Abstract
The ability to cleave DNA is critical to the cellular and pharmacological functions of human type II topoisomerases. However, the low level of cleavage at equilibrium and the tight coupling of the cleavage and ligation reactions make it difficult to characterize the mechanism by which these enzymes cut DNA. Therefore, to establish a system that isolates topoisomerase II-mediated DNA scission from ligation, oligonucleotide substrates were developed that contained a 3’-bridging phosphorothiolate at the scissile bond. Scission of these substrates generates a 3’-terminal –SH moiety that is a poor nucleophile relative to the normal 3’-terminal –OH. Consequently, topoisomerase II cannot efficiently ligate phosphorothiolate substrates once they are cleaved. The characteristics of topoisomerase IIα-mediated cleavage of phosphorothiolate oligonucleotides were identical to those seen with wild-type substrates, except that no ligation was observed. This unidirectional accumulation of cleavage complexes provided critical information regarding coordination of the protomer subunits of topoisomerase IIα and the mechanism of action of topoisomerase II poisons. Results indicate that the two enzyme subunits are partially coordinated and that cleavage at one scissile bond increases cleavage at the other. Furthermore, anticancer drugs such as etoposide and amsacrine that strongly inhibit topoisomerase II-mediated DNA ligation have little effect on the forward scission reaction. In contrast, abasic sites that increase levels of cleavage complexes without affecting ligation stimulate the forward rate of scission. Phosphorothiolate substrates provide significant advantages over traditional “suicide substrates” and should be valuable for future studies on DNA scission and the topoisomerase II-DNA cleavage complex.
Despite the physiological imperative to maintain the genetic material, normal nuclear processes routinely induce topological alterations in DNA that pose a threat to cell survival. For example, recombination generates DNA knots that prevent the separation of the two strands of the double helix and replication produces tangles between daughter chromosomes that impede segregation during mitosis and meiosis (1-4).
The enzymes that remove knots and tangles from the genome are known as type II DNA topoisomerases (3-10). Humans encode two isoforms of the type II enzyme, topoisomerase IIα and topoisomerase IIβ (6, 8-10). Topoisomerase IIα is the isoform whose activity is linked to cell proliferation and is involved in replicative processes (6, 9-13).
Type II topoisomerases resolve knots, tangles, and other topological structures in DNA by passing an intact double helix through a transient double-stranded break that they generate in a separate segment of DNA (6, 9, 10, 14-16). In order to maintain genomic integrity during this process, topoisomerase II forms covalent bonds between active site tyrosyl residues and the 5’-terminal phosphates that are generated during the DNA cleavage reaction (17-19). These covalent enzyme-cleaved DNA intermediates are known as cleavage complexes.
The ability of type II topoisomerases to alter the topological structure of DNA requires these enzymes to generate double-stranded breaks in the genetic material. Thus, while type II topoisomerases are essential to cell survival, they also have the capacity to fragment the genome (6, 9, 10, 14-16). Under normal circumstances, levels of topoisomerase II-DNA cleavage complexes are low and are tolerated by the cell. However, circumstances that shift the DNA cleavage/ligation equilibrium of the enzyme toward the cleaved state can generate permanent double-stranded breaks in DNA, induce recombination/repair events, and in some cases, trigger cell death pathways (6, 10, 16, 20-23).
Beyond its contributions to critical nucleic acid processes, the DNA cleavage/ligation activity of topoisomerase II also plays a fundamental role in both the treatment and development of human malignancies (6, 10, 22, 24-29). A number of widely prescribed anticancer drugs, such as etoposide, kill cells specifically by increasing the concentration of cleavage complexes. To distinguish these agents from catalytic inhibitors of the enzyme, compounds that act by raising levels of cleavage complexes are called topoisomerase II poisons (30). In addition to their role as a target for anticancer drugs, topoisomerase II-generated chromosomal breaks have been linked to specific therapy-related and infant leukemias. These leukemias are characterized by rearrangements involving the mixed-lineage leukemia (MLL) gene at chromosomal band 11q23 (24, 27, 31, 32).
In order to fully understand the basis for topoisomerase II action and the role of these enzymes in both curing and causing cancer, it is essential to understand the mechanism by which they cleave and ligate DNA. Unfortunately, these reactions have been difficult to study for two major reasons. First, the DNA cleavage/ligation equilibrium of mammalian type II topoisomerases lies heavily toward ligation (6, 10, 16-19, 33). Consequently, in unperturbed systems, levels of cleavage complexes generated by type II enzymes, including human topoisomerase IIα, are extremely low (<1%). To overcome this challenge, it is common to include topoisomerase II poisons, such as anticancer drugs (6, 10, 16) or DNA substrates that contain damaged or missing bases, in reaction mixtures (34-37). The use of non-physiological divalent cations such as Ca(II) or Co(II) also has been reported (38, 39). Although all of these conditions shift the cleavage/ligation equilibrium of topoisomerase II toward the cleaved state, they often perturb the specificity of the enzyme. Furthermore, since they do not uncouple the two reaction steps from one another (see below), in most cases they cannot be used to reveal intrinsic properties of the DNA cleavage or ligation reactions.
Second, the DNA cleavage and ligation reactions of type II topoisomerases are tightly coupled (6, 10, 16-18, 38, 40). Because these reactions take place in a highly concerted fashion, it often is difficult to specifically ascribe a given effect to either catalytic event. Several approaches have been employed to separate the DNA cleavage and ligation reactions and study them independently. “Suicide substrates” represent the primary approach that has been used in an attempt to isolate the forward DNA cleavage reaction of topoisomerase II (41-46). These DNA substrates all contain perturbations or non-B-form structures, such as nicks, double strand/single strand junctions, or hairpins. With “normal” intact B-form substrates, both halves of the cleaved DNA intermediate are covalently attached to topoisomerase II. In contrast, when suicide substrates are cut by the enzyme, one of the DNA halves is held in place by non-covalent, rather than covalent, interactions. If the “non-covalent” portion of the cleaved DNA dissociates from topoisomerase II, the enzyme is unable to rejoin the original substrate. This process results in a time-dependent accumulation of cleavage complexes. Although results with suicide substrates often are interpreted as representative of a unidirectional forward cleavage reaction, evidence suggests that topoisomerase II actually establishes a short-lived cleavage/ligation equilibrium with these substrates prior to the dissociation event (44). As a result, some conditions that impair the ability of topoisomerase II to reseal DNA breaks could increase the rate of accumulation of “trapped” cleavage complexes, even though they have no direct effect on the DNA scission reaction. This could lead to erroneous mechanistic conclusions regarding the actions of anticancer drugs or other topoisomerase II poisons.
In contrast to DNA cleavage, a number of approaches have been utilized to isolate the ligation reaction of topoisomerase II (19, 37, 47-51). These include ligation of cleavage complexes generated with suicide substrates to acceptor oligonucleotides, manipulation of the divalent cation, shifting reactions to suboptimal temperatures, or the use of oligonucleotide substrates with activated 5’-terminal phosphates. The last approach has the advantage that it does not require the enzyme to cleave the substrate prior to ligation (50, 51).
The goal of the present study was to uncouple the DNA cleavage reaction of topoisomerase II from ligation. To this end, a novel system was developed that employed oligonucleotide substrates containing a 3’-bridging phosphorothiolate at the scissile bond. Similar DNA substrates containing 5’-bridging phosphorothiolates have been used to provide important mechanistic information about human, bovine, and pox virus type IB topoisomerases as well as bacteriophage lambda integrase (52-56). [5’-Bridging phosphorothiolates were used in these studies because type IB topoisomerases and lambda integrase form covalent links to the 3’-terminal phosphates of cleaved DNA (3, 9, 57, 58), as opposed to the 5’-linkage formed with eukaryotic topoisomerase II (6, 9, 10, 14-18).] 5’-Bridging phosphorothiolate oligonucleotides ligate extremely poorly (rates of ligation mediated by bovine topoisomerase I were <1% those observed for corresponding wild-type oligonucleotides) (54), and have been used in structural studies to trap the cleavage complex formed with human topoisomerase I (59-64).
The use of 3’-bridging phosphorothiolate oligonucleotides to study topoisomerase II mechanism has significant advantages over previous suicide substrates. Phosphorothiolate-containing oligonucleotides are fully double-stranded B-form DNA and no evidence of ligation was observed. Results with human topoisomerase IIα indicate that phosphorothiolate-containing oligonucleotides can be utilized effectively to examine the DNA cleavage reaction of the enzyme in isolation and provide information regarding the mechanism of action of type II topoisomerases and topoisomerase II poisons.
EXPERIMENTAL PROCEDURES
Enzymes
Human topoisomerase IIα and IIβ were expressed in Saccharomyces cerevisiae and purified as described previously (65-67). Yeast (S. cerevisiae) topoisomerase II was isolated as described previously (66, 68).
Preparation of Oligonucleotides
Two 50-base oligonucleotide duplexes were designed using previously identified topoisomerase II cleavage sites from pBR322. These sites correspond to sequences designated as site 1 and site 2 by Fortune et al. (40). Wild-type oligonucleotide sequences were generated using an Applied Biosystems DNA synthesizer. The 50-mer site 1 sequence for the top and bottom strands were 5’–AGCGGTATCAGCTCACTCAAAGGC↓GGTAATACGGTTATCCACAGAATCAG-3’ and 5’–CTGATTCTGTGGATAACCGTAT↓TACCGCCTTTGAGTGAGCTGATACCGCT-3’, respectively (arrow denotes point of cleavage). The 50-mer site 2 top and bottom sequences were 5’-TTGGTATCTGCGCTCTGCTGAAGCC↓AGTTACCTTCGGAAAAAGAGTTGGT-3’ and 5’-ACCAACTCTTTTTCCGAAGGT↓AACTGGCTTCAGCAGAGCGCAGATACCAA-3’, respectively. Nested 40-, 30-, 25-, and 20-mer oligonucleotides were generated by incrementally shortening the 50-mer site 2 sequence from each end and maintaining the central cleavage site. The sequences used for the bottom strand of these shortened oligonucleotides were 5’–AACTCTTTTTCCGAAGGT↓AACTGGCTTCAGCAGAGCGCAG-3’, 5’–TTTTTCCGAAGGT↓AACTGGCTTCAGCAGAG-3’, 5’-TTTTTCCGAAGGT↓AACTGGCTTCAG-3’, and 5’-TTCCGAAGGT↓AACTGGCTTC-3’, respectively. Depending on the experiment, the top strand was either the complementary sequence of the same length or the full-length 50-mer site 2 top strand sequence. In some cases, oligonucleotides contained a tetrahydrofuran abasic site analog in place of a specific nucleotide or a site-specific nick. The positions of these alterations are denoted in the appropriate experiments.
