nonhistone Rubisco MTase (96, 97) (Figure 2). These structures revealed that the SET domain forms a novel β-fold with a series of curved β-strands forming several small sheets, packed together with post-SET, pre-SET, or an additional domain (i-SET) inserted into SET domain.
Figure 2.
Representative examples of SET domain containing structures. (a) Neurospora DIM-5, (b) human SET7/9, (c) S. pombe Clr4, and (d) Rubisco MTase.
The Pre-SET Domain Forms a Triangular Zinc Cluster
The pre-SET domain of the SUV family HKMTs contains nine invariant cysteine residues that arrange into two segments of five and four cysteines separated by various numbers of amino acids (46 in DIM-5 and 28 in Clr4). The nine pre-SET cysteines coordinate three zinc ions to form an equilateral triangular cluster (Figure 1c, left panel). Each zinc ion is coordinated by two unique cysteines (six total) and the remaining three cysteine residues are each shared by two zinc atoms, thus serving as bridges to complete the tetrahedral coordination of the metal atoms. A similar metalthiolate cluster can be found in metallothioneins that are involved in zinc metabolism, zinc transfer, and apoptosis (101). Although the significance of this apparent similarity is unclear, it is intriguing to speculate that the zinc can be transferred from the pre-SET triangular cluster to the post-SET zinc center (see below), analogous to metallothioneins.
The SET Domain Forms a Knot-Like Active Site
The SET domain folds into several small β-sheets that surround a knot-like structure by threading a C terminus through an opening of a short loop formed by a preceding stretch of the sequence (Figure 2). This remarkable knot-like structure brings together the two most-conserved motifs, III and IV, of the SET domain to form an active site in a location immediately next to the methyl-donor-binding pocket and peptide-binding cleft (Figure 1c, middle panel). Of the handful known protein structures that contain a knot, two are AdoMet-related proteins: Escherichia coli AdoMet synthetase (91) and the SPOUT family of RNA MTases (56, 62, 63) (for review on knotted protein structures, see Reference 95).
The Post-SET Zinc-Binding Domain
The post-SET region contains three conserved cysteine residues that are essential for DIM-5 HKMT activity (112). The structure of DIM-5 in a ternary complex with H3 Lys-9 peptide and AdoHcy (113) revealed that the three post-SET cysteines C306, C308, and C313, together with C244 of motif III, tetrahedrally coordinate a zinc ion near the active site (Figure 1c, right panel). Consequently, a narrow channel is formed to accommodate the target lysine side chain (see below).
Close examination of the post-SET region of many SET proteins, including the SUV, SET1, and SET2 families (Figure 1a), suggests that the post-SET metal center observed in DIM-5 is universal among all SET proteins with the cysteine-rich post-SET. For almost all SET proteins, there appears to be an absolute correlation between the presence of the post-SET and a cysteine corresponding to C244 of DIM-5 from the knotted loop formed by the SET signature motif III (Figure 1b). As this metal center is required for enzymatic activity, it represents a good target to design inhibitors that disrupt metal coordination, perhaps analogous to the clinically successful examples of other metalloenzymes such as matrix metalloproteinases (5, 13).
Comparison of the structure of DIM-5 with that of SET7/9 (41, 106) and of Rubisco MTase (96, 97), two SET proteins that do not have a cysteine-rich post-SET domain, reveals a remarkable example of convergent evolution. In particular, like DIM-5, these two enzymes rely on residues C-terminal to the SET domain for the formation of a lysine channel, but do so by packing an α-helix, rather than a metal center, onto the active site (Figure 2b, d).
The Active-Site Channel
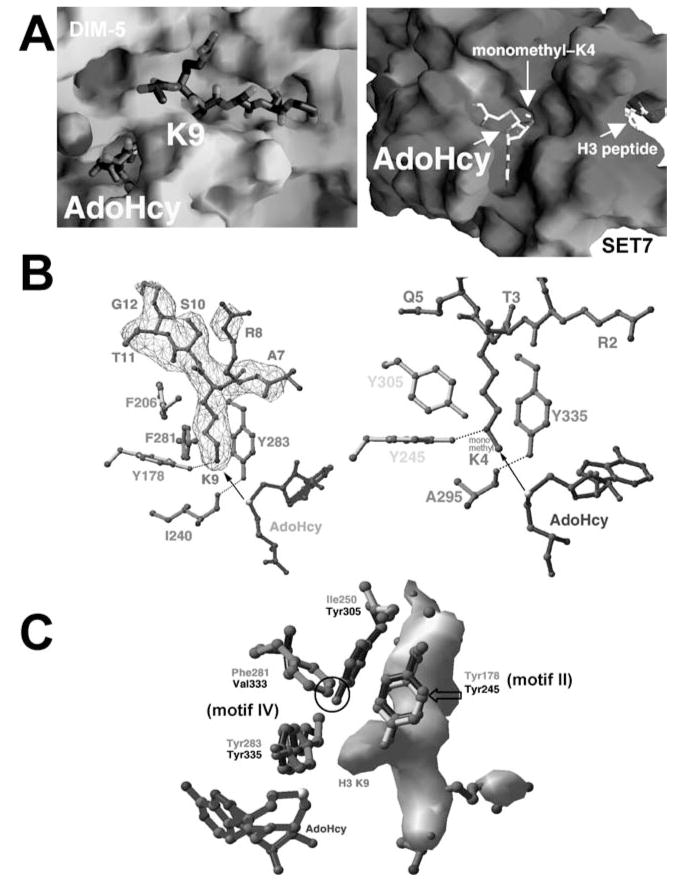
Three ternary structures, SET7/9 in complex with a peptide containing histone H3 Lys-4 (106), DIM-5 in complex with histone H3 Lys-9 peptide (113), and Rubisco MTase in complex with a free lysine (97), revealed the target lysine is inserted into a narrow channel so that the target nitrogen lies in close proximity to the methyl-donor AdoMet at the opposite end of the channel (Figure 3a). The aromatic side chains form the channel wall and make van der Waals contacts to the methylene part of the target lysine side chain (Figure 3b). At the bottom of the channel, the terminal ε-amino group of the substrate lysine hydrogen bonds the hydroxyl of catalytic Tyr of SET motif II (Y178 in DIM-5 and Y245 in SET7/9) and is ~4 Å from the AdoHcy sulfur atom, where the transferable methyl group would be attached on AdoMet.
Figure 3.