Preparation of Phosphorothiolate Oligonucleotides
DNA containing a single 3'-bridging phosphorothiolate linkage at the site of topoisomerase II-mediated cleavage was synthesized using 3'-S-(2-cyanoethyl-N,N-diisopropylphosphorothioamidite)-3'-deoxy-5'-O-(4,4'-dimethoxytrityl)thymidine (69).1 The synthesis of the DNA oligonucleotide was performed on an ABI392 DNA synthesizer (Applied Biosystems) similar to published protocols (70) except that standard activator (0.45M tetrazole in acetonitrile) and low iodine containing oxidizing solution (0.02M I2 in THF/pyridine/water) were used during all coupling and oxidation steps. All reagents for DNA synthesis were purchased from Glen Research. DNA oligonucleotides were deprotected in concentrated ammonia (Aldrich) containing 0.1% β-mercaptoethanol (Aldrich) at room temperature for 48 h, and partially purified by reverse phase HPLC chromatography as described previously (53). Unless stated otherwise, the location of the phosphorothiolate was always on the bottom strand at the scissile bond (denoted by the arrow). Confirmation of the presence of the bridging sulfur was accomplished by treating oligonucleotides with AgNO3 (71) and resolving fragments by gel electrophoresis.
Radioactive Labeling of Oligonucleotides
[γ-32P]ATP (∼5000 Ci/mmol) was obtained from ICN. Single-stranded oligonucleotides were labeled on their 5’ termini using T4 polynucleotide kinase (New England Biolabs). Labeling of 3’ termini with [α-32P]cordycepin (ICN) was performed using terminal deoxynucleotidyl transferase (New England Biolabs). Following labeling and gel purification, complementary oligonucleotides were annealed by incubation at 70 °C for 10 min and cooling to 25 °C.
DNA Cleavage
DNA cleavage assays were carried out by a modification of the procedure of Fortune et al. (40). Unless stated otherwise, oligonucleotide substrates were always 5’ end-labeled. All DNA cleavage reactions with human topoisomerase IIα or IIβ contained 200 nM enzyme and 100 nM double-stranded oligonucleotide in a total of 20 μL of 10 mM Tris-HCl, pH 7.9, 135 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 2.5% glycerol. Reactions with yeast topoisomerase II contained 200 nM enzyme and 100 nM double-stranded oligonucleotide in a total of 20 μL of 10 mM Tris-HCl, pH 7.9, 35 mM KCl, 100 mM NaCl, 5 mM MgCl2, 0.1 mM Na2EDTA, and 2.5% glycerol. Reactions were initiated by the addition of enzyme and were incubated at 37 °C (human) or 28 °C (yeast). In some cases, assay mixtures included 50 μM etoposide or amsacrine. DNA cleavage products were trapped by the addition of 2 μL of 10% SDS followed by 2 μL of 250 mM NaEDTA, pH 8.0. As a test of the reversibility of topoisomerase II-mediated cleavage, 2 μL of 250 mM NaEDTA, pH 8.0, or 2 μL of 5 M NaCl were added and samples were incubated at 37 °C for 5 min prior to the addition of SDS. Proteinase K (2 μL of 0.8 mg/mL) was added to digest the enzyme, and oligonucleotides were precipitated with ethanol. Cleavage products were resolved by electrophoresis in a 14% denaturing polyacrylamide gel. To inhibit oxidation of cleaved oligonucleotides containing 3’-terminal –SH moieties and the formation of multimers in the gel, 100 mM DTT was added to the sample loading buffer. DNA cleavage products were visualized and quantified using a Bio-Rad Molecular Imager.
DNA Ligation
DNA ligation assays were carried out by a modification of the procedure of Kingma et al. (35). DNA cleavage/ligation equilibria were established as described for human topoisomerase IIα in the previous section except that 5 mM CaCl2 replaced the MgCl2 in the cleavage buffer. Topoisomerase IIα-DNA cleavage complexes were trapped by the addition of EDTA, pH 8.0, to a final concentration of 6 mM, followed by NaCl to a final concentration of 0.5 M to prevent re-cleavage of the DNA. Ligation was initiated by the addition of MgCl2 to a final concentration of 0.1 mM and terminated by the addition of 2 μL of 10% SDS. Samples were processed and analyzed, and levels of cleavage were quantified as described above. The percent DNA cleavage at time zero was set to 100%, and the rate of ligation was determined by quantifying the loss of cleaved DNA over time.
RESULTS AND DISCUSSION
3’-Bridging Phosphorothiolates
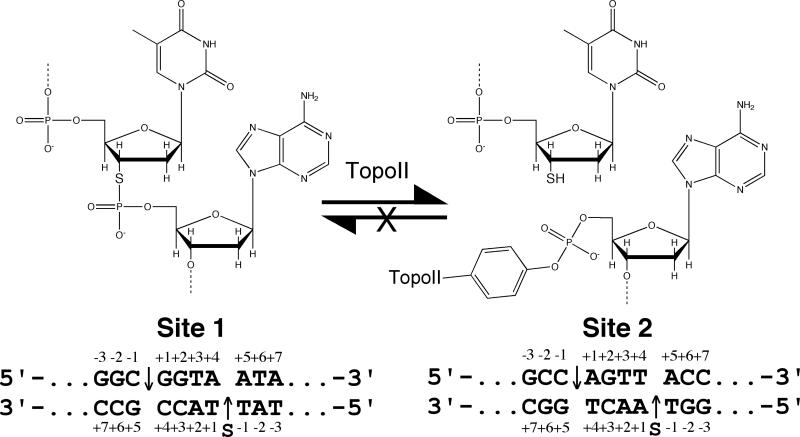
The forward DNA cleavage reaction of type II topoisomerases has been difficult to study because it exists in a tightly coupled equilibrium with the reverse ligation reaction. Furthermore, equilibrium levels of cleavage are low. Therefore, in order to establish a system that isolates topoisomerase II-mediated DNA scission from ligation and allows the unidirectional accumulation of cleavage complexes, double-stranded oligonucleotide substrates that contained a 3’-bridging phosphorothiolate at the scissile bond were synthesized (Figure 1).
Figure 1.
Schematic of topoisomerase II-mediated cleavage of DNA containing a 3’-bridging phosphorothiolate. As diagrammed in the top of the figure, the oxygen at the 3’-bridging position of the scissile bond is replaced with sulfur. Cleavage of this substrate generates a 5’-terminal phosphate that is covalently attached to the enzyme and a 3’-terminal –SH moiety (instead of the –OH group seen with wild-type DNA). In contrast to –OH, the –SH moiety is a poor nucleophile in this context. The sequence of the central portions of site 1 and site 2 are shown at the bottom. The phosphorothiolate modification (denoted “S”) is on the bottom strand of these sequences at the scissile bond (−1/+1 position). Scissile bonds are denoted by arrows.
In contrast to cleavage of wild-type DNA, which generates a 3’-terminal –OH moiety, cleavage of substrates that contain a 3’-bridging phosphorothiolate generates a 3’-terminal –SH group. While the terminal –OH group is a good nucleophile that readily facilitates ligation, the terminal –SH group is very poor nucleophile that does not readily attack the phosphotyrosyl protein-DNA bond (53, 54). Consequently, once phosphorothiolate substrates are cleaved, they should not be able to support appreciable levels of topoisomerase II-mediated DNA ligation.
Cleavage of Oligonucleotide Substrates Containing 3’-Bridging Phosphorothiolates by Human Topoisomerase IIα
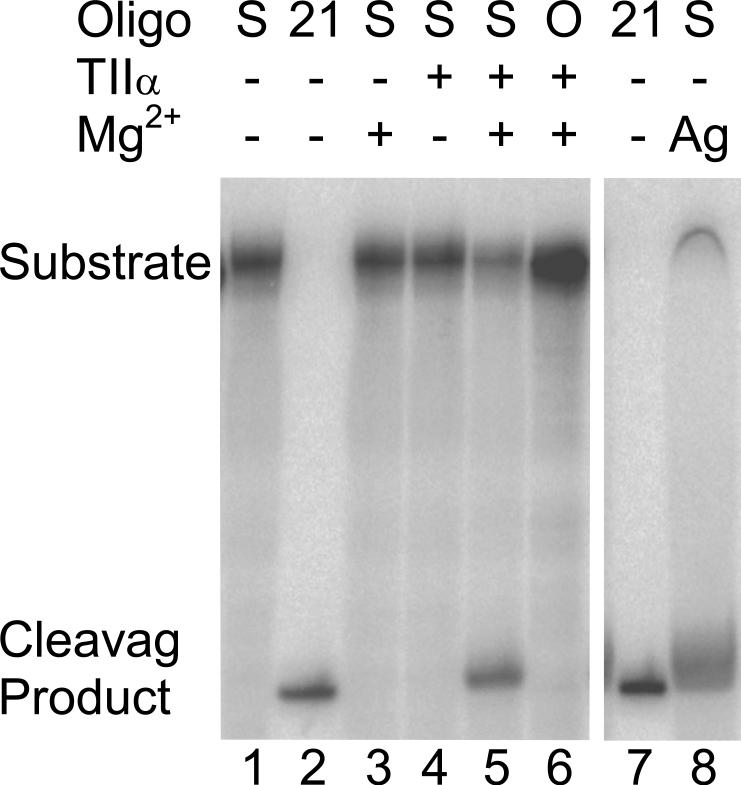
The ability of human topoisomerase IIα to cleave a 5’ end-labeled oligonucleotide substrate (denoted site 2 in Figure 1) containing a 3’-bridging phosphorothiolate at the scissile bond is shown in Figure 2. Scission took place at the predicted scissile bond (lane 5), generating the expected 21-mer product (lane 2). Characteristic of a topoisomerase II-mediated DNA reaction, cleavage required both the human enzyme and a divalent cation (compare lane 5 to lanes 3 and 4). Following a 30-min incubation, cleavage of the phosphorothiolate-containing substrate was dramatically higher than observed with the wild-type phosphate-containing oligonucleotide (compare lanes 5 and 6). Similar results were seen for the site 1 phosphorothiolate oligonucleotide (not shown). Finally, when the site 2 phosphorothiolate oligonucleotide was incubated with AgNO3, which hydrolyzes S-P bonds (71), a 21-mer product resulted (lane 8). This last control demonstrates both the presence of the 3’-bridging phosphorothiolate and its location at the scissile bond.
Figure 2.
Topoisomerase IIα-mediated cleavage of the site 2 oligonucleotide containing a 3’-bridging phosphorothiolate at the scissile bond. An autoradiogram of a polyacrylamide gel is shown. The substrate is denoted at the top (Oligo): S, 3’-bridging phosphorothiolate substrate; O, wild-type substrate; 21, 21-mer marker. The presence of enzyme (TIIα) or divalent cation (Mg2+) is denoted by a plus (+) and the absence by a minus (−). The migration of the 50-mer substrate and the 21-mer cleavage product are denoted on the left side. It is notable that the presence of the 3’-terminal –SH moiety slightly retards the electrophoretic mobility of phosphorothiolate cleavage products as compared to wild-type standards with 3’-terminal –OH groups. The addition of AgNO3 to cleave the S-P bond is denoted by Ag in lane 8. For this and all subsequent figures, the concentrations of topoisomerase IIα and oligonucleotide substrate were 200 nM and 100 nM, respectively. Data are representative of four independent experiments.
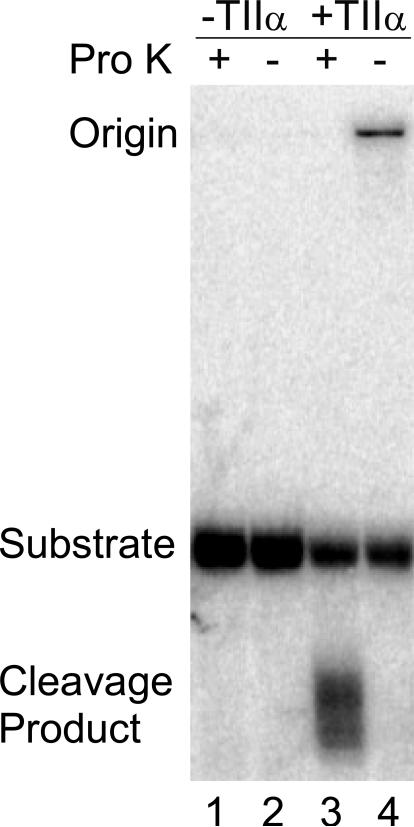
Although cleavage at the 3’-bridging phosphorothiolate requires topoisomerase IIα and Mg(II), it is possible that the environment of the enzyme active site promotes a cleavage event that is independent of the active site tyrosyl residues. A hallmark of topoisomerase II-mediated scission is a covalent linkage between the cleaved DNA and the protein. Therefore, the 3’-bridging phosphorothiolate site 2 substrate was labeled on its 3’-terminus to determine whether the product was covalently linked to topoisomerase IIα (Figure 3). Once again, cleavage was observed only in the presence of enzyme. Following treatment with proteinase K, cleavage products were seen as a diffuse series of bands that ran with a mobility that was faster than that of the substrate band (lane 3). These product bands are indicative of cleaved oligonucleotides that are linked to small peptides that are not completely digested by proteinase K. To confirm the protein-DNA linkage, proteinase K was omitted from the reaction (lane 4). The cleavage product bands disappeared and were replaced by a DNA band that remained at the origin. These results demonstrate that cleavage products generated with a 3’-bridging phosphorothiolate at the scissile bond are covalently attached to topoisomerase IIα. Together with the data shown in Figure 2, these findings provide strong evidence that topoisomerase IIα cleaves the phosphorothiolate oligonucleotide in a manner that is consistent with the normal scission reaction of the enzyme.
Figure 3.
Covalent protein-DNA linkage occurs when the site 2 oligonucleotide containing a 3’-bridging phosphorothiolate is cleaved by topoisomerase IIα. An autoradiogram of a polyacrylamide gel is shown. Substrates were 3’ end-labeled and reacted with topoisomerase IIα. Lanes 1 and 2 were in the absence of enzyme (−TIIα) with (+) or without (−) proteinase K (Pro K). Lane 3 and 4, were in the presence of enzyme (+TIIα) with or without proteinase K. Data are representative of three independent experiments.
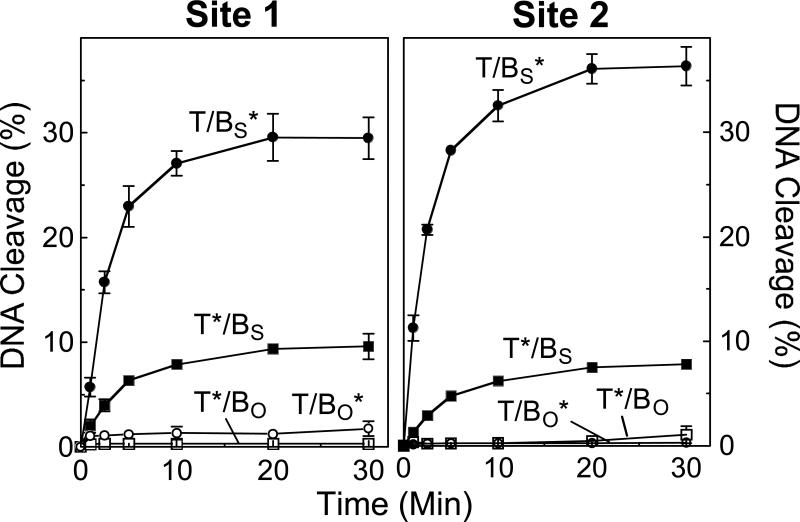
Time courses for the cleavage of site 1 and site 2 by human topoisomerase IIα are shown in Figure 4. Comparable results were observed with both sequences. Two striking features were seen. First, levels of cleavage of the phosphorothiolate substrate were dramatically higher (∼10– to 100–fold at 30 min) than were generated with the comparable wild-type oligonucleotides. Second, while a cleavage/ligation equilibrium was reached rapidly with both wild-type substrates, levels of cleaved phosphorothiolate oligonucleotides continued to accumulate until 20−30 min. Both of these features are characteristic of reactions in which the ability of topoisomerase IIα to ligate cleaved DNA molecules is either blocked or severely compromised. Similar results were seen when the site 2 phosphorothiolate substrate was cleaved by yeast topoisomerase II or human topoisomerase IIβ (shown in Supporting Information Figure S1). Albeit, levels of cleaved products were somewhat lower with these latter two enzymes. Taken together, these findings indicate that 3’-bridging phosphorothiolates can be used with a variety of type II topoisomerases and DNA cleavage sites.
Figure 4.
Time-courses for cleavage of oligonucleotide substrates by topoisomerase IIα. Results for site 1 (left panel) and site 2 (right panel) are shown. The asterisk (*) denotes the 5’ end-labeled strand (top, T; or bottom, B) on which cleavage was monitored. The wild-type duplex containing an O-P scissile bond (subscript O) is denoted by T/BO* for the labeled bottom strand (open circles) and T*/BO for the labeled top strand (open squares). The phosphorothiolate substrate containing an S-P scissile bond (subscript S) is denoted by T/BS* for the labeled phosphorothiolate bottom strand (closed circles) and T*/BS for the labeled wild-type top strand (closed squares). Error bars represent the standard deviation of at least three independent experiments.
Cleavage of 3’-Bridging Phosphorothiolate Substrates is Sequence Specific
Type II topoisomerases cleave DNA with an intrinsic specificity (72). The above experiments demonstrate that human topoisomerase IIα cleaves an S-P linkage when it is situated at the scissile bond of an established topoisomerase II-DNA recognition sequence. However, it is not known whether the ability of the enzyme to cut the S-P linkage requires the bond to be situated at a “normal” cleavage site or is independent of the intrinsic sequence specificity of the protein.
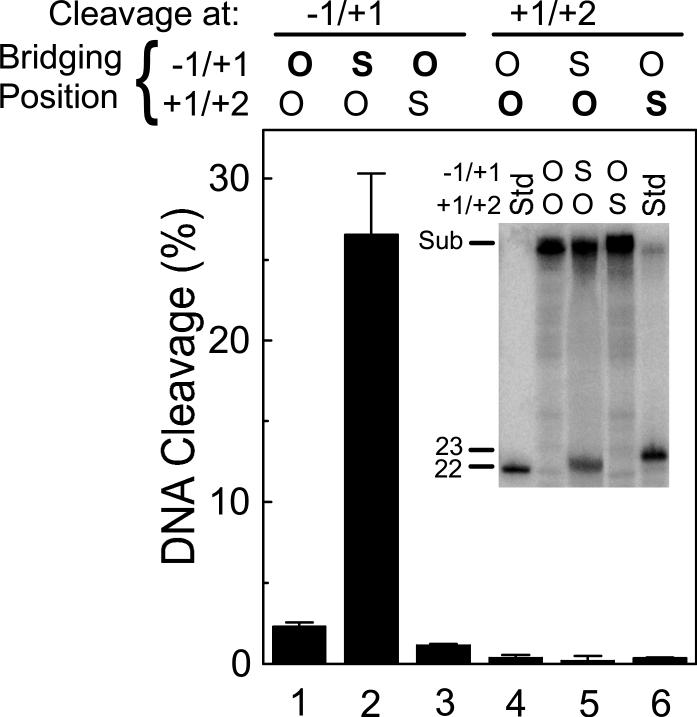
To address this critical issue, the phosphorothiolate of site 1 was moved one position from the natural scissile bond, bridging the −1/+1 nucleotides, to a location bridging the +1/+2 nucleotides (Figure 5). In contrast to the high levels of scission generated when the S-P linkage was situated at the normal −1/+1 position (compare lanes 1 and 2), virtually no cleavage was seen at the S-P bond when it was moved to the +1/+2 position (compare lanes 4 and 6). As a control, to ensure that the oligonucleotide with the +1/+2 S-P bond was capable of being recognized and cleaved by topoisomerase IIα, scission at the −1/+1 position was monitored. While cleavage at the normal scissile bond was decreased somewhat compared to that seen with the completely wild-type oligonucleotide (compare lanes 1 and 3), a significant level of scission still was observed. Taken together, these data demonstrate that the specificity of topoisomerase IIα is not altered by the presence of the 3’-bridging phosphorothiolate.