Active site of SET domain. (a) H3 peptide-binding site in DIM-5 with the target Lys-9 inserted into a channel (PDB 1PEG) (left panel), and the AdoHcy-binding site in SET7/9, located at the opposite end of the target lysine-binding channel (PDB 1O9S) (right panel). (b) The active sites in DIM-5 (PDB 1PEG) (left panel) and SET7/9 (PDB 1MT6) (right panel). The arrow indicates the movement of the methyl group transferred from the AdoMet methylsulfonium group to the target amino group. (c) Structural comparison of active sites in DIM-5 and SET7/9: either two tyrosines and one phenylalanine (DIM-5) or three tyrosines (SET7/9) surrounds the target lysine.
A Tyrosine/Phenylalanine Switch Controls Product Specificity
HKMTs differ both in their substrate specificity for the various acceptor lysines and in their product specificity for the number of methyl groups (one, two or three) they transfer. The Saccharomyces cerevisiae SET1 protein can catalyze di- and trimethylation of H3 Lys-4, and trimethylation of Lys-4 is thought to be present exclusively in active genes (76). Human SET7/9 protein, on the other hand, generates exclusively monomethyl Lys-4 of H3 (106, 113). Furthermore, DIM-5 of N. crassa generates primarily trimethyl Lys-9, which marks chromatin regions for DNA methylation (92). Considering that different methylation products might have different signaling properties (14, 76, 92), it is important to understand the structural basis for this product specificity (113).
DIM-5 and SET7/9 generate distinct products: DIM-5 forms trimethyl-lysine (92, 113) and SET7/9 forms only monomethyl-lysine (106, 113). A likely structural explanation for their different product specificities is that residues in the lysine-binding channel of SET7/9 sterically exclude the target lysine side chain with methyl group(s). Comparison of the two active sites pinpointed the difference to a single amino acid that occupies a structurally similar position in both enzymes (F281 of DIM-5 and Y305 of SET7/9). Although the two residues are not aligned at the primary sequence level, the edge of the F281 phenyl ring in DIM-5 points to the same position as the Y305 hydroxyl in SET7/9, both in close proximity to the terminal ε-amino group of target lysine (Figure 3c). It was hypothesized that the Y305 hydroxyl in SET7/9 may be the source of steric hindrance limiting methylation (113). Therefore, DIM-5 and SET7/9 mutants that swapped these residues were created. Remarkably, this swap almost completely inverts methylation product specificity (Figure 4). Importantly, neither the lysine target specificity (Lys-9 versus Lys-4) nor the overall reaction rate for each enzyme was changed. Thus, DIM-5 was converted from a Lys-9 tri-MTase to a Lys-9 mono/di-MTase by F281Y mutation, while SET7/9 Y305F generated dimethylated instead of monomethylated Lys-4 (Figure 4a, b).
Figure 4.
Mass spectrometry analysis of methylation products for (a) WT DIM-5 and its F281Y variant (113), (b) WT SET7/9 and its Y305F variant (113), and (c) G9a and its F1205Y variant (12a).
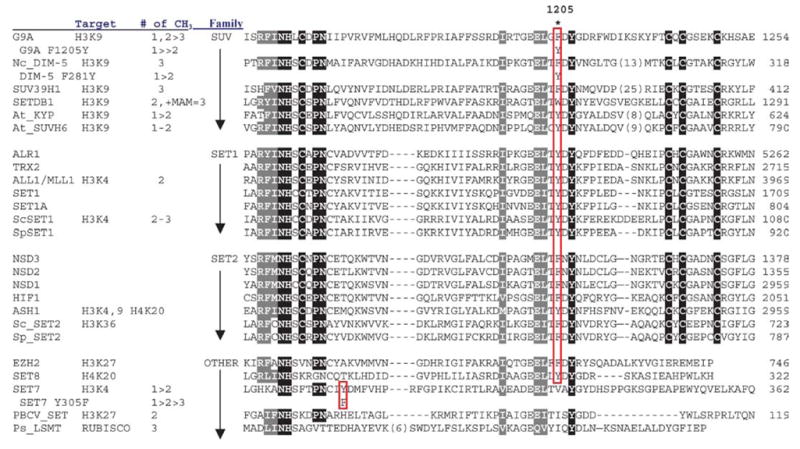
To further test the hypothesis that a single residue in the active site of SET-containing HKMTs, which aligns to F281 in DIM-5 (Table 1), is a major determinant of product specificity, F1205 was replaced with tyrosine (F1205Y) in human G9a, a predominant mammalian H3 Lys-9 HKMT that directs euchromatic mono-and dimethylation (70, 75, 90) but can generate trimethyl-H3K9 in some situations (64). The mutation did not affect the catalytic activity of G9a; however, the reaction by F1205Y stalled at the monomethyl stage (Figure 4c). Thus, the F1205Y mutation changed the product specificity of G9a from a fast mono/di-MTase with a slow tri-MTase activity to a predominantly mono-MTase without affecting overall catalytic activity, analogous to the F281Y mutation for DIM-5 (12a).
TABLE 1.
The locations of the Phe/Tyr switch in SET families
|
Sequence alignment including all HKMTs with known product specificity suggests that the tyrosine/phenylalanine switch rule may be generalized (Table 1). Both Arabidopsis KYP and SUVH6 have a tyrosine and are primarily mono-MTases (29). From a structural perspective, it appears the tyrosine hydroxyl can block substrate lysines with methyl group(s) attached from rotating into a position where they can be further methylated.
Dot1p: Non-SET Domain HKMT
Histone H3 Lys-79 is methylated by Dot1p (21, 43, 61, 100), a protein originally identified as a disruptor of telomeric silencing in S. cerevisiae (83). Methylation of H3 Lys-79 is important for gene silencing and the proper localization of the SIR (silent information regulator) complex in S. cerevisiae (61, 100). A sequence analysis (18) suggested that Dot1p possesses AdoMet-binding motifs characteristic of class-I MTases (78), similar to those in protein arginine MTases that modify arginines on many proteins including histones H3 and H4 (see below). Class-I MTases such as Dot1p are distinct from and do not contain the SET domain. Thus, entirely different structural scaffolding and unrelated local active-site spatial arrangements can catalyze AdoMet-dependent methyl transfer to a protein lysine side chain.
A Conserved Dot1p Core
Yeast Dot1p contains a core region conserved among human, Caenorhabditis elegans, Drosophila, and Anopheles gambiae Dot1p homologues (Figure 5a). The length of these Dot1 proteins varies from 582 amino acids in yeast to 2237 amino acids in Drosophila. The conserved Dot1p core is located at the C terminus in yeast but is at the N terminus in human, C. elegans, Drosophila, and Anopheles gambiae Dot1p homologues.