Figure 5.
Site-specificity of DNA cleavage mediated by topoisomerase IIα is not altered by the location of the 3’-bridging phosphorothiolate. Cleavage was monitored on three versions of the site 1 substrate: wild-type duplex containing an O-P linkage at both the −1/+1 and +1/+2 positions; duplex containing an S-P linkage at the normal scissile bond (−1/+1 position) and an O-P linkage at the +1/+2 position; and duplex containing an O-P linkage at the normal scissile bond (−1/+1 position) and an S-P linkage at the +1/+2 position. Cleavage was quantified at the −1/+1 position (lanes 1−3, 22-mer product) and the +1/+2 position (lanes 4−6, 23-mer product). The nature of the linkage at the bridging position (O-P linkage, O; S-P linkage, S: bolded when being monitored) is denoted at the top of the figure. A representative gel is shown in the inset. The positions of the intact 50-mer substrate (Sub) as well as 22-mer and 23-mer standards (Std) representing cleavage products at the −1/+1 and +1/+2 sites, respectively, are indicated. As noted in Figure 2, the presence of the 3’-terminal –SH moiety slightly retards the electrophoretic mobility of phosphorothiolate cleavage products as compared to wild-type standards with 3’-terminal –OH groups. Error bars represent the standard deviation of three independent experiments.
Cleavage of 3’-Bridging Phosphorothiolates by Topoisomerase IIα Is Not Reversible
Topoisomerase II-mediated cleavage of oligonucleotides that contain a 3’-bridging phosphorothiolate at the scissile bond generate products with a 3’-terminal -SH group (see Figure 1). Previous studies with type I topoisomerases and 5’-bridging phosphorothiolate substrates suggest that the –SH group should not be able to attack the covalent enzyme-DNA phosphotyrosine bond with any level of efficiency (53, 54).
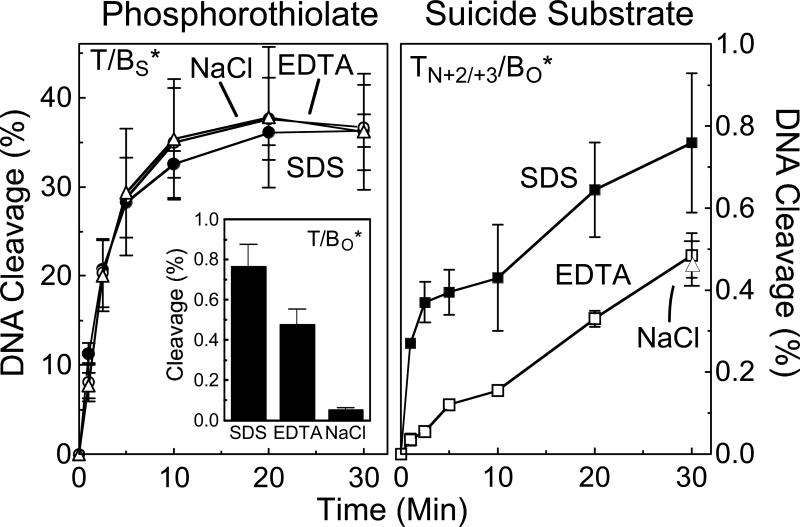
Therefore, to determine whether topoisomerase IIα is able to rejoin cleaved phosphorothiolate oligonucleotides, reactions were terminated with either EDTA or 0.5 M NaCl prior to the addition of SDS. It is believed that Mg(II), which is essential for both cleavage and ligation, can be chelated by EDTA only after ligation has taken place. Consequently, treatment with EDTA allows ligation to take place but prevents re-cleavage (18, 38). Similarly, treatment with high salt allows ligation, but promotes dissociation of topoisomerase II from the DNA once it is no longer covalently attached to the nucleotide substrate. Once again, this leads to a unidirectional closure of the cleaved oligonucleotide (17, 38). As expected, when wild-type site 1 (not shown) or site 2 (Figure 6, left panel inset) oligonucleotide was used as substrate, termination of a 30 min DNA scission reaction with EDTA or NaCl led to a significant reduction in the level of cleaved products. For example, greater than 90% of cleaved oligonucleotides were ligated following a 5-min incubation with 500 mM NaCl.
Figure 6.
Reversibility of DNA cleavage mediated by topoisomerase IIα. Left panel, a time course is shown for cleavage of the site 2 oligonucletide containing a 3’-bridging phosphorothiolate at the scissile bond (T/BS*; see Figure 4 for nomenclature). Reactions were terminated with SDS (closed circles), EDTA (open circles), or NaCl (open triangles). Left panel inset, cleavage of the wild-type site 2 oligonucletide containing an O-P linkage at the scissile bond (T/BO*). A 30-min reaction terminated with SDS, EDTA, or NaCl is shown. Right panel, a time course is shown for cleavage of the site 2 suicide substrate containing a nick (N) between the +2 and +3 positions on the top strand (TN+2/+3/BO*). Reactions were terminated with SDS (closed squares), EDTA (open squares), or NaCl (30-min time point, closed triangle). Error bars represent the standard error of the mean of two independent experiments or the standard deviation of three independent experiments.
In marked contrast, no decrease in cleaved products was seen in parallel reactions that employed either site 1 (not shown) or site 2 (Figure 6, left panel) phosphorothiolate oligonucleotide, even when incubation periods in EDTA or NaCl were extended to 30 min (not shown). Furthermore, time courses for 30-min DNA cleavage reactions terminated with SDS, EDTA, or NaCl were essentially superimposable for these substrates. These findings provide strong evidence that human topoisomerase IIα is incapable of ligating 3’-bridging phosphorothiolate cleavage products with any appreciable level of efficiency. Thus, 3’-bridging phosphorothiolates allow the forward DNA scission reaction of topoisomerase IIα to be studied in isolation from the ligation event.
Cleavage of Suicide Substrates by Topoisomerase IIα Is Reversible
The only other substrates that are believed to promote a unidirectional topoisomerase II-mediated DNA cleavage reaction are known collectively as “suicide substrates.” These systems trap cleavage complexes by utilizing nucleic acid molecules that contain non-contiguous (i.e., nicked) DNA (46), double strand/single strand junctions (42, 43, 45), or unusual secondary structure (i.e., hairpins) (41, 44). When suicide substrates are cut by topoisomerase II, a segment of the cleaved DNA is able to dissociate from the active site, rendering the enzyme unable to ligate the original substrate (43). This process results in a time-dependent accumulation of cleavage complexes.
One obvious advantage of 3’-bridging phosphorothiolates over “traditional” suicide substrates is that the former are composed of intact duplex B-form DNA molecules. More importantly, evidence with a hairpin molecule suggests that topoisomerase II actually establishes a short-lived cleavage/ligation equilibrium with suicide substrates prior to the dissociation event (44). If this is the case, then the use of these substrates to monitor the forward scission reaction may not be appropriate.
Therefore, to compare cleavage of an S-P linkage to that of a traditional suicide substrate, a site 2 oligonucleotide containing a nick between the +2 and +3 positions of the top strand was synthesized. In contrast to scission of the intact wild-type substrate, which reached cleavage/ligation equilibrium within 5 min, cleaved products of the nicked oligonucleotide accumulated throughout the 30 min time course when the reaction was terminated with SDS (Figure 6, right panel). This finding demonstrates that the nicked oligonucleotide acts as a suicide substrate for topoisomerase IIα.
To examine the ability of the human enzyme to ligate the nicked suicide substrate, a parallel time course for scission was terminated with EDTA. As seen in Figure 6 (right panel) cleavage levels were significantly lower in the EDTA-treated reactions. A similar result was observed when a 30 min reaction was terminated by the addition of 0.5 M NaCl (see indicated data point in Figure 6, right panel). These findings indicate that a significant proportion of the initial cleavage complexes formed with the suicide substrate are reversible.
It is notable that the relative fraction of cleavage complexes that was irreversible (i.e., not ligated following treatment with EDTA) rose over time. While levels of irreversible cleavage with the suicide substrate represented only ∼10% of the total scission observed at 1 min, they increased to ∼50% by 20−30 min. This result provides further evidence that suicide substrates (at least the nicked substrate employed in the present study) require a time-dependent conversion from a reversible cleavage complex to an irreversible DNA strand break. Presumably, this conversion reflects the point at which the non-covalent cleavage product dissociates from topoisomerase II.
Thus, while traditional suicide substrates provide an important system to accumulate topoisomerase II-DNA cleavage complexes and to provide starting material for unidirectional ligation assays, they do not segregate the forward enzyme-mediated DNA scission reaction from the cleavage/ligation equilibrium. Consequently, the use of these substrates to analyze the forward cleavage reaction of topoisomerase II may lead to erroneous mechanistic conclusions.
Coordination Between the Two Protomer Subunits of Human Topoisomerase IIα During DNA Cleavage
Although topoisomerase II must generate a double-stranded nucleic acid break in order to carry out the DNA passage reaction, results from numerous laboratories indicate that the two protomer subunits of eukaryotic type II enzymes do not cleave and ligate DNA in a totally concerted fashion. For example, it has long been known that equilibrium levels of DNA strand breaks at the two scissile bonds of a cleavage site (17, 19, 33, 73) and rates of ligation of the two strands are often different (19). Furthermore, alterations of base sequence (or the inclusion of abasic sites) that have a direct affect on rates of ligation of one strand have little effect on rates of ligation of the other strand (37, 50). Finally, changes in base sequence that enhance the ability of etoposide to inhibit ligation at one scissile bond have no significant effect on rates of ligation at the opposite scissile bond (74).
The above notwithstanding, the same alterations in DNA sequence that specifically enhance the ability of etoposide to inhibit ligation at one scissile bond lead to a modest increase in topoisomerase IIα-generated double-stranded breaks at that cleavage site (74). This finding implies that there may be some communication between the protomer subunits of the human enzyme during the DNA cleavage event, even though there appears to be little coordination during ligation.
To address the issue subunit communication, levels of cleavage on the top strand of site 1 and site 2 were monitored in the absence or presence of a 3’-bridging phosphorothiolate on the bottom strand (see Figure 4). As expected, when situated opposite a wild-type O-P scissile bond, rapid DNA cleavage/ligation equilibria characterized by low levels of scission were established on the top strand of both sites. However, when situated opposite an S-P scissile bond that did not allow ligation, levels of cleavage on the top strand of both sites were ∼5– to 7–fold higher as compared to comparable wild-type substrates and rose over the course of the assay. Moreover, the time courses for the accumulation of DNA breaks on the top strands paralleled those seen for the bottom S-P strands.
These results indicate that there is at least some level of coordination between the two protomer active sites of topoisomerase IIα. However, since levels of cleavage on the strand opposite the S-P bond were several–fold lower than observed on the phosphorothiolate-containing strand, it is clear that DNA cleavage by the two protomer subunits does not take place in a completely concerted fashion.
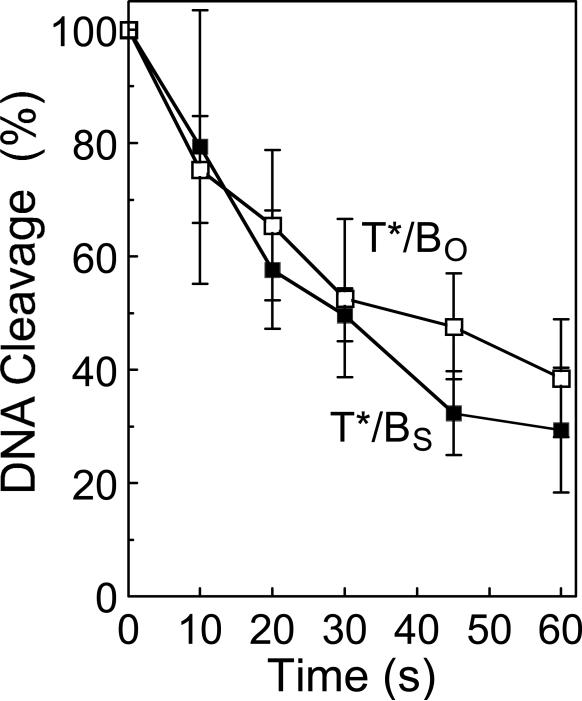
The partial coordination between the protomer subunits of topoisomerase IIα may be explained by at least three independent mechanisms (which are not mutually exclusive). First, cleavage of one scissile bond may decrease the ability of the enzyme to ligate the opposite scissile bond. This possibility seems unlikely, as it would provide no benefit to the catalytic reaction of topoisomerase II. In addition, previous studies have found little coordination between the protomer subunits of topoisomerase IIα during ligation (19, 37, 74). However, as a control, the rate of ligation of the top strand of site 2 was determined when it was situated opposite an O-P or an S-P scissile bond (Figure 7). As predicted, rates of ligation of the two substrates were comparable.
Figure 7.
Time course for topoisomerase IIα-mediated ligation of the wild-type top strand of the site 2 oligonucleotide. Site 2 substrates containing either a wild-type O-P linkage (T*/BO, open squares) or phosphorothiolate S-P linkage (T*/BS, closed squares) at the scissile bond of the bottom strand were employed. The total percent cleavage prior to initiation of ligation was set to 100%. Error bars represent the standard deviation of at least three independent experiments.
Second, trapping a cleavage complex at one scissile bond should increase the concentration of enzyme that contains the opposite scissile bond in its active site. Since levels of cleavage are directly proportional to levels of topoisomerase II-DNA binding, this effect could account for the enhanced scission on the opposite strand. This mechanism requires no alteration in the forward rate of scission and is consistent with the finding that the accumulation of breaks on the top strand of site 1 and site 2 paralleled that seen for the bottom S-P strands (Figure 4).
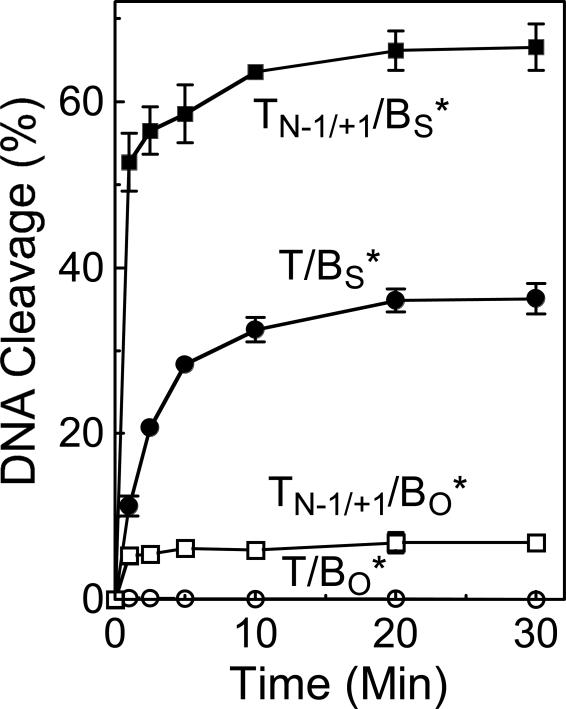
Third, cleavage at one scissile bond may increase the rate of cleavage at the opposite scissile bond. As a first attempt to address this last hypothesis, a site 2 oligonucleotide that contained a nick at the scissile bond (−1/+1 position) of the top strand was employed as a model of a DNA substrate that had already been cut on one of its two strands. It should be noted, however, that the nicked oligonucleotide is not covalently attached to topoisomerase II. Thus, it is unclear whether this substrate adequately mimics an enzyme-DNA cleavage complex in which one of the two strands has been cut by topoisomerase II.
With the above caveat in mind, the ability of topoisomerase IIα to cleave the bottom strand of site 2 was monitored when the top scissile bond was either intact or nicked (Figure 8). The initial experiment utilized a substrate that contained a wild-type O-P linkage at the scissile bond of the bottom strand. Two observations were noted. First, levels of cleavage of the bottom strand increased ∼20–fold when situated opposite a nicked top strand. Second, the concentration of cleavage complexes reached a rapid equilibrium and did not rise over the time course of the experiment. This indicates that unlike the nicked +2/+3 substrate utilized in Figure 6, the nicked scissile bond (−1/+1) oligonucleotide did not act as a suicide substrate for topoisomerase IIα.
Figure 8.
Topoisomerase IIα-mediated cleavage of a substrate containing a nick at one scissile bond. Time courses are shown for the following site 2 substrates: oligonucleotide containing an intact top strand and a wild-type O-P linkage at the scissile bond of the bottom strand (T/BO*, open circles); oligonucleotide containing a nick at the scissile bond (−1/+1 position) of the top strand and a wild-type O-P linkage at the scissile bond of the bottom strand (TN-1/+1/BO*, open squares); oligonucleotide containing an intact top strand and a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (T/BS*, closed circles); and oligonucleotide containing a nick at the scissile bond of the top strand and a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (TN-1/+1/BS*, closed squares). Error bars represent the standard deviation of at least three independent experiments.
To determine whether the enhanced cleavage observed when a wild-type strand was situated opposite a nicked scissile bond reflected an increase in the forward rate of DNA cleavage, the experiment was repeated using a site 2 substrate that contained a 3’-bridging phosphorothiolate at the scissile bond of the bottom strand (Figure 8). When situated opposite the nicked strand, the rate of cleavage of the bottom S-P scissile bond rose considerably. Although rates were too rapid to measure accurately, it appears that they increased at least 5–fold. These data imply that cleavage on one strand by topoisomerase II has the capacity to increase the rate of cleavage on the opposite strand. Whether the increased rate of scission reflects a change in the conformation of the DNA, the enzyme, or a combination of both remains to be determined.
Taken together, the above studies suggest that at least two mechanisms may contribute to the partial coordination between the two subunits of topoisomerase IIα: increased concentration of the second scissile bond in the active site of the enzyme and an enhanced rate of scission of the second strand following cleavage of the first.
Effect of ATP on the Forward DNA Scission Reaction of Human Topoisomerase IIα
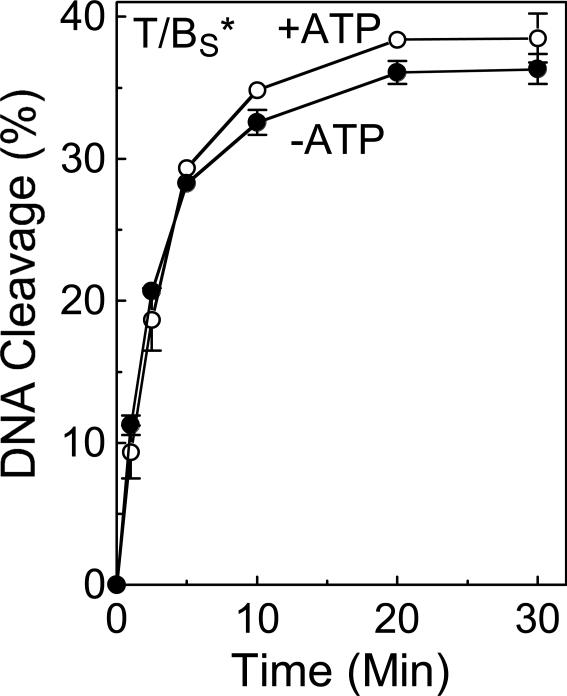
In order for topoisomerase II-mediated DNA strand passage to occur, both strands of the “gate” segment must be cleaved (6, 9, 10, 14-16). ATP binding (one molecule per topoisomerase II protomer) induces opening of the DNA gate (6, 9, 10, 14-16, 75-77), and it is believed that hydrolysis of one of the ATP molecules enhances the rate of strand passage (78). Several laboratories have demonstrated that levels of topoisomerase II-DNA cleavage complexes rise in the presence of the high-energy cofactor (6, 9, 10, 14-16). Although the mechanistic basis for this observation is not known, one study reported that rates of DNA ligation were slowed in the presence of the non-hydrolyzable ATP analog APP(NH)P (48). To determine whether ATP has an effect on the forward DNA scission reaction of human topoisomerase IIα, the ability of the enzyme to cleave the 3’-bridging phosphorothiolate substrate (site 2) in the presence and absence of the nucleotide triphosphate was compared. As seen in Figure 9, ATP had virtually no effect on the rate of DNA scission. Therefore, it is concluded that the increased concentration of cleavage complexes seen in the presence of ATP is not due to an enhanced forward rate of cleavage.