Figure 5.
Dot1p family (non-SETHKMTs). (a) Schematic representation of Dot1 homologues from yeast and human. (b) Dot1 core structure: (left panel) human Dot1L (residues 5–332) in complex with methyl-donor AdoMet (PDB1NW3) and (right panel) yeast Dot1p (residues 176–567) incomplex with reaction by-product AdoHcy (PDB1U2Z). The N-terminal helical domain and the C-terminal catalytic domain are circled. The bound methyl-donor AdoMet in human Dot1L and there action by-product AdoHcy are shown as stick models. The largest conformational difference (indicated by arrows) between the two catalytic domains is the hairpin loop-containing motif VIII forming part of the active site. (c) Five hydrophobic residues of yeast Dot1p form the active-site pocket (the corresponding residue from human Dot1L is in parenthesis) (left panel). The opening of the pocket is approximately 4×5 Å, into which the target lysine could be inserted (middle panel). A surface representation of yeast Dot1p core showing the conserved motifs (X, VI, IV, and VIII) surrounding the active-site pocket, through which only the AdoHcy sulfur atom is visible (right panel). Conserved motifs (I, II, and III) involved in interacting with AdoHcy are buried and invisible from the surface.
The Dot1p conserved core contains an N-terminal helical domain and a seven-stranded catalytic domain (Figure 5b) that harbors the binding site for the methyl-donor and an active-site pocket sided with conserved hydrophobic residues (Figure 5c, left panel). In the N-terminal domain, three helices, together with a hairpin, mimic a classic up-and-down four-helix bundle, at which the hairpin replaces the fourth helix. The C-terminal region forms the catalytic domain with a seven-stranded β-sheet, a characteristic feature of the class-I AdoMet-dependent MTases (11, 78). The Dot1p core contains conserved sequence motifs (X, I, II, III, IV, VI, and VIII) common to class-I MTases (51). Although at the primary sequence level the seven motifs are scattered throughout the conserved core, the crystal structures show that these motifs are clustered together on one surface patch at or near the active site (Figure 5c, right panel). The structure suggests that the conserved Dot1p motifs have functional importance. Aside from being involved in direct interactions with AdoHcy (motif I: D397 and G399 of yeast Dot1p, motif II: E422, and motif III: F460), active-site formation (motif X: G373, motif IV: F481, and motif VIII: W543), and catalysis of methyl transfer (motif IV: N479), many invariant residues (motif I: S400, motif IV: N480, motif VIII: S542) are involved in in-tramolecular interactions that likely confer stability to the molecule, particularly around the methyl-donor binding and active sites.
Functions of Residues Outside of the Dot1p Conserved Core
Dot1p has several unique biochemical properties. (a) Yeast Dot1p and its human homolog Dot1L methylate only nucleosomal substrates, but not free histone H3 protein (21, 43, 61, 100). A stretch of positively charged residues C-terminal to the human Dot1L core or N-terminal to the yeast Dot1p core (Figure 5a) were critical for nucleosome binding and therefore for enzymatic activity (57, 77). (b) Methylation of Lys-79 of H3 in S. cerevisiae requires ubiquitination of Lys-123 of histone H2B in vivo (9, 61). H2B Lys-123 is located on the same nucleosome disk surface, ~30 Å away from the target Lys-79 of histone H3. Contrary to the in vivo data, recombinant yeast Dot1p was active on nucleosomes assembled in vitro from bacterially expressed, recombinant core histones (77), indicating that ubiquitination is not absolutely required for Dot1p activity. Interestingly, a stretch of ~60 amino acids (residues 40–100) of yeast Dot1p has repeated hydrophobic residues every four to five positions and is predicted to form short helices by secondary structure prediction (Figure 5a). This stretch of Dot1p is similar in size to the ~50-amino-acid monoubiquitin-binding domain (CUE) and the ubiquitin-binding UBA domain, both of which have a three-helix bundle structure (17, 33, 60, 72). Dot1p may interact via this region directly with H2B ubiquitinated nucleosome or indirectly through other ubiquitin-binding proteins. Such an interaction could be significant in vivo, recruiting Dot1p to specific high-order chromatin where ubiquitinated histone H2B might serve as a spacer between adjacent nucleosome disk surfaces, allowing Dot1p access to its target lysine (89).
AdoMet/AdoHcy Binding and Processivity
The methyl donor (in the human Dot1L structure) or the methyl-donor product AdoHcy (in the yeast Dot1p structure) is observed at the carboxyl end of the parallel strands of the C-terminal catalytic domain (Figure 5b)—the hallmark of a nucleotide-binding site of the Rossmann fold. The AdoHcy in yeast Dot1p and the AdoMet in human Dot1L are in an extended conformation—most frequently observed in widespread class-I MTases such as the DNA cytosine MTase DNMT2 (19) and the protein arginine MTase PRMT1 (111) (see below). However, the AdoHcy/AdoMet conformation in the Dot1 proteins is significantly different from the folded conformation observed in the SET domain of HKMTs (30, 41, 78, 96, 97, 106, 113). Such different conformations of the cofactor may provide a good target to design inhibitors that are selective for class-I (Dot1p, PRMT1, DNMTs) versus class-V (SET HKMTs) MTases (for the classification of AdoMet-dependent MTases, see Reference 78).
Unlike SET domain proteins such as DIM-5 and SET7/9, in which the bound AdoHcy/AdoMet is largely surface-exposed (see Figure 3a), the bound AdoHcy in yeast Dot1p (Figure 5c, right panel) and the AdoMet in human Dot1L are buried, suggesting that exchange between the methyl-donor AdoMet and the reaction by-product AdoHcy requires the conformational movement of protein. The fact that a mixture of unmodified, mono-, di-, and trimethylated H3 Lys-79 coexists in yeast (100) suggests that the exchange of the reaction product AdoHcy with AdoMet in the closed-lid binding site of Dot1p would require the release of the substrate and therefore should require methyl transfer to proceed distributively. On the other hand, the open-binding pocket in the SET domain protein DIM-5 would permit the exchange of the reaction product AdoHcy with AdoMet without releasing the substrate and therefore should allow methyl transfers to process processively (92, 113). In contrast to the full distribution of methylated products on H3 Lys-79 in yeast, only trimethyl H3 Lys-9 has been detected in Neurospora (92).