Figure 9.
Topoisomerase IIα-mediated DNA cleavage in the presence of ATP. A time course is shown for cleavage of the site 2 substrate containing a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (T/BS*) in the absence (−ATP, closed circles) or presence of ATP (+ATP, open circles). Error bars represent the standard deviation of at least three independent experiments.
Dependence of the Forward Scission Reaction of Topoisomerase IIα on DNA Length
A previous study demonstrated that the ability of human topoisomerase IIα and chlorella virus topoisomerase II to form cleavage complexes was dependent on the length of the DNA substrate (40). Consistent with the footprint of topoisomerase II on DNA (∼28 bp) (79), a minimal requirement for a 20-bp oligonucleotide was noted and substantial scission was not seen until a substrate of ∼40 bp was employed. Levels of cleavage complexes did not correlate with changes in binding affinity (40). Consequently, they must be due to a change in the ability of topoisomerase II to either cleave or ligate short substrates.
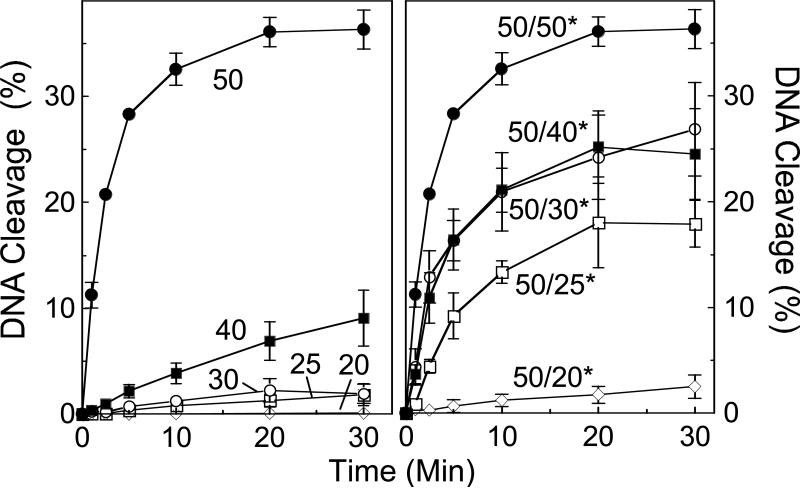
To determine whether the effect of DNA length on cleavage complex formation was due to a decreased ability to cleave short substrates, a series of nested site 2 oligonucleotides with 3’-bridging phosphorothiolates at the scissile bond of the bottom strand was synthesized (Figure 10, left panel). As found previously with wild-type substrates, phosphorothiolate substrates displayed virtually no cleavage with the 20-mer, and low levels were seen with the 25- and 30-mer. A modest rise in cleavage was observed when substrate length was increased to 40 bp, followed by a dramatic rise when it was increased to 50 bp (the length of the parent oligonucleotide). These data indicate that the DNA length dependence of topoisomerase IIα is due to an effect on the forward scission reaction and imply that protein-DNA contacts outside of the active site of the enzyme may be required to promote cleavage.
Figure 10.
Effect of oligonucleotide length on DNA cleavage mediated by topoisomerase IIα. Left panel, time courses are shown for cleavage of the phosphorothiolate-containing bottom strand of nested site 2 substrates. Reactions were monitored with 50-mer (closed circles), 40-mer (closed squares), 30-mer (open circles), 25-mer (open squares), or 20-mer (closed diamonds) duplex substrates. Right panel, time courses are shown for cleavage of the phosphorothiolate-containing bottom strand of nested site 2 substrates containing a 50-mer top strand and a 50-mer (closed circles), 40-mer (closed squares), 30-mer (open circles), 25-mer (open squares), or 20-mer (closed diamonds) bottom strand. Error bars represent the standard deviation of at least three independent experiments.
Although topoisomerase IIα can sense the length of its substrate, only the central portion of the DNA needs to be duplex (Figure 10, right panel). When the nested oligonucleotides containing the S-P linkage at the scissile bond were annealed with the parental wild-type 50-mer top strand, a substantial increase in cleavage was observed as the length of the bottom strand was increased.
Effects of Topoisomerase II Poisons on the Forward DNA Cleavage Reaction of Human Topoisomerase IIα
Topoisomerase II poisons increase the concentration of cleavage complexes by two non-mutually exclusive mechanisms. Anticancer drugs, such as etoposide and amsacrine, strongly inhibit the ability of the enzyme to ligate cleaved DNAs (28, 47, 48, 74, 80, 81). Because of their affect on ligation, it has been assumed that these agents have no significant effect on the forward scission reaction. In contrast, DNA lesions that poison topoisomerase II, such as abasic sites, have little effect on rates of ligation (35, 36, 45, 82, 83). Consequently, it has been assumed that they act primarily by enhancing the forward rate of DNA scission. Unfortunately, it has not been possible to test these assumptions directly. Therefore, the ability of these topoisomerase II poisons to alter the rate of DNA scission was characterized using the phosphorothiolate substrates.
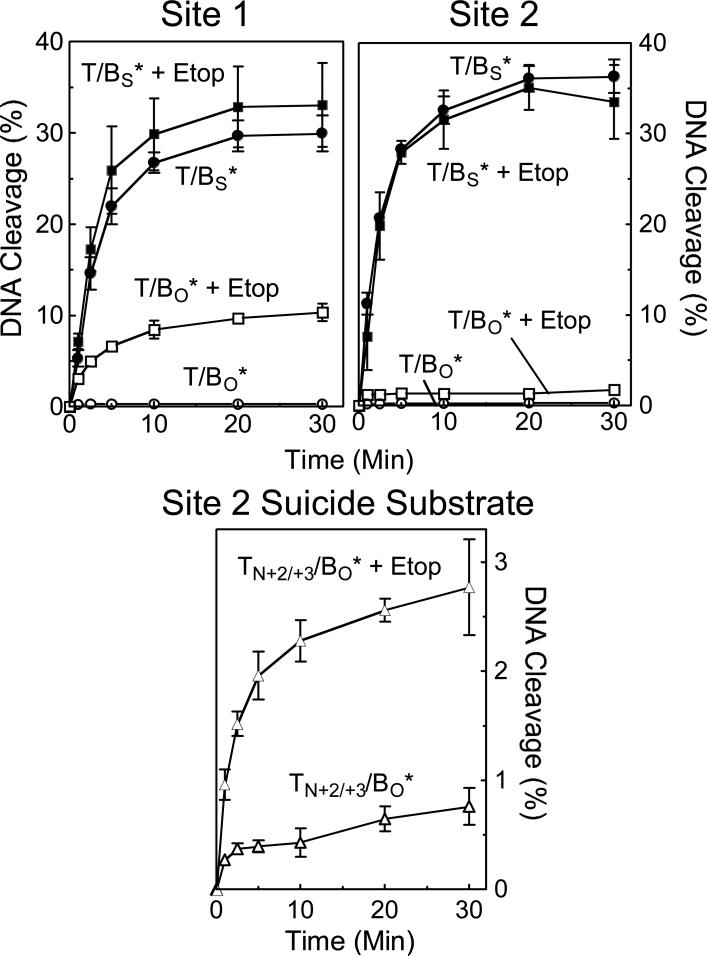
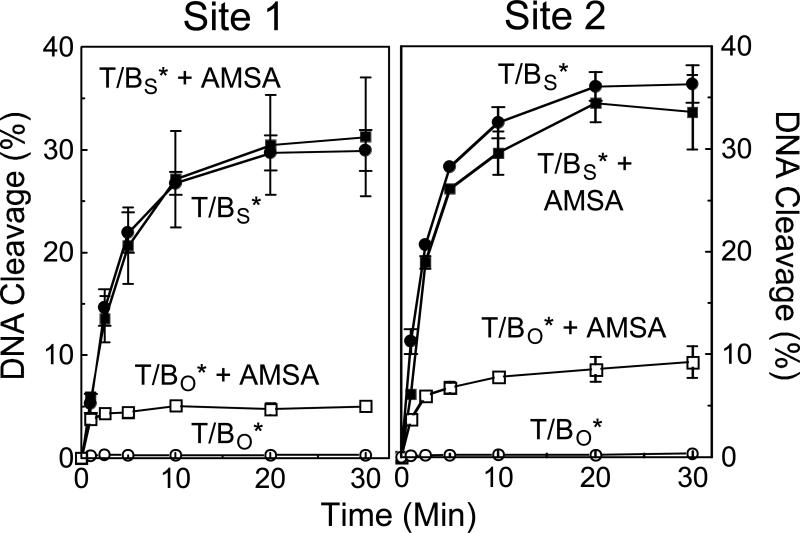
The effects of etoposide and amsacrine on the forward rate of cleavage of site 1 and site 2 are shown in Figures 11 and 12, respectively. In all cases, the drugs increased the concentration of cleavage complexes when wild-type oligonucleotides were employed. However, virtually no increase in the rate of scission was observed when 3’-bridging phosphorothiolate substrates were examined. These data strongly suggest that etoposide and amsacrine increase levels of topoisomerase IIα-DNA cleavage complexes primarily by inhibiting enzyme-mediated ligation.
Figure 11.
Effect of etoposide on the forward rate of topoisomerase IIα-mediated DNA scission. Time courses for cleavage of the bottom strand of the site 1 (left panel) and site 2 (right panel) substrates are shown for oligonucleotides containing a wild-type O-P linkage at the scissile bond of the bottom strand (T/BO*) and oligonucleotides containing a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (T/BS*) in the absence (T/BO*, open circles; T/BS*, closed circles) or presence (T/BO* + Etop, open squares; T/BS* + Etop, closed squares) of 50 μM etoposide. The bottom panel shows time courses for cleavage of the bottom strand of the site 2 suicide substrate containing a nick at the +2/+3 position of the top strand in the absence (TN+2/+3/BO*, open triangles) or presence (TN+2/+3/BO* + Etop, closed triangles) of 50 μM etoposide. Error bars represent the standard error of the mean of two independent experiments or the standard deviations of at least three independent experiments.