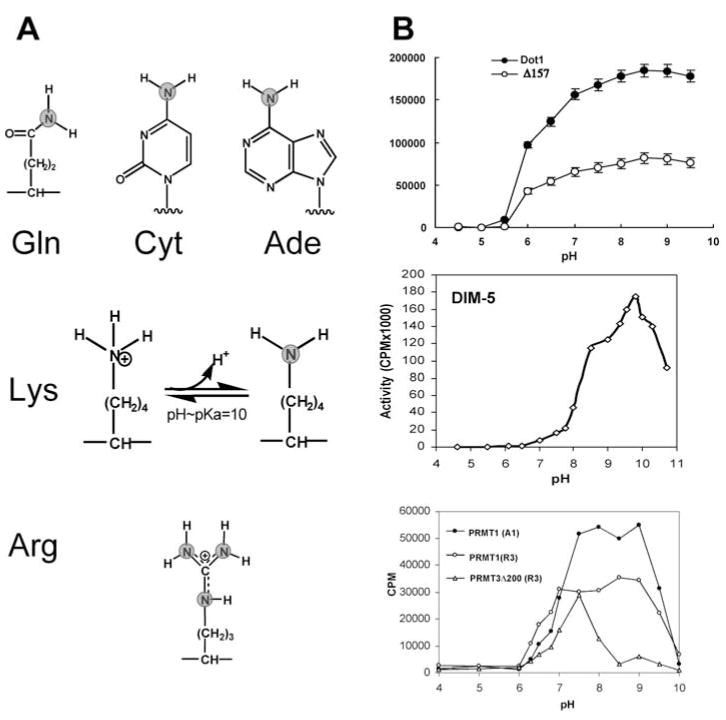
Deprotonation of Target Lysine
The side chain of N479 of yeast Dot1p (the second Asn of NNF motif IV) is the only polar group that protrudes into the active site. Mutations of the equivalent residue in human Dot1L, N241, to D or A abolished activity (57). An asparagine side chain of the so-called NPPY motif is used in the active site of amino MTases of adenine or cytosine in DNA (27, 28) and of protein glutamine MTases (79, 110) (Figure 6a). When AdoHcy is used as an anchoring point, the invariant N479 of yeast Dot1p is superimposable onto the corresponding asparagine in TaqI DNA adenine MTase and HemK protein glutamine MTase (77). This suggests a potential similarity in the catalytic mechanism between Dot1p and class-I amino MTases. In the latter case, the amino group (NH2) that becomes methylated is not charged and is positioned for an in-line attack on AdoMet. However, the amino group ( ) of a lysine side chain is usually positively charged. Under laboratory conditions, yeast Dot1p is active in a broad pH range, from pH 6 to 9.5 (and beyond), with a maximum activity around pH 8.5 (Figure 6b, top panel). This is different from SET-domain-containing HKMTs such as DIM-5 (Figure 6b, middle panel) and SET7, which have a narrower pH range (active at pH 8 or higher) and an unusually high pH optimum (~10) (96, 112). At pH 10, the amino group of the target lysine should be partially deprotonated (Figure 6a, middle panel). Only the deprotonated target lysine has a free lone pair of electrons capable of nucleophilic attack on the AdoMet methyl group. Dot1p must use a different mechanism (or a local microenvironment) to enable the target lysine to be deprotonated.
Figure 6.
(a) Examples of known targets of amino methylation. Only the deprotonated amino group (NH2) has a free lone pair of electrons capable of nucleophilic attack on the AdoMet methyl group. (b) Methyl transfer activities (measured as TCA precipitable counts) as function of pH: (top panel) yeast Dot1p and its N-terminal deletion mutant Δ 157 (77), (middle panel) DIM-5 (112), and (bottom panel) rat PRMT1, rat PRMT3 (full length), and its PRMT core domain (Δ 200) (111).
There are many examples of enzymes that contain lysine residues with significantly depressed pKa values (65). A lysine with a low pKa is generated when a positive charge is immediately proximal to the lysine (104). The proximity of the target lysine to the positively charged methylsulfonium group of AdoMet could have similar effect. A hydrophobic microenvironment is another situation that produces a lowered lysine pKa (66). There are examples of buried lysines with pKa values as low as 6.5 (15). This scenario can best explain the activity of Dot1p over a wide pH range. The active-site pocket sided with all hydrophobic residues (Figure 5c, left panel), in conjunction with a positively charged methylsulfonium group sitting at the bottom of the pocket, is probably essential for lowering the pKa of the ε-amino group of the target lysine so that it can stay in the deprotonated state required for its methylation.
PROTEIN ARGININE METHYLATION
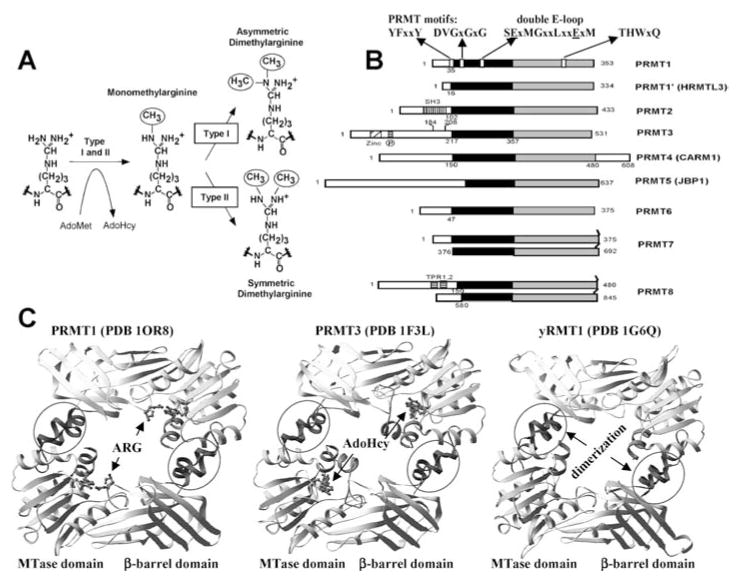
Protein arginine methylation is a common posttranslational modification in eukaryotes. Two major types of protein arginine (R) methyltransferases (PRMTs) transfer the methyl group from AdoMet to the guanidino group of arginines in protein substrates (44). Both catalyze the formation of monomethylarginine, but type I PRMTs also form asymmetric dimethylarginine and type II PRMTs form symmetric dimethylarginine (8) (Figure 7a). Among the known PRMTs, only PRMT5/JBP1 is a type II PRMT (8), which symmetrically dimethylates specific arginines in a few proteins [fewer than 20 proteins have been identified in the past 40 years as containing dimethylated arginine(s) (6)], including myelin basic protein (36), spliceosomal Sm proteins (25), and histones H3 and H4 (66a). PRMT5 was initially identified as a Jak kinase-binding protein (JBP1) (71, 74) and has been found to coexist with substrates in multiprotein complexes (25, 54, 55, 67, 109).
Figure 7.
(a) Two major types of protein arginine methylation. (b) Members of PRMT family. The conserved MTase domain is in black and the unique β-barrel domain to the PRMT family is in gray. The N and C termini of the proteins and the first invariant residue are labeled. (c) Dimer structures of PRMT cores: (left panel) rat PRMT1, (middle panel) rat PRMT3, and (right panel) yeast RMT1/Hmt1.
Multiple PRMT genes are present in eukaryotes from fungi to plants and animals (Figure 7b) (Table 2). For example, seven similar paralogous mammalian PRMT genes have been reported so far: PRMT1 (1, 34, 47), PRMT2 (81), PRMT3 (93), CARM1/PRMT4 (10), JBP1/PRMT5 (45, 71), PRMT6 (24), and PRMT7 (45a, 58). Nine PRMTs are present in the completed Drosophila melanogaster genome (7), and complete PRMT genes from S. pombe, Arabidopsis, and C. elegans have also been identified through genome sequencing projects. In addition, ESTs with strong homology to PRMT1 can also be found in Xenopus, zebrafish, sea urchin, rice, and tomato, indicating that PRMT is a highly conserved family of proteins in eukaryotes. The presence of such a large number of PRMTs may signify the diverse roles they can play.
TABLE 2.
Members of PRMT family
| Human PRMT genes | Presence of PRMT homologous in other organisms | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Activity | Chromosome | EST | Coding Exon | Genomic size (kb) | Accession number | Arabidopsis | Drosophila | C. elegans | S. pombe | S. cerevisiae |
| PRMT1 | +++ | 19q13 | +++ | 9–10 | 10 | CAA71764 | + | + | + | + | + |
| PRMT1′ (HRMTL3) | ? | 12p13 | + | 9 | 52 | AAF91390 | + | − | − | − | − |
| PRMT2 | − | 21q22 | ++ | 10 | 30 | P55345 | − | − | − | − | − |
| PRMT3 | + | 11p15 | + | 13 | 50 | AAH64831 | + | + | − | + | − |
| PRMT4 (CARM1) | + | 19p13 | ++ | 16 | 50 | AL833242 (partial cDNA) | + | + | − | − | − |
| PRMT5 (JBP1) | +(type II) | 14q11 | ++ | 17 | 8.5 | AAF04502 | + | + | + | + | + |
| PRMT6 | + | 1p13 | +/− | 1 | 2.5 | AAK85733 | − | − | − | − | − |
| PRMT7 | + | 16q22 | ++ | 17 | 41 | Q9NVM4 | + | + | + | − | − |
| PRMT8 | ? | 4q31 | + | 10 | 40 | AAH64403 | − | − | + | − | − |
Two well-studied enzymes, PRMT1 and PRMT4/CARM1, methylate histones H3 (3, 10, 50, 80), H4 (88, 102), and H2B (2), in addition to many other substrates. Histone arginine methylation is a component of the histone code that directs a variety of processes involving chromatin (39, 87). For example, methylation of Arg-3 of histone H4 by PRMT1 facilitates H4 acetylation and enhances transcriptional activation by nuclear hormone receptors synergistically with CARM1 (46, 88, 102, 108), in that CARM1 prefers acetylated histone tails in generating H3 Arg-17 methylation (16, 102). In vitro, p53-mediated transcription was stimulated the greatest when all three coactivators (PRMT1, CARM1, and p300) were present, whether added sequentially or at the same time (2). Preincubation of a chromatin template with p53 and PRMT1 significantly stimulated the histone acetyltrans- ferase activity of p300, and similarly, preincubation of the template with p53 and p300 stimulated H3 arginine methylation by CARM1.
PRMT1
PRMT1 is the predominant type I PRMT in mammalian cells, accounting for 85% of cellular PRMT activity (94). It is essential for early postimplantation development, as shown by the embryonic lethality of mouse Prmt1−/− mutants (69). Although PRMT1 is expressed at detectable levels in all tissues examined (47, 69, 81, 93), the expression is highest in developing neural structures in embryos (69), and PRMT1 has been implicated in neuronal differentiation (12). PRMT1 has at least six alternatively spliced transcripts that would produce proteins with an N terminus of 20 to 40 amino acids (81, 111), and these proteins may have different substrate specificities (68).
PRMT1 gene is found in all eukaryotes examined and is highly conserved (Table 2). The sequence identity is over 90% among mammals, zebrafish, and Xenopus, and about 50% even between human and S. cerevisiae. There appears to be another gene (PRMT1′) closely related to PRMT1 genes both in Arabidopsis thaliana and in human (HRMT1L3, AAF91390, on chromosome 12p13), which in each case share 80% amino acid identity with PRMT1. Except for their N termini, the two genes have identical genomic structure: Each pair has eight introns inserted at identical positions, and the locations of seven of those in- trons are also shared between human and A. thaliana. No function about this PRMT1-like gene (PRMT1′) has been reported. Like S. cerevisiae, C. elegans and S. pombe have only one copy of PRMT1 and share some of the splicing sites used by human and A. thaliana. D. melanogaster encodes four to six PRMT proteins of similar size, but of these, only DmPRMT1 (AAF54556) has a high percentage identity with the mammalian PRMT1 (65% versus 15%–35% for the others).
The best-known substrates for PRMT1 are RNA-binding proteins involved in various aspects of RNA processing and/or transport, such as hnRNPs, fibrillarin, nucleolin (26), and poly(A)-binding protein II (84). A growing number of other proteins were found to be substrates of PRMT1, including high-molecular-weight fibroblast growth factor-2 (HMW FGF-2), a nuclear growth factor (38); interleukin enhancer-binding factor 3 (ILF3) (94); STAT1, a transcription factor activated by extracellular signals (59); SPT5, a regulator of transcriptional elongation (40); and histones H4 (88, 102) and H2B (2).
CARM1/PRMT4
PRMT4 was discovered as a transcriptional coactivator-associated arginine (R) MTase (CARM1) (10). CARM1 enhances gene activation by nuclear receptors in a synergistic collaboration with two other classes of coactivators: the p160 coactivators and the protein acetyltransferases p300/CBP (10, 46). CARM1 can methylate specific arginine residues in the N-terminal tail of histone H3 (3, 10, 50, 80).