Figure 12.
Effect of amsacrine on the forward rate of topoisomerase IIα-mediated DNA scission. Time courses for cleavage of the bottom strand of the site 1 (left panel) and site 2 (right panel) substrates are shown for oligonucleotides containing a wild-type O-P linkage at the scissile bond of the bottom strand (T/BO*) and oligonucleotides containing a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (T/BS*) in the absence (T/BO*, open circles; T/BS*, closed circles) or presence (T/BO* + AMSA, open squares; T/BS* + AMSA, closed squares) of 50 μM amsacrine. Error bars represent the standard deviation of at least three independent experiments.
As discussed above, suicide substrates establish a short-lived DNA cleavage/ligation equilibrium before irreversibly trapping cleavage complexes. Consequently, their use as a model for the forward topoisomerase II DNA cleavage reaction was questioned. For example, if etoposide inhibits enzyme-mediated ligation prior to the “suicidal” dissociation of the non-covalent DNA cleavage product, the drug could increase levels of trapped cleavage complexes independent of any effect on the forward rate of scission. To address this critical point, the effect of etoposide on the ability of topoisomerase IIα to cut the nicked suicide substrate described in Figure 6 was characterized (Figure 11, lower panel). Despite the fact that etoposide does not increase the forward rate of DNA scission, a dramatic rise in cleavage complexes was observed when the drug was included in reactions with the suicide substrate. This finding underscores the need to exercise caution when drawing mechanistic conclusions about the forward rate of DNA cleavage based on the results obtained with suicide substrates.
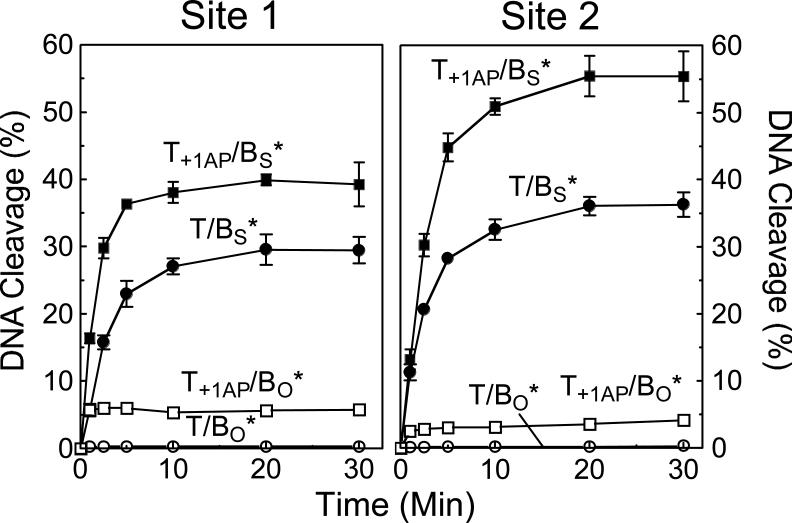
Finally, when abasic sites are situated within the 4-bp region located between the two scissile bonds, their presence raises the concentration of topoisomerase II-DNA cleavage complexes (35, 36, 46, 83). As seen in Figure 13, the inclusion of an abasic site at the +1 position of the top strand of site 1 or site 2 increased levels of cleavage complexes several–fold when substrates with wild-type O-P scissile bonds were used. In contrast to the anticancer drugs, which had little effect on the forward cleavage reaction, the abasic site stimulated the rate of DNA scission of substrates with S-P scissile bonds ∼2– to 3–fold (Figure 13). Similar results were seen with substrates that contained an abasic site at the +2 or +4 position (not shown). These findings confirm the assumption that abasic sites act by stimulating the forward rate of topoisomerase II-mediated DNA scission.
Figure 13.
Effect of an abasic site on the forward rate of topoisomerase IIα-mediated DNA scission. Time courses are shown for the following site 1 (left panel) and site 2 (right panel) substrates: oligonucleotide containing a wild-type top strand and a wild-type O-P linkage at the scissile bond of the bottom strand (T/BO*, open circles); oligonucleotide containing an abasic (AP) site at the +1 position of the top strand and a wild-type O-P linkage at the scissile bond of the bottom strand (T+1AP/BO*, open squares); oligonucleotide containing a wild-type top strand and a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (T/BS*, closed circles); and oligonucleotide containing an abasic site at the +1 position of the top strand and a phosphorothiolate S-P linkage at the scissile bond of the bottom strand (T+1AP/BS*, closed squares). Error bars represent the standard deviation of at least three independent experiments.
Conclusions
Despite the importance of DNA cleavage to the cellular functions of type II topoisomerases and their role as targets for anticancer drugs, the tight coupling of the cleavage/ligation equilibrium has made it difficult to characterize the mechanism by which these enzymes cut the genetic material. In order to uncouple the forward DNA scission reaction of human topoisomerase IIα from the ligation event, a novel system that utilized 3’-bridging phosphorothiolates at the scissile bonds of well-defined topoisomerase II-DNA cleavage sites was developed. Cleavage of these substrates appeared to be irreversible, resulting in a unidirectional enzyme-mediated DNA scission reaction. The use of oligonucleotides with 3’-bridging phosphorothiolates provided critical information on the coordination of the protomer subunits of topoisomerase IIα and the mechanism of action of topoisomerase II poisons. These substrates should be a valuable resource for future studies on DNA scission and the topoisomerase II-DNA cleavage complex.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Omari J. Bandele, Amanda C. Gentry, and Steven L. Pitts for critical reading of the manuscript.
Footnotes
This work was supported by National Institutes of Health research grants GM33944 and GM53960. JED was a trainee under grant T32 CA09592 from the National Institutes of Health.
Due to technical constraints of the synthetic pathway, only the thymidine 3’-bridging phosphorothioamidite was available for the present study. Therefore, all oligonucleotide substrates employed contained thymidine immediately 5’ to the scissile bond (−1 position).
SUPPORTING INFORMATION AVAILABLE
Data showing the ability of eukaryotic type II topoisomerases (comparing yeast topoisomerase II and human topoisomerase IIβ to human topoisomerase IIα) to cleave the phosphorothiolate substrate. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Cozzarelli NR, Wang JC. Cold Spring Harbor: 1990. Cold Spring Harbor Laboratory Press. [Google Scholar]
- 3.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Osheroff N. DNA Topoisomerases. Biochim. Biophys. Acta. 1998:1400. doi: 10.1016/s0167-4781(98)00123-7. [DOI] [PubMed] [Google Scholar]
- 5.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 6.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 7.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 8.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase IIb. BioEssays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 9.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 10.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heck MM, Earnshaw WC. Topoisomerase II: A specific marker for cell proliferation. J. Cell Biol. 1986;103:2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiang YH, Wu HY, Liu LF. Proliferation-dependent regulation of DNA topoisomerase II in cultured human cells. Cancer Res. 1988;48:3230–3235. [PubMed] [Google Scholar]
- 13.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 14.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Quart. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 15.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 16.Wilstermann AM, Osheroff N. Stabilization of eukaryotic topoisomerase IIDNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 17.Liu LF, Rowe TC, Yang L, Tewey KM, Chen GL. Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 1983;258:15365–15370. [PubMed] [Google Scholar]
- 18.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 19.Zechiedrich EL, Christiansen K, Andersen AH, Westergaard O, Osheroff N. Double-stranded DNA cleavage/religation reaction of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim. Biophys. Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 21.Baguley BC, Ferguson LR. Mutagenic properties of topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:213–222. doi: 10.1016/s0167-4781(98)00137-7. [DOI] [PubMed] [Google Scholar]
- 22.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 23.Sordet O, Khan QA, Kohn KW, Pommier Y. Apoptosis induced by topoisomerase inhibitors. Curr. Med. Chem. Anticancer Agents. 2003;3:271–290. doi: 10.2174/1568011033482378. [DOI] [PubMed] [Google Scholar]
- 24.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 25.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr. Med. Chem. Anticancer. Agents. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 26.Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatr. Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 27.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 28.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anticancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 29.Martincic D, Hande KR. Topoisomerase II inhibitors. Cancer Chemother. Biol. Response Modif. 2005;22:101–121. doi: 10.1016/s0921-4410(04)22005-1. [DOI] [PubMed] [Google Scholar]
- 30.Kreuzer KN, Cozzarelli NR. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 1979;140:424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowley JD, Diaz MO, Espinosa R. d., Patel YD, van Melle E, Ziemin S, Taillon-Miller P, Lichter P, Evans GA, Kersey JH, et al. Mapping chromosome band 11q23 in human acute leukemia with biotinylated probes: identification of 11q23 translocation breakpoints with a yeast artificial chromosome. Proc. Natl. Acad. Sci. USA. 1990;87:9358–9362. doi: 10.1073/pnas.87.23.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felix CA, Lange BJ. Leukemia in infants. Oncologist. 1999;4:225–240. [PubMed] [Google Scholar]
- 33.Andersen AH, Christiansen K, Zechiedrich EL, Jensen PS, Osheroff N, Westergaard O. Strand specificity of the topoisomerase II mediated double-stranded DNA cleavage reaction. Biochemistry. 1989;28:6237–6244. doi: 10.1021/bi00441a015. [DOI] [PubMed] [Google Scholar]
- 34.Kingma PS, Osheroff N. The response of eukaryotic topoisomerases to DNA damage. Biochim. Biophys. Acta. 1998;1400:223–232. doi: 10.1016/s0167-4781(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 35.Kingma PS, Osheroff N. Topoisomerase II-mediated DNA cleavage and religation in the absence of base pairing: abasic lesions as a tool to dissect enzyme mechanism. J. Biol. Chem. 1998;273:17999–18002. doi: 10.1074/jbc.273.29.17999. [DOI] [PubMed] [Google Scholar]
- 36.Sabourin M, Osheroff N. Sensitivity of human type II topoisomerases to DNA damage: stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucleic Acids Res. 2000;28:1947–1954. doi: 10.1093/nar/28.9.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilstermann AM, Osheroff N. Positioning the 3'-DNA terminus for topoisomerase II-mediated religation. J. Biol. Chem. 2001;276:17727–17731. doi: 10.1074/jbc.M100197200. [DOI] [PubMed] [Google Scholar]
- 38.Osheroff N, Zechiedrich EL. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 39.Baldwin EL, Byl JA, Osheroff N. Cobalt enhances DNA cleavage mediated by human topoisomerase II alpha in vitro and in cultured cells. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- 40.Fortune JM, Dickey JS, Lavrukhin OV, Van Etten JL, Lloyd RS, Osheroff N. Site-specific DNA cleavage by Chlorella virus topoisomerase II. Biochemistry. 2002;41:11761–11769. doi: 10.1021/bi025802g. [DOI] [PubMed] [Google Scholar]
- 41.Gale KC, Osheroff N. Uncoupling the DNA cleavage and religation activities of topoisomerase II with a single-stranded nucleic acid substrate: evidence for an active enzyme-cleaved DNA intermediate. Biochemistry. 1990;29:9538–9545. doi: 10.1021/bi00493a007. [DOI] [PubMed] [Google Scholar]
- 42.Lund K, Andersen AH, Christiansen K, Svejstrup JQ, Westergaard O. Minimal DNA requirement for topoisomerase II-mediated cleavage in vitro. J. Biol. Chem. 1990;265:13856–13863. [PubMed] [Google Scholar]
- 43.Andersen AH, Sørensen BS, Christiansen K, Svejstrup JQ, Lund K, Westergaard O. Studies of the topoisomerase II-mediated cleavage and religation reactions by use of a suicidal double-stranded DNA substrate. J. Biol. Chem. 1991;266:9203–9210. [PubMed] [Google Scholar]
- 44.Froelich-Ammon SJ, Gale KC, Osheroff N. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 1994;269:7719–7725. [PubMed] [Google Scholar]
- 45.Wang Y, Knudsen BR, Bjergbaek L, Westergaard O, Andersen AH. Stimulated activity of human topoisomerases IIα and IIβ on RNA- containing substrates. J. Biol. Chem. 1999;274:22839–22846. doi: 10.1074/jbc.274.32.22839. [DOI] [PubMed] [Google Scholar]
- 46.Wilstermann AM, Osheroff N. Base excision repair intermediates as topoisomerase II poisons. J. Biol. Chem. 2001;276:46290–46296. doi: 10.1074/jbc.M105733200. [DOI] [PubMed] [Google Scholar]
- 47.Osheroff N. Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry. 1989;28:6157–6160. doi: 10.1021/bi00441a005. [DOI] [PubMed] [Google Scholar]
- 48.Robinson MJ, Osheroff N. Effects of antineoplastic drugs on the post-strand-passage DNA cleavage/religation equilibrium of topoisomerase II. Biochemistry. 1991;30:1807–1813. doi: 10.1021/bi00221a012. [DOI] [PubMed] [Google Scholar]
- 49.Bromberg KD, Osheroff N. DNA cleavage and religation by human topoisomerase IIα at high temperature. Biochemistry. 2001;40:8410–8418. doi: 10.1021/bi010681q. [DOI] [PubMed] [Google Scholar]
- 50.Bromberg KD, Hendricks C, Burgin AB, Osheroff N. Human topoisomerase IIα possesses an intrinsic nucleic acid specificity for DNA ligation. Use of 5' covalently activated oligonucleotide substrates to study enzyme mechanism. J. Biol. Chem. 2002;277:31201–31206. doi: 10.1074/jbc.M204741200. [DOI] [PubMed] [Google Scholar]
- 51.Bromberg KD, Velez-Cruz R, Burgin AB, Osheroff N. DNA ligation catalyzed by human topoisomerase IIα. Biochemistry. 2004;43:13416–13423. doi: 10.1021/bi049420h. [DOI] [PubMed] [Google Scholar]
- 52.Stivers JT, Shuman S, Mildvan AS. Vaccinia DNA topoisomerase I: kinetic evidence for general acid-base catalysis and a conformational step. Biochemistry. 1994;33:15449–15458. doi: 10.1021/bi00255a027. [DOI] [PubMed] [Google Scholar]
- 53.Burgin AB, Jr., Huizenga BN, Nash HA. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 1995;23:2973–2979. doi: 10.1093/nar/23.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henningfeld KA, Arslan T, Hecht SM. Alteration of DNA primary structure by DNA topoisomerase I. Isolation of the covalent topoisomerase I-DNA binary complex in enzymatically competent form. J. Am. Chem. Soc. 1996;118:11701–11714. [Google Scholar]
- 55.Krogh BO, Cheng C, Burgin A, Jr., Shuman S. Melanoplus sanguinipes entomopoxvirus DNA topoisomerase: site-specific DNA transesterification and effects of 5'-bridging phosphorothiolates. Virology. 1999;264:441–451. doi: 10.1006/viro.1999.0022. [DOI] [PubMed] [Google Scholar]
- 56.Burgin AB. Synthesis and use of DNA containing a 5'-bridging phosphorothioate as a suicide substrate for type I DNA topoisomerases. Methods Mol. Biol. 2001;95:119–128. doi: 10.1385/1-59259-057-8:119. [DOI] [PubMed] [Google Scholar]
- 57.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 58.Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 59.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WGJ. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 60.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr., Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chrencik JE, Burgin AB, Pommier Y, Stewart L, Redinbo MR. Structural impact of the leukemia drug 1-beta-D-arabinofuranosylcytosine (Ara-C) on the covalent human topoisomerase I-DNA complex. J. Biol. Chem. 2003;278:12461–12466. doi: 10.1074/jbc.M212930200. [DOI] [PubMed] [Google Scholar]
- 62.Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, Redinbo MR. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 63.Marchand C, Antony S, Kohn KW, Cushman M, Ioanoviciu A, Staker BL, Burgin AB, Stewart L, Pommier Y. A novel norindenoisoquinoline structure reveals a common interfacial inhibitor paradigm for ternary trapping of the topoisomerase IDNA covalent complex. Mol. Cancer Ther. 2006;5:287–295. doi: 10.1158/1535-7163.MCT-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 65.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 66.Elsea SH, Hsiung Y, Nitiss JL, Osheroff N. A yeast type II topoisomerase selected for resistance to quinolones. Mutation of histidine 1012 to tyrosine confers resistance to nonintercalative drugs but hypersensitivity to ellipticine. J. Biol. Chem. 1995;270:1913–1920. doi: 10.1074/jbc.270.4.1913. [DOI] [PubMed] [Google Scholar]
- 67.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIa and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 68.Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti M-A, Patchan MW, Thompson RB, Osheroff N. Topoisomerase II•Etoposide Interactions Direct the Formation of Drug-Induced Enzyme-DNA Cleavage Complexes. J. Biol. Chem. 1996;271:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 69.Cosstick R, Vyle JS. Solid phase synthesis of oligonucleotides containing 3'-thiothymidine. Tetrahedron Lett. 1989;30:4693–4696. [Google Scholar]
- 70.Sabbagh G, Fettes KJ, Gosain R, O'Neil IA, Cosstick R. Synthesis of phosphorothioamidites derived from 3'-thio-3'-deoxythymidine and 3'-thio-2',3'-dideoxycytidine and the automated synthesis of oligodeoxynucleotides containing a 3'-S-phosphorothiolate linkage. Nucleic Acids Res. 2004;32:495–501. doi: 10.1093/nar/gkh189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sontheimer EJ. Bridging sulfur substitutions in the analysis of pre-mRNA splicing. Methods. 1999;18:29–37. doi: 10.1006/meth.1999.0754. [DOI] [PubMed] [Google Scholar]
- 72.Capranico G, Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
- 73.Muller MT, Spitzner JR, DiDonato JA, Mehta VB, Tsutsui K, Tsutsui K. Single-strand DNA cleavages by eukaryotic topoisomerase II. Biochemistry. 1988;27:8369–8379. doi: 10.1021/bi00422a012. [DOI] [PubMed] [Google Scholar]
- 74.Bromberg KD, Burgin AB, Osheroff N. A two-drug model for etoposide action against human topoisomerase IIα. J. Biol. Chem. 2003;278:7406–7412. doi: 10.1074/jbc.M212056200. [DOI] [PubMed] [Google Scholar]
- 75.Osheroff N. Eukaryotic topoisomerase II. Characterization of enzyme turnover. J. Biol. Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 76.Lindsley JE, Wang JC. Study of allosteric communication between protomers by immunotagging. Nature. 1993;361:749–750. doi: 10.1038/361749a0. [DOI] [PubMed] [Google Scholar]
- 77.Smiley RD, Collins TR, Hammes GG, Hsieh TS. Single-molecule measurements of the opening and closing of the DNA gate by eukaryotic topoisomerase II. Proc. Natl. Acad. Sci. USA. 2007;104:4840–4845. doi: 10.1073/pnas.0700342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee MP, Sander M, Hsieh T. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. J. Biol. Chem. 1989;264:21779–21787. [PubMed] [Google Scholar]
- 80.Sorensen BS, Sinding J, Andersen AH, Alsner J, Jensen PB, Westergaard O. Mode of action of topoisomerase II-targeting agents at a specific DNA sequence. Uncoupling the DNA binding, cleavage and religation events. J. Mol. Biol. 1992;228:778–786. doi: 10.1016/0022-2836(92)90863-f. [DOI] [PubMed] [Google Scholar]
- 81.Byl JA, Cline SD, Utsugi T, Kobunai T, Yamada Y, Osheroff N. DNA topoisomerase II as the target for the anticancer drug TOP-53: mechanistic basis for drug action. Biochemistry. 2001;40:712–718. doi: 10.1021/bi0021838. [DOI] [PubMed] [Google Scholar]
- 82.Cline SD, Osheroff N. Cytosine arabinoside lesions are position-specific topoisomerase II poisons and stimulate DNA cleavage mediated by human type II enzymes. J. Biol. Chem. 1999;274:29740–29743. doi: 10.1074/jbc.274.42.29740. [DOI] [PubMed] [Google Scholar]
- 83.Velez-Cruz R, Riggins JN, Daniels JS, Cai H, Guengerich FP, Marnett LJ, Osheroff N. Exocyclic DNA lesions stimulate DNA cleavage mediated by human topoisomerase IIα in vitro and in cultured cells. Biochemistry. 2005;44:3972–3981. doi: 10.1021/bi0478289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.