Both CARM1 as well as PRMT1 act in concert with the acetyltransferase CBP/p300, along with the p160 coactivator family to enhance transcription from hormone-responsive promoters (85, 86). Similarly, CARM1 and PRMT1 act as coactivators in the tumor suppressor protein p53-mediated transcription, via direct interactions with p53 and its associated coactivator partner p300 (2). These results provide compelling evidence that the histones are relevant targets for CARM1, PRMT1, and p300, and that the resulting histone modifications are directly important for transcription.
A Conserved PRMT Core
The PRMT proteins vary in length from 348 amino acids in S. cerevisiae RMT to 608 amino acids in CARM1, but they all contain a conserved core region of approximately 310 amino acids (Figure 7b). The sequences beyond the conserved PRMT core region are all N-terminal additions; however, CARM1 also has a C-terminal addition. The size of the N-terminal additions varies from ~20 amino acids in S. cerevisiae RMT1 to 200 amino acids in PRMT3. The varied N termini could subject each PRMT to a different regulation. An interesting feature of PRMT7 (58), and PRMT8 (Figure 7b) (Table 2) is that they seem to have arisen from a gene duplication event and contain two conserved core regions, each with a putative AdoMet-binding motif.
Structure of the Conserved PRMT Core
Three crystal structures of PRMTs are currently available: rat PRMT1 (amino acids 41–353) (111), the rat PRMT3 catalytic core (amino acids 208–528) (114), and yeast RMT1/Hmt1 (amino acids 30–348) (103). These structures reflect a striking structural conservation of the PRMT catalytic core (Figure 7c). The overall monomeric structure of the PRMT core can be divided into three parts: an MTase domain, a β-barrel, and a dimerization arm. The MTase domain has the consensus fold conserved in a class-I AdoMet-dependent MTase that harbors an AdoMet-binding site (11, 78). The β-barrel domain is unique to the PRMT family (114).
PRMT Dimerization is Essential for AdoMet Binding and Enzymatic Activity
An identical hydrophobic dimer interface is observed in PRMT1 (111), the PRMT3 core (114), and yeast RMT1/Hmt1 (103) (Figure 7c), despite different crystallization conditions, space groups, and cell dimensions. This observation supports the notion that dimer formation is a conserved feature in the PRMT family (114). A mutant of yeast RMT1/Hmt1 that replaces the dimerization arm with alanines resulted in the loss of dimer formation and methylation activity (105). The mutant PRMT1 ΔARM lacks the entire dimerization arm (residues 188–222), elutes as a monomer on a gel filtration column, and completely lacks enzymatic activity, most likely because it is unable to bind the AdoMet cofactor, as determined by UV cross-linking experiments (111). In the crystal structure, the dimer interface forms between the arm and the outer surface of the AdoMet-binding site (Figure 7c). It is conceivable that dimererization is required to engage the residues in the AdoMet-binding site in a manner in which they can interact with AdoMet properly. Interestingly, the higher order oligomerization of PRMT1 (47, 111) does not occur in the absence of dimerization (i.e., in the case of ΔARM) (111).
Another potential function of the conserved PRMT dimer might be to allow processive production of the final methylation product, asymmetric dimethylarginine. PRMT substrates isolated in vivo are usually completely or nearly completely dimethylated (37, 38, 48, 49, 84). In vitro, PRMT6 forms dimethylarginine in a processive manner (24). It is conceivable that a ring-like dimer could allow the product of the first methylation reaction, monomethylarginine, to enter the active site of the second molecule of the dimer without releasing the substrate from the ring or replenishing the methyl-donor.
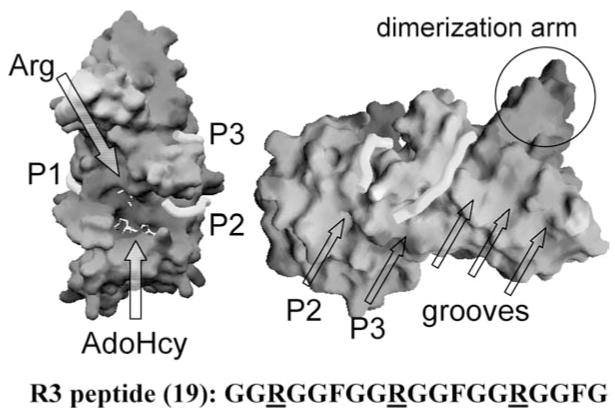
Multiple-Substrate-Binding Grooves
Most PRMT1 substrates contain glycine- and arginine-rich sequences that include multiple arginines in RGG context (26, 38, 84). Peptides that contain three copies of the consensus RGG repeat sequence (R3) were cocrystallized with PRMT1 (111). Three peptide-binding grooves were identified (Figure 8), which probably represent a mixture of binding modes of the R3 peptide, which contains three potential methylation targets at position 3, 9, and 15. Additional acidic grooves running parallel to site P3 were identified (Figure 8). These grooves could form additional binding sites for protein substrates with more RGG repeats.
Figure 8.
Peptide-binding grooves (P1, P2, and P3) in the structure of ternary complex of PRMT1-AdoHcy-R3 peptide (sequence shown at the bottom): solvent-accessible molecular surface with bound AdoHcy and arginine shown as stick models and indicated by the arrows (left panel). If the central Arg-9 were the target bound in the active site, connecting peptide-binding sites P1 and P2 would cover the active site and the entire length of the peptide. When the end arginine (either Arg-3 or Arg-15) is bound in the active site, connection of peptide-binding sites P2 and P3 would account for the length of the whole peptide (right panel). Site P3 corresponds to one of the grooves perpendicular to the strands of the β-barrel domain.
Asymmetric and Symmetric Dimethylarginines
The target arginine is situated in a deep acidic pocket between the MTase domain and the β-barrel domain (Figure 8). The residues that make up the active site are conserved across the PRMT family, and a “double-E” hairpin loop (Figure 7b) contributes most of the residues in the active-site pocket. Two invariant glutamates (E144 and E153 of PRMT1 and E326 and E335 of PRMT3) are used to neutralize the positive charge on the substrate guanidino group: The interaction with E153 of PRMT1 (or E335 of PRMT3) redistributes the positive charge on the guanidino group toward one amino group while leaving a lone pair of electrons on the other amino group to attack the cationic methylsulfonium moiety of AdoMet (114). The corresponding mutant in CARM1/PRMT4 (E267Q) has been used to demonstrate that the MTase activity of CARM1 was required for synergy among nuclear receptor coactivators (46).
The three solved PRMT structures, rat PRMT1, rat PRMT3, and yeast RMT1, are all type I enzymes. Interestingly, all the PRMTs except mammalian PRMT5 (or yeast Hsl7) contain an active-site methionine, the last residue of the double-E loop (amino acid 155 for PRMT1, amino acid 337 for PRMT3, and amino acid 143 for yeast RMT1), which has been proposed to exclude binding of monomethylated arginine in a conformation that would allow its symmetric methylation (114). However, in both PRMT5 (amino acid 446) and Hsl7 (amino acid 474), the residue corresponding to M155 of PRMT1 is serine. The smaller bulk of the side chain of this residue may allow for symmetric dimethyl arginine formation by the type II enzymes PRMT5 and possibly Hsl7 (see figure 7 of Reference 8).
FUTURE PROSPECTS
AdoMet-dependent MTases are involved in biosynthesis, signal transduction, protein repair, chromatin regulation, and gene silencing. Methylation substrates range in size from arsenite through DNA and proteins, and the atomic targets can be carbon, oxygen, nitrogen, sulfur, or even halides. Methylation can function as a reversible signal, as in the case of O(xygen)-methylation, in which the side chain carboxyl groups of glutamate residues or the C-terminal carboxyl groups are reversibly methylated. However, it is unclear whether the N(itrogen)-methylations (of arginine, lysine, glutamine, asparagine, histidine residues, and the amino group at the N terminus) are reversible in the cell or the N-methylation function is a more permanent modification that affects the activity or surface hydrophobicity of a substrate. Recently, a human nuclear peptidyl arginine deiminase, PAD4, has been shown to antagonize methylation on the arginine residues by converting arginine to citrulline (13a, 102a), and a human nuclear amine oxidase, LSD1, functions as a histone di/monomethyl-lysine demethylase via an oxidation reaction (81a). Despite recent advances in identifying MTases, we still know little about what regulates their activities or determines their specificity. This is evident by a recent report that SET7/9 activity is not limited to histones; it also methylates the tumor suppressor p53 (11a). With the increasing interest in protein (histone) methylation as a mechanism for gene regulation, we will undoubtedly discover other exciting roles for MTases and the cellular processes that they direct.
Acknowledgments
We wish to thank most warmly the members of our laboratory and our collaborators (Drs. Eric U. Selker, Mike R. Stallcup, Yoichi Shinkai) whose hard work was responsible for much of the protein methyltransferase story. Work in our laboratories is supported in part by grants from the National Institute of Health (GM49245 and GM61355 to X.Z. and X.C.) and training grant T32 GM08367 (to R.E.C). X.C. is a Georgia Research Alliance Eminent Scholar. Color versions of Figures 1–8 have been provided as Supplemental Material (follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org).
LITERATURE CITED
- 1.Abramovich C, Yakobson B, Chebath J, Revel M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–66. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–48. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–33. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode W, Fernandez-Catalan C, Tschesche H, Grams F, Nagase H, Maskos K. Structural properties of matrix metalloproteinases. Cell Mol Life Sci. 1999;55:639–52. doi: 10.1007/s000180050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–30. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Boulanger MC, Miranda TB, Clarke S, Di Fruscio M, Suter B, et al. Characterization of the Drosophila protein arginine methyltransferases DART1 and DART4. Biochem J. 2004;379:283–89. doi: 10.1042/BJ20031176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, et al. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–76. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 9.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, et al. Gene silencing: transhistone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, Ma H, Hong H, Koh SS, Huang SM, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–77. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 11.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyl-transferases and base flipping. Nucleic Acids Res. 2001;29:3784–95. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–60. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 12.Cimato TR, Tang J, Xu Y, Guarnaccia C, Herschman HR, et al. Nerve growth factor-mediated increases in protein methylation occur predominantly at type I arginine methylation sites and involve protein arginine methyltransferase 1. J Neurosci Res. 2002;67:435–42. doi: 10.1002/jnr.10123. [DOI] [PubMed] [Google Scholar]
- 12a.Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–70. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–92. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 13a.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 15.Dao-Pin S, Anderson DE, Baase WA, Dahlquist FW, Matthews BW. Structural and thermodynamic consequences of burying a charged residue within the hydrophobic core of T4 lysozyme. Biochemistry. 1991;30:11521–29. doi: 10.1021/bi00113a006. [DOI] [PubMed] [Google Scholar]
- 16.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–97. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 17.Dieckmann T, Withers-Ward ES, Jarosinski MA, Liu CF, Chen IS, Feigon J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat Struct Biol. 1998;5:1042–47. doi: 10.1038/4220. [DOI] [PubMed] [Google Scholar]
- 18.Dlakic M. Chromatin silencing protein and pachytene checkpoint regulator Dot1p has a methyltransferase fold. Trends Biochem Sci. 2001;26:405–7. doi: 10.1016/s0968-0004(01)01856-4. [DOI] [PubMed] [Google Scholar]
- 19.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439–48. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutnall RN. Cracking the histone code: one, two, three methyls, you’re out! . Mol Cell. 2003;12:3–4. doi: 10.1016/s1097-2765(03)00282-x. [DOI] [PubMed] [Google Scholar]
- 21.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–58. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 22.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–79. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 23.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–43. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 25.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 27.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyl-transferase M. TaqI in complex with DNA and a cofactor analog. Nat Struct Biol. 2001;8:121–25. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 28.Gong W, O’Gara M, Blumenthal RM, Cheng X. Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–15. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–15. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs SA, Harp JM, Devarakonda S, Kim Y, Rastinejad F, Khorasanizadeh S. The active site of the SET domain is constructed on a knot. Nat Struct Biol. 2002;9:833–38. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 32.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, et al. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–30. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 34.Katsanis N, Yaspo ML, Fisher EM. Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mamm Genome. 1997;8:526–29. doi: 10.1007/s003359900491. [DOI] [PubMed] [Google Scholar]
- 35.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–72. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Lim IK, Park GH, Paik WK. Biological methylation of myelin basic protein: enzymology and biological significance. Int J Biochem Cell Biol. 1997;29:743–51. doi: 10.1016/s1357-2725(97)00009-5. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Merrill BM, Rajpurohit R, Kumar A, Stone KL, et al. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry. 1997;36:5185–92. doi: 10.1021/bi9625509. [DOI] [PubMed] [Google Scholar]
- 38.Klein S, Carroll JA, Chen Y, Henry MF, Henry PA, et al. Biochemical analysis of the arginine methylation of high molecular weight fibroblast growth factor-2. J Biol Chem. 2000;275:3150–57. doi: 10.1074/jbc.275.5.3150. [DOI] [PubMed] [Google Scholar]
- 39.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 40.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11:1055–66. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 41.Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, et al. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–24. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 43.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–24. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 44.Lee HW, Kim S, Paik WK. S-adenosylmethionine: protein-arginine methyltransferase. Purification and mechanism of the enzyme. Biochemistry. 1977;16:78–85. doi: 10.1021/bi00620a013. [DOI] [PubMed] [Google Scholar]
- 45.Lee JH, Cook JR, Pollack BP, Kinzy TG, Norris D, Pestka S. Hsl7p, the yeast homologue of human JBP1, is a protein methyltransferase. Biochem Biophys Res Commun. 2000;274:105–11. doi: 10.1006/bbrc.2000.3049. [DOI] [PubMed] [Google Scholar]
- 45a.Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson S, et al. PRMT7: a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005;280:3656–64. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 46.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621–32. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–44. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 48.Lischwe MA, Cook RG, Ahn YS, Yeoman LC, Busch H. Clustering of glycine and NG, NG-dimethylarginine in nucleolar protein C23. Biochemistry. 1985;24:6025–28. doi: 10.1021/bi00343a001. [DOI] [PubMed] [Google Scholar]
- 49.Lischwe MA, Ochs RL, Reddy R, Cook RG, Yeoman LC, et al. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG, NG-dimethylarginine. J Biol Chem. 1985;260:14304–10. [PubMed] [Google Scholar]
- 50.Ma H, Baumann CT, Li H, Strahl BD, Rice R, et al. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11:1981–85. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 51.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–32. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 52.Manzur KL, Farooq A, Zeng L, Plotnikova O, Koch AW, et al. A dimeric viral SET domain methyltrans-ferase specific to Lys27 of histone H3. Nat Struct Biol. 2003;10:187–96. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 53.Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 54.Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–94. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 55.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–63. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michel G, Sauve V, Larocque R, Li Y, Matte A, Cygler M. The structure of the RlmB 23S rRNA methyl-transferase reveals a new methyltransferase fold with a unique knot. Structure. 2002;10:1303–15. doi: 10.1016/s0969-2126(02)00852-3. [DOI] [PubMed] [Google Scholar]
- 57.Min J, Zhang X, Cheng X, Grewal SI, Xu RM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9:828–32. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 58.Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004;279:22902–7. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- 59.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–41. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 60.Mueller TD, Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J Mol Biol. 2002;319:1243–55. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 61.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–27. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nureki O, Shirouzu M, Hashimoto K, Ishitani R, Terada T, et al. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr D. 2002;58:1129–37. doi: 10.1107/s0907444902006601. [DOI] [PubMed] [Google Scholar]
- 63.Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, et al. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure. 2004;12:593–602. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Osipovich O, Milley R, Meade A, Tachibana M, Shinkai Y, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol. 2004;5:309–16. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 65.Paetzel M, Dalbey RE. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem Sci. 1997;22:28–31. doi: 10.1016/s0968-0004(96)10065-7. [DOI] [PubMed] [Google Scholar]
- 66.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–90. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 66a.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–87. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pawlak MR, Banik-Maiti S, Pietenpol JA, Ruley HE. Protein arginine methyltransferase I: substrate specificity and role in hnRNP assembly. J Cell Biochem. 2002;87:394–407. doi: 10.1002/jcb.10307. [DOI] [PubMed] [Google Scholar]
- 69.Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20:4859–69. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 71.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyl-transferase activity. J Biol Chem. 1999;274:31531–42. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 72.Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–20. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- 73.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–99. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 74.Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276:11393–401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- 75.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–98. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 76.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 77.Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine79 methyltransferase. J Biol Chem. 2004;279:43294–306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–35. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schubert HL, Phillips JD, Hill CP. Structures along the catalytic pathway of PrmC/HemK, an N5-glutamine AdoMet-dependent methyltransferase. Biochemistry. 2003;42:5592–99. doi: 10.1021/bi034026p. [DOI] [PubMed] [Google Scholar]
- 80.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–56. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 81.Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, Henry MF. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–40. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 81a.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 82.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–32. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, et al. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–34. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 85.Stallcup MR. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–20. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 86.Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85:139–45. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- 87.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 88.Strahl BD, Briggs SD, Brame CJ, Cald-well JA, Koh SS, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 89.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 90.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takusagawa F, Kamitori S. A real knot in protein. J Am Chem Soc. 1996;118:8945–46. [Google Scholar]
- 92.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, et al. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 93.Tang J, Gary JD, Clarke S, Herschman HR. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–45. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 94.Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyl-transferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275:19866–76. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- 95.Taylor WR, Xiao B, Gamblin SJ, Lin K. A knot or not a knot? SETting the record ‘straight’ on proteins. Comput Biol Chem. 2003;27:11–15. doi: 10.1016/s1476-9271(02)00099-3. [DOI] [PubMed] [Google Scholar]
- 96.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 97.Trievel RC, Flynn EM, Houtz RL, Hurley JH. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol. 2003;10:545–52. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 98.Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 99.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–91. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 100.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–56. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 101.Vasak M, Hasler DW. Metallothioneins: new functional and structural insights. Curr Opin Chem Biol. 2000;4:177–83. doi: 10.1016/s1367-5931(00)00082-x. [DOI] [PubMed] [Google Scholar]
- 102.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–57. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 102a.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 103.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat Struct Biol. 2000;7:1165–71. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 104.Westheimer FH. Coincidences, decarboxylation, and electrostatic effects. Tetrahedron. 1995;51:3–20. [Google Scholar]
- 105.Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–15. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 106.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–56. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 107.Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Curr Opin Struct Biol. 2003;13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 108.Xu W, Chen H, Du K, Asahara H, Tini M, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–11. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 109.Yanagida M, Hayano T, Yamauchi Y, Shinkawa T, Natsume T, et al. Human fibrillarin forms a sub-complex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins alpha 3 and beta 1 that is independent of its association with preribosomal ribonucleoprotein complexes. J Biol Chem. 2004;279:1607–14. doi: 10.1074/jbc.M305604200. [DOI] [PubMed] [Google Scholar]
- 110.Yang Z, Shipman L, Zhang M, Anton BP, Roberts RJ, Cheng X. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J Mol Biol. 2004;340:695–706. doi: 10.1016/j.jmb.2004.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–20. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, et al. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–27. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–85. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19:3509–19. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]